Abstract
The apoptotic protein Hrk is expressed in hematopoietic progenitors after growth factor deprivation. Here we identify a silencer sequence in the 3′ untranslated region of the hrk gene that binds to the transcriptional repressor DREAM in interleukin-3 (IL-3)-dependent hematopoietic progenitor cells, and abrogates the expression of reporter genes when located downstream of the open reading frame. In addition, the binding of DREAM to the hrk gene is reduced or eliminated when cells are cultured in the absence of IL-3 or treated with a calcium ionophore or a phosphatidylinositol 3-kinase-specific inhibitor, suggesting that both calcium mobilization and phosphorylation can regulate the transcriptional activity of DREAM. Furthermore, we have shown that DREAM is phosphorylated by a phosphatidylinositol 3-kinase-dependent, but Akt-independent pathway. In all cases, loss of the DREAM–DNA binding complex was correlated with increased levels of Hrk and apoptosis. These data suggest that IL-3 may trigger the activation of DREAM through different signaling pathways, which in turn binds to a silencer sequence in the hrk gene and blocks transcription, avoiding inappropriate cell death in hematopoietic progenitors.
Keywords: apoptosis/DREAM/hrk/transcription
Introduction
Apoptotic cell death plays a critical role in the development of hematopoietic progenitors. It is well documented that both growth factors and stromal cell interactions maintain survival of progenitor cells within the bone marrow, and that the absence or reduction of these survival signals induces cells to undergo apoptosis (Williams et al., 1990; Yu et al., 1993).
In hematopoietic progenitors, several mechanisms have been proposed to explain how growth factors regulate survival through Bcl-2 family members. Several hematopoietins, including interleukin-3 (IL-3) and erythropoietin, have been shown to maintain the protein levels of Bcl-2, Bcl-xL, Mcl-1, Bax, Bad and Bak in both cell lines and primary myeloid progenitors (Packham et al., 1998; Sanz et al., 2000). In addition, growth factors can promote survival through post-translational modifications such as phosphorylation of Bad via activation of Akt kinase (del Peso et al., 1997). We have shown previously that the apoptotic protein Hrk, a recently described BH3-only member of the Bcl-2 family (Imaizumi et al., 1997; Inohara et al., 1997), is tightly controlled at the transcriptional level in hematopoietic progenitors (Sanz et al., 2000). The expression of Hrk is undetectable in cells cultured with growth factors, but is specifically upregulated upon growth factor deprivation, and this pattern of expression correlates with induction of apoptosis. Furthermore, ectopic expression of Hrk induced cell death of hematopoietic progenitors cultured with IL-3, indicating that the presence of this protein is sufficient to induce apoptosis.
Identifying the factors and signaling pathways that regulate the levels of Bcl-2 family members is important in understanding better the response of cells to apoptotic stimuli such as chemotherapeutic drugs. Some of the key transcriptional activators of anti-apoptotic members have been identified (Lee et al., 1999; Tamatani et al., 1999; Lord et al., 2000). For example, it has been described that Stats and NFκB factors transactivate the bcl-x gene in a variety of cell systems (Socolovsky et al., 1999; Chen et al., 2000), and that anti-apoptotic NFκB signaling can be activated by the phosphatidylinositol 3 (PI 3)-kinase/Akt pathway (Romashkova and Makarov, 1999). In contrast, few data exist regarding negative regulatory factors for apoptotic members of the Bcl-2 family, although transcriptional repression of strong apoptosis inducers would be a safeguard mechanism to avoid inappropriate cell death in normal hematopoietic progenitors. We report here the identification of a downstream regulatory element (DRE) sequence in the 3′ untranslated region (UTR) of the hrk gene. In IL-3-dependent hematopoietic progenitor cell lines, this sequence binds to a calcium-binding protein, DREAM, which functions as a transcriptional repressor (Carrion et al., 1999). Furthermore, the DREAM-binding sequence (DRE-hrk) abrogates the expression of reporter genes when located downstream of the open reading frame. In addition, we show that the activity of DREAM can be regulated by calcium mobilization and also by direct phosphorylation through a PI 3-kinase-dependent, but Akt-independent pathway, which may allow this repressor to bind DNA and inhibit transcription of hrk in the presence of IL-3.
Results
The hrk gene contains a DRE that binds to the transcriptional repressor DREAM in hematopoietic progenitors
We have shown previously that Hrk is not expressed in viable hematopoietic progenitors, but it is specifically induced at the mRNA and protein level after growth factor deprivation (Sanz et al., 2000). Since Hrk is sufficient to induce apoptosis in hematopoietic progenitor cells, we argued that its expression should be tightly controlled. Thus, we searched for regulatory elements in the mouse hrk gene and found a sequence in the 3′ UTR with significant homology to a gene silencer sequence named DRE (downstream regulatory element) located in the 5′ UTR of the prodynorphin gene (Carrion et al., 1998). As the DRE site binds to a repressor protein, DREAM (Carrion et al., 1999), we analyzed first the capacity of the sequence found in hrk to bind to recombinant human DREAM prepared from Escherichia coli. As shown in Figure 1, recombinant DREAM was able to bind to a radiolabeled DRE sequence from the hrk gene (DRE-hrk) as measured in electrophoretic mobility shift assays (EMSAs). Two specific DRE-hrk-retarded bands are competed by an excess (5- and 50-fold) of unlabeled probe. However, unlabeled irrelevant probes, including a random sequence of the same size and nucleotide composition, failed to compete off the DRE-hrk probe (data not shown). The presence of two bands is likely to be the result of either partial digestion or aggregated forms of the recombinant protein. As a control of binding specificity, recombinant DREAM was shown to bind to the wild-type (wt) DRE sequence of the prodynorphin gene (DRE-PD) but not to a mutated sequence (mut) (Figure 1).
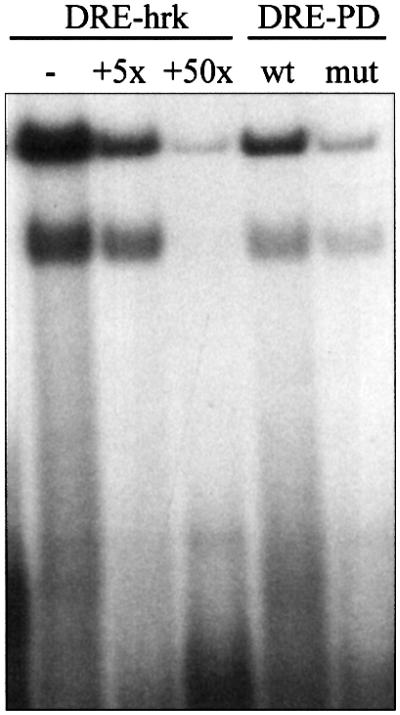
Fig. 1. The DRE sequence of the hrk gene binds to recombinant DREAM. Radiolabeled DRE-hrk, or wild-type (wt) and mutated (mut) DRE-PD probes were incubated with recombinant human DREAM, and the formation of the binding complex was analyzed by EMSA. For competition experiments, DREAM was pre-incubated with 5- and 50-fold molar excess of unlabeled DRE-hrk probe.
Next, we analyzed the expression of DREAM in two IL-3-dependent hematopoietic progenitor cell lines, FL5.12 and FDCP-Mix, previously shown to upregulate Hrk after IL-3 deprivation (Sanz et al., 2000). Nuclear extracts from these cells were prepared and used in southwestern analysis with a radiolabeled DRE-hrk probe. As shown in Figure 2A, this probe reproducibly bound to two proteins of 34 and 110 kDa, which correspond to a monomer and a tetramer of DREAM, as previously characterized in another cell system (Carrion et al., 1999). These DNA–protein complexes were inhibited upon competition with an excess of unlabeled probe, but were not modified by an excess of unlabeled irrelevant probe (data not shown). The expression of DREAM in FL5.12 and FDCP-Mix cells was confirmed further by western blotting with a specific antibody, and as shown in Figure 2B, both monomeric and tetrameric forms of DREAM were detected.
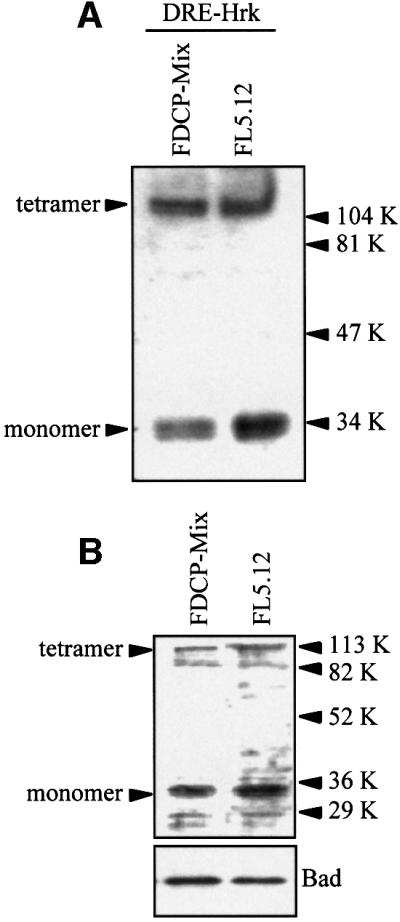
Fig. 2. DREAM is expressed in FDCP-Mix and FL5.12 hematopoietic progenitor cells. (A) Nuclear extracts were analyzed by southwestern blotting using a radiolabeled DRE-hrk probe. Arrowheads indicate the monomeric and tetrameric DREAM proteins. (B) The expression of DREAM protein was analyzed by western blotting with a specific goat antibody.
The DRE sequence of the hrk gene represses the expression of reporter genes
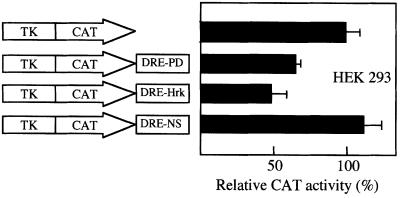
To examine the transcriptional-regulatory activity of the DRE-hrk sequence, we performed transient transfection assays with the heterologous promoter construct pCMV-EGFP containing either a DRE-hrk sequence or an irrelevant, nonsense sequence of the same size and nucleotide composition (DRE-NS) at the 3′ end of the EGFP cDNA. When we examined the FDCP-Mix cells 24 h after transfection with DRE-NS, 17.6% of the cells expressed EGFP, as determined by flow cytometry analysis (Figure 3). In contrast, cells transfected with the DRE-hrk-containing plasmid failed to express the EGFP protein (0.2%) showing the transcriptional repressor activity of this sequence. Since Hrk is expressed following IL-3 withdrawal, we tried to re-induce the expression of EGFP by incubating the transfected cells in the absence of IL-3. However, very little or no expression of EGFP protein was observed after growth factor deprivation (data not shown), most likely due to the inability of FDCP-Mix cells to activate the transcription of EGFP under a strong apopto tic stimulus. To confirm further the repressor activity of DRE-hrk, HEK293 cells were transiently transfected with a different reporter construct, pTK-CAT, containing either a DRE-hrk sequence, a DRE sequence of the prodynorphin gene (DRE-PD) or an irrelevant sequence (DRE-NS) at the 3′ end of the CAT cDNA. Since HEK293 cells have very little or no endogenous DREAM (Carrion et al., 1999), the reporter plasmids were co-transfected with pCDNA3 containing the DREAM cDNA. As shown in Figure 4, the presence of DRE-hrk in the construct reduced the CAT activity by half [48.2 ± 9.7% (mean ± SD)] with respect to the pTK-CAT plasmid. In addition, CAT activity was also reduced when cells were transfected with the DRE-PD-containing plasmid, although to a lower extent (66.1 ± 3.6%), and no reduction in CAT activity was detected when DRE-NS was present in the construct. Therefore, these data show that DRE-hrk is able to reduce or abrogate the expression of heterologous genes when located at the 3′ end of the transcription unit.
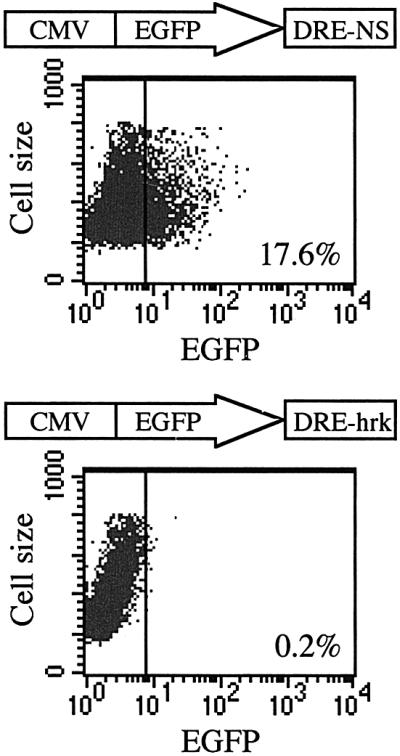
Fig. 3. The expression of EGFP is blocked when DRE-hrk is inserted downstream of the reporter cDNA. Flow cytometry analysis of FDCP-Mix cells after 24 h of transfection with the reporter plasmid pCMV-EGFP containing the DREAM binding sequence of the hrk gene (DRE-hrk) or a random, nonsense sequence of the same size and nucleotide composition (DRE-NS). DNA sequences were inserted downstream of the EGFP open reading frame. Dot plots are from a representative experiment (n = 3).
Fig. 4. The expression of CAT is inhibited when DRE-hrk is located at the 3′ end of the reporter cDNA. The reporter plasmid pTK-CAT containing the DREAM binding sequence of the hrk gene (DRE-hrk), or a random, nonsense sequence of the same size and nucleotide composition (DRE-NS), or the DRE sequence of the prodynorphin gene (DRE-PD), was introduced into HEK 293 cells. After 24 h of transfection, CAT activity was determined relative to that of the pTK-CAT reporter plasmid. Data (mean ± SD) were collected from three independent transfections.
Increase of cytoplasmic calcium and the absence of IL-3 inhibit binding of DREAM to the DRE-hrk sequence and induce Hrk expression
Since the transcriptional repression of DREAM has been shown to be regulated by calcium in human embryonic kidney 293 cells, we asked whether calcium mobilization could affect binding to DRE-hrk in the hematopoietic cell lines. FDCP-Mix and FL5.12 cells were cultured in the absence of IL-3, a well characterized stimulus for the induction of Hrk, or treated with the calcium ionophore ionomycin, and then nuclear extracts were prepared and analyzed in an EMSA (Figure 5). We showed first that the treatment of cells with IL-3 resulted in the formation of a DNA–protein complex when using a DRE-hrk probe, which was reduced after 6 h (not shown), and virtually lost after 24 h of IL-3 deprivation, indicating that DREAM was activated in response to IL-3. In addition, treatment of FDCP-Mix cells with ionomycin for 24 h significantly reduced the DREAM–DNA binding complex. Interest ingly, ionomycin failed to affect the formation of the complex in FL5.12 cells in the same time period, although it was significantly decreased at longer incubation times (48 h) (not shown). In order to correlate the formation of DREAM–DNA binding complex with the expression of the hrk gene, we analyzed the levels of Hrk protein under the same culture conditions. As expected, the presence of IL-3 inhibited the expression of Hrk, which was upregulated after growth factor withdrawal (Figure 5). Consistent with the different DREAM–DNA binding pattern observed in the two cell lines, treatment with ionomycin for 24 h induced the expression of Hrk in FDCP-Mix but not FL5.12 cells, although an incubation time of 48 h significantly upregulated Hrk in FL5.12 (not shown). Consistent with previously described data (Sanz et al., 2000), expression of Hrk correlated with induction of apoptosis in both cell lines (data not shown). Interestingly, none of the treatments modified significantly the protein (Figure 5) and mRNA (data not shown) levels of DREAM, suggesting that the activity of DREAM is not transcriptionally regulated.
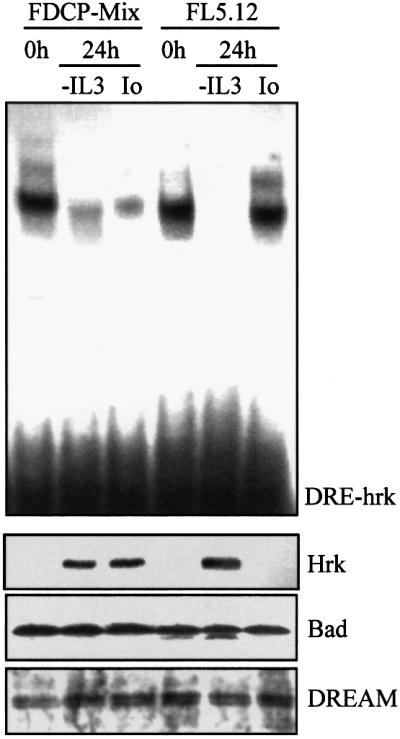
Fig. 5. Treatment with ionomycin blocks binding of DREAM to the DRE-hrk sequence and induces Hrk protein in FDCP-Mix myeloid progenitors. Cells were cultured in the absence of IL-3 or treated with ionomycin for 24 h, and then analyzed for the formation of the DREAM–DNA complex by EMSA using radiolabeled DRE-hrk (top), and for the expression of Hrk, Bad and DREAM proteins by western blotting (bottom).
DREAM is activated through a PI 3-kinase-dependent pathway
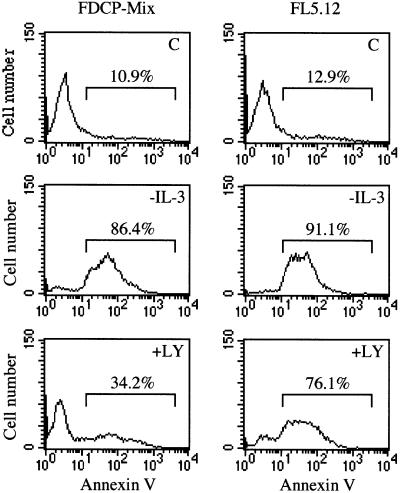
PI 3-kinase is recruited and activated during the intracellular signal transduction of many receptors and has been implicated in the signaling of survival factors such as IL-3 (Yao and Cooper, 1995; Franke et al., 1997). Since calcium mobilization does not seem to regulate efficiently the hrk-binding capacity of DREAM in FL5.12 cells, we studied whether a PI 3-kinase-dependent pathway could contribute to the formation of the binding complex. FDCP-Mix and FL5.12 cells were incubated for 24 h with LY294002, a specific inhibitor of PI 3-kinase, and the DNA binding activity of DREAM was assessed by EMSA. As shown in Figure 6, LY294002 reduced binding to DRE-hrk in both cell lines. Interestingly, while the binding complex was only slightly reduced in FDCP-Mix, it almost disappeared in FL5.12 cells. This pattern of DREAM– DNA complex formation was consistent with the expression of Hrk protein, which was slightly induced in FDCP-Mix and clearly upregulated in FL5.12 cells, further indicating the Hrk repressor activity of DREAM. As previously shown in cells treated with ionomycin, the expression of DREAM remained constant following treatment with LY294002 (Figure 6). Moreover, a clear correlation was observed between the upregulated expression of Hrk and the increase in the number of apoptotic cells as determined by flow cytometry analysis with annexin V (Figure 7). By 24 h of treatment with LY294002, 34.2% of FDCP-Mix cells were annexin V positive, whereas most of the FL5.12 cells (76.1%) were apoptotic within the same time period. Similar results were obtained when cells were treated with wortmannin, another specific inhibitor of PI 3-kinase (data not shown). As expected, in the absence of IL-3 most of the cells (∼90%) were apoptotic (Figure 7).
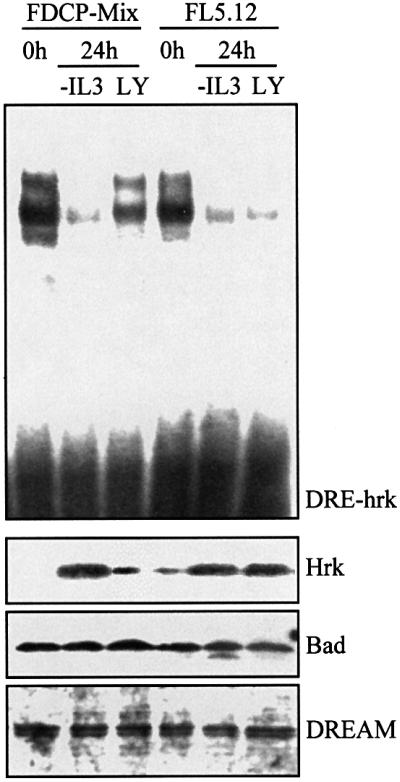
Fig. 6. Inactivation of PI 3-kinase inhibits binding of DREAM to DRE-hrk and induces Hrk protein in FL5.12 lymphoid progenitors. FDCP-Mix and FL5.12 cells were cultured in the absence of IL-3 or treated with LY294002 for 24 h. An EMSA was performed using a radiolabeled DRE-hrk probe to analyze the formation of the DREAM–DNA complex (top). The expression of Hrk, Bad and DREAM proteins was analyzed by western blotting (bottom).
Fig. 7. Inactivation of PI 3-kinase induces apoptotic cell death. FDCP-Mix and FL5.12 cells were cultured in the absence of IL-3 or treated with LY294002 for 24 h, and then analyzed by flow cytometry with fluorescein isothiocyanate (FITC)-labeled annexin V. Numbers above the selected regions indicate the percentage of apoptotic cells.
DREAM is phosphorylated by a PI 3-kinase-dependent and Akt-independent pathway
These results prompted us to analyze whether DREAM was phosphorylated in response to IL-3. In these experiments, FL5.12 cells were stimulated with IL-3 in the presence or absence of LY294002, and the phosphorylation of endogenous DREAM was assayed in DREAM immunoprecipitates. As shown in Figure 8, stimulation with IL-3 induced phosphorylation of DREAM, which was abrogated when cells were treated with the PI 3-kinase inhibitor (Figure 8). Thus, IL-3 induced DREAM phosphorylation through a PI 3-kinase-dependent pathway in hematopoietic progenitor cell lines, which may allow DREAM to bind to the hrk gene.
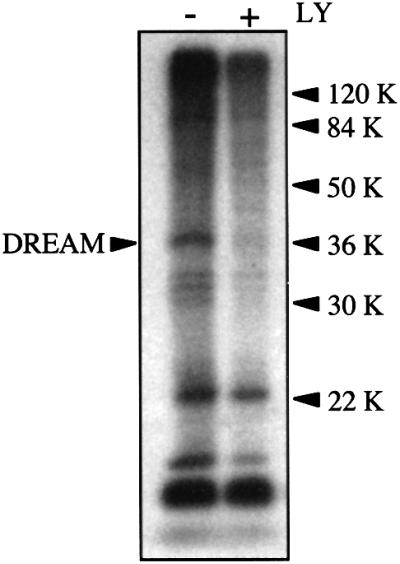
Fig. 8. DREAM is phosphorylated by a PI 3-kinase-dependent pathway. FL5.12 cells were cultured in the presence of [32P]ortho phosphate with or without LY294002. After labeling, cells were lysed and DREAM was immunoprecipitated using a specific polyclonal antibody and protein A/G–agarose beads. Phosphorylation of DREAM was determined by autoradiography.
Because Akt plays an important role in anti-apoptotic PI 3-kinase signaling (del Peso et al., 1997), we studied whether this serine/threonine kinase was able to phosphorylate DREAM. However, we could not find a consensus phosphorylation site for Akt within the DREAM amino acid sequence, making this protein an unlikely candidate for Akt phosphorylation. To determine experimentally whether Akt could phosphorylate DREAM, recombinant DREAM was incubated with Akt immunoprecipitates from FDCP-Mix and FL5.12 cells, and consistent with the previous finding, Akt failed to phosphorylate DREAM (data not shown).
Discussion
Since the expression of Hrk is sufficient to induce apoptosis in hematopoietic progenitors, it is likely that this gene is tightly controlled at the transcriptional level to avoid inappropriate cell death within the hematopoietic compartment. Here we have shown the presence of a DREAM binding sequence in the 3′ UTR of hrk mRNA, DRE-hrk. This sequence specifically binds to the transcriptional repressor DREAM in cells cultured with IL-3, and this binding is lost in the absence of growth factor, suggesting the existence of a DREAM regulatory pathway triggered by IL-3. A DREAM binding sequence has been described recently as a gene silencer located in the 5′ UTR of the human prodynorphin gene (Carrion et al., 1998). Interestingly, our results show that the DREAM binding sequence found in hrk inhibits transcription when located downstream of the open reading frame. DNA sequences that act as silencers when located in the 3′ region of the transcription unit have been described for a number of genes, including the serine protease inhibitor 2.3 and neuronal genes (Paul et al., 1998; Thiel et al., 1998), but not for genes involved in the execution of apoptosis. Thus, the mechanism of transcriptional inhibition described here represents a novel regulatory mechanism for pro-apoptotic members of the Bcl-2 family that is triggered by IL-3 in hematopoietic progenitors. It is interesting that IL-3 also induces the expression of the anti-apoptotic protein Bcl-xL through activation of transcriptional factors such as Stat5 and Stat3 (Catlett-Falcone et al., 1999; Kieslinger et al., 2000), which indicates that IL-3 activates a transcriptional pathway that leads to the expression of an anti-apoptotic protein, and also a repressor pathway that blocks the expression of a highly lethal protein. It has been described previously that Bax, an apoptosis-promoting member of the Bcl-2 gene family, can be transcriptionally repressed by the Gfi-1 proto-oncogene through direct binding to the bax promoter in IL-2-dependent T-cell lines (Grimes et al., 1996). However, in contrast to Hrk, Bax is normally expressed in most hematopoietic cells, including T cells (Grimes et al., 1996; Sanz et al., 2000). Thus, although transcriptional repression of Bax may contribute to balance the ratio between pro-apoptotic and anti-apoptotic members of the Bcl-2 family, the expression of Bax is not sufficient to induce apoptosis. To this end, it has been shown that hematopoietic and non-hematopoietic cells can be stably transfected with Bax (Kobayashi et al., 1998; Sawada et al., 2000). In contrast, we have been unable to obtain cells stably overexpressing Hrk (Sanz et al., 2000; our unpublished data).
We have also shown that the hrk transcriptional repressor can be regulated by different signaling pathways, including phosphorylation and calcium mobilization. Binding of DREAM to the DRE-hrk sequence can be inhibited in FDCP-Mix cells following treatment with ionomycin, whereas the binding complex is impaired in FL5.12 cells treated with PI 3-kinase inhibitors. It has been shown that when intracellular calcium concentration rises, DREAM binds to calcium and releases the DRE sequence, permitting a higher level of promoter activity (Carrion et al., 1999). Thus, an increase in cytosolic calcium, which has been implicated in apoptosis (McConkey et al., 1989; Caron-Leslie et al., 1991), may enable the expression of Hrk through inactivation of its transcriptional repressor. In addition, phosphorylation might play an important role in the activation of DREAM in hematopoietic progenitors. Unlike other transcriptional repressors (ERF, Bcl-6 and HES-1) that have been shown to be inactivated by phosphorylation (Strom et al., 1997; Niu et al., 1998; Le Gallic et al., 1999), DREAM seems to exert its repressor activity when it is phosphorylated by a PI 3-kinase-dependent pathway. Thus, mitogenic signals may inactivate transcriptional repressors (ERF, HES-1) that control genes involved in cell growth, whereas the same or other mitogenic signals may activate transcriptional repressors (DREAM) that block the expression of cell death genes. It is not clear why DREAM is preferentially regulated by calcium in FDCP-Mix myeloid progenitors and by phosphorylation in FL5.12 pre-B cells. A likely explanation is that different apoptotic members of the Bcl-2 family, mainly the BH3-only proteins, are required to execute particular death responses in individual cell types. Consistent with this is the observation that pre-B cells, as well as pre-T cells, from mice deficient in the BH3-only protein Bim are refractory to apoptosis induced by calcium flux, indicating that Bim must be the dominant transducer of this cytotoxic signal in lymphocytes (Bouillet et al., 1999). In this regard, it would be interesting to analyze the expression of Hrk in Bim-deficient lymphoid cells treated with calcium mobilization agents and other apoptotic stimuli in order to shed light on the participation of Hrk in the survival/apoptosis of this cell lineage.
In conclusion, several BH3-only proteins are expressed in viable cells but are maintained in a latent state until activated by apoptotic signals (Li et al., 1998; Puthalakath et al., 1999). By contrast, Hrk is not expressed in viable cells and is induced only after an apoptotic stimulus. We describe here a transcriptional repression mechanism triggered by IL-3 that at least may contribute to block the expression of Hrk and avoid apoptosis in hematopoietic progenitor cell lines. Further studies will need to address the physiological relevance of Hrk transcriptional regulation within the hematopoietic system. It will be of special interest to find other target genes of DREAM that may be involved in proliferation and survival.
Materials and methods
Cell culture
FL5.12 and FDCP-Mix cell lines were grown in RPMI 1640 medium (Seromed Biochrom KG, Berlin, Germany), supplemented with 10% fetal calf serum (FCS) and 10% Wehi3B culture supernatant as an IL-3 source. Human embryonic kidney 293 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS. When indicated, cell lines were treated with 20 µM LY294002 (Sigma, St Louis, MO), a specific inhibitor of PI 3-kinase, or 1 µM ionomycin (SIGMA), a carrier of calcium across membranes. The apoptotic cells were detected with annexin V labeled with FITC (Pharmingen, San Diego, CA) by flow cytometry.
Gene reporter assays
A double-stranded oligonucleotide from the 3′ UTR of the hrk gene (DRE-hrk; 5′-TCGAGTTAACGAAACACAGACAGAGGAAGCCCCTCGGGAG-3′) or a nonsense double-stranded sequence of the same size and nucleotide composition (DRE-NS; 5′-TCGAGTTAACGGACCAAAAAGCAGCAGAGACACTCGCGGG-3′) containing a XhoI site at 5′ was cloned into the XhoI and Bst1107I sites of pCMV-EGFP vector (Clontech, Palo Alto, CA). Cloning was confirmed by linearization of the construct with HpaI, incorporated as a unique site at the 5′ end of the insert, and dideoxy sequencing. FDCP-Mix cells (5 × 106) were transfected by electroporation as previously described (Sanz et al., 2000). After 24 h of transfection, cells were analyzed for expression of green fluorescent protein by flow cytometry using a FACScan analyzer (Becton Dickinson, San Jose, CA). The reporter plasmids pTK-CAT containing the sequences described above or the DREAM binding sequence of the prodynorphin gene at the 3′ end of the CAT cDNA were transfected into 293 cells by calcium phosphate precipitation as previously described (Carrion et al., 1999). Briefly, a mixture of DNA containing 3 µg of reporter plasmid and DREAM cDNA cloned in pCDNA3, 1 µg of β-galactosidase expression vector (Pharmacia, Uppsala, Sweden) and 6 µg of carrier plasmid DNA was coprecipitated with calcium phosphate and added to the 293 cells. After 12 h, cells were washed and left for an additional period of 24 h with fresh medium. The chloramphenicol acetyltransferase (CAT) activity was assayed in 100 µg of protein extract and normalized with respect to β-galactosidase activity.
Protein analysis
The expression of Hrk was determined by western blotting as previously described (Silva et al., 1996). Blots were incubated with rabbit antibodies against Hrk (Sanz et al., 2000) and Bad (Santa Cruz Biotechnology, Santa Cruz, CA), or goat antibodies against DREAM (Santa Cruz Biotechnology), and then incubated with secondary antibodies conjugated to alkaline phosphatase (Tropix, Bedford, MA). Bound antibodies were detected by a chemiluminescence system (Tropix).
Southwestern analysis was carried out as described previously (Carrion et al., 1998). Briefly, nuclear proteins (50 µg) were resolved in SDS–10% polyacrylamide gels and transferred to nitrocellulose membranes. The blots were renatured in phosphate-buffered saline, blocked with 3% non-fat dry milk, and then incubated for 2 h in the presence of 32P-labeled double-stranded DRE-hrk oligonucleotide (106 c.p.m./ml). After washing to remove the unbound probe, blots were exposed for autoradiography.
In vivo phosphorylation assay
FDCP-Mix cells were starved of FCS for 12 h, cultured for 1 h in phosphate-free RPMI containing 20 µM LY294002, and then incubated in the presence of [32P]orthophosphate (100 µCi/ml) for 2 h. Cells were lysed and endogenous DREAM was immunoprecipitated with peptide-specific DREAM polyclonal antibody (J.R.Naranjo, B.Mellstrom and W.A.Link, in preparation). Immunocomplexes were collected with protein A–Sepharose, resolved by 9% SDS–PAGE and analyzed by autoradiography.
Electrophoretic mobility shift assays
Cells were cultured for different periods of time in the presence or absence of ionomycin or LY294002, and were then lysed as previously described (Silva et al., 1999). In brief, nuclear extracts (5 µg of total protein) were incubated with 32P-labeled double-stranded DRE-hrk oligonucleotide, and then run on a 5% non-denaturing polyacrylamide gel. Gels were dried and visualized by autoradiography. In some experiments, 50 ng of recombinant DREAM were incubated with the labeled DRE-hrk probe or a labeled probe from the promoter of the prodynorphin gene (DRE-PD), containing a wild-type or a mutated DRE sequence (Carrion et al., 1999). For competition assays, the recombinant protein was pre-incubated with 5- and 50-fold molar excess of unlabeled DRE-hrk probe.
Acknowledgments
Acknowledgements
The authors thank Dr Adalberto Benito for helpful discussion and Drs J.P.Alvar and L.Krammer for help with the preparation of DREAM antibodies. This work was supported by Comision Interministerial de Ciencia y Tecnologia Grants No. SAF99–0139 to J.L.F.-L. and SAF98-0055 to J.R.N. and by a grant from the ‘Fundacion Marcelino Botin’ (proyecto Terapia Genica) to J.L.F.-L. C.S. is a recipient of a postdoctoral fellowship from the ‘Fundacion Marques de Valdecilla’.
References
- Bouillet P., Metcalf,D., Huang,D.C., Tarlinton,D.M., Kay,T.W., Kontgen,F., Adams,J.M. and Strasser,A. (1999) Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science, 286, 1735–1738. [DOI] [PubMed] [Google Scholar]
- Caron-Leslie L.A. and Cidlowski,J.A. (1991) Similar actions of glucocorticoids and calcium on the regulation of apoptosis in S49 cells. Mol. Endocrinol., 5, 1169–1179. [DOI] [PubMed] [Google Scholar]
- Carrion A.M., Mellstrom,B. and Naranjo,J.R. (1998) Protein kinase A-dependent derepression of the human prodynorphin gene via differential binding to an intragenic silencer element. Mol. Cell. Biol., 18, 6921–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion A.M., Link,W.A., Ledo,F., Mellstrom,B. and Naranjo,J.R. (1999) DREAM is a calcium-regulated transcriptional repressor. Nature, 398, 80–84. [DOI] [PubMed] [Google Scholar]
- Catlett-Falcone R. et al. (1999) Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity, 10, 105–115. [DOI] [PubMed] [Google Scholar]
- Chen C., Edelstein,L.C. and Gelinas,C. (2000). The Rel/NFκB family directly activates expression of the apoptosis inhibitor Bcl-xL. Mol. Cell. Biol., 20, 2687–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Peso L., Gonzalez-Garcia,M., Page,C., Herrera,R. and Nuñez,G. (1997) Interleukin-3-induced phosphorylation of Bad through the protein kinase Akt. Science, 278, 687–689. [DOI] [PubMed] [Google Scholar]
- Franke T.F., Kaplan,D.R. and Cantley,L.C. (1997) PI3K: downstream AKTion blocks apoptosis. Cell, 88, 435–437. [DOI] [PubMed] [Google Scholar]
- Grimes H.L., Gilks,C.B., Chan,T., Porter,S. and Tsichlis,P.N. (1996) The Gfi-1 protooncoprotein represses Bax expression and inhibits T-cell death. Proc. Natl Acad. Sci. USA, 93, 14569–14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi K., Tsuda,M., Imai,Y., Wanaka,A., Takagi,T. and Tohyama,M. (1997) Molecular cloning of a novel polypeptide, DP5, induced during programmed neuronal death. J. Biol. Chem., 272, 18842–18848. [DOI] [PubMed] [Google Scholar]
- Inohara N., Ding,L., Chen,S. and Nuñez,G. (1997) harakiri, a novel regulator of cell death, encodes a protein that activates apoptosis and interacts selectively with survival-promoting proteins Bcl-2 and Bcl-xL. EMBO J., 16, 1686–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieslinger M., Woldman,I., Moriggl,R., Hofman,J., Marine,J.C., Ihle,J.N., Beug,H. and Decker,T. (2000) Anti-apoptotic activity of Stat5 required during terminal stages of myeloid differentiation. Genes Dev., 14, 232–244. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Ruan,S., Clodi,K., Kliche,K.O., Shiku,H., Andreeff,M. and Zhang,W. (1998) Overexpression of Bax gene sensitizes K562 erythroleukemia cells to apoptosis induced by selective chemotherapeutic agents. Oncogene, 16, 1587–1591. [DOI] [PubMed] [Google Scholar]
- Lee H.H., Dadgostar,H., Cheng,Q., Shu,J. and Cheng,G. (1999) NF-κB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl Acad. Sci. USA, 96, 9136–9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gallic L., Sgouras,D., Beal,G. and Mavrothalassitis,G. (1999) Transcriptional repressor ERF is a Ras/mitogen-activated protein kinase target that regulates cellular proliferation. Mol. Cell. Biol., 19, 4121–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhu,H., Xu,C.J. and Yuan,J. (1998) Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell, 94, 491–501. [DOI] [PubMed] [Google Scholar]
- Lord J.D., McIntosh,B.C., Greenberg,P.D. and Nelson,B.H. (2000) The IL-2 receptor promotes lymphocyte proliferation and induction of the c-myc, bcl-2, and bcl-x genes through the trans-activation domain of Stat5. J. Immunol., 164, 2533–2541. [DOI] [PubMed] [Google Scholar]
- McConkey D.J., Nicotera,P., Hartzell,P., Bellomo,G., Wyllie,A.H. and Orrenius,S. (1989) Glucocorticoids activate a suicide process in thymocytes through an elevation of cytosolic calcium concentration. Arch. Biochem. Biophys., 269, 365–370. [DOI] [PubMed] [Google Scholar]
- Niu H., Ye,B.H. and Dalla-Favera,R. (1998) Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the Bcl-6 transcription factor. Genes Dev., 12, 1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packham G. et al. (1998) Selective regulation of Bcl-xL by a Jak kinase-dependent pathway is bypassed in murine hematopoietic malignancies. Genes Dev., 12, 2475–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul C., Simar-Blanchet,A.E., Ro,H.S. and La Cam,A. (1998) Characterization of three transcriptional repressor sites within the 3′ untranslated region of the rat serine protease inhibitor 2.3 gene. Eur. J. Biochem., 254, 538–546. [DOI] [PubMed] [Google Scholar]
- Puthalakath H., Huang,D.C., O’Reilly,L.A., King,S.M. and Strasser,A. (1999) The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell, 3, 287–296. [DOI] [PubMed] [Google Scholar]
- Romashkova J.A. and Makarov,S.S. (1999) NFκB is a target of Akt in anti-apoptotic PDGF signaling. Nature, 401, 86–89. [DOI] [PubMed] [Google Scholar]
- Sanz C., Benito,A., Inohara,N., Ekhterae,D., Nuñez,G. and Fernandez-Luna,J.L. (2000) Specific and rapid induction of the proapoptotic protein Hrk after growth factor withdrawal in hematopoietic progenitor cells. Blood, 95, 2742–2747. [PubMed] [Google Scholar]
- Sawada M., Nakashima,S., Banno,Y., Yamakawa,H., Takenaka,K., Shinoda,J., Nishimura,Y., Sakai,N. and Nozawa,Y. (2000) Influence of Bax or Bcl-2 overexpression on the ceramide-dependent apoptotic pathway in glioma cells. Oncogene, 19, 3508–3520. [DOI] [PubMed] [Google Scholar]
- Silva M., Grillot,D., Benito,A., Richard,C., Nuñez,G. and Fernandez-Luna,J.L. (1996) Erythropoietin can promote erythroid progenitor survival by repressing apoptosis through Bcl-xL and Bcl-2. Blood, 88, 1576–1582. [PubMed] [Google Scholar]
- Silva M., Benito,A., Sanz,C., Prosper,F., Ekhterae,G., Nuñez,G. and Fernandez-Luna,J.L. (1999) Erythropoietin can induce the expression of Bcl-xL through Stat5 in erythropoietin-dependent progenitor cell lines. J. Biol. Chem., 274, 22165–22169. [DOI] [PubMed] [Google Scholar]
- Socolovsky M., Fallo,A., Wang,S., Brugnara,C. and Lodish,H.F. (1999) Fetal anemia and apoptosis of red cell progenitors in Stat5a–/–/5b–/– mice: a direct role for Stat5 in Bcl-xL induction. Cell, 98, 181–191. [DOI] [PubMed] [Google Scholar]
- Strom A., Castella,P., Rockwood,J., Wagner,J. and Caudy,M. (1997) Mediation of NGF signaling by post-translational inhibition of HES-1, a basic helix–loop–helix repressor of neuronal differentiation. Genes Dev., 11, 3168–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamatani M., Che,Y.H., Matsuzaki,H., Ogawa,S., Okado,H., Miyake,S., Mizuno,T. and Tohyama,M. (1999) Tumor necrosis factor induces Bcl-2 and Bcl-x expression through NfκB activation in primary hippocampal neurons. J. Biol. Chem., 274, 8531–8538. [DOI] [PubMed] [Google Scholar]
- Thiel G., Lietz,M. and Cramer,M. (1998) Biological activity and modular structure of RE-1-silencing transcription factor (REST), a repressor of neuronal genes. J. Biol. Chem., 273, 26891–26899. [DOI] [PubMed] [Google Scholar]
- Williams G.T., Smith,C.A., Spooncer,E., Dexter,T.M. and Taylor,D.R. (1990) Hemopoietic colony stimulating factors promote cell survival by suppressing apoptosis. Nature, 343, 76–79. [DOI] [PubMed] [Google Scholar]
- Yao R. and Cooper,G.M. (1995) Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science, 267, 2003–2006. [DOI] [PubMed] [Google Scholar]
- Yu H., Bauer,B., Lipke,G.K., Phillips,R.L. and Van Zant,G. (1993) Apoptosis and hematopoiesis in murine fetal liver. Blood, 81, 373–384. [PubMed] [Google Scholar]