Abstract
Bovine spongiform encephalopathy (BSE) and its human equivalent, variant Creutzfeldt–Jakob disease (vCJD), are caused by the same strain of infectious agent, which is similar to, but distinct from, >20 strains of their sheep scrapie homologue. A better understanding of the molecular strain determinants could be obtained from cells in monoculture than from whole animal studies where different cell targeting is commonly a strain-related feature. Although a few cell types can be infected with different strains, the phenotypes of the emergent strains have not been studied. We have cured the scrapie-infected, clonal SMB cell line with pentosan sulfate, stably re-infected it with a different strain of scrapie and shown that biological properties and prion protein profiles characteristic of each original strain are propagated faithfully in this single non-neuronal cell type. These findings attest to the fact that scrapie strain determinants are stable and host-independent in isolated cells.
Keywords: cultured cell/prion disease/prion protein isoforms/scrapie strain/transmissible spongiform encephalopathy
Introduction
The prion concept holds that a phenotype can be transmitted across generations using a protein as the carrier of information rather than a nucleic acid (Prusiner, 1982). It originated in the field of the transmissible spongiform encephalopathies (TSEs) which are progressive neurodegenerative diseases such as scrapie of sheep and goats, Creutzfeldt–Jakob disease (CJD) of humans and bovine spongiform encephalopathy (BSE), and has been extended recently to explain non-Mendelian inheritance of certain traits in yeast and fungi (Masison, 2000). The existence of several distinct strains of mammalian agent has been recognized for several years and these strains were first defined by two principal criteria, their incubation times in Sincs7 and Sincp7 inbred mice and their heterozygote cross, and the types and patterns of brain lesions they induce at the terminal stage of disease in these mice (Dickinson et al., 1986). The prion protein (PrP) is encoded at the Sinc (or Prn-i) locus, and one (or both) of two amino acid differences in the mouse protein encoded by the s7 or p7 alleles is the molecular basis of these host differences in survival time (Moore et al., 1998). Although genetic elements other than the protein-coding element within the Sinc locus can influence the brain pathology following infection with a particular strain (Moore et al., 1998), distinct features can be seen which are obviously determined by the strain of inoculated agent independently of the host species (Fraser, 1976; Bruce, 1993).
The conversion of normal PrP (PrPC) to a protease-resistant isoform, PrPSc, is a key event in the pathogenesis of scrapie and all other prion diseases, and PrPSc, according to prion theory, is the sole component of the infectious particle (Prusiner, 1998) so molecular differences in PrPSc must form the basis of the different phenotypes of CJD and scrapie. It has been speculated that variation in protease-resistant fragment size or conformation and the degree of its asparagine-linked glycosylation (Bessen and Marsh, 1994; Collinge et al., 1996; Telling et al., 1996; Caughey et al., 1998; Parchi et al., 1998) form the basis of agent strain differences. Such physicochemical differences could ‘translate’ into differences in the survival times and pathology seen with various strains as a consequence of differing rates of production, accumulation and clearance of the prion protein by different cell types within the brain (Safar et al., 1998). It follows that cell-specific variation in conformation and/or degree of glycosylation of PrP would be the effectors of this strain propagation (Weissmann, 1991). Separating cause of infection from its pathological by-products has been hindered by the need to study strain effects in the whole organism: for example, it has been impossible to determine if a single PrPSc glycoform—one molecular signature of a particular strain (Aguzzi and Weissmann, 1997)—can target only one specific subset of nerve cells in the brain capable of producing only that glycoform (DeArmond et al., 1997, 1999). To investigate these processes requires the stable infection of a single cell type with more than one strain of scrapie.
The best defined scrapie strains have been isolated and passaged in mice (Dickinson et al., 1986); therefore, to avoid interpretational difficulties arising from inter- species transmission, a mouse cell line would be optimal. However, persistent scrapie infection can be established in only a very few cell lines in vitro. Scrapie-infected mouse C1300 neuroblastoma-derived cell lines ScMNB (Race et al., 1987) or ScN2a (Butler et al., 1988) have been valuable in defining events in PrPSc production (Race et al., 1988; Taraboulos et al., 1990, 1992; Caughey et al., 1991; McKinley et al., 1991; Vey et al., 1996; Naslavsky et al., 1997) but the wild-type cells have only a low susceptibility to in vitro challenge with scrapie. The cell line we chose for these experiments was SMB (Clarke and Haig, 1970) which was established originally by culture from a brain taken from a mouse clinically affected by the Chandler scrapie isolate and shown to be a cell line of mesodermal origin (Haig and Clarke, 1971). We reasoned that since SMB cells possessed a high degree of competence in propagating the original infection, a non-infected SMB variant might exhibit equal competence at hosting a newly introduced, different scrapie strain.
Results
Curing SMB with pentosan sulfate
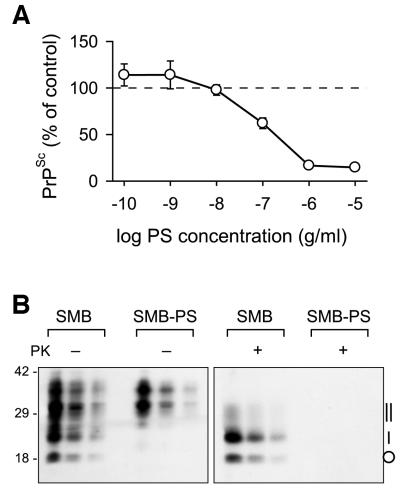
In preliminary experiments, when we examined clonally derived colonies of SMB for PrPSc, a surrogate marker for infectivity, no PrPSc-negative colonies were identified. We therefore investigated making an uninfected variant by treating the SMB cell line with pentosan sulfate (PS) to cure it of the infection with Chandler scrapie. PS is one of several polyanionic compounds capable of altering the scrapie incubation period in vivo (Diringer and Ehlers, 1991) and has been shown to curtail PrPSc production and replication of infectivity in ScMNB cells (Caughey and Raymond, 1993; Priola and Caughey, 1994). When grown for a single passage in the presence of PS at a range of concentrations up to 1 × 10–5 g/ml, the growth of SMB cells was unaffected, but PrPSc accumulation was inhibited in a dose-dependent manner (Figure 1A). The concentration required to inhibit PrPSc accumulation by 50% (IC50) was 1.5 × 10–7 g/ml. Even at the highest concentrations of PS tested, there was a residual PrPSc content of ∼12–14% of the control value, which is in accord with previous work showing that PS effectively inhibits de novo accumulation of PrPSc but has little effect on reducing the amount of pre-existing PrPSc in cells (Caughey and Raymond, 1993).
Fig. 1. PrPSc expression in SMB cells was cured by pentosan sulfate. (A) The inhibition by PS of PrPSc accumulation in SMB cells is dose dependent. Cultures of SMB cells were grown for 7 days in the continuous presence of the indicated concentrations of PS, after which time PrPSc levels were measured by quantitative dot-blotting. The points are the means (±SEM) from four experiments in which each data point was triplicated. (B) Western blot analysis of the PrP in SMB and the pentosan sulfate-treated SMB-PS cells. Proteins in post-nuclear cell extracts prepared either with (+) or without (–) proteinase K (PK) treatment, were precipitated with methanol/chloroform prior to electrophoresis. Successive lanes in each triplet of lanes represents a 5-fold dilution series of the previous sample. Molecular weight standards (kDa) are indicated on the left and the position of PK-treated PrPSc isoforms on the right: non-glycosylated (circle), monoglycosylated (single vertical bar) and diglycosylated (double vertical bar).
To effect a permanent cure, SMB cells were grown in the continuous presence of 1 × 10–6 g/ml PS for seven passages so that all pre-formed accumulated PrPSc was effectively removed through dilution. With an ∼8-fold increase in cell number at each pass, PrPSc quickly became undetectable, the seven passages representing a dilution of some 2 × 107 in the initial amount of PrPSc. PrPSc subsequently has remained undetectable in this PS-treated cell line, designated SMB-PS, maintained in the absence of PS for >50 passages.
Western blot analysis of total protein extracts of SMB and SMB-PS cells showed a complex pattern of PrP isoforms (Figure 1B). In SMB cells, the pattern was typical of a mixture of PrPSc and PrPC. In SMB-PS cells, a much simpler pattern of PrP isoforms existed composed predominantly of 35–37 kDa isoforms, most probably full-length diglycosylated PrPC, 30–33 kDa isoforms which were probably a mixture of full-length monoglycosylated and truncated diglycosylated PrPC (Chen et al., 1995), with very little evidence of non-glycosylated PrPC (26 kDa). Treatment with proteinase K (PK) produced a subset of proteinase-resistant bands from SMB cells typical of PrPSc, but resulted in the degradation of all the PrP in SMB-PS samples.
To confirm that the PrPSc-cured cells had also been cured of infectious Chandler agent, the SMB-PS cell line was tested for scrapie infectivity. Of 18 C57BL mice inoculated intracerebrally with SMB-PS cells, 11 survived to the end of the experiment and were culled without clinical signs at 796 days post-inoculation; the other seven mice were killed as a result of contracting inter-current diseases (on days 370, 377, 473, 604, 628, 673 and 790). In the entire group of 18 (both inter-current and survivors), there were no symptomatic cases of scrapie, no post-mortem histopathological signs of scrapie and no western blot-detectable PrPSc in any of the brains. The survival curve for SMB-PS-inoculated mice (not shown) is not significantly different from that obtained for sham-inoculated controls but it is significantly longer than for mice inoculated with an equivalent number of SMB cells (see below), and is consistent with SMB-PS being cured of infectivity.
Re-infection of the cured SMB-PS cell
Two sources of scrapie have been particularly important in the derivation of individual scrapie strains, the ‘drowsy goat’ source and the Cheviot sheep-passaged SSBP/1 source (Dickinson et al., 1986). It was the drowsy goat source that first gave clues to the existence of different scrapie strains (Pattison and Millson, 1961; Pattison, 1972), was the first to be transmitted to mice (Chandler, 1961) and which was also the source of the SMB cell infection. Strains isolated from the drowsy goat source are ‘Chandler’, widely distributed and, after derivation and cloning, known as 139A or RML, and 79A which are all unique to this source (Dickinson et al., 1986). Among the several strains derived from SSBP/1 is 22F, and its very distinctive biological phenotype makes it ideal for comparison with the original infection of the SMB cell (Dickinson et al., 1986; Bruce, 1993).
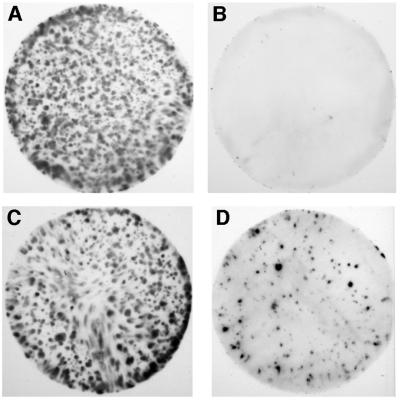
To survey the susceptibility of SMB-PS cells to in vitro infection using the 22F strain, we employed a colony-lift blotting procedure. The SMB cell is phagocytic (Haig and Clarke, 1971) and this gross screening method allowed us to introduce very large dilution factors post-challenge, favouring the subsequent detection of PrPSc synthesized de novo and discriminating it from any residual PrPSc of the inoculum taken up by the cells. Challenge of SMB-PS cells with 22F-infected brain homogenate resulted in consistently high stimulation of PrPSc (24.8% of the colonies, n = 4 experiments) while in SMB-PS exposed to normal mouse brain homogenate, PrPSc remained undetectable (<0.05%, n = 7) (Figure 2). Using 139A or 79A brain homogenates in SMB-PS cell challenges also gave rise to PrPSc-expressing colonies (11 and 27%, respectively) while controls involving challenge of N2a cells with these three strains gave nothing above background (data not shown). A further control applying the hamster-adapted scrapie (263K) on SMB-PS cells also failed to stimulate PrPSc-positive colonies. We did not vary the infectious titre used for these challenges, nor was each source titred prior to use. Based on typical clinical mouse brains from these scrapie strains containing 3 × 108 LD50 units per gram, our challenge would employ ∼50 LD50 units per cell. Hamster 263K clinical brain usually has a higher titre containing between 1 × 109 and 1 × 1010 LD50 units per gram.
Fig. 2. Scrapie brain but not normal brain stimulates PrPSc synthesis after in vitro challenge. SMB-PS cells were exposed to a homogenate of either normal mouse brain (A and B) or 22F-infected brain (C and D); the donor mice were of the VM(Sincs7) strain. Filters from identical cultures were subjected to development with anti-PrP antibody (IA8) either without proteinase K (PK) treatment, giving the total clone density (A and C), or after treatment with 100 µg/ml PK (B and D), showing only those clones containing PrPSc.
Given that a substantial proportion of 22F-challenged clones of SMB-PS could be stimulated to express PrPSc, we proceeded to repeat 22F challenges of SMB-PS cells and, instead of the destructive colony-lift procedure, selected 40 colonies at random and picked them using cloning rings. The expansion of these colonies from 6.4 mm diameter wells to 60 mm diameter dishes was done over six passages and, in those lines where sufficient cells had grown to permit western blot analysis on a portion of each culture, seven of these remained positive for PrPSc expression. Importantly, in these seven lines, the synthesis of PrPSc appeared to be stable through multiple successive passages.
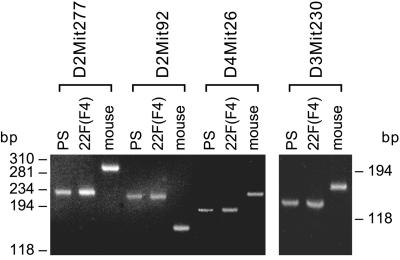
Since SMB cells were originally grown from dispersed cells from whole mouse brain, it was relevant to confirm that these clones originated from 22F-infected SMB-PS rather than from outgrowth of surviving cells in the 22F scrapie brain homogenate. The founder SMB (and therefore SMB-PS) cells were derived from the Compton White mouse strain while the 22F scrapie brain homogenate was obtained from a VM(Sincs7) congenic mouse. These two mouse strains and their derivative cells have the same PrP genotype but can be distinguished readily by microsatellite genetic analysis. Of the eight microsatellite loci tested, four were informative since they generated PCR products of the same size from DNA isolated from SMB-PS and 22F-challenged SMB-PS cells (Figure 3) cells, but of a distinctly different size to that produced on DNA from the VM(Sincs7) congenic mouse. The PrPSc-positive 22F-challenged SMB-PS lines were therefore derived from SMB-PS cells and did not originate from the mouse donor of the 22F infection.
Fig. 3. The same lineage of SMB-PS and 22F-infected cells was determined by microsatellite analysis. PCR-microsatellite analysis using the indicated primers was performed on DNA from SMB-PS cells (PS), 22F-challenged SMB-PS cells, clone F4 [22F (F4)] and the VM(Sincs7) mouse (mouse), the 22F-infected donor. Size calibration in base pairs (bp) is shown to left and right.
Comparison of strain characteristics
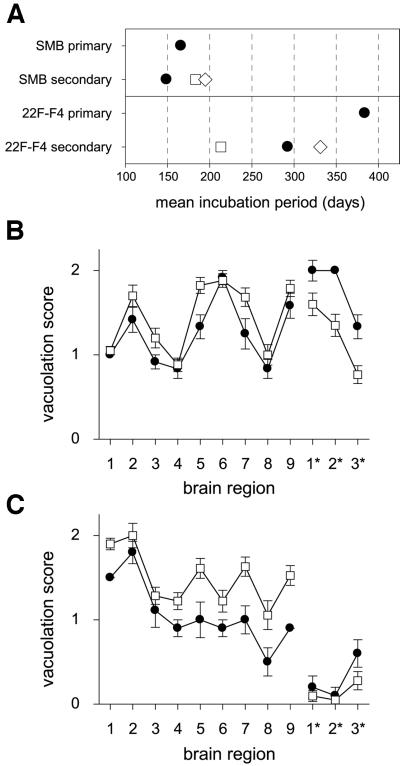
The biological phenotype (incubation period and lesion profile) of the infectious agent in one of the re-infected lines, SMB[22F]F4, was determined at the 14th passage after its infection and compared with that of the infectious agent present in the original parent SMB cell. Clinical signs characteristic of scrapie developed in all the mice inoculated with SMB or SMB[22F]F4 cells, and confirmation of a scrapie infection in every recipient was obtained by standard histological procedures and by western blot analysis of PrPSc. Consistent with their respective strain properties, the incubation period for mice injected with the SMB cell homogenate (infected with the Chandler strain; 166 ± 1.6 days) was shorter than for mice injected with the SMB[22F]F4 cell homogenate (infected with 22F; 384 ± 9.5 days) (Figure 4A). Although the lengths of these incubation periods cannot be compared directly because the infectious titres were not measured by serial dilution, the incubation periods of SMB[22F]F4-inoculated mice stretch beyond the survival time of animals infected with a limiting dose of the Chandler/SMB agent (250 days maximum; data not shown). To obtain the optimal differentiation of the biological scrapie strain characteristics of the two cell types, brain material from this primary transmission was inoculated into mice of all three Sinc genotypes constituting a secondary transmission experiment.
Fig. 4. Strain typing of cell-derived scrapie revealed distinctive characteristics. (A) Comparative ranking of the scrapie incubation periods in mice at the primary and secondary transmissions. Primary transmissions were established by injecting 7.5 × 104 cell equivalents intracerebrally into mice, and secondary transmissions were made by injecting 0.03 ml of a 10–2 homogenate of the brain taken from a single primary transmission mouse once it had reached the clinical stage of the disease. Strains of mice used were C57BL (Sincs7; filled circles), VM (Sincp7; open squares) and their F1 cross (Sincs7p7; open diamonds). Each point is the mean of 12 mice (primary transmissions) or 20–24 mice (secondary transmissions); the error bars fall within the boundaries of the symbols and are therefore not seen. (B and C) Lesion profiles generated by the primary (filled circles) and secondary transmission (open squares) of cell-borne scrapie; from SMB cells (B) or from SMB[22F]F4 cells (C). In both (B) and (C), each point is the mean score at the specified brain region of between 12 and 22 animals, the bars are 1 SEM. Grey matter regions scored: 1, dorsal medulla; 2, cerebellar cortex; 3, superior colliculus; 4, hypothalamus; 5, medial thalamus; 6, hippocampus; 7, septum; 8, thalamic cortex; 9, forebrain cortex. White matter regions scored: 1*, cerebellar white matter; 2*, mesencephalic tegmentum; 3*, pyramidal tract.
In the secondary transmission from SMB cells, the rank order of incubation periods (Figure 4A) was that expected for the 139A strain. The shortening of the incubation period by 17 days in the C57BL mouse between the primary and secondary transmissions is entirely consistent with a higher infectious dose in brain than in cultured cells. The rank order of incubation periods resulting from the secondary transmission from SMB[22F]F4 cells was entirely characteristic of the 22F scrapie strain and revealed the singularly most informative feature of this scrapie strain; of all the commonly Sincs7-passaged strains, 22F is unique in that it effects a shorter incubation period in Sincp7 than in Sincs7 recipients (Bruce et al., 1992). There was a 91 day difference between the incubation periods of the C57BL mouse in the primary and secondary transmissions, too large to be accounted for entirely by dosage but which could be accounted for by the passage history of the 22F source infectious brain that we used. Strain 22F arises when the Sincp7-derived scrapie strain 22A is passed in Sincs7 mice; over the course of three serial passages, the incubation period in C57BL mice shortens from ∼460 days to ∼290 days (Bruce et al., 1992), at which it is stabilized. Our 22F infectious source was 22A that had been passaged once in a Sincs7 mouse and would therefore give C57BL incubation periods intermediate on this scale. It therefore raises the intriguing possibility that the 22F in SMB[22F]F4 cells has been ‘frozen’ with a phenotype intermediate between early 22F and stable 22F. Irrespect ive of this incubation period phenomenon, its identification as 22F from the ranking of incubation periods is unassailable.
Our conclusion that the scrapie strains in SMB and SMB[22F]F4 cells were different from each other was reinforced by histopathological analysis of the brains of recipient mice. The patterns of brain pathology of C57BL mice infected with the two cell lines were clearly distinct at both primary and secondary transmission levels. The vacuolation lesion profiles generated in mice inoculated with SMB cells (Figure 4B) were largely characteristic of the Chandler strain and were substantially different from those generated by inoculation with the re-infected SMB[22F]F4 clone which were characteristic of 22F-infected mice (Figure 4C). Most striking were the differences in the three white matter areas, which were almost devoid of lesions following inoculation with SMB[22F]F4, whereas the high vacuolation scores in these white matter tracts following SMB infection were entirely characteristic of the Chandler scrapie isolate and its derivative 139A strain in C57BL mice. With both strains, there were minor differences in the severity of vacuolation between primary and secondary transmission, an effect that can be related to initial dose, but in both strains the specific trends were constant between first and second pass.
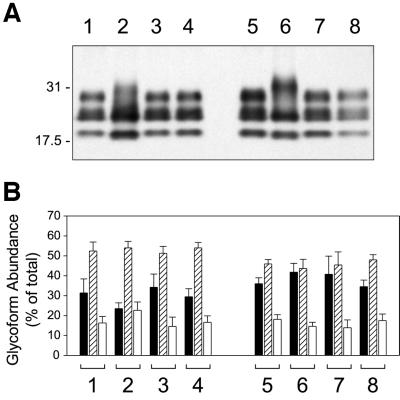
A more recently described scrapie strain-related characteristic is the relative abundance and molecular masses of the three PrPSc isoforms, the so-called PrP glycoform profile. Western blot analyses of PK-treated PrPSc isolated from the two cell lines indicated that there was a major difference in the relative amounts of the diglycosyl form of PrPSc from SMB and SMB[22F]F4 cells (Figure 5A). SMB cell PrPSc always contained less diglycosyl PrP than that of SMB[22F]F4 cells, distinct patterns that were entirely stable through many passes; the PrPSc western blot profiles used to construct the densitometric data (Figure 5B) were collected intermittently from cells kept in continuous culture for >1 year (>50 passages). The ratio diglycosyl/monoglycosyl PrPSc (D/M) from SMB cells was 0.46 ± 0.19 (mean ± SD; n = 11) compared with that from SMB[22F]F4 of 0.98 ± 0.31 (n = 14), marking the significance of these observations (ANOVA, P <0.05). A low D/M was not only evident for SMB cells but also for standard 139A (and 79A, not shown) brain PrPSc and also in the SMB cell primary and secondary transmission mice (Figure 5B, groups 1, 3 and 4). Compared with these Chandler-infected cells and brains, PrPSc from standard 22F brain, SMB[22F]F4 cells and the 22F cell-transmitted brains (Figure 5B, groups 5–8) all had higher D/M ratios. Whereas the PrPSc glycoform profiles of the two cell lines were visually distinctive (Figure 5A, lanes 2 and 6), the differences between the brain samples were not so easily distinguished without densitometry, which together with the natural variations of the brain glycoform samples reinforces earlier findings that scrapie strains with diverse biological phenotypes are not necessarily endowed with very different PrPSc glycoform profiles (Somerville et al., 1997).
Fig. 5. PrPSc isoform analysis of infected cells revealed distinct differences both between strains and between cells and brains. (A) Western blotting was used to compare PK-treated PrPSc derived from mouse brain with that from SMB and SMB[22F]F4 cells. The lanes were loaded as follows: lane 1, 139A reference mouse brain sample; lane 2, SMB cells, ∼8 × 105 cell equivalents; lane 3, mouse brain from primary transmission of SMB cells; lane 4, mouse brain from secondary transmission of SMB cells; lane 5, 22F reference mouse brain sample; lane 6, SMB[22F]F4 cells, ∼3 × 105 cell equivalents; lane 7, mouse brain from primary transmission of SMB[22F]F4 cells; and lane 8, mouse brain from secondary transmission of SMB[22F]F4 cells. Each brain sample was ∼40 µg wet weight of tissue. Molecular weight standards (kDa) are marked on the left. (B) Densitometric analysis constructed from a number of western-blotted samples developed with the IA8 antiserum. Grouped bars depicting the diglycosyl (filled bar), monoglycosyl (hatched bar) and unglycosylated (open bar) PrPSc isoforms are numbered the same as in (A). The columns are the mean of between six and 12 brains (samples 1, 3–5, 7 and 8) or from 11 or 14 cell samples (samples 2 and 6, respectively) collected intermittently over a 50 passage series; the bars represent 1 SD.
A feature common to PrPSc isolated from both cell lines was that the apparent relative molecular weights of the unglycosylated and diglycosylated PrP isoforms were different from those in all of the brain-derived PrPSc samples. Cell-derived unglycosylated PrP migrated faster than the brain-derived isoform by some 100–150 Da, while the diglycosyl isoform from the cells was both more disperse and of different mean molecular weight, higher than the respective brain isoform by some 1.5–2.0 kDa. The reason for these disparities is not evident from simple one-dimensional SDS–PAGE analysis but, since such differences have been described in other studies as being due to conformational differences, will bear further investigation.
Discussion
SMB cells were isolated as an infected cell line; therefore, there was no uninfected line of comparable cells to use as experimental controls. The persistence of scrapie infectivity in SMB through >150 passages (Clarke, 1979), with a constant infectious titre, was indicative of a high degree of competence as a host cell for scrapie. To exploit this competence in a study of scrapie strain determinants, we would first need to isolate a revertant, scrapie-free subline of SMB, but our search for subclones spontaneously ‘cured’ of PrPSc was unsuccessful. PS enabled us to redress this as it proved to be effective at inhibiting PrPSc accumulation, though when compared with the IC50 of 1 × 10–9 g/ml obtained with ScMNB cells (Priola and Caughey, 1994) SMB cells were far less sensitive to PS than the neuroblastoma line. There is no clear reason for this difference in sensitivity, but we can speculate that fundamental differences in cell type might be responsible. The mode of action of PS is uncertain although it is known to bind directly to isolated PrPC (Caughey et al., 1994) and recombinant PrP (Brimacombe et al., 1999) and to cause the subcellular re-distribution of PrPC expressed in mouse neuroblastoma cells (Shyng et al., 1995), so a direct interaction of PS with PrP during the curing process is implicated. Since polyanionic glycosaminoglycans such as PS can bind to numerous cell surface ligands, in particular to components of the extracellular matrix, differences in levels of expression of such ligands between SMB and neuroblastoma cells might serve to sequester PS to a greater extent and thereby modify the amount of polyanion available for binding PrP.
It is probable that the susceptibility of a cell to infection with a TSE agent and its ability to sustain the replication of that infection are governed by separate factors. There are several reports of the ability to infect different types of cell with scrapie in vitro but because SMB had not been raised in this way originally we had to determine its sensitivity to in vitro infection directly. Our finding, therefore, that >20% of 22F-challenged SMB-PS cells growing out as clonal colonies had been stimulated to express PrPSc was encouraging. In contrast, the wild-type N2a neuroblastoma-derived cells had only a low frequency of PrPSc stimulation, consistent with reports that maximally ∼1% of cells became infected when challenged with similar infectious doses by the in vitro challenge route (Butler et al., 1988; Race et al., 1988). Two recent reports both employing N2a and GT-1 cells, the latter is an immortalized mouse hypothalamic cell line that is sensitive to infection with scrapie (Schätzl et al., 1997), have concluded that either overexpression of wild-type PrPC (Nishida et al., 2000) or simply pre-selection of cells intrinsically more sensitive to infection (Bosque and Prusiner, 2000) can directly increase the success rate of in vitro infection.
There were two reasons why attaining sustainable infection was one of our aims. The first was simply that any trace of infectivity from the inoculum must be removed by dilution before attempting transmissions to mice. The second was that we wanted a model of infection that is entirely self-sustaining. Cell-free conversion (Kocisko et al., 1994) of PrPC to PrPSc following their mixing in vitro is well established and it has also been demonstrated that PrP-associated strain-dependent criteria can be transmitted in this way (Bessen et al., 1995). It is possible, therefore, that the application of exogenous PrPSc to PrPC-bearing cells might convert PrPC upon contact whether or not this conversion would ultimately become established as a sustained process within the cell. Studying de novo PrPSc synthesis close to the time of challenge of the cells with exogenous PrPSc, or at any time prior to the complete removal by dilution of the PrPSc contained in the challenge material, might therefore be more analogous to studying cell-free conversion and less relevant to the study of the cellular components required for sustainable TSE replication. The successful challenge of SMB-PS cells with infectious scrapie confirmed our hypothesis that this cell type is a competent scrapie host. Our observation that PrPSc expression remained constant over >50 passages following infection with 22F without requiring subcloning indicates the inherent stability of this cell system.
Our demonstration that highly specific strain-related biological phenotypic properties of both the Chandler and 22F strains can be propagated reliably in a single cell type in vitro goes beyond the initial level of study of different scrapie strains in culture, which has been the infection of cells. Rubenstein et al. (1994) showed that the rat PC12 cell line could be infected by different scrapie strains and that resultant alterations in metabolic functions might be strain specific. However, scrapie strain properties of the emergent PC12 cultures were not reported. More recently, various scrapie strains have been used to challenge N2a or GT-1 cell-based models, and some degree of PrPSc glycoform differences has been reported (Nishida et al., 2000). The fact that our different strain-infected SMB-type cells express scrapie strain-related PrPSc glycoforms suggests that this will be a useful way to provide early indication of strain-specific reproduction prior to the more lengthy but requisite biological strain phenotyping.
The introduction of the 22F strain has been particularly informative in this study. Comparison of scrapie strains by incubation period alone is not ideal, particularly in the primary transmission from the cell where infectious titre is always relatively low. The specific ranking of incubation period given by 22F in mice of the three Sinc genotypes is unique in this context. Not only will these cells provide a useful substrate for the study of these scrapie strains in particular and strain-related phenomena in general, but they also signify that other disease-specific TSE cultured cell models can be generated.
Materials and methods
Cell culture
SMB cells were restored from a stock frozen in 10% (v/v) dimethylsulfoxide (DMSO) at the 74th passage. They were grown in tissue culture plastic dishes in Medium 199 supplemented with 10% newborn bovine serum and 5% fetal bovine serum of Australian origin at 33°C in an atmosphere of 5% CO2 in air. Medium was changed every 3rd or 4th day, and every 7th–10th day the confluent cells were passaged using 0.05% trypsin, 0.002% EDTA, at a split ratio of between 3 and 5. Although suffering a poor cloning efficiency, cloning was successful either by limiting dilution or by low density seeding into 90 mm culture plates with selection using cloning rings. One typical clone was selected for all the subsequent work, clone SMB.s15 (referred to throughout as SMB); it exhibited a population doubling time of 33 h at 33°C. A report that scrapie infection of cells in vitro was generally more productive when cultured at a lower temperature led us to use 33°C (Taraboulos et al., 1990).
Quantitative PrPSc dot-blot assay
Cells growing in 24-well cluster plates were extracted with 0.1 ml of lysis buffer [LyB; 10 mM Tris–HCl pH 7.6, 100 mM NaCl, 10 mM EDTA, 0.5% (v/v) NP-40, 0.5% (w/v) sodium deoxycholate], centrifuged (1000 g, 5 min) and the post-nuclear supernatant applied under gentle vacuum to a nitrocellulose membrane (0.45 µm; BA85; Schleicher and Schuell) via a 96-well dot-blot manifold. The membrane was air-dried thoroughly before application of the in situ processing to discriminate PrPSc from PrPC (adapted from Taraboulos et al., 1990). Briefly, the membranes were immersed in a 75 µg/ml solution of PK in 20 mM Tris–HCl-buffered saline (TBS) for 60 min at 37°C, stopped with 1 mM phenylmethylsulfonyl fluoride (PMSF), washed extensively with TBS, then immersed in 3.0 M guanidine thiocyanate in TBS for 10 min at room temperature to denature the residual PrPSc and expose epitopes. After further extensive washing, the membrane was blocked in 5% fat-free milk powder in phosphate-buffered saline (PBS) then processed with rabbit polyclonal anti-PrP antiserum IB3 or IA8 and developed using ECL (Amersham Pharmacia Biotech). Relative quantitation was made by normalizing loading to cellular protein, measured by detergent-compatible Lowry assay (DC Assay; Bio-Rad) using bovine serum albumin (BSA) as standard, and densitometric analysis of multiple unsaturated films using Phoretix Array 2 software (Non-Linear Dynamics, Newcastle-upon-Tyne), results being normalized to a fixed number of untreated SMB cells.
In vitro scrapie infection and colony-lift blotting
Cultures of 5 × 104 SMB-PS cells in 0.4 ml of medium seeded in a single well of a 24-well plate and allowed to establish for 24 h were exposed to 0.1 ml of brain homogenate [10% (w/v) in PBS] of normal mouse brain or scrapie-infected brain in the 33°C incubator. After 4 h, this was supplemented with an additional 2 ml of complete medium and incubation continued overnight. They were washed extensively with PBS then grown until confluent, at which time they were trypsinized and plated into 10 cm diameter plates at low density and grown on for 4–6 weeks to allow growth of discrete colonies. To obtain a gross numerical analysis of PrPSc-producing colonies, albeit not a true estimate of the frequency of infection because of their low cloning efficiency, these plates were subjected to a colony-lift procedure utilizing PVDF filters soaked in LyB, based on that described by Taraboulos et al. (1990). Except that the filters were not air-dried and that we used 100 µg/ml PK to differentiate PrPC from PrPSc, the blot development procedure was similar to dot-blot processing (above). Colonies in the central 85% of the diameter of the filters (so avoiding edge effects) were enumerated with the aid of SigmaScan (v2.5) software (SPSS Inc.). Alternatively, suitably isolated colonies were selected using cloning rings and expanded gradually by continuing culture.
Infectious material and lesion profiling
Infectious mouse scrapie brain material was derived by serial passage of scrapie strains of known lineage by 0.02 ml intracerebral injection under halothane anaesthesia of a 10% (w/v) homogenate in saline of brain from a clinically infected mouse. When determined to be in the clinical phase of the disease, brain tissue was dissected out and stored frozen at –80°C. The source of 22F used was a single pass of cloned strain 22A in a VM(Sincs7) mouse which converts 22A irreversibly to 22F.
To challenge mice directly from cultured cells, cultures from two or three dishes at 80–90% confluence were washed twice with PBS then scraped into PBS, pooled and put through one rapid freeze–thaw cycle to disrupt them; cells from identical cultures grown in parallel were trypsinized and counted in a haemacytometer chamber to establish numbers. Ethanol (2.3 vols) was added to the freeze–thaw homogenate which was then stored at 4°C for 24 h to (nominally) kill adventitious agents. The precipitate was collected and washed twice with PBS by 10 000 g centrifugation, finally being resuspended using cup-horn sonication (Heat Systems) and thorough homogenization in sterile 0.9% (w/v) saline at 2.5 × 106 cell equivalents per ml. Mice under halothane anaesthesia were injected intracerebrally with 30 µl (7.5 × 104 cell equivalents) of this homogenate. Mice continued to be monitored for clinical signs of scrapie; after reaching a pre-defined clinical end point, or surviving to ∼800 days post-inoculation, they were killed and their brains rapidly dissected out. Brains were sectioned coronally and one part fixed with formaldehyde prior to histological analysis of the pathological lesion profile (Fraser, 1976) while the remainder was snap-frozen and stored at –80°C for later biochemical analysis or for serial subpassage in mice as described above. All animal work was conducted strictly according to local and national guidelines.
Microsatellite analysis
This was done essentially by standard methods (Hearne et al., 1992). Genomic DNA prepared from cells or from brain tissue using a commercial kit (Qiagen) was used to prime single-stranded length polymorphism (SSLP; microsatellite) PCRs using eight sets of Map-Pair primer pairs (Research Genetics Inc.): D1Mit47, D2Mit92, D2Mit277, D3Mit230, D4Mit26, D7Mit259, D13Mit202 and D17Mit93. We used Tth DNA polymerase (Roche Molecular Biochemicals) in conditions otherwise specified by the primer pair vendor. After 25 cycles of amplification, the DNA products were separated on 2.0 or 2.5% Metaphor agarose (FMC Ltd.) gels in TAE buffer and stained with ethidium bromide. Since these primer pairs were chosen at random, only four pairs gave meaningful data, while the other four gave bands of identical size irrespective of the target DNA.
Ultracentrifugal preparation of PrPSc for glycoform analysis
Cells growing in 60 mm culture plates were rinsed twice with PBS then scraped from the plate in 1.0 ml of LyB and nuclei and cytoskeletal remnants were removed by centrifugation at 2000 g for 5 min. Brain tissue was homogenized at 5% (w/v) in LyB and centrifuged in the same way. The post-nuclear extract of either type of sample was adjusted to 1 mg/ml protein (DC assay; Bio-Rad) and PK added to 25 µg/ml before 37°C, 60 min incubation after which PMSF was added (1 mM) to inhibit PK. The sample was layered over a 0.25 ml of 20% sucrose, 1 mM PMSF cushion in a conical polyallomer ultracentrifuge tube (sample volume was adjusted to fit by addition of LyB containing 1 mM PMSF) before centrifugation [300 000 g(max), 10°C, 60 min] in a SW50.1 rotor (Beckman). The pellet and ∼0.1 ml of cushion were resuspended in 2 ml of water by sonication (cup-horn) then centrifuged [541 000 g(max), 10°C, 15 min] in a polycarbonate tube in a TLA-100.3 rotor (Beckman). The PrPSc-containing pellet was drained thoroughly before solubilization for electrophoresis.
Western blot analysis
Proteins that were obtained by methanol/chloroform precipitation (Wessel and Flugge, 1984) of post-nuclear LyB detergent extracts of cells or by ultracentrifugation were dispersed by sonication (cup-horn) in a modified electrophoresis sample buffer (Laemmli, 1970) containing 4% (w/v) SDS, 5% (v/v) 2-mercaptoethanol, 62.5 mM Tris–HCl pH 6.8, 10% (w/v) sucrose and dissolved by heating at 100°C for 5 min. Proteins were separated on 12.5 or 15% polyacrylamide gels in Tris-glycine-SDS buffer and electrophoretically transferred to PVDF membranes (Immobilon P, Millipore) in 25 mM Tris, 192 mM glycine, 0.05% SDS. Membranes were blocked with 5% fat-free milk powder in PBS (MPBS) for a minimum of 1 h. Polyclonal anti-PrP antiserum IA8 diluted 1:5000 in MPBS was used as primary antibody; blots were developed with ECL reagents (Amersham-Pharmacia Biotech). Non-saturating films were scanned (Epson GT9600 in transmission mode), then calibrated and analysed using Phoretix 1D v4 software (Non-Linear Dynamics Ltd).
Acknowledgments
Acknowledgements
We thank our many colleagues at IAH-Compton and IAH-Edinburgh for valuable discussion and also for the supply of anti-PrP sera, especially Christine Farquhar, Robert Somerville and James Hope, also to Kate Fergusson, Louise Gibbard and Natasha Fielder for excellent technical assistance. This work was funded by Biotechnology and Biological Sciences Research Council and, additionally, at IAH-Edinburgh, by the Medical Research Council of the UK.
References
- Aguzzi A. and Weissmann,C. (1997) Prion research: the next frontiers. Nature, 389, 795–798. [DOI] [PubMed] [Google Scholar]
- Bessen R.A. and Marsh,R.F. (1994) Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol., 68, 7859–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen R.A., Kocisko,D.A., Raymond,G.J., Nandan,S., Lansbury,P.T. and Caughey,B. (1995) Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature, 375, 698–700. [DOI] [PubMed] [Google Scholar]
- Bosque P.J. and Prusiner,S.B. (2000) Cultured cell sublines highly susceptible to prion infection. J. Virol., 74, 4377–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe D.B., Bennett,A.D., Wusteman,F.S., Gill,A.C., Dann,J.C. and Bostock,C.J. (1999) Characterization and polyanion-binding properties of purified recombinant protein. Biochem. J., 342, 605–613. [PMC free article] [PubMed] [Google Scholar]
- Bruce M.E. (1993) Scrapie strain variation and mutation. Br. Med. Bull., 49, 822–838. [DOI] [PubMed] [Google Scholar]
- Bruce M.E., Fraser,H., McBride,P.A., Scott,J.R. and Dickinson,A.G. (1992) The basis of strain variation in scrapie. In Prusiner,S.B., Collinge,J., Powell,J. and Anderton,B. (eds), Prion Diseases of Humans and Animals. Ellis-Horwood, Chichester, pp. 497–508.
- Butler D.A., Scott,M.R.D., Bockman,J.M., Borchelt,D.R., Taraboulos,A., Hsiao,K.K., Kingsbury,D.T. and Prusiner,S.B. (1988) Scrapie-infected murine neuroblastoma cells produce protease-resistant prion proteins. J. Virol., 62, 1558–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B. and Raymond,G.J. (1993) Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J. Virol., 67, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B., Raymond,G.J., Ernst,D. and Race,R.E. (1991) N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s). J. Virol., 65, 6597–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B., Brown,K., Raymond,G.J., Katzenstein,G.E. and Thresher,W. (1994) Binding of the protease-sensitive form of prion protein PrP to sulfated glycosaminoglycan and Congo red. J. Virol., 68, 2135–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B., Raymond,G.J. and Bessen,R.A. (1998) Strain-dependent differences in β-sheet conformations of abnormal prion protein. J. Biol. Chem., 273, 32230–32235. [DOI] [PubMed] [Google Scholar]
- Chandler R.L. (1961) Encephalopathy in mice produced with scrapie brain material. Lancet, i, 1378–1379. [DOI] [PubMed] [Google Scholar]
- Chen S.G., Teplow,D.B., Parchi,P., Teller,J.K., Gambetti,P. and Autiliogambetti,L. (1995) Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem., 270, 19173–19180. [DOI] [PubMed] [Google Scholar]
- Clarke M.C. (1979) Infection of cell cultures with scrapie agent. In Prusiner,S.B. and Hadlow,W.J. (eds), Slow Transmissible Diseases of the Nervous System. Academic Press, New York, NY, pp. 225–233.
- Clarke M.C. and Haig,D.A. (1970) Evidence for the multiplication of scrapie agent in cell culture. Nature, 225, 100–101. [DOI] [PubMed] [Google Scholar]
- Collinge J., Sidle,K.C.L., Meads,J., Ironside,J.W. and Hill,A.F. (1996) Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature, 383, 685–690. [DOI] [PubMed] [Google Scholar]
- DeArmond S.J. et al. (1997) Selective neuronal targeting in prion disease. Neuron, 19, 1337–1348. [DOI] [PubMed] [Google Scholar]
- DeArmond S.J., Qiu,Y., Sanchez,H., Spilman,P.R., Ninchak-Casey,A., Alonso,D. and Daggett,V. (1999) PrPC glycoform heterogeneity as a function of brain region: implications for selective targeting of neurons by prion strains. J. Neuropathol. Exp. Neurol., 58, 1000–1009. [DOI] [PubMed] [Google Scholar]
- Dickinson A.W., Outram,G.W., Taylor,D.M. and Foster,J.D. (1986) Further evidence that scrapie agent has an independent genome. In Court,L.A., Dormont,D., Brown,P. and Kingsbury,D.T. (eds), Unconventional Virus Diseases of the Central Nervous System. Commisariat à l’Energie Atomique, Paris, France, pp. 446–460.
- Diringer H. and Ehlers,B. (1991) Chemoprophylaxis of scrapie in mice. J. Gen. Virol., 72, 457–460. [DOI] [PubMed] [Google Scholar]
- Fraser H. (1976) The pathology of natural and experimental scrapie. In Kimberlin,R.H. (ed.), Slow Virus Diseases of Animals and Man. North-Holland, Amsterdam, The Netherlands, pp. 267–305.
- Haig D.A. and Clarke,M.C. (1971) Multiplication of the scrapie agent. Nature, 234, 106–107. [DOI] [PubMed] [Google Scholar]
- Hearne C.M., Ghosh,S. and Todd,J.A. (1992) Microsatellites for linkage analysis of genetic traits. Trends Genet., 8, 288–294. [DOI] [PubMed] [Google Scholar]
- Kocisko D.A., Come,J.H., Priola,S.A., Chesebro,B., Raymond,G.J., Lansbury,P.T. and Caughey,B. (1994) Cell-free formation of protease-resistant prion protein. Nature, 370, 471–474. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Masison D.C. (2000) Expanding the prion model for the yeast [PSI+] element. Trends Microbiol., 8, 1–2. [DOI] [PubMed] [Google Scholar]
- McKinley M.P., Taraboulos,A., Kenaga,L., Serban,D., Stieber,A., DeArmond,S.J., Prusiner,S.B. and Gonatas,N. (1991) Ultrastructural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cells. Lab. Invest., 65, 622–630. [PubMed] [Google Scholar]
- Moore R.C., Hope,J., McBride,P.A., McConnell,I., Selfridge,J., Melton,D.W. and Manson,J.C. (1998) Mice with gene targeted prion protein alterations show that Prnp, Sinc and Prni are congruent. Nature Genet., 18, 118–125. [DOI] [PubMed] [Google Scholar]
- Naslavsky N., Stein,R., Yanai,A., Friedlander,G. and Taraboulos,A. (1997) Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J. Biol. Chem., 272, 6324–6331. [DOI] [PubMed] [Google Scholar]
- Nishida N., Harris,D.A., Vilette,D., Laude,H., Frobert,Y., Grassi,J., Casanova,D., Milhavet,O. and Lehmann,S. (2000) Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol., 74, 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchi P. et al. (1998) Different patterns of truncated prion protein fragments correlate with distinct phenotypes in P102L Gerstmann–Straussler–Scheinker disease. Proc. Natl Acad. Sci. USA, 95, 8322–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison I.H. (1972) Scrapie—a personal view. J. Clin. Pathol. Suppl. (R. Coll. Pathol.), 6, 110–114. [PMC free article] [PubMed] [Google Scholar]
- Pattison I.H. and Millson,G.C. (1961) Scrapie produced experimentally in goats with special reference to the clinical syndrome. J. Comp. Pathol., 71, 101–108. [DOI] [PubMed] [Google Scholar]
- Priola S.A. and Caughey,B. (1994) Inhibition of scrapie-associated PrP accumulation—probing the role of glycosaminoglycans in amyloidogenesis. Mol. Neurobiol., 8, 113–120. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B. (1982) Novel proteinaceous infectious particles cause scrapie. Science, 216, 136–144. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B. (1998) Prions. Proc. Natl Acad. Sci. USA, 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race R.E., Fadness,L.H. and Chesebro,B. (1987) Characterisation of scrapie infection in mouse neuroblastoma cells. J. Gen. Virol., 68, 1391–1399. [DOI] [PubMed] [Google Scholar]
- Race R.E., Caughey,B., Graham,K., Ernst,D. and Chesebro,B. (1988) Analysis of frequency of infection, specific infectivity, and prion protein biosynthesis in scrapie-infected neuroblastoma cell clones. J. Virol., 62, 2845–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein R., Deng,H., Race,R., Ju,W., Scalici,C., Papini,M., Rubenstein,A., Kascsak,R. and Carp,R. (1994) Scrapie strain infection in vitro induces changes in neuronal cells. Mol. Neurobiol., 8, 129–138. [DOI] [PubMed] [Google Scholar]
- Safar J., Wille,H., Itri,V., Groth,D., Serban,H., Torchia,M., Cohen,F.E. and Prusiner,S.B. (1998) Eight prion strains have PrPSc molecules with different conformations. Nature Med., 4, 1157–1165. [DOI] [PubMed] [Google Scholar]
- Schätzl H.M., Laszlo,L., Holtzman,D.M., Tatzelt,J., DeArmond,S.J., Weiner,R.I., Mobley,W.C. and Prusiner,S.B. (1997) A hypothalamic neuronal cell line persistently infected with scrapie prions exhibits apoptosis. J. Virol., 71, 8821–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyng S.L., Lehmann,S., Moulder,K.L. and Harris,D.A. (1995) Sulfated glycans stimulate endocytosis of the cellular isoform of the prion protein, PrPC in cultured-cells. J. Biol. Chem., 270, 30221–30229. [DOI] [PubMed] [Google Scholar]
- Somerville R.A., Chong,A., Mulqueen,O.U., Birkett,C.R., Wood,S.C.E.R. and Hope,J. (1997) Biochemical typing of scrapie strains. Nature, 386, 564. [DOI] [PubMed] [Google Scholar]
- Taraboulos A., Serban,D. and Prusiner,S.B. (1990) Scrapie prion proteins accumulate in the cytoplasm of persistently infected cultured cells. J. Cell Biol., 110, 2117–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Raeber,A.J., Borchelt,D.R., Serban,D. and Prusiner,S.B. (1992) Synthesis and trafficking of prion proteins in cultured cells. Mol. Biol. Cell, 3, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telling G.C. et al. (1996) Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science, 274, 2079–2082. [DOI] [PubMed] [Google Scholar]
- Vey M., Pilkuhn,S., Wille,H., Nixon,R., DeArmond,S.J., Smart,E.J., Anderson,R.W., Taraboulos,A. and Prusiner,S.B. (1996) Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc. Natl Acad. Sci. USA, 93, 14945–14949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C. (1991) A ‘unified theory’ of prion propagation. Nature, 352, 679–683. [DOI] [PubMed] [Google Scholar]
- Wessel D. and Flugge,U.I. (1984) A method for the quantitative recovery of protein in dilute-solution in the presence of detergents and lipids. Anal. Biochem., 138, 141–143. [DOI] [PubMed] [Google Scholar]