Abstract
IFN-γ induction of the CIITA (class II transactivator) promoter (pIV) requires Brahma-related gene 1 (BRG1), a chromatin-remodeling enzyme. However, the events that lead to pIV activation are only partially understood, and the point at which BRG1 acts is unknown. The first IFN-γ-induced event triggers nuclear translocation of STAT1 (signal transducer and activator of transcription 1), which binds IFN-γ-responsive promoters. BRG1 is recruited after activator binding at several other inducible loci, and STAT family members are known to bind BRG1, suggesting that BRG1 might act downstream of STAT1. Here, we delineate a comprehensive view of factor assembly and detailed histone modifications at pIV and show that all events, even STAT1 binding, require BRG1 at CIITA pIV and other IFN-γ target promoters. Recruitment of IFN-stimulated gene factor-3 (ISGF3) [STAT1/STAT2/IFN regulatory factor 9 (IRF9)] to several IFN-α-responsive promoters is also BRG1-dependent. In contrast, constitutive BRG1 association at IFN targets is STAT1-independent. Furthermore, BRG1 is required for IFN-induced restriction enzyme and DNase I accessibility at promoters. Thus, BRG1 has an apical role in cytokine-induced promoter assembly, acting upstream of STAT complexes at multiple IFN target loci.
Keywords: BRG1-associated factor, chromatin, interferon, SWI/SNF
Two classes of enzymes remodel chromatin: ATP-dependent complexes and enzymes that covalently modify histones (1, 2). SWI/SNF is the first identified complex of the former class and is powered by one of two related ATPases, Brahma-related gene 1 (BRG1) and human Brahma (hBRM) (3). Each contributes to separate complexes with overlapping subunit composition and both distinct and common gene targets (4, 5). Covalent histone modification is catalyzed by enzymes such as histone acetyl transferases (HATs) and histone methyl transferases (HMTs). Lysine acetylation is typically associated with gene activation whereas methylation is linked to both gene induction and repression depending on the residue affected and the number of methylated amino groups (6, 7).
SWI/SNF has a key role in tumor suppression because BRG1 heterozygous mice develop cancer and some human tumors lack BRG1 and hBRM (8, 9). The role of BRG1 in neoplasia has been linked to regulation of cell cycle and differentiation (9). We exposed a third possibility by showing that SWI/SNF is required for induction of some IFN-γ-responsive genes (10). Subsequently, BRG1 was shown to be critical for induction of some IFN-α targets (11, 12). Thus, SWI/SNF has a major role in the immune response (reviewed in ref. 3). Mice lacking the IFN-γ receptor or STAT1 (signal transducer and activator of transcription 1), a critical mediator of IFN-γ signaling, develop tumors, indicating that this pathway is critical in immune surveillance (13). Inactivating SWI/SNF may be another way tumor cells evade this defense against cancer.
A multistep cascade of events precedes transcription by RNA polymerase II (Pol II). Chromatin immunoprecipitation (ChIP) is used to study the order of recruitment of proteins to regulatory elements in vivo (14). In yeast, ChIP showed that SWI/SNF is recruited after Swi5 activator binds the inducible homothallism (HO) endonuclease promoter, and genetics coupled with ChIP established that Swi5 binds independent of SWI/SNF (15–18). Such studies are hindered in mammals by the lack of appropriate mutations. However, in vitro chromatin reconstitution studies reveal that SWI/SNF is required downstream of enhanceosome formation at the virus-responsive IFN-β promoter (19). Temporal analyses of factor assembly at other loci also suggest that SWI/SNF action is a secondary event (20–22).
IFN signaling triggers phosphorylation and nuclear translocation of STAT proteins. IFN-γ induces STAT1 homodimerization whereas IFN-α induces formation of a STAT1/STAT2 heterodimer, which interacts with IFN regulatory factor 9 (IRF9) (p48) to generate the IFN-stimulated gene factor-3 (ISGF3) trimer (23, 24). STAT1 or ISGF3 complexes target the IFN-γ-activated sequence (GAS) or the IFN-α stimulated regulatory element (ISRE), respectively, and activate many target genes (25).
A subset of IFN-inducible genes is BRG1-dependent. BRG1 is required for the IFN-γ induction of guanylate-binding protein 1 (GBP1) and class II transactivator (CIITA), the master regulator of MHC class II gene induction, whereas IRF1 induction is BRG1-independent (10). Similarly, IFN-α induction of 9-27, IFN-inducible protein 27 (IFI27), and GBP1 requires BRG1, whereas induction of 6-16 and ISGF3G is BRG1-independent (11, 12). Consistent with the notion that BRG1 acts downstream of STAT recruitment, it is inducibly targeted to the IFN-γ-responsive CIITA promoter and, reminiscent of the Swi5-SWI/SNF interaction in yeast, IFN-α-activated STAT2 binds BRG1 (10, 11). Notably, BRG1 is constitutively present at a subset of IFN-α promoters (11, 12), but whether it influences STAT recruitment at all these targets or is required after STAT binding is unclear. Moreover, it is unclear whether STAT1 complexes play a role in the constitutive recruitment of BRG1. This issue is key given that STATs can reach the nucleus even in the absence of IFN signaling (26, 27).
In most cell types, IFN-γ induction of CIITA is mediated by promoter IV (pIV), one of four alternative promoters (28). pIII is somewhat IFN-γ-responsive, although its major role is to drive constitutive expression in antigen-presenting cells (28, 29). IFN-γ induces binding of the activators STAT1, upstream stimulatory factor 1 (USF1), and IRF1, as well as histone acetylation at pIV in vivo (30). Here, we expand insight into this cascade and show that IFN-γ stimulates biphasic formation of a STAT1-USF1/c-MYC-IRF1 complex, accompanied by histone methylation, HAT-recruitment, acetylation of specific histone residues, and recruitment of Pol II. Critically, unlike homothallism endonuclease, where the apical activator Swi5 binds independent of SWI/SNF, STAT1 binding to CIITA pIV and several other IFN-γ target promoters requires BRG1. IFN-γ induced chromatin remodeling is also BRG1-dependent. This predominance is duplicated at multiple SWI/SNF-dependent IFN-α-inducible loci where ISGF3 recruitment also requires BRG1. In contrast, constitutive recruitment of BRG1 to IFN target genes does not require STAT1. Cytokine-responsive genes are the first identified targets where SWI/SNF is required for the primary step in promoter assembly.
Methods
Cell Culture. HeLa-ini1-11 (HeLa11), SW13, 2fTGH, and U3A cells were grown as described (10, 31, 32). Cells were treated with 300 units/ml of human IFN-γ (PHC4834, BioSource International, Camarillo, CA) or 1,000 units/ml IFN-α (PHC4841, BioSource International).
Adenoviruses. Adenoviral vectors were based on pAdlox (33). Construction details and complete sequences are available on request. Vectors were used to generate adenovirus as described (33). Each virus was plaque purified to remove contaminating normal adenovirus. Virus was amplified in the 293-derived Cre8 cell line (33).
RNA Extraction and Real-Time Quantitative PCR (qPCR). RNA extraction and reverse transcription were as described (10). qPCR used primers in the last exon of each gene (Table 1, which is published as supporting information on the PNAS web site). qPCR was performed by using an Applied Biosystems PRISM 7900HT in duplicate with SYBR Green PCR master mix (Applied Biosystems) according to the manufacturer's instructions. PCR consisted of 40 cycles of 95°C for 15 s and 55°C for 30 s. A final cycle (95°C, 15 s, 60°C) generated a dissociation curve to confirm a single product. The cycle quantity required to reach a threshold in the linear range (Qt) was determined and compared with a standard curve for each primer set generated by five 3-fold dilutions of genomic DNA samples of known concentration. Values were normalized to β-actin.
ChIP. ChIP DNA was prepared as described (34) and subjected to qPCR as above by using primers in Table 1. Copy number was calculated from Qt values as above. The amount of DNA precipitated by a control GAL4 antibody was subtracted, and the percent ChIP DNA relative to input was calculated (for an antibody list, see Table 2, which is published as supporting information on the PNAS web site). Statistically significant differences were assessed by using analysis of variance (ANOVA) with an ad hoc Tukey's test. ChIP efficiency can vary among antibodies. However, analysis of maximally induced IFN-responsive genes (6 h post-IFN) revealed that histone ChIPs are most efficient (≈2–10% of input chromatin), followed by DNA-bound factors (Pol II, STAT1, STAT2, IRF1, IRF9, c-Myc, USF1: ≈0.5–2% of input), followed by cofactors recruited by protein–protein interactions [BRG1, CREB-binding protein (CBP), p300: 0.1–0.5% of input]. These data imply that, rather than differences in antibody affinity, the primary determinant of ChIP efficiency, at least for factors studied here, may be the potential for cross-linking to DNA.
Restriction Enzyme and DNase I Accessibility Assays. Restriction enzyme and DNase I accessibility experiments were performed as described (10). Extracted DNA was subjected to qPCR as above by using primers in Table 1.
Western Blotting. Western blotting was performed as described (35).
Results
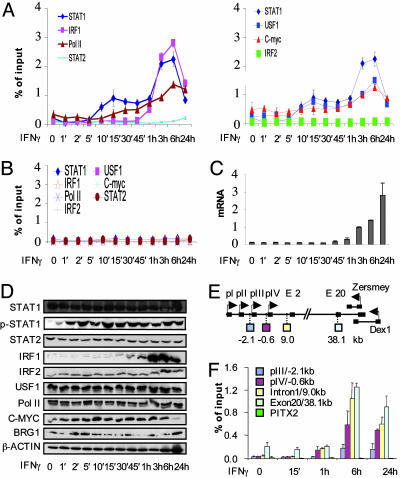
Biphasic Formation of a Putative Enhanceosome at CIITA pIV. To understand the timing of BRG1 action, we initially focused on CIITA pIV and first deduced when activators and Pol II bind after IFN-γ-induction. pIV contains sites for STAT1 and USF-1 homodimers, and IRF1/2 homo/heterodimers (28). USF1 and c-MYC share related DNA-binding motifs; thus, we also tested whether c-MYC might bind pIV. We also checked for STAT2 recruitment because the STAT1/STAT2 heterodimer, typically induced by IFN-α, may also be activated by IFN-γ (36).
ChIP assays revealed that STAT1 and USF1 bound pIV within 10 min of IFN-γ treatment (Fig. 1A). By 30 min, at the initial peak of STAT1/USF1 binding, a small increase in Pol II was observed at pIV, and, by 45 min, mRNA was evident (Fig. 1C). IRF1 binding was detected 1 h post-IFN-γ (Fig. 1A) coincident with its expression (Fig. 1D), and induced a stable STAT1/USF1/IRF1 complex, further recruitment of Pol II, and an exponential increase in mRNA (Fig. 1 A and C). In vitro gel shifts argued against a role for c-MYC in CIITA regulation (37), but in vivo detectable levels were observed at the promoter before IFN-γ treatment, and both USF1 and c-MYC binding increased after formation of the putative enhanceosome (Fig. 1A). None of the activators or Pol II was detected at the irrelevant paired-like homeodomain transcription factor 2 (PITX2) promoter (Fig. 1B), and neither STAT2 nor IRF2 was recruited to pIV or the irrelevant control locus (Fig. 1 A and B). All proteins were expressed at every time point, with the expected exception of IRF1, which is induced by STAT1 (Fig. 1D). STAT1 was rapidly phosphorylated within 5 min of IFN-γ stimulation (Fig. 1D).
Fig. 1.
Biphasic promoter assembly at CIITA pIV. (A) HeLa 11 cells were exposed to IFN-γ for the indicated times. ChIP assays were performed with the indicated antibodies. DNA was analyzed by qPCR by using CIITA pIV primers. Results are on two graphs, with the STAT1 data on both to facilitate comparison. (B) ChIP DNA from A was analyzed by qPCR by using PITX2 control primers. (C) RNA from cells treated as in A was analyzed by qPCR to monitor CIITA induction. Levels are in arbitrary units. (D) Lysate from cells treated as in A was analyzed by Western blot by using the indicated antibodies. (E) The CIITA locus. Exons are shown as black boxes. pI, pII, pIII, and pIV are alternate promoters (see introduction). pIV is the major IFN-γ-responsive promoter. The regions amplified for F are shown below the CIITA gene. Two genes, DEX1 and ZERSMEY, overlap the end of CIITA and are on the opposite strand. (F) Pol II elongation correlates with promoter assembly and dissolution. ChIP assays were performed with phospho-ser2-Pol II antibodies, and the DNA was analyzed by qPCR by using the indicated CIITA or control PITX2 primers. Above-background signal with exon 20 primers is due to constitutive low-level expression of inverse transcripts (see E). Graphs in this and all subsequent figures show the average of three separate experiments, each performed in duplicate ±SD.
Thus, IFN-γ-induced activator recruitment to pIV is biphasic: STAT1 and USF-1 associate first, but maximal Pol II binding and significant mRNA expression correlate with assembly of the complete STAT1-USF1/c-MYC-IRF1 complex.
CIITA Promoter Assembly and Dissolution Correlates with Pol II Activity. Peak complex formation on pIV at 6 h was followed by disassembly at 24 h (Fig. 1A). mRNA continued to accumulate (Fig. 1C), but steady state reflects the combination of new and prior transcripts rather than active transcription. To determine whether promoter assembly and disassembly correlated with Pol II activity, we measured elongating Pol II present along the CIITA locus with an antibody that recognizes phosphorylated serine 2 (S2) of the C-terminal domain of the large subunit (38). Active Pol II was detected at pIV, intron 1, and exon 20 1 h after IFN-γ treatment, peaked at 6 h, and diminished by 24 h (Fig. 1F). Delayed low level Pol II activity at pIII (Fig. 1F) is consistent with other delayed events at that upstream location (see Fig. 2). Pol II was not detected at the control PITX2 locus (Fig. 1F). Thus, Pol II activity at the CIITA locus correlated precisely with promoter assembly and disassembly.
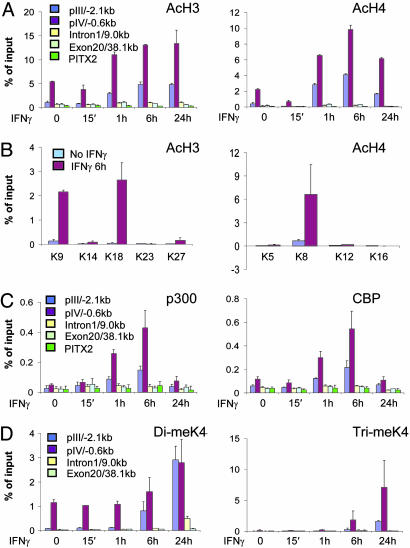
Fig. 2.
The CIITA histone code. HeLa 11 cells were exposed to IFN-γ for the indicated times. ChIP assays were performed with antibodies against tetraacetylated histone H3 and diacetylated histone H4 (A), individual acetylated histone H3 and H4 residues (B), p300 or CBP (C), and di- or trimethylated lysine 4 of histone H3 (D). DNA was analyzed by qPCR by using the indicated CIITA or control PITX2 primers.
The CIITA Histone Code. IFN-γ induces histone H3/H4 acetylation at pIV (30), but the HATs involved, the residues affected, the role of methylation and the extent of histone modifications across the CIITA locus are unknown. STAT1, c-MYC, and IRF1 bind CBP/p300 HATs (39–41); thus, we hypothesized that these cofactors may acetylate histones at pIV. Antibodies that bind multiple acetylated sites on H3 (K9 and/or K14)or H4 (K5, K8, K12, and/or K16) and antibodies for CBP and p300 were used in ChIP assays with chromatin from cells treated with IFN-γ for 0 min, 15 min, 1 h, 6 h, and 24 h (Fig. 2). Before IFN-γ treatment, acetylation was already higher at pIV than elsewhere in the CIITA locus or at the silent PITX2 control (P < 0.05, Fig. 2A). IFN-γ-induced H3/H4 acetylation centered on pIV nucleosomes (Fig. 2A). Acetylation rose at 1 h and peaked 6 h after IFN-γ treatment, mirroring promoter assembly (compare Fig. 1A).
Next, ChIPs were performed with antibodies against individual acetylated residues. After IFN-γ treatment, acetylated H3-K9 and K18 and H4-K8, but not H3-K14, K23 or K27, or H4-K5, K12 or K16 were detected at pIV (Fig. 2B). These modifications may be mediated by CBP/p300 because recruitment of these HATs, but not Gcn5 (general control of amino acid synthesis 5) or p300/CBP-associated factor (PCAF), was detected after IFN-γ treatment (Fig. 2C and data not shown).
H3-K4 tri-methylation is linked to activation whereas H3-K9 tri-methylation is linked to silencing (42–44). ChIP did not detect H3-trimeK9 at pIV, suggesting that it does not silence CIITA in the absence of IFN-γ (data not shown). In contrast, H3-dimeK4, like acetylation, was detected at the silent promoter (P < 0.05) and increased 6 h after IFN-γ treatment (Fig. 2D). Induced dimethylation was also detected 2.1 kb upstream of pIV at pIII, but the response lagged several hours behind pIV (Fig. 2D). Low and delayed H3-dimeK4 were also induced in intron 1 by 24 h, but only background levels were seen at exon 20 (Fig. 2D). Unlike H3-dimeK4, H3-trimeK4 was very low at the silent promoter but was induced after IFN-γ treatment (Fig. 2D). This modification was more localized because only low levels were observed upstream and no trimethylation was detected downstream of pIV (Fig. 2D). Thus, CIITA induction is linked to multiple specific histone modifications centered on pIV that are synchronized with promoter assembly.
Constitutive Recruitment of BRG1 at pIV. To determine when BRG1 binds the CIITA promoter, time course ChIP assays were performed with HeLa 11 chromatin. Importantly, BRG1 was present at pIV in uninduced cells at levels above neighboring upstream and downstream locations (P < 0.05, Fig. 3A). IFN-γ increased the amount of BRG1 above constitutive levels, rising at 1 h, peaking at 6 h, and falling by 24 h (Fig. 3A). Thus, BRG1 shows both constitutive and induced binding to pIV. In contrast, negligible levels of STAT1 were found at pIV before IFN-γ treatment (Fig. 1A).
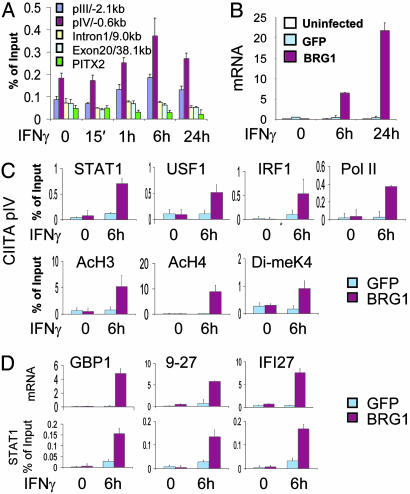
Fig. 3.
BRG1-dependent STAT1 binding at IFN-γ-responsive promoters. (A) BRG1 recruitment at CIITA pIV. HeLa 11 cells were exposed to human IFN-γ for the indicated times, ChIP assays were performed with BRG1 antibody, and DNA was analyzed by qPCR by using the indicated CIITA or control PITX2 primers. (B) BRG1-dependent CIITA expression. qPCR was used to assess the level of CIITA mRNA in SW13 cells that were left uninfected or transduced with GFP or BRG1 adenovirus and treated with IFN-γ for the indicated times. (C) BRG1-dependent events at pIV. SW13 cells were transduced with Ad-FG or Ad-FG-BRG and left untreated or exposed to IFN-γ for 6 h. ChIP assays were performed with the indicated antibodies and isolated DNA analyzed by qPCR by using CIITA pIV primers. (D) BRG1-dependent STAT1 binding at multiple IFN-γ targets. SW13 cells were treated as in C, and mRNA levels were assessed by qPCR. STAT1 ChIP assays were performed, and enriched DNA fragments were quantified by using primers for the indicated promoters. In C and D, negligible binding was detected by ChIP at the 3′ end of each gene or at the PITX2 promoter (data not shown). mRNA data in B and D are shown as arbitrary units.
Predominant Role for BRG1 in Factor Assembly at CIITA pIV. Having established a thorough view of constitutive and induced events at pIV, we asked which steps require BRG1. Because STAT1 mediates IFN-γ signaling, because BRG1 binds STATs, and because BRG1 typically acts downstream of activators, we expected STAT1 binding to be BRG1-independent. BRG1-deficient SW13 cells were transduced with an adenovirus vector that we developed (Ad-FG-BRG1) expressing BRG1 fused to an N-terminal Flag-GFP tag, or a control virus (Ad-FG), expressing Flag-tagged GFP. More than 90% of the cells were transduced after 24 h (data not shown), and Western blotting confirmed expression of Flag-tagged GFP and GFP-BRG1 (Fig. 7A, which is published as supporting information on the PNAS web site). BRG1 levels in Ad-FG-BRG1-transduced SW13 cells were comparable with the endogenous levels in HeLa 11 cells (Fig. 7 B and C). Cells were harvested either without treatment or after exposure to IFN-γ, and qPCR showed that CIITA induction was rescued by BRG1 (Fig. 3B). ChIP assays revealed that, as seen in HeLa cells, high levels of STAT1, USF1, IRF1, and Pol II recruitment and histone H3/H4 acetylation and H3 methylation were observed at pIV 6 h in IFN-γ-treated SW13 cells reconstituted with BRG1 (Fig. 3C). In contrast, Pol II recruitment, histone modification, and formation of the putative enhanceosome were blocked in BRG1-deficient Ad-FG-transduced cells (Fig. 3C). Most importantly, STAT1 recruitment was also blocked in the absence of BRG1 (Fig. 3C). Similar results were obtained 30 min after IFN-γ induction (data not shown), indicating that even the initial phase of STAT1 binding before IRF1 recruitment was blocked. IFN-γ-induced recruitment of STAT1 to several BRG1-independent target promoters (ISFG3G, IRF1, and 6-16) was similar in BRG1 reconstituted and control cells (Fig. 8, which is published as supporting information on the PNAS web site), indicating that the absence of BRG1, rather than impaired STAT1 function, blocks factor assembly at pIV.
Apical Role for BRG1 at Multiple IFN-γ-Responsive Promoters. We next determined whether BRG1 also acts before STAT1 at other BRG1-dependent IFN-γ-responsive targets. IFN-γ induction of GBP1, 9-27, and IFI27 mRNA was BRG1-dependent (Fig. 3D). Moreover, both STAT1 recruitment and histone acetylation at these targets required BRG1 (Fig. 3D and Fig. 9, which is published as supporting information on the PNAS web site). Thus, BRG1 is critical for STAT1 binding and subsequent events at multiple IFN-γ-responsive genes.
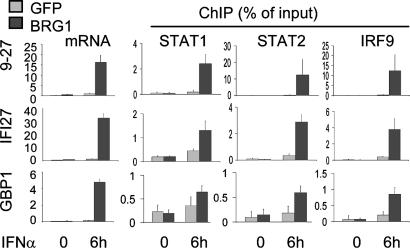
Predominant Role for BRG1 at IFN-α-Induced Promoters. BRG1 is also critical for induction of some IFN-α-responsive genes (11, 12). Thus, we asked whether BRG1 was required for recruitment of the trimeric STAT1/STAT2/IRF9 ISGF3 complex to the SWI/SNF-dependent IFN-α target genes. As expected (11, 12), BRG1 was necessary for IFN-α induction of 9-27, IFI27, and GBP1 (Fig. 4) but not 6-16 or ISGF3G mRNAs (Fig. 10, which is published as supporting information on the PNAS web site). Recruitment of STAT1, STAT2, and IRF9 to 9-27, IFI27, and GBP1 promoters required BRG1 (Fig. 4), whereas binding to 6-16 or ISGF3G was BRG1-independent (Fig. 10). Thus, again, BRG1 absence does not impair STAT activity, per se, but blocks access to specific genes. Downstream events at IFN-α-responsive promoters, such as H3/H4 acetylation and H3 methylation, showed the same BRG1 dependency as ISGF3 binding (Fig. 11, which is published as supporting information on the PNAS web site). IFIT1 and IFITM3 induction show partial requirement for SWI/SNF (Fig. 12A, which is published as supporting information on the PNAS web site), and here recruitment of ISGF3 was partially blocked in the absence of BRG1 (Fig. 12A), which resulted in partial acetylation of promoter-associated nucleosomes (Fig. 12B). Thus, the degree to which BRG1 is required for gene induction correlates with the level to which ISGF3 depends on BRG1 for access to target promoters.
Fig. 4.
BRG1-dependent ISFG3 recruitment. SW13 cells were transduced with Ad-FG or Ad-FG-BRG and left untreated or exposed to IFN-α for 6 h. qPCR was used to assess the mRNA level of indicated genes (levels in arbitrary units). ChIP assays were used to assess the level of ISGF3 components at these promoters. Control PCRs using primers at downstream locations were negative (data not shown).
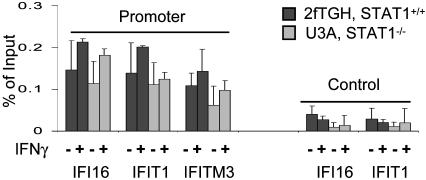
BRG1 Recruitment Does Not Require STAT Complexes. The above data show that STAT recruitment to IFN targets is BRG1-dependent but do not resolve whether BRG1 recruitment requires STAT. Thus, we compared the amount of constitutive BRG1 present at IFN-responsive promoters in STAT1-expressing 2fTGH cells and STAT1-deficient U3A cells. STAT1 is essential for both IFN-γ (STAT1 dimer) and IFN-α (ISGF3 trimer) signaling. Notably, STAT1 absence did not affect constitutive BRG1 association at several promoters (Fig. 5). IFN-γ treatment did not significantly enhance BRG1 recruitment at these promoters (Fig. 5). Thus, although recruitment of IFN-induced STAT complexes is BRG1-dependent, BRG1 recruitment is STAT-independent.
Fig. 5.
Constitutive recruitment of BRG1 at IFN targets is STAT1-independent. 2fTGH or derivative STAT1-deficient U3A cells were left untreated or exposed to IFN-γ for 6 h, and BRG1 ChIP assays were performed. Control PCRs used primers from exon 9 of IFI16 or the last exon of IFIT1.
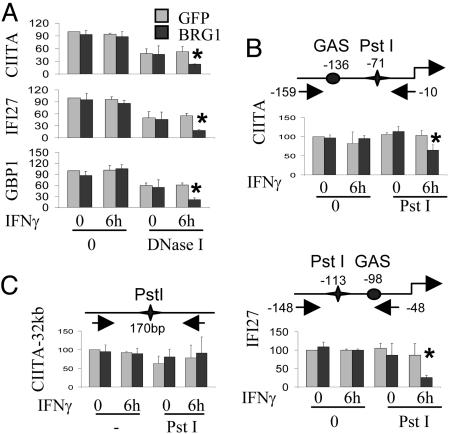
BRG1-Dependent Chromatin Remodeling at IFN Target Promoters. Previously, we used DNase I and restriction enzyme (RE) digestion to show that BRG1 increases chromatin accessibility at CIITA pIV after IFN-γ treatment (10). To confirm and extend this analysis, we used qPCR coupled with DNase I and RE digestion to quantify accessibility at CIITA, IFI27, and/or GBP1 promoters in the presence or absence of BRG1 and IFN-γ. GFP- or BRG1-expressing SW13 cells were left untreated or exposed to IFN-γ and nuclei incubated with DNase I, or PstI, which cuts near the GAS element in the CIITA and IFI27 promoters. Levels of intact DNA were determined by qPCR. In GFP-transduced cells, promoter accessibility was identical plus or minus IFN-γ (Fig. 6 A and B). In contrast, BRG1 markedly increased accessibility, either to DNase I or REs in the presence of IFN-γ (Fig. 6 A and B). BRG1 did not affect baseline accessibility in the absence of IFN-γ (Fig. 6 A and B). Similar results were observed upon IFN-α induction of IFI27 (data not shown). Enzyme accessibility at a control locus 32 kb upstream of CIITA was identical in untreated or IFN-γ-treated cells transduced with GFP or BRG1 (Fig. 5C). These data suggest that BRG1 may not alter baseline chromatin accessibility, at least as assessed by DNase I or PstI digests, but is essential for IFN-induced chromatin remodeling.
Fig. 6.
IFN-γ-induced chromatin remodeling requires BRG1. SW13 cells transduced with Ad-FG or Ad-FG-BRG1 were left untreated or exposed to IFN-γ; then, nuclei were prepared and exposed to 2.5 Krunitz units/100 μl DNaseI for 3 min (A) or 50 units/350 μl of PstI for 15 min (B and C). DNA was purified and analyzed by qPCR by using primers that flank the GAS elements in the indicated promoters (A and B) or a control region 32 kb upstream of CIITA IV (C). The schematic diagrams show the position of GAS elements, PstI sites, and primers (arrows). Results are the percentages of the level of intact DNA in GFP-transduced cells that were not exposed to IFN-γ and are the average of three independent experiments ±SD. Asterisks indicate a significant increase in accessibility (P < 0.05).
Discussion
SWI/SNF Acts Upstream of STATs. SWI/SNF recruitment and/or action is secondary to one or more activators in many gene induction cascades (15–22). Constitutive BRG1 binding has been reported at some IFN-α-inducible targets (11, 12), but it was not clear whether BRG1 was required for STAT recruitment at all these targets, or whether it acts up or downstream of STAT1 at IFN-γ targets. It was also unknown whether BRG1 recruitment required STAT binding at either IFN-α or -γ target genes. It is well known that STAT1 translates IFN-γ signaling to the nucleus, and, because STATs interact with BRG1 (11, 45), it seemed logical that recruitment of STAT1 at CIITA pIV might supercede either BRG1 recruitment and/or the requirement for BRG1 activity. Thus, one possibility was that SWI/SNF-dependent inducible IFN targets would behave like the homothallism endonuclease gene, where the Swi5 activator (analogous to STAT) binds independent of SWI/SNF (15, 16). Instead, STAT1-recruitment to CIITA pIV and several other IFN-γ responsive promoters (GBP1, 9-27, and IFI27) was BRG1-dependent. Moreover, recruitment of the ISGF3 trimer to IFN-α-inducible genes (GBP1, 9-27, IFI27) was also BRG1-dependent. BRG1-dependent IFN-inducible promoters exhibited constitutive BRG1 binding, which was independent of the presence of functional STAT complexes. Thus, whereas recruitment of either IFN-γ-activated STAT1-dimers or IFN-α-activated ISGF3 requires BRG1, BRG1 recruitment does not require either complex. Cytokine-induced genes are the first targets identified at which BRG1 action supercedes recruitment of the primary activator. These data underscore the importance of SWI/SNF in the immune response.
The mechanism by which BRG1 enhances STAT recruitment is unclear. The idea that BRG1 acts indirectly seems unlikely given the direct constitutive association with IFN-responsive promoters. BRG1 was critical for IFN-γ-induced DNase I and RE accessibility at three promoters we tested (CIITA, GBP1, and IFI27) but did not increase baseline accessibility in the absence of IFN-γ. DNase I and RE accessibility assays may miss subtle BRG1-dependent variations in chromatin structure at quiescent promoters. Alternatively, BRG1 may facilitate STAT recruitment by acting as an interaction surface for STAT proteins. BRG1 ATPase activity is crucial for IFN-induced chromatin remodeling and gene induction (10), so, if this protein is a STAT interaction surface, the latter is not its only role. An intriguing possibility is that BRG1 may modify chromatin at more distant sites that influence STAT1 promoter binding. Indeed, we have evidence that there may be long-range effects at the CIITA locus (Z.N., Z.X., T.Y., and R.B., unpublished results). It will be important to determine the role of BRG1 in this context.
The Ordered Cascade at CIITA pIV. Induction of CIITA pIV is a biphasic event involving multiple factors and specific histone modifications. Before activation, low levels of c-MYC, HATs, BRG1, and Pol II, as well as various histone modifications, are present at pIV. Ten to 45 min after IFN-γ exposure, STAT1 and USF1 bind cooperatively to the GAS and E-box elements, resulting in some increase in Pol II. Formation of a stable promoter complex and maximal Pol II binding require IRF1, which is expressed and recruited 1 h after IFN-γ treatment. Promoter assembly peaks after 6 h and drops by 24 h. The creation and dissolution of this structure coincide with the rise and fall of HAT recruitment and histone modifications, as well as elevated levels of BRG1. Promoter assembly and disassembly also correlate with the presence and loss of active Pol II traversing the CIITA locus. In addition to STAT1 recruitment, all downstream events are BRG1-dependent. Similarly, at IFN-α-inducible promoters, induction of histone acetylation and methylation were blocked in the absence of BRG1.
Apart from elucidating the position of BRG1 in IFN signaling, our work expands insight into pIV regulation in many ways. First, histone acetylation was detected on H3-K9 and K18, and on H4-K8, but not on H3-K14, on K23 or K27, or on H4-K5, K12, or K16. Second, CBP and p300, but not GCN5 or CBP-associated factor (PCAF), likely mediate site-specific acetylation, although we do not have functional proof that this is the case. Multiple distinct activation domains may combine to create an ideal interaction surface for CBP/p300, akin to the IFN-β promoter (40). Indeed, STAT1, IRF1, and c-MYC interact with CBP/p300 (39–41). Third, elevated diand trimethylation of histone H3-K4 were also linked to pIV induction. Fourth, whereas histone modifications associated with gene activation or transcriptional competency can range over tens of kb and encompass several inactive and active genes (44, 46, 47), the histone modifications we detected at pIV were confined, suggesting that localized acetylation and methylation may be characteristic of genes that are rapidly induced and silenced (reviewed in ref. 6). Methylation of H3-K4 has been linked to elongation in yeast (48–50), but we did not observe this modification in intragenic CIITA regions at intron 1 or exon 20. This modification does not seem to be required for transcription through most of the ≈55-kb CIITA locus. Fifth, above-background levels of both acetylation and methylation were detected at the silent promoter. Yeast studies suggest that trimethylated H3-K4 is a marker for active loci, including both promoters and transcribed regions, and that dimethyl H3-K4 marks regions that are poised but not necessarily active (42, 50), which is consistent with our observations at pIV. Sixth, despite links between IFN-γ and STAT2 activation (36), and between IRF2 and CIITA induction (51), our ChIP analyses argue against a direct role for these proteins in promoter assembly at least in the cells we studied. Finally, previous in vitro gel shift analysis excluded a role for c-MYC in regulating CIITA (37), but data here show that c-MYC associates with pIV in vivo. These data clarify the series of events that precede and mediate CIITA induction.
How Is BRG1 Recruited? Our data argue against the idea that STAT complexes play a role in constitutive BRG1 recruitment to IFN targets, placing the focus on various alternatives. SWI/SNF is targeted to the IFN-β promoter by specific interaction with H4-AcK8 (52), a modification we observed at CIITA pIV. SWI/SNF components have been detected in a multiprotein complex that contains both histone methyl transferases and HATs (53); thus, it is intriguing that CBP and p300, like BRG1, are present at low levels at the uninduced pIV. DNA-binding proteins may also help tether BRG1 and HATs to the silent pIV. SWI/SNF and CBP/p300 interact with c-MYC and Pol II, and, in untreated cells, each of these factors is present at pIV at detectable levels. Finally, Sp1 binds BRG1 and recruits SWI/SNF to the IFN-α-responsive IFITM3 locus (12). These mechanisms are not mutually exclusive and may cooperate to tether BRG1 to IFN target genes independent of STAT proteins.
Supplementary Material
Acknowledgments
We thank Dr. G. Stark (Cleveland Clinic Foundation, Cleveland) for U3A cells. E.K. was supported by a Vision Science Research Program (VSRP) fellowship. Z.N. was supported by an Ontario Graduate Studentship, by fellowships from the VSRP and the Frank Fletcher Memorial Fund, and by a Dr. R. Dittakavi & Dr. P. Rao Graduate Award. This work was supported by the National Cancer Institute of Canada with funds from the Canadian Cancer Society.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HAT, histone acetyl transferase; STAT, signal transducer and activator of transcription; Pol II, polymerase II; ChIP, chromatin immunoprecipitation; IRF, IFN regulatory factor; ISGF, IFN-stimulated gene factor; GAS, IFN-γ-activated sequence; CIITA, class II transactivator; pIV, promoter IV; USF, upstream stimulatory factor; qPCR, real-time quantitative PCR; CBP, CREB-binding protein; RE, restriction enzyme; BRG1, Brahma-related gene 1; GBP1, guanylate-binding protein 1; IFI27, IFN-inducible protein 27; PITX2, paired-like homeodomain transciption factor 2.
References
- 1.Becker, P. B. & Horz, W. (2002) Annu. Rev. Biochem. 71, 247-273. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074-1080. [DOI] [PubMed] [Google Scholar]
- 3.Chi, T. (2004) Nat. Rev. Immunol. 4, 965-977. [DOI] [PubMed] [Google Scholar]
- 4.Wang, W., Xue, Y., Zhou, S., Kuo, A., Cairns, B. R. & Crabtree, G. R. (1996) Genes Dev. 10, 2117-2130. [DOI] [PubMed] [Google Scholar]
- 5.Kadam, S. & Emerson, B. M. (2003) Mol. Cell 11, 377-389. [DOI] [PubMed] [Google Scholar]
- 6.Turner, B. M. (2000) BioEssays 22, 836-845. [DOI] [PubMed] [Google Scholar]
- 7.Fischle, W., Wang, Y. & Allis, C. D. (2003) Curr. Opin. Cell Biol. 15, 172-183. [DOI] [PubMed] [Google Scholar]
- 8.Bultman, S., Gebuhr, T., Yee, D., La Mantia, C., Nicholson, J., Gilliam, A., Randazzo, F., Metzger, D., Chambon, P., Crabtree, G. & Magnuson, T. (2000) Mol. Cell 6, 1287-1295. [DOI] [PubMed] [Google Scholar]
- 9.Klochendler-Yeivin, A., Muchardt, C. & Yaniv, M. (2002) Curr. Opin. Genet. Dev. 12, 73-79. [DOI] [PubMed] [Google Scholar]
- 10.Pattenden, S. G., Klose, R., Karaskov, E. & Bremner, R. (2002) EMBO J. 21, 1978-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, M., Qian, F., Hu, Y., Ang, C., Li, Z. & Wen, Z. (2002) Nat. Cell. Biol. 4, 774-781. [DOI] [PubMed] [Google Scholar]
- 12.Liu, H., Kang, H., Liu, R., Chen, X. & Zhao, K. (2002) Mol. Cell Biol. 22, 6471-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, G. P., Bruce, A. T., Ikeda, H., Old, L. J. & Schreiber, R. D. (2002) Nat. Immunol. 3, 991-998. [DOI] [PubMed] [Google Scholar]
- 14.Cosma, M. P. (2002) Mol. Cell 10, 227-236. [DOI] [PubMed] [Google Scholar]
- 15.Cosma, M. P., Tanaka, T. & Nasmyth, K. (1999) Cell 97, 299-311. [DOI] [PubMed] [Google Scholar]
- 16.Krebs, J. E., Kuo, M. H., Allis, C. D. & Peterson, C. L. (1999) Genes Dev. 13, 1412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhoite, L. T., Yu, Y. & Stillman, D. J. (2001) Genes Dev. 15, 2457-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosma, M. P., Panizza, S. & Nasmyth, K. (2001) Mol. Cell 7, 1213-1220. [DOI] [PubMed] [Google Scholar]
- 19.Agalioti, T., Lomvardas, S., Parekh, B., Yie, J., Maniatis, T. & Thanos, D. (2000) Cell 103, 667-678. [DOI] [PubMed] [Google Scholar]
- 20.DiRenzo, J., Shang, Y., Phelan, M., Sif, S., Myers, M., Kingston, R. & Brown, M. (2000) Mol. Cell. Biol. 20, 7541-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martens, J. H., Verlaan, M., Kalkhoven, E. & Zantema, A. (2003) Mol. Cell. Biol. 23, 1808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salma, N., Xiao, H., Mueller, E. & Imbalzano, A. N. (2004) Mol. Cell. Biol. 24, 4651-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boehm, U., Klamp, T., Groot, M. & Howard, J. C. (1997) Annu. Rev. Immunol. 15, 749-795. [DOI] [PubMed] [Google Scholar]
- 24.Reith, W. & Mach, B. (2001) Annu. Rev. Immunol. 19, 331-373. [DOI] [PubMed] [Google Scholar]
- 25.Aaronson, D. S. & Horvath, C. M. (2002) Science 296, 1653-1655. [DOI] [PubMed] [Google Scholar]
- 26.Chatterjee-Kishore, M., Wright, K. L., Ting, J. P. & Stark, G. R. (2000) EMBO J. 19, 4111-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer, T., Marg, A., Lemke, P., Wiesner, B. & Vinkemeier, U. (2003) Genes Dev. 17, 1992-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhlethaler-Mottet, A., Otten, L. A., Steimle, V. & Mach, B. (1997) EMBO J. 16, 2851-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piskurich, J. F., Linhoff, M. W., Wang, Y. & Ting, J. P. (1999) Mol. Cell. Biol. 19, 431-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris, A. C., Beresford, G. W., Mooney, M. R. & Boss, J. M. (2002) Mol. Cell. Biol. 22, 4781-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sif, S., Stukenberg, P. T., Kirschner, M. W. & Kingston, R. E. (1998) Genes Dev. 12, 2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKendry, R., John, J., Flavell, D., Muller, M., Kerr, I. M. & Stark, G. R. (1991) Proc. Natl. Acad. Sci. USA 88, 11455-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardy, S., Kitamura, M., Harris-Stansil, T., Dai, Y. & Phipps, M. L. (1997) J. Virol. 71, 1842-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells, J. & Farnham, P. J. (2002) Methods 26, 48-56. [DOI] [PubMed] [Google Scholar]
- 35.Dorval, K. M., Bobechko, B. P., Ahmad, K. F. & Bremner, R. (2005) J. Biol. Chem. 280, 10100-10108. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto, M., Tanaka, N., Harada, H., Kimura, T., Yokochi, T., Kitagawa, M., Schindler, C. & Taniguchi, T. (1999) Biol. Chem. 380, 699-703. [DOI] [PubMed] [Google Scholar]
- 37.Muhlethaler-Mottet, A., Di Berardino, W., Otten, L. A. & Mach, B. (1998) Immunity 8, 157-166. [DOI] [PubMed] [Google Scholar]
- 38.Komarnitsky, P., Cho, E. J. & Buratowski, S. (2000) Genes Dev. 14, 2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, J. J., Vinkemeier, U., Gu, W., Chakravarti, D., Horvath, C. M. & Darnell, J. E., Jr. (1996) Proc. Natl. Acad. Sci. USA 93, 15092-15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merika, M., Williams, A. J., Chen, G., Collins, T. & Thanos, D. (1998) Mol. Cell 1, 277-287. [DOI] [PubMed] [Google Scholar]
- 41.Vervoorts, J., Luscher-Firzlaff, J. M., Rottmann, S., Lilischkis, R., Walsemann, G., Dohmann, K., Austen, M. & Luscher, B. (2003) EMBO Rep. 4, 484-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., Schreiber, S. L., Mellor, J. & Kouzarides, T. (2002) Nature 419, 407-411. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D. & Grewal, S. I. (2001) Science 292, 110-113. [DOI] [PubMed] [Google Scholar]
- 44.Litt, M. D., Simpson, M., Gaszner, M., Allis, C. D. & Felsenfeld, G. (2001) Science 293, 2453-2455. [DOI] [PubMed] [Google Scholar]
- 45.Giraud, S., Hurlstone, A., Avril, S. & Coqueret, O. (2004) Oncogene 23, 7391-7398. [DOI] [PubMed] [Google Scholar]
- 46.Schubeler, D., Francastel, C., Cimbora, D. M., Reik, A., Martin, D. I. & Groudine, M. (2000) Genes Dev. 14, 940-950. [PMC free article] [PubMed] [Google Scholar]
- 47.Noma, K., Allis, C. D. & Grewal, S. I. (2001) Science 293, 1150-1155. [DOI] [PubMed] [Google Scholar]
- 48.Krogan, N. J., Kim, M., Tong, A., Golshani, A., Cagney, G., Canadien, V., Richards, D. P., Beattie, B. K., Emili, A., Boone, C., et al. (2003) Mol. Cell. Biol. 23, 4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krogan, N. J., Dover, J., Wood, A., Schneider, J., Heidt, J., Boateng, M. A., Dean, K., Ryan, O. W., Golshani, A., Johnston, M., et al. (2003) Mol. Cell 11, 721-729. [DOI] [PubMed] [Google Scholar]
- 50.Ng, H. H., Robert, F., Young, R. A. & Struhl, K. (2003) Mol. Cell 11, 709-719. [DOI] [PubMed] [Google Scholar]
- 51.Xi, H., Goodwin, B., Shepherd, A. T. & Blanck, G. (2001) Oncogene 20, 4219-4227. [DOI] [PubMed] [Google Scholar]
- 52.Agalioti, T., Chen, G. & Thanos, D. (2002) Cell 111, 381-392. [DOI] [PubMed] [Google Scholar]
- 53.Nakamura, T., Mori, T., Tada, S., Krajewski, W., Rozovskaia, T., Wassell, R., Dubois, G., Mazo, A., Croce, C. M. & Canaani, E. (2002) Mol. Cell 10, 1119-1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.