Abstract
The genome of Saccharomyces cerevisiae encodes three close homologues of the Escherichia coli 2′-O-rRNA methyltransferase FtsJ/RrmJ, designated Trm7p, Spb1p and Mrm2p. We present evidence that Trm7p methylates the 2′-O-ribose of nucleotides at positions 32 and 34 of the tRNA anticodon loop, both in vivo and in vitro. In a trm7Δ strain, which is viable but grows slowly, translation is impaired, thus indicating that these tRNA modifications could be important for translation efficiency. We discuss the emergence of a family of three 2′-O-RNA methyltransferases in Eukaryota and one in Prokaryota from a common ancestor. We propose that each eukaryotic enzyme is located in a different cell compartment, in which it would methylate a different RNA that can adopt a very similar secondary structure.
Keywords: maturation/modification/RNA binding protein/RNA methyltransferase/three-dimensional structure
Introduction
One of the long-term goals of modern biology is to predict the function of a protein by analysing its amino acid sequence and, whenever possible, by inspection of its three-dimensional architecture (Burley, 2000). This can be achieved by a combination of structural genomics and bioinformatics approaches, followed by testing the predicted function in vivo and in vitro. With the completion of the sequencing projects for a growing number of organisms, it is now possible to carry out genome-wide searches for genes encoding putative proteins that share sequence motifs and thus potentially belong to the same family (Goffeau et al., 1996; Koonin et al., 1998). For instance, methyltransferases (MTases) that use S-adenosyl-l-methionine (AdoMet) as a cofactor exhibit conserved motifs that map onto the catalytic face of the common fold (Fauman et al., 1999). Recently, a structural analysis has revealed that the heat shock protein FtsJ/RrmJ of Escherichia coli exhibits the typical MTase fold (Bügl et al., 2000). It has been demonstrated that FtsJ/RrmJ actually catalyses the formation of Um2552, which is located within a loop of five nucleotides present on domain V of the peptidyl transferase centre of the 23S rRNA (Caldas et al., 2000a). In vitro, FtsJ/RrmJ is able to methylate the 23S rRNA when it is assembled into 50S ribosomal subunits, and it can also catalyse the formation of 2′-O-methylribose in E.coli tRNA, although the modified nucleotide(s) has not yet been mapped (Bügl et al., 2000).
In yeast, almost all types of RNA molecules contain 2′-O-methylnucleotides: tRNA, snRNA, small nucleolar RNA (snoRNA) and rRNA (Rozenski et al., 1999). Although transcription of these various RNAs takes place either in the nucleus (and the nucleolus) or in mitochondria, RNA maturation can be completed sequentially in every cellular compartment. For instance, several tRNA modifications occur after removal of the intron, probably after the tRNA has been transported to the cytoplasm (reviewed by Grosjean et al., 1997). Since the formation of methylnucleotides in eukaryotes takes place in different compartments, it could either be achieved by the same enzymes distributed in various compartments or catalysed by different enzymes located in the nucleus, the cytoplasm and the mitochondria.
Remarkably, the yeast genome contains three open reading frames (ORFs) that exhibit significant sequence similarity to E.coli FtsJ/RrmJ: YBR061c (hereafter referred to as TRM7; see below), YGL136c (MRM2) and YCL054w (SPB1). Spb1p is a putative MTase from the yeast nucleolus that is involved in 60S ribosomal subunit synthesis (Pintard et al., 2000). In a separate report, we demonstrated that Mrm2p is a mitochondrial site-specific 2′-O-ribose MTase that catalyses the formation of Um2791 on the peptidyl transferase centre of the 21S rRNA (Pintard et al., 2002). Here, we report the results of experimental and bioinformatic analysis of Trm7p, which suggest that it is a new cytoplasmic 2′-O-RNA MTase.
In yeast tRNAs, 2′-O-methylriboses have been found at positions 4, 18, 32, 34 and 44 (Grosjean et al., 1995; Sprinzl et al., 1998). Although certain modifications occur on the pre-tRNA in the nucleus, methylation at positions 18, 32, 34 and 44 appears to occur after the removal of the intron (reviewed by Grosjean et al., 1997). So far, Trm3p is the only putative yeast 2′-O-tRNA MTase that has been described. It is required for the site-specific formation of Gm at position 18 in the D-loop of intron-less tRNA (Cavaillé et al., 1999). Two of the remaining four 2′-O-methylnucleotides are located within the anticodon loop, at positions 32 and 34, in tRNALeu,Phe,Trp. Modification of position 34, the wobble nucleotide of the anticodon, has been reported to be important for translation efficiency and/or fidelity (Curran, 1998; Satoh et al., 2000). We report here that Trm7p is a yeast MTase that catalyses the formation of 2′-O-methylribose at positions 32 and 34 in the anticodon loop of various yeast tRNAs. We propose that the three proteins Trm7p, Spb1p and Mrm2p form a subclass of 2′-O-ribose MTases that have emerged from a single common ancestor and have co-evolved with their three different RNA substrates, located in the cytoplasm, the nucleolus and the mitochondria, respectively.
Results
Trm7p belongs to a family of putative MTases from Eukaryota that are structurally related to the 2′-O-rRNA MTase FtsJ/RrmJ from E.coli
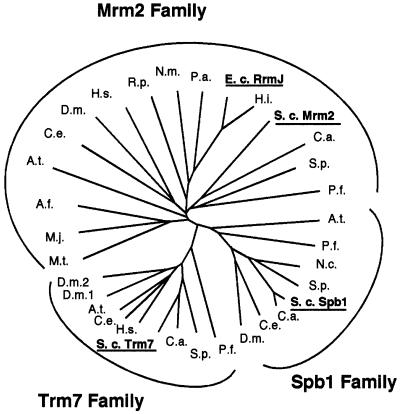
By searching the yeast genome, we identified three proteins exhibiting similarity to the bacterial 2′-O-rRNA MTase FtsJ/RrmJ (Caldas et al., 2000a,b): Spb1p (Pintard et al., 2000), Mrm2p (Pintard et al., 2002) and Ybr061c (referred to here as Trm7p; see below). We undertook the bioinformatic analysis to predict the structure of the three yeast proteins based on the data available for other MTases. The E.coli FtsJ/RrmJ sequence was used as a query in a PSI-BLAST search (Altschul et al., 1997) of the non-redundant amino acid sequence database at NCBI (http://www.ncbi.nlm.nih.gov/) with a stringent expectation value cut-off of 10–10. The search converged in the fifth iteration, yielding the alignment of highly conserved core segments. Sequences of 32 putative homologues of FtsJ/RrmJ were chosen and realigned to the PSI-BLAST core profile using Clustal_X (Thompson et al., 1997). A distance-based phylogeny was reconstructed according to the neighbour-joining method (Saitou and Nei, 1987), revealing three evolutionary lineages with representatives from yeast that were named the Trm7, Spb1 and Mrm2 subfamilies (Figure 1). Mrm2 groups together with the FtsJ/RrmJ family members from the Eubacteria and Archaea, suggesting that its ancestor has been brought into the eukaryotic cell via the mitochondrial endosymbiont. The topology of the tree suggests that a gene duplication event leading to Trm7 and Spb1 occurred in a common ancestor of the Eukaryota.
Fig. 1. Phylogenetic tree analysis reveals three putative orthologous lineages: the Trm7, Spb1 and Mrm2 families. The Mrm2 family is represented by Mrm2p from S.cerevisiae (P53123), FtsJ/RrmJ from E.coli (AAC76211) and other prokaryotic proteins [M.t., Methanobacterium thermoautotrophicum (G69103); M.j., Methanococcus jannaschii (Q58771); A.f., Archaeglobus fulgidus (O28228); R.p., Rickettsia prowazekii (Q9ZE00); N.m., Neisseria meningitidis (AAF41212); P.a., Pseudomonas aeruginosa (AAG08138); H.i., Haemophilus influenzae (P45162)] and eukaryotic proteins [A.t., Arabidopsis thaliana (CAB69851); C.e., Caenorhabditis elegans (O62251); D.m., Drosophila melanogaster (Q9VDT6); H.s., Homo sapiens (AAF22488); C.a., Candida albicans (unfinished sequence from the Stanford Genome Technology Center, SGTC, http://sequence-www.stanford.edu); S.p., Schizosaccharomyces pombe (P78860); and P.f., Plasmodium falciparum 3D7 (unfinished sequence from the P.falciparum Genome Project–PlasmoDB, http://www.plasmodb.org)]. The Spb1 family is represented by Spb1p from S.cerevisiae (CAA42391) and seven other members [A.t., A.thaliana (CAB69851); P.f., P.falciparum (unfinished sequence from the PlasmoDB); N.c., Neurospora crassa (CAB88626); S.p., S.pombe (T37754); C.a., C.albicans (unfinished sequence from the SGTC); C.e., C.elegans (AAF39868); and D.m., D.melanogaster (AAF48557)]. The Trm7 family is represented by Trm7p from S.cerevisiae (CAA85004) and eight other members [P.f., P.falciparum (unfinished sequence from the PlasmoDB); S.p., S.pombe (Z98980); C.a., C.albicans (unfinished sequence from the SGTC); H.s., H.sapiens (HSA005892); C.e., C.elegans (Q22031); A.t., A.thaliana (CAB69851); and D.m.1. and D.m.2., D.melanogaster (AAF55380 and AAF55857)].
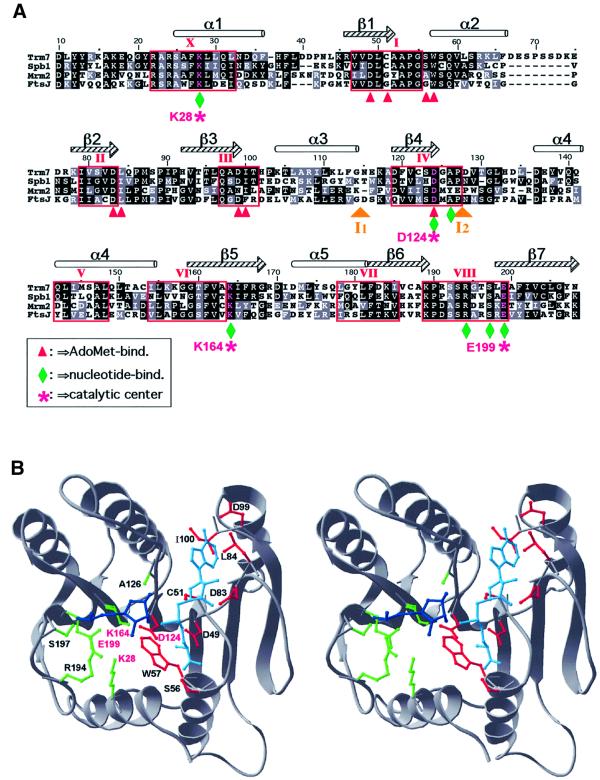
The three-dimensional structures of the three putative MTases from yeast were homology modelled using Modeller (Sali and Blundell, 1993) based on the refined sequence alignment with FtsJ/RrmJ (Figure 2). The stereochemistry and pseudoenergetic features of the models passed all tests implemented in ProsaII (Z-scores ≤ –7.7; Sippl, 1993). Figure 2A shows the key elements of the predicted AdoMet- and ribose-binding sites of Trm7p, including the predicted catalytic tetrad of two basic and two acidic side chains (K28, D124, K164 and E199, each marked by an asterisk) conserved in many 2′-O-ribose MTases exhibiting the common ‘MTase fold’ (Bujnicki and Rychlewski, 2001). All three putative MTases from yeast and their orthologues (including E.coli FtsJ/RrmJ) exhibit striking conservation of the predicted active site and its neighbourhood, suggesting that their target must be similar.
Fig. 2. Three-dimensional structure prediction for Trm7p. (A) Alignment of the AdoMet-binding domain of the three yeast proteins (Trm7p, Spb1p and Mrm2p) with the E.coli protein FtsJ/RrmJ. Identical and chemically equivalent residues are highlighted in black and grey, respectively. Secondary structural elements are indicated above the alignment (α-helices as cylinders and β-strands as arrows). Conserved motifs have been labelled according to the nomenclature proposed by Posfai et al. (1988). Predicted interactions with AdoMet and the methylated nucleotide are designated by red arrowheads and green diamonds, respectively. The predicted catalytic tetrad K–D–K–E is labelled with purple stars. Two insertions in Mrm2p [I1 (58 amino acids) and I2 (21 amino acids)] have been omitted for the sake of clarity. (B) The stereogram of the Trm7p model in cartoon representation. The AdoMet moiety is shown in cyan, and the ribose and the phosphate group of the nucleotide to be methylated are shown in blue. The predicted binding and catalytic residues are shown in the wireframe representation, with colour coding analogous to that in (A).
In the proposed model of interaction with an RNA substrate (Figure 2B), we docked the methylated ribose based on the superposition of the active sites of FtsJ/RrmJ and the yeast proteins with the coordinates of vaccinia mRNA cap-I 2′-O-MTase (Hodel et al., 1998). Although we cannot predict with confidence all specific interactions of the enzyme with the entire RNA molecule, we predict that the solvent-exposed side chains of residues S197 and R194 interact specifically with the phosphate group of the methylated nucleotide. Coordinates and additional structural representations are available (http://www.crbm.cnrs-mop.fr/∼lapeyre/extpages/structure.html).
Trm7p is able to bind tritiated AdoMet in vitro
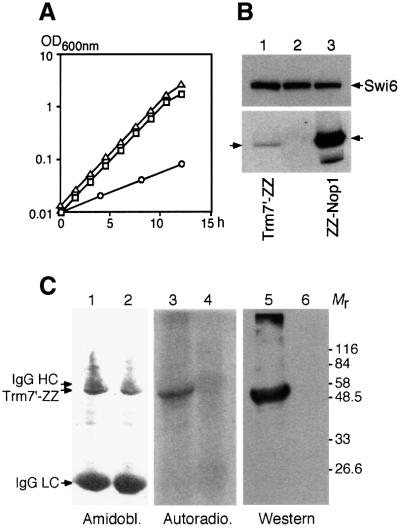
To challenge the model presented above, we tested the ability of a recombinant tagged Trm7p to bind AdoMet in vitro, as described previously for Spb1p (Pintard et al., 2000). Expression of Trm7′-ZZp (see Materials and methods) was able to complement the growth defect of a trm7Δ strain, thus demonstrating that the fusion protein was functional (Figure 3A). The level of expression of Trm7′-ZZp (Mr = 49 700) was compared with the level of the abundant ZZ-Nop1p by western blot analysis (Figure 3B). Signals obtained after serial dilutions indicate that Trm7p is ∼200 times less abundant in the cell than Nop1p, an observation that is in agreement with the codon adaptation index calculated for the two proteins (0.143 and 0.492, respectively; Sharp and Li, 1987).
Fig. 3. Trm7p binds tritiated AdoMet in vitro. (A) The doubling time was determined for various strains grown in YPD at 30°C. Open squares, wild-type strain (BMA64); open circles, trm7Δ strain (YBL4409); open triangles, trm7Δ transformed with a centromeric plasmid expressing the TRM7′-ZZ gene (YBL4494). (B) Relative expression of Trm7p. Cells expressing various tagged proteins were probed by western blotting using mouse IgG coupled to peroxidase (lower panel). Lane 1, Trm7′-ZZp (arrow on the left, strain YBL4494); lane 2, no tagged protein (BMA64); lane 3, ZZ-Nop1p (arrow on the right points to the major band, and a degradation product is detected below). Similar amounts of protein were loaded in each lane, as demonstrated using anti-Swi6p antibodies (upper panel). (C) Trm7′-ZZp was overexpressed from the GAL1-10 promoter (YBL4502), immunoprecipitated and incubated with 5 µCi of [3H]AdoMet before being UV crosslinked. Radiolabelled complexes were then denatured, separated by 10% SDS–PAGE and transferred to nitrocellulose. The membrane was first stained with amidoblack to localize the heavy and light chains of immunoglobulins used during the immunoprecipitation reaction (lanes 1 and 2). This membrane was then autoradiographed for 4 weeks at –70°C with an intensifying screen (lanes 3 and 4). Finally, the same membrane was analysed by western blotting to reveal Trm7′-ZZp (lanes 5 and 6). Lanes 1, 3 and 5, extracts prepared from the GAL::TRM7′-ZZ strain. Lanes 2, 4 and 6, extracts from an untagged strain as a control. Mr, molecular weight markers in thousands.
Affinity-purified overexpressed recombinant Trm7′- ZZp was clearly capable of binding tritiated AdoMet in vitro (Figure 3C, lane 3). This binding strictly depends on UV irradiation, as is also the case for Spb1p (Pintard, 2000), thus demonstrating that labelling of the proteins was not due to alkylation by free methyl groups that could arise after degradation of AdoMet. Large amounts of immunoglobulins present in the reaction were not crosslinked to AdoMet under these conditions (Figure 3C, lane 1). Similarly, no signal was detected when bovine serum albumin or recombinant nucleolar Gar1p was used as control (data not shown), thus demonstrating the specificity of the reaction. This result strongly supports the view that Trm7p is a new MTase from Saccharomyces cerevisiae.
Deletion of TRM7 impairs mRNA translation
Sequence similarity existing between Trm7p and Spb1p led us to investigate whether Trm7p could also be a nucleolar protein. However, immunofluorescence studies performed with three different tagged proteins gave signals that were indistinguishable from the background noise. Intense signals were obtained only when TRM7 was overexpressed from the GAL1-10 promoter, revealing the protein throughout the cytoplasm (data not shown). Unlike Spb1p, Trm7p does not contain a nuclear localization signal, and PSORT software (Nakai and Kanehisa, 1992) predicts a cytoplasmic location for Trm7p (score = 0.650). Although we cannot definitely ascertain the location of the protein, since it was detectable only when overexpressed in the cell, our observations, combined with the PSORT prediction, support the view that Trm7 is a cytoplasmic protein.
To understand better the role of Trm7p, we further analysed the phenotype of the trm7Δ strain. Incorporation of labelled methionine revealed that protein synthesis is reduced to 30% in the deleted strain, compared with an isogenic wild-type strain. This result was confirmed by the low level of polysomes detected in the mutant, compared with a wild-type strain, when extracts were fractionated on sucrose gradients (Figure 4A). We also noticed that the trm7Δ strain is highly sensitive to paromomycin, an antibiotic of the aminoglycoside family that impairs translation by increasing codon misreading in Prokaryota and Eukaryota (Figure 4B) (Chernoff et al., 1994). Taken together, these results suggests that Trm7p is a cytoplasmic protein that plays a role in mRNA translation.
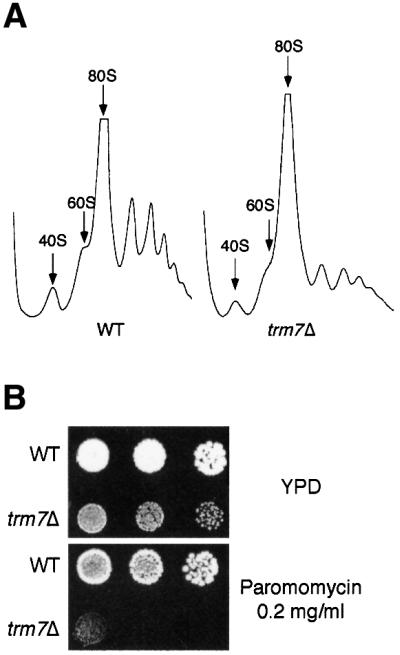
Fig. 4. Translation is impaired in a trm7Δ strain. (A) Polysome profile analysis. Cellular extracts were fractionated onto 15–50% linear sucrose gradients. A continuous record of the absorbance at 254 nm for each gradient is presented, with the top of the gradient on the left. The arrows indicate the peaks for the 40S, 60S and 80S subunits. Extracts were prepared from the strains as indicated: WT, wild type (BMA64); trm7Δ (YBL4409). (B) The trm7Δ strain is sensitive to the translation inhibitor paromomycin. The wild-type strain (WT, BMA64) and the trm7Δ strain (YBL4409) were grown in YPD until mid-log phase; then 5 µl of 5-fold serial dilutions were spotted onto YPD plates containing (or not) 0.2 mg/ml paromomycin as indicated, and strains were grown for 3 days at 30°C.
Trm7p is required for tRNA 2′-O-ribose methylation in vivo
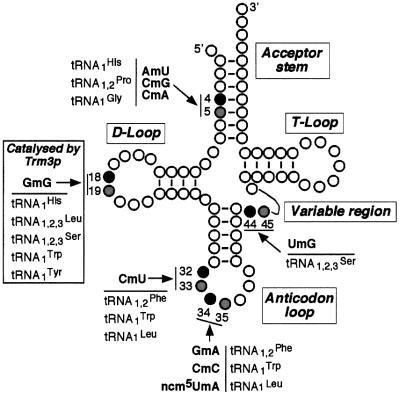
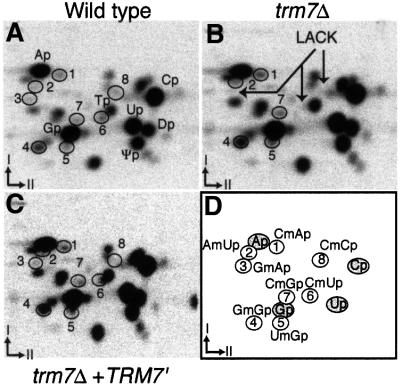
The close structural similarity existing between Trm7p and the E.coli rRNA MTase FtsJ/RrmJ suggests that Trm7p could be a new yeast 2′-O-RNA MTase. In vitro, FtsJ/RrmJ is also able to methylate tRNA (Bügl et al., 2000). This result, taken together with our observation that Trm7p is likely to be a cytoplasmic MTase that plays a role in mRNA translation, prompted us to test whether Trm7p was involved in 2′-O-ribose methylation of a nucleotide within a loop of yeast tRNA. Positions 32 and 34 in the anticodon loop represented good candidates that were tested further (Figure 5). Interestingly, when position 32 of a given tRNA is 2′-O-methylated, the wobble position 34 of the same tRNA is also 2′-O-methylated. To identify the modified nucleotides, we used the property that RNase T2 cleaves all phosphodiester linkages between nucleotides, except those that are 2′-O-methylated. Since the 3′-neighbouring nucleotides of each type of 2′-O-methyl derivative in the different isoacceptor tRNA are different, analysis of the dinucleotides produced by RNase T2 digestion reveals the identity of the modified nucleotide. The result of RNase T2 digestion of total tRNA extracted from various strains grown in the presence of inorganic [32P]orthophosphate, followed by two-dimensional thin-layer chromatography (TLC), is presented in Figure 6A–C. As expected, each tRNA digest revealed a complex pattern of radiolabelled spots, of which the four canonical 3′-monophosphate nucleosides Ap, Cp, Gp and Up are the most abundant. Spots corresponding to ribothymidine monophosphate (Tp), dihydrouridine monophosphate (Dp) and pseudouridine monophosphate (Ψp), which are present in all naturally occurring tRNA in yeast, are also clearly seen. Other modified nucleotides that are present only in certain tRNA yield spots that are less intense but still easily detected on the autoradiogram. Three spots (3, 6 and 8) of the wild-type strain (Figure 6A) are missing in the trm7Δ strain (indicated by arrows in Figure 6B). Spot 3 corresponds to GmAp (positions 34–35 in tRNAPhe), spot 6 corresponds to CmUp (positions 32–33 in tRNAPhe,Trp,Leu) and spot 8 corresponds to CmCp (positions 34–35 in tRNATrp). Two of these three radioactive spots are clearly visible in the trm7Δ strain transformed with the centromeric plasmid harbouring a synthetic TRM7′ gene (Figure 6C). Of note is the presence in all the chromatograms of the radioactive spots corresponding to CmAp (spot 1, from positions 4–5 in major tRNAGly), AmUp (spot 2, from positions 4–5 in tRNAHis), GmGp (spot 4, from positions 18–19 in several yeast tRNAs), UmGp (spot 5, from positions 44–45 in all tRNASer) and CmGp (spot 7, from positions 4–5 in major tRNAPro). The spot corresponding to the dinucleotide ncm5UmAp (positions 34–35 in minor tRNALeu) was not identified in this experiment, probably because it was in amounts too low to be detected and/or it co-migrates with another radioactive spot on the TLC plates.
Fig. 5. Position of the five 2′-O-methylnucleotides in the yeast cytoplasmic tRNA and its major domains. Positions in black indicate the site of methylation in tRNA containing 2′-O-methylnucleotide, and positions in grey correspond to its 3′-neighbouring nucleotide. The indicated dinucleotides correspond to those obtained after complete digestion of the tRNA by RNase T2.
Fig. 6. Position 32 of several tRNAs and position 34 of tRNAPhe and tRNATrp are not 2′-O-methylated in a trm7Δ strain. Autoradiograms of selected two-dimensional TLC of modified mono- and dinucleotides after RNase T2 digestion of various 32P-labelled tRNA recovered from cells that were grown in the presence of [32P]orthophosphate. Hydrolysate of total tRNA from (A) wild-type strain (BMA64α), (B) trm7Δ strain (YBL4409) and (C) trm7Δ strain transformed with a plasmid harbouring the TRM7′ gene (YBL4363) have been analysed with the N/R chromatographic solvent system. (D) A reference map indicating the location of the nucleotides of interest. Arrows point to the spots that disappear in the disrupted strain.
Purification of wild-type and mutant recombinant Trm7 proteins
As another means to demonstrate the catalytic activity of Trm7p, we mutated the AdoMet-binding domain of Trm7p to abolish cofactor binding by this enzyme. Based on the homology model of Trm7p and analysis of the FtsJ/RrmJ structure, D49 of Trm7p has been predicted to make an essential water-mediated interaction with AdoMet, the lack of which would interfere with cofactor binding. Therefore, a mutation was introduced into the TRM7 gene to change this aspartic residue into an alanine (→Trm7[D49A]). In vivo, a plasmid expressing the recombinant Trm7 wild-type protein tagged with six histidines (Trm7-H6p) was able to complement the growth defect of a trm7Δ strain, whereas expression of the mutant Trm7[D49A]-H6p in a trm7Δ strain did not restore wild-type growth (data not shown). Then, to assert unambiguously that the MTase activity was borne by Trm7p itself, the two recombinant and mutant proteins were affinity purified (Figure 7) and their tRNA MTase activity was tested in vitro. Few minor contaminants were detected by Coomassie Blue staining, the most abundant of which was also present in a fraction prepared from a trm7Δ strain expressing no recombinant His-tagged protein (Figure 7, lane 7).
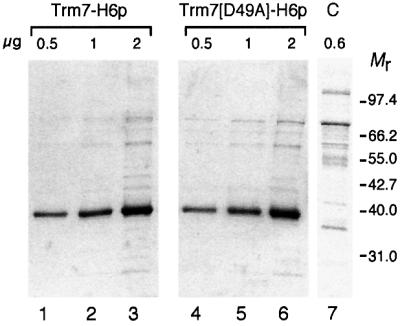
Fig. 7. Purification of the His-tagged wild-type and mutant recombinant Trm7 proteins. Affinity-purified wild-type and D49A mutant Trm7p were analysed by 10% SDS–PAGE. To assess the purity of the proteins, various amounts were loaded (∼0.5, 1 and 2 µg, from left to right, as indicated). Lanes 1–3, wild-type Trm7-H6p; lanes 4–6, D49A mutant Trm7-H6p; lane 7, control (C) from an untagged strain. Mr, molecular weight markers in thousands.
Trm7p is required for tRNA 2′-O-ribose methylation in vitro
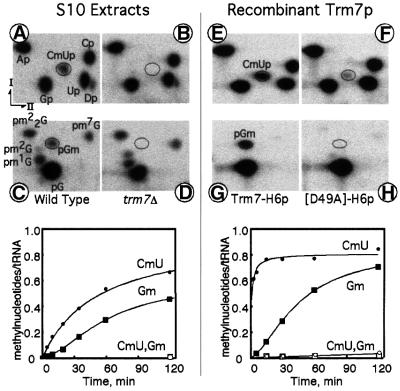
To test in vitro for AdoMet-dependent MTase activity at positions 32 and 34, a synthetic tRNA substrate was prepared. Intron-less yeast tRNAPhe was transcribed in vitro using [α-32P]GTP as a radioactive precursor. This permits, using the same transcript, the detection of the presence of either CmU32p after RNase T2 digestion or 32pGm after nuclease P1 digestion. When this substrate was incubated with a S10 cellular extract prepared from a wild-type strain, enzymatic formation of both Cm32 and Gm34 was detected, although a small but reproducible lag phase in the formation of Gm34 was visible (Figure 8, left part). Formation of Cm32 and Gm34 was completely abolished in an S10 extract from a trm7Δ yeast strain (Figure 8B and D). Similar results were obtained with an intron-less yeast tRNATrp transcript (data not shown). Several other radioactive spots were generated by the S10 extract from the wild-type strain, which were also present in the trm7Δ strain extract: Dp from position 17, in addition to the expected CmUp after T2 RNase digestion; and pm2G, pm22G, pm1G and pm7G from positions 10, 26, 37 and 46, respectively, in addition to pGm after nuclease P1 digestion (Figure 8A and C). This result demonstrates that only the formation of Cm32 and Gm34 is abolished in the trm7Δ strain. Moreover, enzymatic formation of Cm32 and Gm34 was restored when the S10 extract was prepared from a trm7Δ strain transformed with a centromeric plasmid expressing the TRM7 gene (data not shown).
Fig. 8. Affinity-purified Trm7p is able to methylate positions 32 and 34 of tRNAPhe in vitro. Upper part: autoradiograms of selected two- dimensional TLC of modified mono- and dinucleotides after RNase T2 (A, B, E and F) or nuclease P1 (C, D, G and H) digestion of synthetic intron-less [α-32P]GTP-labelled tRNAPhe initially incubated either with S10 cell extracts or with purified recombinant proteins. Left four panels: tRNA incubated for 2 h at 30°C with 2 mg/ml protein from an S10 cell extract prepared from a wild-type strain (BMA64α) (A and C) or from a trm7Δ strain (YBL4409) (B and D). Right four panels: tRNA incubated as above at 30°C with 10 µg/ml purified Trm7-H6p (E and G) or with D49A mutant Trm7-H6p (F and H). Lower part: time-course formation of Cm32 (detected as CmU) and Gm34 on synthetic intron-less [α-32P]GTP-labelled tRNAPhe incubated at 30°C either with S10 extracts (left panel) or with recombinant proteins (right panel). Molar ratios of methylated nucleotides versus substrate tRNA plotted as a function of incubation time in minutes (methylnucleotides/tRNA). Left panel: S10 extracts prepared from a wild-type yeast (filled circles, CmU; filled squares, Gm) or from a trm7Δ mutant strain that does not give rise to any detectable modification (open circles and open squares, respectively). Right panel: purified recombinant wild-type Trm7-H6p (filled circles, CmU; filled squares, Gm) or with purified mutant Trm7[D49A]-H6p (open circles and open squares, respectively).
The same synthetic tRNA substrate was then incubated with the recombinant proteins, either the wild-type or the D49A mutant Trm7p, and analysed as above for the presence of modified nucleotides. The wild-type protein was clearly capable of driving the formation of both Cm32 and Gm34, although the rate of Cm32 formation was ∼10 times faster than the rate of Gm34 formation in intron-less tRNAPhe, and a slight but reproducible lag phase exists for the enzymatic formation of Gm34 (Figure 8E and G, and see the corresponding kinetics below). It is noteworthy that no other spots corresponding to other modified mono- or dinucleotides were detected when using the recombinant protein, thus demonstrating that the affinity-purified Trm7p was not contaminated by other tRNA modification enzymes detectable in our assay. Moreover, mutation D49A abolishes almost completely the formation of Cm32, and no activity was detected for G34 (Figure 8F and H, and see the corresponding kinetics below).
From these results obtained in vitro with cellular extracts and recombinant proteins, together with those obtained in vivo, after analysing naturally occurring tRNA from wild-type and trm7Δ mutant yeast strains (Figure 6), we conclude that TRM7 codes for a MTase that catalyses the formation of 2′-O-methylnucleotides at positions 32 and 34 of the yeast tRNAPhe, tRNATrp and, possibly, tRNALeu.
Discussion
tRNAs contain the highest number of modified nucleotides of all cellular RNAs (Björk, 1995). Several enzymes responsible for the post-transcriptional methylation of tRNAs have now been characterized and named Trm (for tRNA methyltransferase). In S.cerevisiae, Trm1p and Trm2p are base MTases that catalyse the site-specific formation of m22G26 and m5U54, respectively (Ellis et al., 1986; Nordlund et al., 2000); Trm3p is likely to be the site-specific Gm18 2′-O-ribose MTase (Cavaillé et al., 1999); Trm4p is a multisite-specific m5C methylase (Motorin and Grosjean, 1999); Trm5p catalyses the formation of m1G37 and m1I37 (Björk et al., 2001); and Trm6 (Gcd10p/Gcd14p) is a two-subunit complex required for the formation of m1A58 (Anderson et al., 2000).
In this study, we report the characterization of a new multisite-specific enzyme from yeast that catalyses the formation of 2′-O-methylribose in the tRNA anticodon loop. According to the current nomenclature for tRNA modification enzymes, we propose the name Trm7p for this new enzyme. This proposition is based on the facts that: (i) Trm7p is an AdoMet-binding protein likely to be located in the cytoplasm; (ii) deletion of the TRM7 gene leads to a low rate of protein synthesis and sensitivity to the translation inhibitor paromomycin; (iii) T2 RNase hydrolysates of total tRNA extracted from the trm7Δ strain reveal a lack of dinucleotide CmU (corresponding to positions 32 and 33 of tRNAPhe, tRNATrp and tRNALeu), as well as GmA and CmC (corresponding to positions 34 and 35 of tRNAPhe and tRNATrp, respectively); (iv) extracts prepared from a trm7Δ strain can no longer methylate positions 32 and 34 in tRNAPhe and tRNATrp in vitro; (v) the wild-type TRM7 gene expressed from a centromeric vector in a trm7Δ strain restores the enzymatic formation of Cm32 and Gm34 both in vivo and in vitro; (vi) affinity-purified recombinant Trm7p catalyses in vitro the formation of Cm32 and Gm34 in tRNAPhe; and (vii) a single point mutation (D49A) within the AdoMet-binding motif of Trm7p abolishes almost completely the tRNA MTase activity in vitro and, in vivo, expression of this construction in a trm7Δ strain is unable to restore wild-type growth. Taken together, these results demonstrate that Trm7p is the RNA MTase that catalyses the formation of 2′-O-methylribose at positions 32 and 34 of the anticodon loop in several yeast tRNAs.
Trm7p belongs to a group of three yeast proteins (Trm7p, Spb1p and Mrm2p) that exhibit sequence similarity to the E.coli rRNA MTase FtsJ/RrmJ. Bioinformatic analysis allowed the delineation of solvent-exposed features that are common to all these proteins and the categorization of the conserved residues into three different groups (Figure 2A and B). The first group that is conserved in most MTases is involved in the recognition of the AdoMet cofactor. This prediction was experimentally confirmed for position 49, since the mutation D49A was sufficient to abolish almost completely the MTase activity of Trm7p. The second group of residues (K28, D124, K164 and E199) corresponds to the predicted active site conserved between different RNA 2′-O-ribose MTases (Bujnicki and Rychlewski, 2001). Finally, we noticed a group of 14 residues that are perfectly conserved between E.coli FtsJ/RrmJ and the three yeast proteins (Figure 2A), suggesting that the four proteins could share other characteristics in addition to being specific for ribose in RNA.
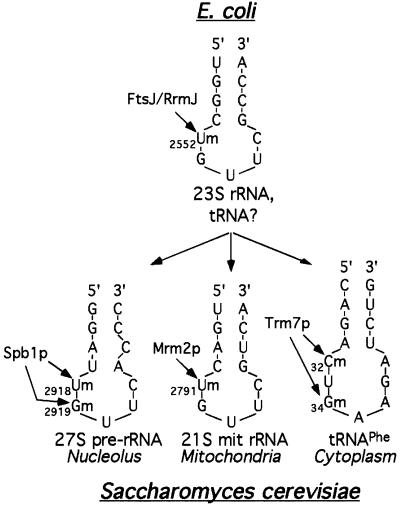
In prokaryotes, a unique enzyme, FtsJ/RrmJ, methylates the peptidyl transferase centre of the large rRNA and possibly also certain tRNAs (Bügl et al., 2000). Its rRNA target has a peculiar structure that is composed of a hairpin and a loop of five nucleotides, with the modified nucleotide being located at the 5′ end of the loop (Figure 9, top). Interestingly, in yeast this structure is remarkably conserved in the mitochondrial 21S rRNA, in the 25S rRNA and, to a lesser extent, in the tRNA anticodon loop (Figure 9, bottom). We have demonstrated recently that Mrm2p is a mitochondrial protein that catalyses the formation of Um2791 of the peptidyl transferase centre of the 21S rRNA, at a position equivalent to U2552 of the E.coli 23S rRNA (Pintard et al., 2002). Spb1p is a nucleolar protein required for 25S rRNA synthesis. Owing to its structural similarity with FtsJ/RrmJ, we hypothesize that Spb1p is responsible for the methylation of U2918 (and possibly G2919), which is equivalent to U2552 of the E.coli 23S rRNA. When compared with the three rRNA loops, the tRNA anticodon loop seems to be slightly different, with a longer loop of seven nucleotides instead of five. However, the target nucleotide to be modified (C32) is also located at the 5′ end of the loop, and the seven-nucleotide-long anticodon loop can be reduced to five nucleotides, due to a base-pairing between positions 32 and 38 (Auffinger and Westhof, 2001), thus rendering the tRNA structure even more similar to the rRNA ones. Therefore, we propose that the three yeast proteins Trm7p, Spb1p and Mrm2p have evolved from a common ancestor to function in three different cell compartments, i.e. the cytoplasm, the nucleolus and the mitochondria, where they each recognize a similar hairpin/loop RNA structure (Figure 9). Interestingly, the enzymes of the FtsJ/RrmJ family exhibit no significant similarity to other known yeast 2′-O-ribose MTases, such as Trm3p or Mrm1p/Pet56p. These observations suggest the existence of distinct subfamilies of enzymes that originated at an early point in evolution and then co-evolved with their target RNA, independently of the other classes of 2′-O-ribose MTases.
Fig. 9. Schematic representation of the secondary structure of the target sites for each of the three enzymes that belong to the 2′-O-RNA MTase family. Only the 2′-O-ribose methylations discussed in this report are represented. Top: location of U2552 in the hairpin/loop of domain V of the peptidyl transferase centre of the E.coli 23S rRNA. In addition, FtsJ/RrmJ can also methylate tRNA in vitro; however, the position of the methyl-nucleotides has not yet been determined (Bügl et al., 2000). Bottom, left: location of Um2918 and Gm2919 in the hairpin/loop of the peptidyl transferase centre of the nuclear 27S pre-rRNA. Bottom, middle: location of Um2791 in the hairpin/loop of the 21S mitochondrial rRNA. Bottom, right: location of C32 and G34 in the anticodon loop of tRNAPhe. The modified nucleotides are indicated by arrows, with their known (FtsJ/RrmJ, Trm7p, Mrm2p) or putative (Spb1p) modification enzymes.
Data presented here, together with data published previously, indicate that methylation at positions 32 and 34 exhibits certain similarities but also some significant differences. Amongst the similarities, in yeast the two modifications are catalysed by the same enzyme, Trm7p. Also, both modifications occur at a late stage, after the removal of the intron (reviewed by Grosjean et al., 1997), and they do not depend on other tRNA modifications, since affinity-purified Trm7p modifies efficiently a synthetic intron-less tRNAPhe in vitro. As we detected Trm7p in the cytoplasm, it is likely that methylation of the anticodon loop takes place in the cytoplasm on spliced tRNA. Among the differences in the formation of Cm32 and Gm34, first we observed a lag in the formation of Gm34, as compared with Cm32, on tRNAPhe in vitro (Figure 8; see also Jiang et al., 1997); secondly, point mutations that affect the three-dimensional structure of the tRNAPhe molecule have little effect on the formation of Cm32 but almost completely prevent the formation of Gm34 (H.G. and F.L., unpublished).
It has been shown that the hypomodified anticodon loop is conformationally more flexible than the modified one present in the mature tRNA (Agris, 1996; Sundaram et al., 2000). We attempted to dock the tRNAPhe structure (Shi and Moore, 2000) to the Trm7 model using the VP39 MTase–mRNA complex (Hodel et al., 1998) as a guide, by superposition of the corresponding protein structures and the C32 and G34 ribose rings onto the 2′-O-methylated ribose of mRNA. Docking of C32, whose 2′-O-methylated group is buried in the tRNAPhe interior, resulted in extensive steric clashes between tRNAPhe and the Trm7 model (data not shown). However, docking of G34 revealed striking complementarity between the positively charged groove on the Trm7p surface and the tRNAPhe molecule (http://www.crbm.cnrs-mop.fr/∼lapeyre/ext pages/structure.html). This suggests that G34 fits into the active site of Trm7p if tRNAPhe assumes the crystal-like structure, while the conformation of the anticodon loop needs to be altered in order to present C32 to the Trm7p active site. A nucleotide flipping mechanism for tRNA modification has been demonstrated recently in the case of E.coli TruB, which catalyses the formation of pseudouridine-55 in the T-loop of tRNA (Hoang and Ferre-D’Amare, 2001). The flipping mechanism seems quite attractive for C32 methylation by the Trm7p MTase, since it is common in the reaction catalysed by DNA MTases, which are structurally and evolutionarily related to Trm7p (reviewed by Cheng and Roberts, 2001). Based on the aforementioned experimental results and the docking model, we hypothesize that C32 is methylated in the conformationally flexible, hypomodified anticodon hairpin, where its 2′-hydroxyl can be presented to the active site of the enzyme, whereas formation of Gm34 occurs preferably in a rigid, hypermodified loop. It remains to be determined whether the potential of G34 to be methylated is increased only by Cm32 or also by modified nucleotides at other positions of the anticodon loop (m1G/Wye37, Psi-39 and m5C40; Grosjean et al., 1990; Jiang et al., 1997), as suggested by the results obtained in vitro with affinity-purified Trm7p.
Materials and methods
Microbiological methods and recombinant DNA work
The strains used in this study (Table I) were constructed from strain BMA64 (Baudin et al., 1993). Disruption of the TRM7 gene was performed by using a PCR-based strategy with oligonucleotides OBL57 (AAAACAACAGTAGATGACTACAAGGCAAGCTTAAAAAGAAAGTATTCAAGCTGATGCGGTATTTTCTCCT)/OBL58 (ATAATGTAGTGACTTATTTAATTGTTTGATTAGAGGTAGTAATATTGCAACGGGTGTTGGCGGGTGTC) to amplify a TRP1 cassette (Baudin et al., 1993). A synthetic TRM7 gene bearing two unique restriction sites at the initiation and stop codons was constructed as follows. The 5′ UTR, the ORF and 3′ UTR of TRM7 were PCR amplified with oligonucleotides OBL59 (GCTGCAGAAAGACACCAAACCTAAGTA)/OBL60 (GGGGATCCGGCCGACCATCTTGAATACTTTCTT), OBL63 (CGGATCCTCTAGACCCGGGTTGATTGCAATATTACTACCTC)/OBL64 (CGGTACCGCAGATGTTTTAAGAGACGAG) and OBL61 (CCCCGGCCGTGGTAAGAGCAGCAAAGAT)/OBL62 (GCTCTAGAACTGATCTAGTGAGTTTCC), respectively, using yeast genomic DNA as a template. The three PCR products were gel purified, digested with PstI and EagI (for the 5′ UTR), EagI and XbaI (for the ORF), and XbaI and KpnI (for the 3′ UTR), and inserted between the PstI and KpnI sites of Ycplac22 (Sikorski and Hieter, 1989) to give plasmid pBL579, which encodes a slightly modified Trm7p (denoted Trm7′p), with three additional residues at the N-terminus of the protein (VGR) and four at the C-terminus (LDPG). Tagging Trm7′p with two IgG-binding domains of Staphylococcus aureus protein A (Trm7′-ZZp) was performed by PCR amplification of a ZZ fragment with oligonucleotides OBL144 (TCCCCCGGGGAAGCTGGAGCTCAAAACC)/OBL145 (AGGCCCGGGTTGGTTGACTTCCCCG) and plasmid pBS1173 as a template (Puig et al., 1998). pBL598 was obtained by ligating this fragment digested with SmaI into the same site of pBL579. A DNA fragment containing the GAL1-10 promoter was obtained by PCR using oligonucleotides OBL147 (GCTGCAGCGACGGCCAGTGAATTCG)/OBL148 (CCCCGGCCGACCATGGATCCGGGGTTTTTTCTCC) and pDK376 as a template (Kressler et al., 1999). This fragment was digested with PstI and EagI, and used to replace the TRM7 promoter in pBL598 to yield pBL601. Another recombinant gene encoding a Trm7p derivative tagged with six histidines at the C-terminus (Trm7-H6p) was constructed as follows. Oligonucleotides OBL157 (AAGGATCCATGGGTAAGAGCAGCAAAGATAAAAG)/OBL158 (AAGAATTCTTTAGTGGTGGTGGTGGTGGTGAACTGATCTAGTGAGTTTCCCGCTCC) were used to amplify a BamHI–EcoRI cassette, which was then introduced into the same sites of YeDP60, a vector shown to achieve a high level expression of cytochrome P-450 under the control of the GAL1-10 promoter (Urban et al., 1990) and which contains the two selection markers URA3 and ADE2. The resulting plasmid was called pBL606 and introduced into the trm7Δ strain YBL4409 to yield strain YBL4537. In order to disrupt the interaction of Trm7p with AdoMet, a mutation was introduced using an ExSite PCR-based kit (Stratagene) to change an aspartate residue into an alanine residue (position 49 of the wild-type protein) with oligonucleotides OBL159 (CTTAAAAAGAGTTGTAGCTTTGTGTGCAGCACCAGG)/OBL160 (CCTGGTGCTGCACACAAAGCTACAACTCTTTTTAAG). The resulting plasmid, called pBL607, was then transformed into strain YBL4409 to give strain YBL4538.
Table I. Strains used in this study.
| Strain | Genotypea | Plasmid | Main property | Origin |
|---|---|---|---|---|
| BMA64 | MATa/α | wild type | Baudin et al. (1993) | |
| YBL4409 | MATα trm7Δ::TRP1 | trm7Δ | this work | |
| YBL4363 | MATα trm7Δ::TRP1 | pBL579 | +TRM7′ | this work |
| YBL4494 | MATα trm7Δ::TRP1 | pBL598 | +TRM7′-ZZ | this work |
| YBL4502 | MATa/α trm7Δ::TRP1 | pBL601 | +GAL::TRM7′-ZZ | this work |
| YBL4518 | MATα trm7Δ::TRP1 | YeDP60 | trm7Δ | this work |
| YBL4537 | MATα trm7Δ::TRP1 | pBL606 | +GAL::TRM7-H6 | this work |
| YBL4538 | MATα trm7Δ::TRP1 | pBL607 | +GAL::TRM7D49A-H6 | this work |
| ZZ-Nop1p | MATα nop1Δ::HIS3 | pZZ-Nop1p | Gautier et al. (1997) |
aAll strains were ade2, his3, leu2, trp1-Δ, ura3, can1-100 unless otherwise specified.
Protein analysis
Protein extraction, SDS–PAGE and western blotting were carried out as described previously (Pintard et al., 2000). Antibodies were used at the following dilutions: anti-Swi6p, 1/100 000; secondary antibodies coupled to horseradish peroxidase (Sigma), 1/5000 (anti-mouse) or 1/10 000 (anti-rabbit). Immunoprecipitation of Trm7′-ZZp was performed using 5 mg of native protein extract prepared from YBL4502 and 20 µl of IgG–Sepharose beads (Pharmacia). Recombinant Trm7-H6p and Trm7[D49A]-H6p were affinity purified using Ni-NTA–Superflow beads (Qiagen). Eluted proteins were dialysed against 100 mM Tris–HCl pH 8, 100 mM Na acetate, 50% glycerol and 1 mM DTT. A 1 l cell culture yields ∼0.5 mg of purified protein using 150 µl of Superflow beads. Polysomes and yeast S10 extract from strain BMA64α were prepared as described previously (Jiang et al., 1997; Pintard et al., 2000).
32P-labelling of tRNA and analysis of modified nucleotides
Cells (0.25 OD600 nm) were labelled with 300 µCi of [32P]orthophosphate (Amersham, UK) for 38 h at 30°C. Total [32P]tRNA was phenol extracted and purified by gel electrophoresis as described previously (Grosjean et al., 1990). In vitro synthesis of [32P]tRNA transcripts was as described previously (Jiang et al., 1997). [32P]tRNA was digested into 3′-phosphate nucleosides by RNase T2 (Sigma). Similarly, synthetic tRNA transcripts radiolabelled with [α-32P]GMP and modified in vitro as described previously (Jiang et al., 1997) with S10 cellular extracts or recombinant purified proteins were digested with RNase T2 or to 5′-phosphate nucleosides with nuclease P1 (Sigma). Hydrolysates were analysed by TLC with solvent system N/R as described previously (Jiang et al., 1997).
Acknowledgments
Acknowledgements
We thank O.Uhlenbeck for the p67YF0 plasmid. This work was supported by the CNRS and by grants from the Ligue contre le Cancer, the Fondation pour la Recherche Médicale (to B.L.) and the Association pour la Recherche sur le Cancer (ARC5914 to B.L. and ARC5297 to H.G.). L.P. and F.L. were supported by fellowships from the MENESR and by the Association pour la Recherche sur le Cancer. J.M.B. was supported by the Polish State Committee for Scientific Research Grant 8T11F01019 and by BioInfoBank.
References
- Agris P.F. (1996) The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol., 53, 79–129. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J., Phan,L. and Hinnebusch,A.G. (2000) The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P. and Westhof,E. (2001) An extended structural signature for the tRNA anticodon loop. RNA, 7, 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos,O., Denouel,A., Lacroute,F. and Cullin,C. (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk G.R. (1995) Biosynthesis and function of modified nucleosides. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis and Function. American Society of Microbiology Press, Washington, DC, pp. 165–206.
- Björk G.R., Jacobsson,K., Nilsson,K., Johansson,M.J., Byström,A.S. and Persson,O.P. (2001) A primordial tRNA modification required for the evolution of life? EMBO J., 20, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bügl H., Fauman,E.B., Staker,B.L., Zheng,F., Kushner,S.R., Saper,M.A., Bardwell,J.C. and Jakob,U. (2000) RNA methylation under heat shock control. Mol. Cell, 6, 349–360. [DOI] [PubMed] [Google Scholar]
- Bujnicki J.M. and Rychlewski,L. (2001) Reassignment of specificities of two cap methyltransferase domains in the reovirus λ2 protein. Genome Biol., 2, RESEARCH 0038.1–0038.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K. (2000) An overview of structural genomics. Nature Struct. Biol. Suppl., 7, 932–934. [DOI] [PubMed] [Google Scholar]
- Caldas T., Binet,E., Bouloc,P., Costa,A., Desgres,J. and Richarme,G. (2000a) The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23S ribosomal RNA methyltransferase. J. Biol. Chem., 275, 16414–16419. [DOI] [PubMed] [Google Scholar]
- Caldas T., Binet,E., Bouloc,P. and Richarme,G. (2000b) Translational defects of Escherichia coli mutants deficient in the Um2552 23S ribosomal RNA methyltransferase RrmJ/FTS. Biochem. Biophys. Res. Commun., 271, 714–718. [DOI] [PubMed] [Google Scholar]
- Cavaillé J., Chetouani,F. and Bachellerie,J.P. (1999) The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-O-ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA, 5, 66–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X. and Roberts,R.J. (2001) AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res., 29, 3784–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y.O., Vincent,A. and Liebman,S.W. (1994) Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J., 13, 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J.F. (1998) Modified nucleosides in translation. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. American Society for Microbiology, Washington, DC, pp. 493–516.
- Ellis S.R., Morales,M.J., Li,J.M., Hopper,A.K. and Martin,N.C. (1986) Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J. Biol. Chem., 261, 9703–9709. [PubMed] [Google Scholar]
- Fauman E.B., Blumenthal,R.M. and Cheng,X. (1999) Structure and function of AdoMet-dependent methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific, Singapore, pp. 1–38.
- Gautier T., Bergès,T., Tollervey,D. and Hurt,E. (1997) Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol., 17, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A. et al. (1996) Life with 6000 genes. Science, 274, 546, 563–547. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Droogmans,L., Giegé,R. and Uhlenbeck,O.C. (1990) Guanosine modifications in runoff transcripts of synthetic transfer RNAPhe genes microinjected into Xenopus oocytes. Biochim. Biophys. Acta, 1050, 267–273. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Sprinzl,M. and Steinberg,S. (1995) Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie, 77, 139–141. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Szweykowska-Kulinska,Z., Motorin,Y., Fasiolo,F. and Simos,G. (1997) Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: a review. Biochimie, 79, 293–302. [DOI] [PubMed] [Google Scholar]
- Hoang C. and Ferre-D’Amare,A.R. (2001) Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell, 107, 929–939. [DOI] [PubMed] [Google Scholar]
- Hodel A.E., Gershon,P.D. and Quiocho,F.A. (1998) Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol. Cell, 1, 443–447. [DOI] [PubMed] [Google Scholar]
- Jiang H.Q., Motorin,Y., Jin,Y.X. and Grosjean,H. (1997) Pleiotropic effects of intron removal on base modification pattern of yeast tRNAPhe: an in vitro study. Nucleic Acids Res., 25, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Tatusov,R.L. and Galperin,M.Y. (1998) Beyond complete genomes: from sequence to structure and function. Curr. Opin. Struct. Biol., 8, 355–363. [DOI] [PubMed] [Google Scholar]
- Kressler D., Rojo,M., Linder,P. and de la Cruz,J. (1999) Spb1p is a putative methyltransferase required for 60S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Nucleic Acids Res., 27, 4598–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y. and Grosjean,H. (1999) Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA, 5, 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K. and Kanehisa,M. (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics, 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund M.E., Johansson,J.O., von Pawel-Rammingen,U. and Bystrom,A.S. (2000) Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA, 6, 844–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L. (2000) Spb1p is a putative methylase required for mRNA maturation. PhD Thesis. University of Montpellier I, Medical School, Montpellier, France.
- Pintard L., Kressler,D. and Lapeyre,B. (2000) Spb1p is a yeast nucleolar protein associated with Nop1p and Nop58p that is able to bind S-adenosyl-l-methionine in vitro. Mol. Cell. Biol., 20, 1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L., Bujnicki,J., Lapeyre,B. and Bonnerot,C. (2002) MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J., 21, 1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai J., Bhagwat,A.S. and Roberts,R.J. (1988) Sequence motifs specific for cytosine methyltransferases. Gene, 74, 261–265. [DOI] [PubMed] [Google Scholar]
- Puig O., Rutz,B., Luukkonen,B.G., Kandels-Lewis,S., Bragado-Nilsson,E. and Séraphin,B. (1998) New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast, 14, 1139–1146. [DOI] [PubMed] [Google Scholar]
- Rozenski J., Crain,P.F. and McCloskey,J.A. (1999) The RNA Modification Database: 1999 update. Nucleic Acids Res., 27, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N. and Nei,M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sali A. and Blundell,T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Satoh A., Takai,K., Ouchi,R., Yokoyama,S. and Takaku,H. (2000) Effects of anticodon 2′-O-methylations on tRNA codon recognition in an Escherichia coli cell-free translation. RNA, 6, 680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M. and Li,W.H. (1987) The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H. and Moore,P.B. (2000) The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA, 6, 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl M.J. (1993) Recognition of errors in three-dimensional structures of proteins. Proteins, 17, 355–362. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M., Durant,P.C. and Davis,D.R. (2000) Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry, 39, 12575–12584. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P., Cullin,C. and Pompon,D. (1990) Maximizing the expression of mammalian cytochrome P-450 monooxygenase activities in yeast cells. Biochimie, 72, 463–472. [DOI] [PubMed] [Google Scholar]