Abstract
The mouse mammary tumor virus (MMTV) promoter is induced by glucocorticoid hormone via the glucocorticoid receptor (GR). The hormone-triggered effects on MMTV transcription and chromatin structure were studied in Xenopus oocytes. We previously showed that the nucleosomes organizing the MMTV promoter became translationally positioned upon hormone induction. A single GR-binding site was necessary and sufficient for the chromatin events to occur, while transcription and basal promoter elements were dispensable. Here we show that addition of the hormone antagonists RU486 or RU43044 to the previously hormone-induced MMTV promoter results in cessation of transcription and loss of chromatin remodeling and nucleosome positioning. In vivo footprinting demonstrated agonist- and RU486-induced GR binding to its DNA response element (GRE), while the other antagonist, RU43044, did not promote GR–GRE interaction. These results demonstrate that induction and maintenance of nucleosome positioning is an active process that requires constant ‘pressure’ of agonist–GR-recruited chromatin-modifying factor(s) rather than GR–DNA binding itself.
Keywords: chromatin remodeling/hormone antagonist/MMTV promoter/nucleosome positioning/Xenopus oocyte
Introduction
The genetic material in eukaryotic cells is organized as nucleosomes consisting of 146 bp of DNA wrapped around a histone octamer (Luger et al., 1997) and internucleosomal linker DNA, which is associated with one molecule of linker histone (Wolffe and Guschin, 2000). In the living cell, the nucleosomes form a higher order structure, the 30 nm fiber. The structure of this chromatin fiber is highly dynamic (Wolffe and Guschin, 2000). Several enzyme complexes have evolved whose function is to modify the repressive chromatin structure (Kingston and Narlikar, 1999). These complexes can be divided into two different groups: one that acts by an energy-dependent nucleosome disruption, the SWI/SNF and ISWI group (Peterson and Workman, 2000), and the other that acts by chemical modification of the N-terminal histone tails (Strahl and Allis, 2000). The enzymes that modify chromatin structure are likely to participate in all cellular processes where the DNA is involved. The emerging picture of metazoan gene regulation is that during cellular differentiation, chromatin-modifying processes are sorting genes into active or inactive domains (El-Osta and Wolffe, 2000). Gene activation is mediated by specific DNA-binding proteins that recruit chromatin-modifying enzymes and other molecular machines to their cognate sites (Lemon and Tjian, 2000; Strahl and Allis, 2000).
Nucleosomes, at least at certain genomic loci, are positioned non-randomly with respect to DNA sequence, and this phenomenon is known as translational nucleosome positioning (Simpson, 1991). Many factors may contribute to the establishment of nucleosome positioning in vivo. The influence of the DNA sequence itself is perhaps the best experimentally documented factor. In many cases, the positioning of nucleosomes in vitro was shown to be similar to the dominant positioning found in vivo (Fragoso et al., 1995; Roberts et al., 1995; Shim et al., 1998; Agalioti et al., 2000). In addition to the DNA sequence, other factors, such as DNA-binding protein(s), may contribute to the establishment of nucleosome positioning (Simpson, 1991; Lu et al., 1994; Cirillo and Zaret, 1999).
The hormone-inducible mouse mammary tumor virus (MMTV) promoter has offered a useful model for studies on the relationship between chromatin structure and transcription (Beato and Eisfeld, 1997; Hager, 2000). The MMTV promoter is silent in the absence of hormone (i.e. agonist) and is induced rapidly to high transcription levels by glucocorticoid hormones as well as progestins and androgens. Each hormone manifests its biological effects by interacting with its intracellular receptor, e.g. the glucocorticoid receptor (GR), which is located in the cytoplasm. Hormone-bound GR is then translocated to the nucleus, where it binds to its cognate DNA acceptor site (Becker et al., 1986; McNally et al., 2000), i.e. the glucocorticoid response elements (GREs), and triggers transcription. This induction event involves the disruption of the nucleosome structure in the vicinity of the GREs, the promoter–enhancer segment between –200 and –50 with respect to the transcription initiation site (+1) (Zaret and Yamamoto, 1984; Fragoso et al., 1998). Studies in tissue culture cells showed this DNA segment to be organized in a positioned nucleosome designated as nucleosome B (Richard-Foy and Hager, 1987; Truss et al., 1995), which is located within an array of six positioned nucleosomes (A–F) covering the entire MMTV long terminal repeat (LTR) of 1.2 kb. The discovery of translationally positioned nucleosomes within the MMTV LTR prior to hormone induction has led to the assumption that it may be of functional importance.
The reconstitution of glucocorticoid-induced MMTV transcription in Xenopus oocytes showed that hormone induction results in dramatic changes in chromatin structure in the vicinity of the GREs (Belikov et al., 2000). In contrast to previous studies, the nucleosomes were positioned randomly prior to hormone activation, and it was the hormone induction event itself that resulted in establishment of an array of translationally positioned nucleosomes over the MMTV LTR. Furthermore, a single high affinity GR-binding site was necessary and sufficient for this structural transition to occur. Both basal promoter elements and ongoing transcription were dispensable. Previous in vitro nucleosome reconstitution experiments showed that the DNA segment corresponding to the MMTV B-nucleosome was able to position a histone octamer translationally either by itself (Perlmann and Wrange, 1988; Pina et al., 1990) or in combination with the A-nucleosome DNA segment, thus forming a dinucleosome (Archer et al., 1991; Flaus and Richmond, 1998), or minichromosomes (Venditti et al., 1998).
We wondered whether the chromatin assembly process in tissue culture cells might be more prone to DNAsequence-directed nucleosome positioning than Xenopus oocytes. Could it be that the nucleosomes in Xenopus oocytes require help to be pushed into the DNA sequence-directed translational position? If this was the case, then hormone-induced nucleosome positioning of the activated MMTV promoter in Xenopus oocytes, which shows positioning identical to that observed in tissue culture cells, would also result in a maintained pattern of positioned nucleosomes after transcription repression. We hypothesized that pushing the nucleosomes into an energetically more favorable position, as determined by the DNA sequence of the MMTV LTR, would also make them stay in this position after turning off transcription. This was addressed experimentally by monitoring chromatin transitions during hormone induction followed by repression of MMTV transcription.
The process of hormone activation of the responsive genes can be inhibited by steroid antagonists at various steps, including the binding of hormone to the receptor, binding of liganded receptor to its cognate DNA target, activation of transcription, etc. (Cadepond et al., 1997). All currently available glucocorticoid hormone antagonists are derivatives of testosterone and fall into two classes based on their ability to promote binding of liganded receptor to DNA. Type I antagonists, such as RU43044, do not promote DNA binding (Bocquel et al., 1993). Type II antagonists, such as RU486 (Mifepristone™), stimulate high affinity binding of the progesterone receptor to the MMTV GRE (Mymryk and Archer, 1995).
Here we show that the addition of glucocorticoid antagonists, either RU486 or RU43044, to Xenopus oocytes previously treated with a glucocorticoid agonist results in inhibition of the GR–agonist-dependent transcription. This inhibition is accompanied by a parallel reversal of chromatin remodeling and a transition of nucleosome positioning from a precise translational pattern to a more random pattern. Furthermore, a detectable chromatin remodeling event, including translational nucleosome positioning, was already generated at 1–2% of maximally achieved MMTV transcription. In vivo footprinting by dimethylsulfate (DMS) methylation protection showed that the agonist triamcinolone acetonide (TA)–GR complex binds avidly to the MMTV GRE, while the type II RU486–GR complex binds weakly and the type I RU43044–GR complex did not bind at all. We conclude that activation-induced nucleosome positioning is reversible and strictly related to promoter activity. Furthermore, the agonist-induced chromatin remodeling and nucleosome positioning is not caused by GR–DNA binding itself but is due to agonist-dependent recruitment of chromatin-modifying factor(s).
Results
Glucocorticoid antagonists RU486 and RU43044 block glucocorticoid-induced transcription of the MMTV promoter
Previous studies have shown that Xenopus oocytes are well suited to address questions on chromatin structure and gene regulation (Belikov et al., 2000; Urnov et al., 2000, and references therein). The MMTV promoter (Figure 1) was injected into the nucleus as a single-stranded (ss) DNA construct. Second strand synthesis coupled to chromatin assembly is completed within 3 h after injection (Almouzni and Wolffe, 1993). GR was delivered to the Xenopus oocyte via cytoplasmic injection of in vitro transcribed rat GR mRNA. The addition of glucocorticoid hormone, either the potent synthetic agonist triamcinolone acetonide (TA) or the weaker natural agonist corticosterone (Cort), to these oocytes resulted in a robust hormone- and receptor-dependent activation of MMTV transcription and concomitant chromatin remodeling of the MMTV promoter (Belikov et al., 2000 and data not shown). In a search for a strategy to reversibly induce and then inhibit MMTV transcription, we found that hormone withdrawal did not reduce MMTV transcription significantly within the 2–3 days life span of an injected oocyte (data not shown). This is most probably a consequence of the large pool of lipophilic hormone trapped in the interior of the large oocyte; its diameter is ∼1.2 mm.
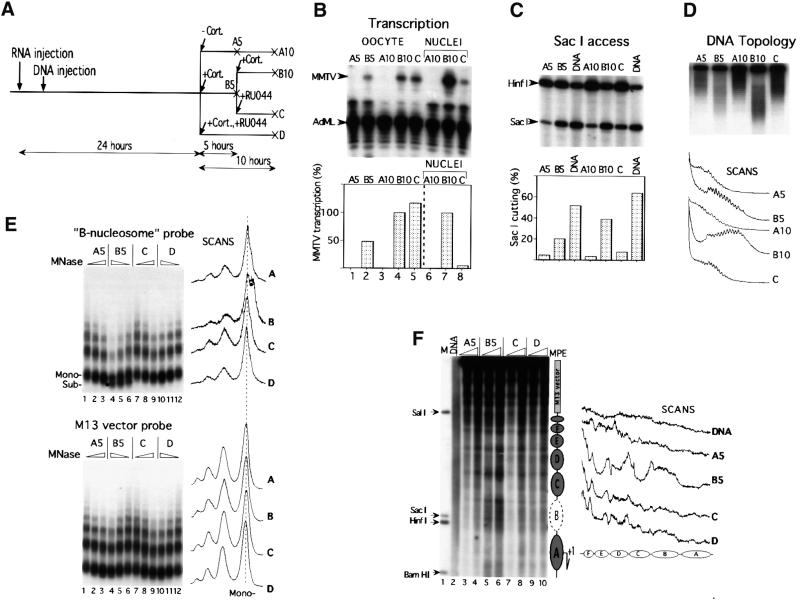
Fig. 1. The reporter DNA construct, the pMTV:M13 used for injection with the primers used for primer extension analysis of the SacI in situ accessibility assay and DMS methylation protection (solid black arrows), and the restriction enzyme cleavage sites that are referred to in the text. White boxes designate GRE hexanucleotide elements I–IV; the black box (Truss et al., 1995) designates an NF1 site; light gray boxes designate OCT1 sites; and the dark gray box designates the TATA sequence. The B-nucleosome probe used in MNase experiments is shown below.
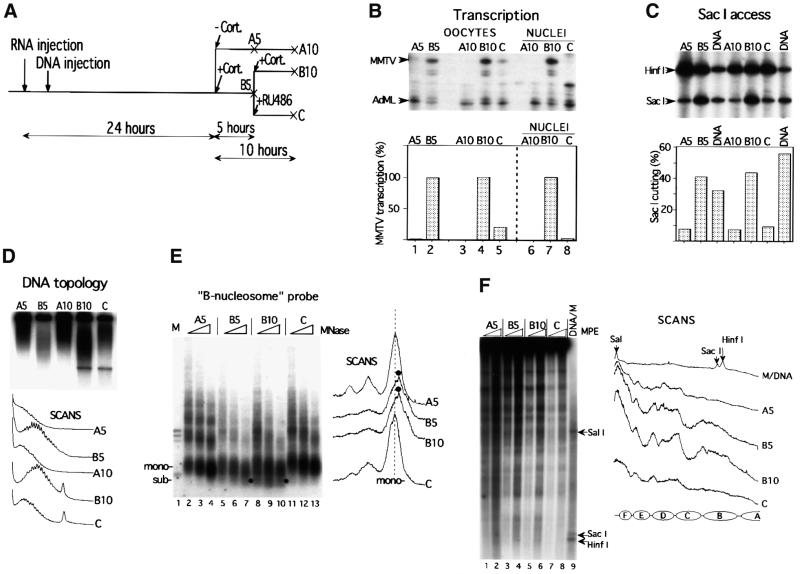
The GR antagonists RU486 and RU43044 were then evaluated for their capacity to mediate transcriptional repression. RU486 (Mifepristone™) is one of the most studied and clinically important progesterone antagonists (Cadepond et al., 1997). RU486 was also defined as a type II anti-glucocorticoid for its ability to bind to GR and promote its binding to DNA (Webster et al., 1988), whereas RU43044 was classified as a type I anti-glucocorticoid because of its inability to promote GR binding to DNA (Bocquel et al., 1993). Initial hormone titration experiments showed that a 5 h incubation with 50 nM of the potent synthetic glucocorticoid agonist TA could not be inhibited efficiently by the consecutive addition of a 1000-fold excess of either of the antagonists used here over a prolonged time (data not shown). This problem was solved by the use of the natural glucocorticoid agonist Cort, which has a 10-fold lower affinity for GR than synthetic glucocorticoid agonists dexamethasone and TA (Rousseau and Schmit, 1977). The experimental design is outlined in Figures 2A and 3A for the RU43044 and RU486 experiments, respectively. Oocytes were first injected with 10 ng of synthetic GR mRNA into the cytoplasm, and 2–5 h later 1 ng of ssMMTV reporter DNA was injected into the oocyte nuclei (Belikov et al., 2000). The next day, 100 nM Cort was added (path B) or not added (path A) to the oocytes. In some experiments, 100 nM Cort was added together with a 50 µM concentration of antagonist (path D) to control its capacity to inhibit transcription (cf. Figure 2A). After a 5 h incubation, the pool of Cort-treated oocytes (B5) was divided into two groups. One group of oocytes was incubated further for another 5 h with 100 nM Cort (B10). The other group of oocytes was rinsed and transferred to medium containing the antagonist, either RU43044 (Figure 2) or RU486 (Figure 3), and also incubated for another 5 h (path C). Antagonists were added in 50 µM concentration, i.e. a 500-fold excess over the originally used agonist, in order to displace the agonist from GR. Total RNA was isolated from groups of oocytes at 5 and 10 h and analyzed for MMTV transcription by S1 nuclease protection assay using the constitutively expressed adenovirus major late (AdML) promoter as a reference (Figures 2B and 3B).
Fig. 2. RU43044 repressed agonist-driven MMTV transcription and reversed chromatin remodeling and nucleosome positioning. (A) Schematic representation of the experimental design. A 100 nM concentration of Cort and 50 µM RU43044 (RU044) were added at the indicated times. A = control; B = agonist; C = antagonist added after agonist; D = agonist + antagonist added at the same time. The 5 and 10 designate hours ± hormone incubation for A and B incubations. (B) Transcription analysis by S1 nuclease protection of MMTV and AdML RNA. Effect of addition of RU43044 on MMTV transcription analyzed in whole oocytes (lanes 1–5) and in manually isolated nuclei (lanes 6–8), six oocytes per group. The diagram below shows quantification of radioactivity. One oocyte equivalent per lane was present in lanes 1–5 and six nuclei per lane in 6–8. B10 is set to 100%. (C) SacI accessibility assay, six oocytes per group. Arrows show specific bands generated by SacI and HinfI as developed by primer extension. DNA shows the analysis with a naked DNA control. The staple diagram below shows the quantification; SacI cutting is given as a percentage of total DNA. (D) Effect of addition of RU43044 on DNA topology of MMTV minichromosomes extracted from the Xenopus oocytes. Radioactivity scans of the lanes are shown below. (E) Effect of addition of RU43044 on nucleosomal array organization. Groups of 10 oocytes were injected and treated with hormone agonist/antagonist as indicated. Oocytes were harvested and digested with increasing amounts of MNase. DNA was resolved in an agarose gel, transferred and hybridized with a labeled MMTV promoter probe encompassing region –192/–100 (B-nucleosome probe) and then washed and rehybridized with an M13 vector probe (below). The black dot shows the subnucleosomal DNA fragment of ∼120 bp revealed only after hybridization with the specific probe. Radioactivity scans of lanes with corresponding maximum MNase concentration are shown to the right. (F) Effect of addition of RU43044 on nucleosome positioning over the MMTV promoter. Groups of seven oocytes, injected and treated as indicated, were digested with MPE for 3 and 10 min. Isolated DNA was digested with EcoRV (+425), resolved on an agarose gel, blotted and hybridized with a random-primed labeled fragment adjacent to the EcoRV site (EcoRV–SacI fragment). M, internal molecular weight markers; DNA, naked dsMMTV DNA digested with MPE. To the right is a schematic summary of MPE cuts with putative nucleosome positions. Radioactivity scans of corresponding lanes are shown to the right.
Fig. 3. RU486 represses agonist-driven MMTV transcription and reverses chromatin remodeling and nucleosome positioning. (A–F) As described in Figure 2 legend.
Addition of hormone resulted in induction of transcription from the MMTV promoter (compare lanes A5 and A10 with B5 and B10 in Figures 2B and 3B). Concomitant addition of the agonist and a 500-fold excess of either of the antagonists gave no transcriptional stimulation (data not shown), thus showing full antagonistic capacity. Agonist incubation for 5 h followed by a switch to antagonist incubation had only a partial and variable effect on the cellular MMTV RNA as analyzed from whole oocytes (compare lanes B and C in Figure 2B and 3B). Oocyte cytoplasm inevitably contains RNA accumulated from the previous 5 h transcription driven by the agonist. In order to monitor the ongoing transcription, we manually isolated oocyte nuclei and determined their nuclear RNA content. This analysis showed a dramatic decline in the level of MMTV transcription after addition of antagonists, down to 6 and 2% of the agonist control for RU43044 and RU486, respectively (compare B10 with C in ‘Nuclei’ in Figures 2B and 3B). However, the oocyte nuclei may still contain MMTV RNA from previous transcription events and/or traces of agonist that may still be bound to GR and drive transcription. We conclude that the antagonists, RU486 and RU43044, added either sequentially or concomitantly (data not shown) with hormone could inhibit hormone-induced transcription.
Agonist-induced chromatin remodeling is reversed by antagonist-driven transcription repression
Several assays were used to monitor the changes in chromatin structure in the course of transcription repression by hormone antagonists. A restriction enzyme accessibility assay utilized SacI, which cleaves the MMTV promoter at position –108 in the bottom strand, i.e. within the cluster of GRE sites (Figure 1). Consistent with previous results (Belikov et al., 2000), inefficient cleavage by SacI was seen in the absence of hormone as compared with the naked DNA control (see Figures 2C and 3C, compare lanes A5 and A10 with DNA control lanes). However, the efficiency of SacI cleavage was increased 4.3- and 5.5-fold, respectively, upon 5 h incubation with Cort (compare lanes A5 with B5 in Figures 2C and 3C). The consecutive addition of antagonist, RU43044 or RU486, to the oocyte medium reversed the SacI accessibility to a similar level as seen in the absence of hormone (compare lanes A5 and A10 with lane C in Figures 2C and 3C). Compared with the SacI accessibility after 5 h of agonist treatment (lane B5), the consecutive 5 h of antagonist incubation (lane C) rendered a 62 and a 77% reduction for RU43044 and RU486, respectively (Figures 2C and 3C).
A supercoiling assay was used to monitor the effects on DNA topology. This assay is based on the observation that each nucleosome organized on a circular DNA molecule induces one negative superhelical turn (Germond et al., 1975), and that the DNA topoisomers can be separated on chloroquine-containing agarose gels. Disruption of the nucleosome structure leads to alterations in DNA topology even if histones are not physically removed from the DNA (Norton et al., 1989). A nucleosomal repeat of 190 bp infers that the 10.25 kb reporter construct organizes ∼54 nucleosomes. Hormone-induced transcription of the MMTV promoter led to changes in the DNA topology that are equivalent to a loss of 7–8 nucleosomes, corresponding to ∼14% of the negative superhelicity (compare lanes A10 and B10 in Figures 2D and 3D). These changes in topology are independent of ongoing transcription as revealed by experiments with α-amanitin (data not shown). Addition of antagonist after 5 h of agonist incubation almost completely reversed the agonist-mediated changes, thus resulting in the topology pattern of inactive chromatin (compare lanes A5 and A10 with lane C in Figures 2D and 3D).
Micrococcal nuclease (MNase) digestion was used to evaluate the antagonist-induced chromatin effects. The electrophoretically separated DNA fragments after MNase digestion were blotted to a filter. This was hybridized with a –192 to –100 probe (B-nucleosome probe), which covers the GRE cluster and harbors the region of strong nucleosome remodeling (Zaret and Yamamoto, 1984; Richard-Foy and Hager, 1987; Truss et al., 1995; Belikov et al., 2000). As can be seen in Figure 2E (compare lanes 3 and 4, upper panel) and Figure 3E (compare lanes 4 and 7), hormone activation led to a drastic increase in MNase cutting in the vicinity of the probed region and in the generation of an unusual subnucleosomal particle of ∼120 bp (labeled with a black dot in the corresponding lane and scan in Figure 2E upper panel, and Figure 3E) (Belikov et al., 2000). This subnucleosomal particle is not present in the vector DNA sequences, as revealed by rehybridization of the same filter with a vector probe (Figure 2E, lower panel). The addition of antagonist (lanes C) to the oocytes after 5 h of agonist incubation resulted in a nucleosomal pattern almost identical to that of inactive chromatin (compare lanes A5 with C and D in the upper panel of Figure 2E and lanes A5 and C in Figure 3E). We conclude that sequential induction and repression of MMTV transcription is accompanied by parallel and reversible changes in chromatin structure.
Addition of antagonist leads to loss of the translational nucleosome positioning
A key question in the antagonist reversibility approach was whether the hormone-induced translational nucleosome positioning would remain after transcription shut off. The analysis of nucleosome positioning was performed with the chemical nuclease methidiumpropyl-EDTA (MPE) due to problems with DNA sequence-specific cutting with MNase in the MMTV promoter (Richard-Foy and Hager, 1987). In agreement with our previous results (Belikov et al., 2000), hormone activation led to the establishment of precise translational nucleosome positioning despite the lack of significant positioning in the inactive state (compare A5 lanes with B5 lanes in Figure 2F and 3F). On the other hand, the concomitant addition of hormone agonist and antagonist RU43044 did not result in any major changes in the nucleosomal pattern (Figure 2F, compare lanes A and D). The sequential incubation of oocytes, first with agonist and then with antagonist, resulted in almost complete loss of translational nucleosome positioning (compare scans for lanes B5 and C in Figures 2F and 3F). However, the nucleosome pattern over the MMTV promoter was not completely returned to the inactive pattern (compare scans through lanes A5 and C in Figures 2F and 3F). Could it be that the residual transcription that was still observed after addition of antagonists (see Figures 2B and 3B) is responsible for the traces of an active nucleosome pattern observed after transcription inhibition by addition of antagonists? This was addressed by monitoring the chromatin remodeling at a low level of MMTV transcription.
Nucleosome positioning is a sensitive marker for hormone-induced transcription
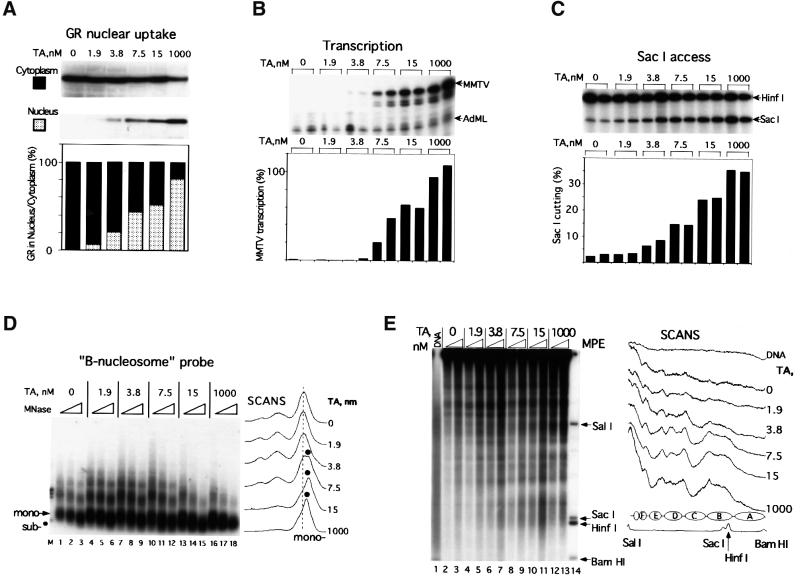
A gradual increase in transcriptional activity from the MMTV promoter was achieved by a stepwise increase of glucocorticoid hormone in the oocyte medium. Non-liganded GR stays exclusively in the cytoplasm (Picard and Yamamoto, 1987) and this is also the case in Xenopus (Figure 4A). The hormone-induced nuclear translocation was analyzed by incubating GR mRNA-injected oocytes with increasing concentrations of hormone (TA) followed by manual dissection of oocyte nuclei and immunoblot analysis of GR content in the nuclei and cytoplasm. This showed a direct correlation between hormone concentration and nuclear translocation (Figure 4A). The nuclear GR translocation was accompanied by a gradual increase in the transcription activity of the MMTV promoter (Figure 4B). Transcription was not detected at 1.9 nM TA and was barely detectable at 3.8 nM, which rendered ∼1.6% of full transcription activity. Half-maximal transcription was obtained at ∼10 nM TA (estimated from a linear plot of data in Figure 4B; Belikov et al., 2000).
Fig. 4. Increasing hormone (TA) concentration induces GR nuclear translocation, MMTV transcription, chromatin remodeling and nucleosome positioning. (A) Nuclear translocation of GR. Oocytes were injected with GR mRNA and pMTV:M13 ssDNA. On the next day, hormone (TA) at the indicated concentrations (nM) was added; after 8 h, oocytes were harvested, manually dissected in groups of five and GR content was evaluated separately in nucleus and cytoplasm by immunoblotting. Relative cytosol/nuclear amounts are given below. (B) Effect of hormone concentration on MMTV transcription assayed by S1 nuclease protection. Six oocytes per group were homogenized and analyzed separately for each TA concentration. (C) Effect of hormone concentration on SacI accessibility assay of MMTV DNA. Six oocytes per group were analyzed for each hormone concentration. See legend of Figure 2C for details. (D) Effect of hormone concentration on nucleosomal array organization in the MMTV promoter. Groups of 10 oocytes were MNase digested and DNA was processed as indicated in Figure 2E. (E) Effect of hormone concentration on nucleosome positioning over the MMTV promoter. Groups of seven oocytes were homogenized and digested with MPE and analyzed as indicted in Figure 2F. M, internal molecular weight markers; DNA, naked MMTV dsDNA digested with MPE. To the right is a schematic summary of MPE cuts with putative nucleosome positions. Radioactivity scans are shown to the right.
The correlation between chromatin remodeling and transcription was monitored by SacI accessibility and MNase digestion. A 2.9-fold increase in SacI cleavage was obtained at 3.8 nM TA as compared with the non-hormone-treated control (Figure 4C). In accordance with the transcription data, the SacI accessibility reached half-maximal level at ∼10 nM TA. The supercoiling assay for circular DNA was also used to monitor the changes in chromatin structure. We observed almost no difference in the topoisomer profile at a 0–3.8 nM interval of hormone, whereas at 7.5 nM hormone a change in topoisomer distribution towards less negative supercoiling was evident (data not shown).
MNase digestion with subsequent visualization of the digestion products by hybridization to a B-nucleosome probe was used to monitor nucleosome structure and remodeling within the GRE segment. Nucleosomal organization within the GRE region remained almost unchanged at the low transcription levels generated by addition of up to 3.8 nM hormone (compare panels ‘0’, ‘1.9’ and ‘3.8’ in Figure 4D). However, a further increase in transcription activity by elevating the concentration of hormone led to increased sensitivity to MNase and the gradual appearance of a subnucleosomal particle of ∼120 bp. At 15 nM TA, the mononucleosome was substituted almost exclusively by the subnucleosome particle (see panel ‘15’ in Figure 4D). A further increase in transcription rate did not induce any changes in the MNase pattern (compare panels ‘15’ and ‘1000’ in Figure 4D).
The development of nucleosome positioning at increasing hormone concentration was addressed by MPE digestion. This showed a significant reorganization of the nucleosomes from an almost random nucleosome pattern seen at lower hormone concentrations towards a translationally positioned pattern clearly detectable already at 3.8 nM TA (Figure 4E, see scans on the right). We conclude that even at very low transcriptional activity, ∼1–2% of maximum level, we detect significant nuclear translocation of GR, a distinctly increased chromatin remodeling and the formation of translational nucleosome positioning.
TA and RU486 but not RU43044 trigger GR binding to DNA in vivo
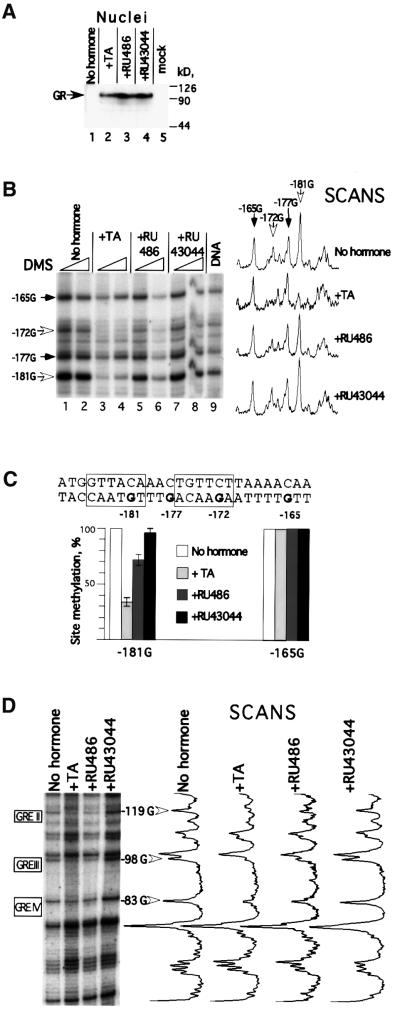
As demonstrated above, GR was localized exclusively in the cytoplasm in the absence of hormone, and ∼80% of total cellular GR was translocated to the nucleus at saturating amounts of hormone agonist (Figure 4A). The addition of antagonists, RU486 or RU43044, also resulted in GR translocation to the nucleus to a similar extent (Figure 5A), in agreement with previous work (Hache et al., 1999).
Fig. 5. Agonist- and antagonist-induced nuclear uptake and GR–GRE binding. (A) Nuclear translocation of hormone/antagonists-liganded GR. Oocytes were injected with GR mRNA and the next day treated with agonist (1 µM TA) or antagonists (50 µM RU486 or RU43044), respectively. Eight hours later, oocytes, five per group, were harvested and GR content in manually dissected nuclei was evaluated by immunoblotting. Mock designates non-injected oocytes. (B) DMS methylation protection over the GRE I segment. Oocytes in groups of five were treated with DMS (see Materials and methods). The methylation pattern was developed by primer (–291/–265) extension. Corresponding Gs are indicated with arrows (white arrows indicate protected guanosines). Radioactivity scans of lanes corresponding to the maximum DMS concentration are shown to the right. (C) The sequence of the GRE I segment with the boxed GR-binding sites and the mean value of the quantification of two separate DMS protection experiments. The –165 G was used as internal standard. (D) DMS methylation protection over the GRE II–IV segments. The methylation pattern was developed by primer (+92/+65) extension. Protected Gs within corresponding GREs are indicated with white arrows. Radioactivity scans of lanes are shown to the right.
The presence of proteins bound to DNA in vivo can be analyzed by DMS methylation (Truss et al., 1995), and specific cleavage of DNA by alkali (Maxam and Gilbert, 1977). This method allows detection of DNA–protein interactions at the N7 position of guanines in the major groove and at the N3 position of adenines via the minor groove of DNA (Maxam and Gilbert, 1977). Here it was used to monitor GR–DNA interactions in the MMTV promoter. The specific cleavage sites caused by methylated DNA were detected by primer extension. The methylation pattern of guanine residues observed for non-hormone-treated oocytes was virtually identical to that obtained for naked DNA (Figure 5B, compare lanes 1 and 2 with lane 9, and data not shown). This indicates that no proteins are bound to this segment of the MMTV promoter via the major groove in the non-induced state. Addition of hormone resulted in a drastic reduction in DMS methylation over GRE I. We observed 35% DMS methylation of the guanosine at position –181 as compared with untreated oocytes (Figure 5B, compare lanes 1 and 2 with lanes 3 and 4; see also Figure 5C). As demonstrated by the radioactivity scans (Figure 5B, right), only guanosines –181 and –172 located within GRE I (marked by boxes in Figure 5C) were protected from DMS methylation, while there was no protection of guanosines –177 and –165, which are located between the GR-binding sites and outside of GRE I, respectively (Figure 5C). This shows that the monitored pattern is specific and in agreement with the previously described GR-dependent methylation interference pattern in vitro (Eriksson and Wrange, 1990). Addition of the hormone antagonists RU486 or RU43044 to oocyte media resulted in transcription cessation and reversal of the agonist-induced chromatin remodeling to a similar extent for both antagonists (see Figures 2 and 3). However, the effect of the two antagonists on GR–DNA binding was quite different. The RU43044-liganded GR complex did not result in any significant protection from DMS methylation, suggesting that it did not promote any specific DNA binding (see corresponding lanes and scans in Figure 5B and C). In contrast, addition of RU486 resulted in significantly lower levels of methylation of guanosine residues over the GR-binding site, ∼70% of that observed in non-induced promoter. Hence, RU486 did not render methylation protection as strong as that detected for the MMTV promoter induced by the agonist TA (see Figure 5C). Both the TA-dependent and RU486-dependent GRE methylation protection was reproducible and was also seen at the three proximal GREs (GRE II–IV) within the MMTV LTR (Figure 5D). We conclude that both TA and RU486 promote GR binding to the GRE while RU43044 does not.
Discussion
Our results show for the first time that the glucocorticoid agonist-induced translational nucleosome positioning of the MMTV promoter is reversible and requires the continuous presence of the GR–agonist complex.
DNA sequence in the MMTV promoter does not direct translational nucleosome positioning in Xenopus oocytes
Previous restriction enzyme mapping of the borders of the dinucleosome covering the MMTV GRE area demonstrated that nucleosomes become translationally positioned upon hormone induction (Belikov et al., 2000). This argues strongly in favor of the hormone-induced MPE digestion pattern being caused by nucleosome positioning. Our previous conclusion that nucleosome positioning is not a prerequisite for MMTV induction but rather is a consequence of hormone activation (Belikov et al., 2000) is confirmed and extended by the fact that the translational nucleosome positioning was reversible and strictly correlated to the hormone-induced transcription. A similar correlation between transcription and nucleosome positioning has been observed previously for the serum albumin enhancer (McPherson et al., 1993) and the –3.9 kb chicken lysozyme enhancer (Huber et al., 1996). Our results show that the nucleosome positioning of the MMTV promoter belongs to the same class of chromatin arrangements, i.e. an activation-dependent nucleosome positioning.
Many in vitro studies had demonstrated that nucleosomes reconstituted on MMTV DNA, either on small DNA fragments forming mono- or dinucleosomes (Perlmann and Wrange, 1988; Pina et al., 1990; Flaus and Richmond, 1998), or on minichromosomes (Venditti et al., 1998), tend to be positioned translationally similarly to the in vivo positioning. For this reason, we assumed that if nucleosomes were forced into the preferred translational positioning in Xenopus oocytes, then they would also stay in this position after antagonist-driven repression. Our results show, however, that this is not the case. Instead the nucleosomes became disordered and adopted the random pattern of the non-induced promoter. We conclude that the in vitro detectable, DNA sequence-directed nucleosome positioning is not able to determine the nucleosome position in vivo.
In contrast to our results in Xenopus oocytes, previous work on the MMTV promoter in tissue culture cells demonstrated a similar nucleosome-positioning pattern in either the presence or the absence of exogenously added hormone, together with local hormone-dependent chromatin remodeling. The latter was often revealed as a DNase I-hypersensitive site (Richard-Foy and Hager, 1987; Truss et al., 1995; Fragoso et al., 1998). However, the MMTV expression in the absence of hormone is significant in tissue culture cells (cf. Truss et al., 1995). As shown here, 2% of the maximally achieved transcription is sufficient to detect translational nucleosome positioning (Figure 4B and F). This finding may explain the difference in nucleosome pattern of the non-hormone-treated MMTV promoter in tissue culture cells as compared with Xenopus oocytes. The often observed high level of MMTV transcription in the absence of exogenous hormone in tissue culture cells might be explained by the presence of endogenous hormone(s) that may be trapped in the lipophilic membrane compartments of the cells and/or bound to serum proteins in the tissue culture medium. This small amount of hormone may be enough to trigger the chromatin transition into a positioned nucleosome array.
The antagonist-induced loss of nucleosome positioning demonstrates the in vivo dynamics of the chromatin fiber, which is overriding the MMTV DNA translational nucleosome-positioning signals detected in vitro (Perlmann and Wrange, 1988; Flaus and Richmond, 1998; Venditti et al., 1998). Based on these findings, we propose that establishment of translational nucleosome positioning in the MMTV LTR is an active process that requires the participation of factor(s) other than just the DNA sequence. However, the absence of translational positioning in the inactive MMTV promoter does not exclude a preferred rotational setting on the nucleosome surface. In fact, in situ hydroxyradical footprinting suggests that this is the case at least for the B-nucleosome area (S.Belikov and Ö.Wrange, unpublished). Never theless, we can not exclude that additional factor(s), which may be absent in Xenopus oocytes, might direct nucleosomes into a preferred translational positioning in other cell types.
The mechanism of chromatin remodeling and nucleosome positioning
The distinct GR footprint shown here in oocytes reveals one advantage of this experimental system. Gross amounts of GR are expressed in the Xenopus oocyte by GR mRNA injection (up to ∼0.5 pmol GR/oocyte; T.Klenka and Ö.Wrange, unpublished). The injected 2–10 × 108 gene copies of the MMTV promoter are involved concomitantly in hormone-triggered chromatin remodeling (Belikov et al., 2000).
To the best of our knowledge, we present the first direct evidence, based on DMS in vivo footprinting, showing that the hormone antagonist RU486 does promote GR–GRE binding. We revealed specific TA–GR binding and RU486–GR binding to all GREs (GRE I–IV; Figure 1) of the MMTV LTR but no detectable binding by the RU43044–GR complex (Figure 5). The weaker protection from DMS methylation by the RU486–GR complex as compared with the TA–GR complex might reflect lower ligand–receptor affinity, thus resulting in a faster off rate from the DNA. Alternatively, it might be due to an antagonist-induced difference in spacing or steric arrangement of the GR homodimer, thus rendering a weaker DNA binding affinity. RU486 was shown previously to induce progesterone receptor (PR) binding to its response element (PRE) in vitro by gel retardation experiments (Klein-Hitpass et al., 1991) and in vivo based on competition studies (Guiochon-Mantel et al., 1988) and its effect on chromatin remodeling and nuclear factor 1 loading in the MMTV promoter (Mymryk and Archer, 1995). On the other hand, Truss et al. (1994) showed that RU486 inhibits PR–DNA interaction at the MMTV promoter, and Becker et al. (1986) reported that RU486 did not promote GR binding at the tyrosine aminotransferase gene.
Previous work and results presented here define RU486 as a type II antagonist, i.e. it promotes DNA binding. The other antagonist, RU43044, previously was described not to induce DNA binding and is hence referred to as a type I antagonist (Bocquel et al., 1993). We could not detect any difference in antagonist-induced nuclear translocation when comparing with the agonist. However, only the agonists TA and Cort cause a distinct chromatin remodeling and induce nucleosome positioning. Importantly, there was no detectable difference in the chromatin pattern of the type I and type II antagonists despite the fact that RU486 induced GR–GRE binding and RU43044 did not. This shows that GR binding per se is not causing any detectable chromatin changes. We showed before that a single GR-binding site (GRE I) is necessary and sufficient for chromatin remodeling and nucleosome positioning to occur (Belikov et al., 2000). Both transcription and DNA-binding sites for NF1, OCT1 and TFIID were dispensable. Taken together, this suggests that other factors are attracted to the MMTV promoter by the GR–agonist complex and mediate the chromatin remodeling and nucleosome-positioning events. There are several candidate coactivators that might contribute to these chromatin effects. Both GRIP1 (Hong et al., 1996) and the SWI/SNF subunit BRG1 (Fryer and Archer, 1998) were shown to interact with GR in an agonist-dependent fashion. We could not detect any GR–BRG1 interaction in oocytes by co-immunoprecipitation (IP) (B.Gelius and Ö.Wrange, unpublished; see also DiRenzo et al., 2000). This should be addressed further by chromatin IP analysis (Shang et al., 2000).
We show that the ‘pressure’ from the agonist–GR-recruited chromatin remodeling machinery maintains the precisely organized structure of the active promoter. When this pressure is released by addition of antagonist, the nucleosomes return to the randomized state. We suggest that this may be of general importance for the understanding of chromatin dynamics in the eukaryotic cell. We speculate that the highly sensitive switch from a random to a positioned nucleosome pattern already at a low level of transcription provides a memory effect or a pre-setting that shortens the time to acquire full gene activation. This sensitive chromatin patterning process may be reminiscent of the early steps in development of tissue-specific gene expression, as characterized in the serum albumin enhancer (Cirillo and Zaret, 1999) and the –3.9 kb lysozyme enhancer (Huber et al., 1996).
Materials and methods
DNA and plasmids
Construction of the MMTV reporter and the plasmid designated for in vitro transcription of rat GR mRNA was described previously (Belikov et al., 2000).
Oocyte microinjection and maintenance
Xenopus laevis oocytes were prepared and collagenized as described (Robinett and Dunaway, 1999). In a typical experiment, 10 ng of in vitro transcribed GR RNA (mMessage mMachine™; Ambion) were injected into the oocyte cytoplasm followed 2–6 h later by intranuclear injection of 1 ng of pMTV:M13 ssDNA and 0.35–0.5 ng of pAdML dsDNA, the latter as a transcription reference (Ohlsson and Edlund, 1986). For in vivo footprinting, 5 ng of pMTV:M13 ssDNA and no pAdML were injected. Microinjections were done using a Nanoliter 2000™ (World Precision instruments, Inc.). The indicated amounts of steroid hormones were added the following day. Antagonists RU486 (Sigma and kindly donated by Exelgyn) and RU43044 (kindly donated by Dr F.Nique, Hoescht Marion Rousell) were prepared as a 5 × 10–2 M solution in dimethylsulfoxide (DMSO) and added to oocyte media. TA and Cort (Sigma) were dissolved in ethanol as 10–2 M concentration. All incubations contained the same concentrations of DMSO and ethanol. The experiments presented have been performed twice or more with reproducible results.
RNA analysis and immunoblotting
RNA analysis and immunoblotting using a mouse monoclonal antibody against rat GR (MA1-510; Affinity Bioreagents Inc.) were performed as previously described (Gelius et al., 1999). Immunoblots were quantified using a Fuji LAS-1000 Chemiluminiscence detector system.
Chromatin analysis
MNase digestion, the SacI accessibility assay and in situ cleavage by MPE were carried out as described previously (Belikov et al., 2000). The supercoiling assay was carried out as described previously (Clark and Wolffe, 1991) with a chloroquine concentration of 60 µg/ml. 32P radioactivity scans and quantifications were carried out with a Fuji Bio-Imaging analyzer BAS-2500 using the Image Gauge V3.3 software.
DMS in vivo footprinting
DMS was added to injected oocytes in groups of five in OR-2 solution (Robinett and Dunaway, 1999) to achieve a final concentration of 0.3 and 0.6% (v/v). Incubation was performed at 20°C for 10 min. Next, oocytes were rinsed quickly with OR-2 solution, homogenized in 200 µl of OR-2 solution containing 1 M β-mercaptoethanol, and then an equal volume of 2× STOP solution was added to render a final concentration of 0.5% SDS and 10 mM EDTA. DNA was purified as described (Belikov et al., 2000). DNA was dissolved in 20 µl of 5 mM HEPES pH 7.8, heated at 90°C for 15 min followed by addition of 2 µl of 1 M NaOH with subsequent incubation at 95°C for 30 min, and then neutralized with 2 µl of 1 M acetic acid and ethanol precipitated (Maxam and Gilbert, 1977). The DMS-induced cleavage pattern was visualized by primer extension as described previously (Belikov et al., 2000).
Acknowledgments
Acknowledgements
We gratefully acknowledge Ulla Björk for skillful technical assistance, Kyle Sousa for comments on the manuscript, Dr E.E.Baulieu for help with RU43044, and Dr F.Nique for providing RU43044 from Hoescht Marion Roussel Inc. RU486 (Mifepristone™) was kindly provided by Dr R.Sitruk-Ware, Roussel Uclaf and Exelgyn. This work was supported by the Swedish Cancer Foundation (project 2222-B00-16XCC) also supporting a PhD student salary to B.G., and by the Royal Swedish Academy of Sciences (12682). A guest research fellowship by STINT (98/793) supported S.B. Project support was also provided by the European Commission, TMR, to Ö.W. (Network Contract ERBFMRXCT-98-0191).
References
- Agalioti T., Lomvardas,S., Parekh,B., Yie,J., Maniatis,T., Thanos,D., El-Osta,A. and Wolffe,A.P. (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell, 103, 667–678. [DOI] [PubMed] [Google Scholar]
- Almouzni G. and Wolffe,A.P. (1993) Replication-coupled chromatin assembly is required for repression of basal transcription in vivo. Genes Dev., 7, 2033–2047. [DOI] [PubMed] [Google Scholar]
- Archer T.K., Cordingley,M.G., Wolford,R.G. and Hager,G.L. (1991) Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol. Cell. Biol., 11, 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M. and Eisfeld,K. (1997) Transcription factor access to chromatin. Nucleic Acids Res., 25, 3559–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P.B., Gloss,B., Schmid,W., Strahle,U. and Schutz,G. (1986) In vivo protein–DNA interactions in a glucocorticoid response element require the presence of the hormone. Nature, 324, 686–688. [DOI] [PubMed] [Google Scholar]
- Belikov S., Gelius,B., Almouzni,G. and Wrange,Ö. (2000) Hormone activation induces nucleosome positioning in vivo. EMBO J., 19, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquel M.T., Ji,J., Ylikomi,T., Benhamou,B., Vergezac,A., Chambon,P. and Gronemeyer,H. (1993) Type II antagonists impair the DNA binding of steroid hormone receptors without affecting dimerization. J. Steroid Biochem. Mol. Biol., 45, 205–215. [DOI] [PubMed] [Google Scholar]
- Cadepond F., Ulmann,A. and Baulieu,E.E. (1997) RU486 (mifepristone): mechanisms of action and clinical uses. Annu. Rev. Med., 48, 129–156. [DOI] [PubMed] [Google Scholar]
- Cirillo L.A. and Zaret,K.S. (1999) An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell, 4, 961–969. [DOI] [PubMed] [Google Scholar]
- Clark J.D. and Wolffe,A.P. (1991) Superhelical stress and nucleosome-mediated repression of 5S RNA transcription in vitro. EMBO J., 10, 3419–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRenzo J., Shang,Y., Phelan,M., Sif,S., Myers,M., Kingston,R. and Brown,M. (2000) BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol., 20, 7541–7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Osta A. and Wolffe,A.P. (2000) DNA methylation and histone deacetylation in the control of gene expression: basic biochemistry to human development and disease. Gene Expr., 9, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P. and Wrange,Ö. (1990) Protein–protein contacts in the glucocorticoid receptor homodimer influence its DNA binding properties. J. Biol. Chem., 265, 3535–3542. [PubMed] [Google Scholar]
- Flaus A. and Richmond,T.J. (1998) Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol., 275, 427–441. [DOI] [PubMed] [Google Scholar]
- Fragoso G., John,S., Roberts,M.S. and Hager,G.L. (1995) Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev., 9, 1933–1947. [DOI] [PubMed] [Google Scholar]
- Fragoso G., Pennie,W., John,S. and Hager,G.L. (1998) The position and length of the steroid dependent hypersensitive region in the MMTV LTR is invariant despite multiple nucleosome B frames. Mol. Cell. Biol., 18, 3633–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C.J. and Archer,T.K. (1998) Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature, 393, 88–91. [DOI] [PubMed] [Google Scholar]
- Gelius B., Wade,P., Wolffe,A.P., Wrange,Ö. and Östlund Farrants,A.-K. (1999) Characterization of a chromatin remodeling activity in Xenopus oocytes. Eur. J. Biochem., 262, 426–434. [DOI] [PubMed] [Google Scholar]
- Germond J.E., Hirt,B., Oudet,P., Gross-Bellark,M. and Chambon,P. (1975) Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc. Natl Acad. Sci. USA, 72, 1843–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiochon-Mantel A., Loosfelt,H., Ragot,T., Bailly,A., Atger,M., Misrahi,M., Perricaudet,M. and Milgrom,E. (1988) Receptors bound to antiprogestin from abortive complexes with hormone responsive elements. Nature, 336, 695–698. [DOI] [PubMed] [Google Scholar]
- Hache R.J., Tse,R., Reich,T., Savory,J.G. and Lefebvre,Y.A. (1999) Nucleocytoplasmic trafficking of steroid-free glucocorticoid receptor. J. Biol. Chem., 274, 1432–1439. [DOI] [PubMed] [Google Scholar]
- Hager G.L. (2000) Understanding nuclear receptor function: from DNA to chromatin to the interphase nucleus. Prog. Nucleic Acid Res. Mol. Biol., 66, 279–305. [DOI] [PubMed] [Google Scholar]
- Hong H., Kohli,K., Trivedi,A., Johnson,D.L. and Stallcup,M.R. (1996) GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl Acad. Sci. USA, 93, 4948–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M.C., Kruger,G. and Bonifer,C. (1996) Genomic position effects lead to an inefficient reorganization of nucleosomes in the 5′-regulatory region of the chicken lysozyme locus in transgenic mice. Nucleic Acids Res., 24, 1443–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Klein-Hitpass L., Cato,A.C., Henderson,D. and Ryffel,G.U. (1991) Two types of antiprogestins identified by their differential action in transcriptionally active extracts from T47D cells. Nucleic Acids Res., 19, 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B. and Tjian,R. (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev., 14, 2551–2569. [DOI] [PubMed] [Google Scholar]
- Lu Q., Wallrath,L.L. and Elgin,S.C. (1994) Nucleosome positioning and gene regulation. J. Cell. Biochem., 55, 83–92. [DOI] [PubMed] [Google Scholar]
- Luger K., Mäder,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Maxam A.M. and Gilbert,W. (1977) A new method for sequencing DNA. Proc. Natl Acad. Sci. USA, 74, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally J.G., Muller,W.G., Walker,D., Wolford,R. and Hager,G.L. (2000) The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science, 287, 1262–1265. [DOI] [PubMed] [Google Scholar]
- McPherson C.E., Shim,E.-Y., Friedman,D.S. and Zaret,K.S. (1993) An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell, 75, 387–398. [DOI] [PubMed] [Google Scholar]
- Mymryk J.S. and Archer,T.K. (1995) Dissection of progesterone receptor mediated chromatin remodeling and transcriptional activation in vivo. Genes Dev., 9, 1366–1376. [DOI] [PubMed] [Google Scholar]
- Norton V.G., Imai,B.S., Yau,P. and Bradbury,E.M. (1989) Histone acetylation reduces nucleosome core particle linking number change. Cell, 57, 449–457. [DOI] [PubMed] [Google Scholar]
- Ohlsson H. and Edlund,T. (1986) Sequence-specific interactions of nuclear factors with insulin gene enhancer. Cell, 45, 35–44. [DOI] [PubMed] [Google Scholar]
- Perlmann T. and Wrange,Ö. (1988) Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J., 7, 3073–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L. and Workman,J.L. (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev., 10, 187–192. [DOI] [PubMed] [Google Scholar]
- Picard D. and Yamamoto,K.R. (1987) Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J., 6, 3333–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B., Bruggemeier,U. and Beato,M. (1990) Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell, 60, 719–731. [DOI] [PubMed] [Google Scholar]
- Richard-Foy H. and Hager,G.L. (1987) Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J., 6, 2321–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M.S., Fragoso,G. and Hager,G.L. (1995) Nucleosomes reconstituted in vitro on mouse mammary tumor virus B region DNA occupy multiple translational and rotational frames. Biochemistry, 34, 12470–12480. [DOI] [PubMed] [Google Scholar]
- Robinett C.C. and Dunaway,M. (1999) Modeling transcriptional regulation using microinjection into Xenopus oocytes. Methods, 17, 151–160. [DOI] [PubMed] [Google Scholar]
- Rousseau G.G. and Schmit,J.P. (1977) Structure–activity relationships for glucocorticoids. I. Determination of receptor binding and biological activity. J. Steroid Biochem., 8, 911–919. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Shim E.Y., Woodcock,C. and Zaret,K.S. (1998) Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev., 12, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R.T. (1991) Nucleosome positioning: occurrence, mechanisms and functional consequences. Prog. Nucleic Acid Res. Mol. Biol., 40, 143–184. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Truss M., Bartsch,J. and Beato,M. (1994) Antiprogestins prevent progesterone receptor binding to hormone responsive elements in vivo. Proc. Natl Acad. Sci. USA, 91, 11333–11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truss M., Bartsch,J., Schulbert,A., Hache,R.J.G. and Beato,M. (1995) Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J., 14, 1737–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov F.D., Yee,J., Sachs,L., Collingwood,T.N., Bauer,A., Beug,H., Shi,Y.B. and Wolffe,A.P. (2000) Targeting of N-CoR and histone deacetylase 3 by the oncoprotein v-erbA yields a chromatin infrastructure-dependent transcriptional repression pathway. EMBO J., 19, 4074–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditti P., Di Croce,L., Kauer,M., Blank,T., Becker,P.B. and Beato,M. (1998) Assembly of MMTV promoter minichromosomes with positioned nucleosomes precludes NF1 access but not restriction enzyme cleavage. Nucleic Acids Res., 26, 3657–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N.J., Green,S., Jin,J.R. and Chambon,P. (1988) The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell, 54, 199–207. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. and Guschin,D. (2000) Review: chromatin structural features and targets that regulate transcription. J. Struct. Biol., 129, 102–122. [DOI] [PubMed] [Google Scholar]
- Zaret K.S. and Yamamoto,K.R. (1984) Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell, 38, 29–38. [DOI] [PubMed] [Google Scholar]