Introduction
In order for the genome to be faithfully maintained, chromosomal DNA must be precisely replicated and segregated in each cell cycle. Over the last decade an enormous amount has been learned about how this is achieved. Much of the progress has come from genetic analysis in yeast. However, biochemical analysis of cell cycle events using extracts prepared from eggs of the South African clawed toad Xenopus laevis has also played an important role. Although there are differences in the detailed regulation of the yeast and frog cell cycles, the basic cell cycle machinery appears to have been conserved throughout evolution. The results obtained in these model organisms, therefore, seem likely to be generally applicable to the cell cycles of all eukaryotes.
Xenopus eggs and egg extracts
The fertilized Xenopus egg undergoes 12 synchronous rounds of cell division in ∼8 h. These cell divisions take place in the absence of growth, subdividing the large (∼1 mm diameter) single-celled egg into ∼4000 smaller cells. Transcription does not occur during these early embryonic divisions, although translation of pre-existing maternal mRNA continues. Most of the proteins required for cell cycle progression are pre-formed in the egg, and the continuing translation of a single protein (cyclin B) can support passage through the whole cell cycle (Murray and Kirschner, 1989). The stockpile of cell cycle proteins present in the Xenopus egg became exploitable by biochemical means following the development of cell-free extracts that support all the nuclear events of the early embryonic cell division cycle (Lohka and Masui, 1983). Gentle lysis associated with minimal dilution of the cytoplasm yields a ‘low speed supernatant’ that maintains all the cell cycle activities present in the intact egg. These ‘low speed supernatants’ support precise rounds of DNA replication on exogenously added DNA templates, and like the intact egg, only re-replicate DNA after passage through mitosis (Blow and Laskey, 1986, 1988). The initiation of DNA replication is dependent on the template DNA being assembled into a functional nucleus, whilst re-replication is dependent on nuclear disassembly. In recent years, the molecular basis of these dependencies has become clearer.
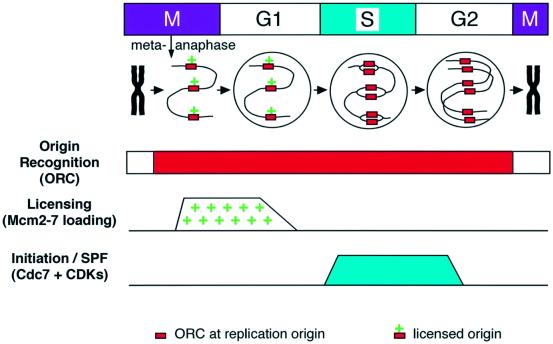
Precise chromosome duplication can be thought of as being dependent on three distinct systems. Figure 1 shows a small segment of chromosomal DNA passing through the cell cycle. The origin recognition system is responsible for positioning replication origins at appropriate sites on the DNA. If any pair of adjacent replication origins were too far apart, the replication forks initiated from them would be unable to replicate all the intervening DNA before the cell passes through mitosis, and the chromosome would be broken. The origin recognition complex (ORC) is specifically recruited to replication origins and is required for origins to function. In the early Xenopus embryo, ORC remains bound to origins throughout interphase (Carpenter et al., 1996; Rowles et al., 1996, 1999). The second system, the replication licensing system, is responsible for ensuring that these origins fire only once in a single cell cycle (Blow and Laskey, 1988). It is activated on exit from metaphase, and supports the loading of Mcm(2–7) complexes onto origins, which ‘licenses’ them for replication in the coming S phase. The third system is S phase-promoting factor (SPF), which provides the trigger for replication forks to initiate at licensed origins. SPF consists of at least two activities: the Cdc7 protein kinase and S phase-promoting cyclin-dependent kinases (CDKs). As each origin initiates, its Mcm(2–7) proteins are displaced from it, thereby ensuring that the origin fires only once in each cell cycle. So long as the licensing system cannot re-license origins once SPF has been activated, no sections of DNA will be replicated more than once. This review will describe current understanding of these three systems in the early Xenopus embryo.
Fig. 1. Three systems required for the precise duplication of chromosomal DNA replication. Top panel, a small segment of DNA carrying three replication origins passing through the cell cycle. Red boxes denote ORC and a plus denotes a licensed origin. The metaphase chromosome decondenses, assembles licensed origins, is assembled into a nucleus, replicates, and then recondenses. Below are the activities of the origin recognition system, the licensing system and SPF during the course of the cell cycle. See text for further details.
The origin recognition system
Replication origins in yeast and in mammalian somatic cells are located at particular DNA sequences (reviewed in DePamphilis, 1999). In Xenopus eggs and egg extracts, however, replication origins are positioned randomly with respect to DNA sequence (Hyrien and Méchali, 1992, 1993; Mahbubani et al., 1992). This conclusion is consistent with earlier experiments showing that no special DNA sequences are required for DNA molecules to be replicated in Xenopus eggs (Harland and Laskey, 1980). However, when transcription starts later in embryonic development, origins become restricted to specific DNA sequences as is seen in somatic cells of other eukaryotes (Hyrien et al., 1995).
Replication forks in the Xenopus embryo move at ∼10 nt/s (Mahbubani et al., 1992), so the two replication forks initiated from a single bi-directional origin can replicate no more than ∼25 kb in each ∼20 min S phase. If origins in the early embryo are positioned randomly with respect to DNA sequence, how then does the early embryo ensure that no two adjacent origins are >25 kb apart? A completely random distribution of origins would give a geometric distribution of inter-origin distances with a significant number >25 kb. In fact, replication origins are not randomly distributed in the Xenopus early embryo, but instead are roughly spaced every 5–15 kb along the chromosomal DNA (Blow et al., 2001). A similar periodic spacing of replication origins has been observed in the Drosophila early embryo (Blumenthal et al., 1974). Since the Drosophila and Xenopus early embryos are both transcriptionally quiescent, this may represent an adaptation for optimizing DNA replication under circumstances where transcription does not occur. The periodic spacing of replication origins can ensure complete chromosome replication by removing the risk of generating excessively large inter-origin distances without using many more origins than are actually needed. It is possible that regularly spaced origins represent a default state that will also be found in the transcriptionally quiescent chromatin of somatic cells.
It seems likely that the periodic spacing of replication origins is mediated by periodic binding of ORC along the chromosomal DNA. Sperm nuclei incubated in Xenopus egg extract becomes saturated with one copy of ORC per 8–15 kb DNA (Rowles et al., 1996). When the quantity of ORC is limited, the spacing between origins increases and the subsequent replication rate drops (Rowles et al., 1999; Blow et al., 2001). It would be interesting to understand how this presumed periodic spacing of ORC is achieved. Two different models can be envisaged. One possibility is that chromosomal DNA is organized into a regular structure, independent of DNA sequence, that provides periodic ORC binding sites. An attractive candidate for such a repeating structure might be the organization of DNA into chromosomal loops. Alternatively, ORC might play an active role in establishing origin spacing, so that once one molecule of ORC has bound to a particular site on the DNA, it sets up an ‘exclusion zone’ where other ORCs cannot bind.
The replication licensing system
Origins are ‘licensed’ for a single round of replication by binding the Mcm(2–7) proteins (Chong et al., 1995; Kubota et al., 1995, 1997; Thömmes et al., 1997). A number of different complexes between these six proteins have been described (reviewed in Kearsey and Labib, 1998), which probably represent different intermediates in a complex assembly/disassembly pathway (Prokhorova and Blow, 2000). Although various Mcm complexes can bind to chromatin, only the Mcm(2–7) heterohexamer (containing one each of Mcm2, 3, 4, 5, 6 and 7) supports functional licensing (Prokhorova and Blow, 2000). Ten to 20 copies of Mcm(2–7) are loaded onto each replication origin in Xenopus extracts (as also occurs in yeast and mammalian cells), although maximal replication rates are still observed when this is reduced to only ∼2 copies per origin (Mahbubani et al., 1997). While Mcm(2–7) are essential for DNA replication, their precise role remains unclear. They possess DNA helicase activity, and one plausible idea is that they act as a helicase to unwind DNA ahead of the replication fork (reviewed in Labib and Diffley, 2001). This would also provide an explanation for how Mcm(2–7) are displaced from each origin as it initiates, an essential part of the licensing model (Figure 1).
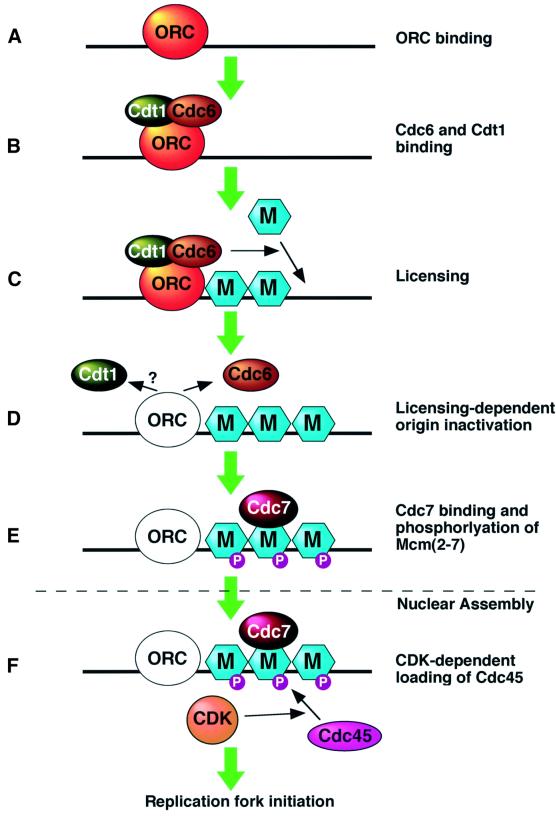
The licensing system must be inactivated before the start of S phase so that re-replication of DNA does not occur. However, Mcm(2–7) levels and activity do not appear to fluctuate significantly during the cell cycle (Mahbubani et al., 1997; Kearsey and Labib, 1998). Instead licensing is regulated by other proteins that are required for Mcm(2–7) to be loaded onto origins. The presence of ORC on the chromatin is essential for origin licensing to occur (Figure 2A) (Coleman et al., 1996; Romanowski et al., 1996; Rowles et al., 1996). Once ORC is bound, two further proteins called Cdc6 and Cdt1 (also known as RLF-B) are then required for licensing (Coleman et al., 1996; Maiorano et al., 2000; Nishitani et al., 2000; Tada et al., 2001). Both Cdc6 and Cdt1 bind to the ORC-containing chromatin, probably forming a complex (Figure 2B). Licensing can then occur, which results in multiple copies of the Mcm(2–7) heterohexamer being loaded onto chromatin (Figure 2C). Mcm(2–7) can bind to chromatin independently from the other proteins since ORC, Cdc6 and Cdt1 can be removed from chromatin whilst leaving functional Mcm(2–7) still bound (Donovan et al., 1997; Hua and Newport, 1998; Rowles et al., 1999; Maiorano et al., 2000). However, once licensing is complete, the quantity of chromatin-bound Cdc6 drops, a process called ‘licensing-dependent origin activation’ (Figure 2D) (Rowles et al., 1999; Jares and Blow, 2000). At the same time, the association of ORC with DNA changes, so that it binds less tightly to the DNA. Since ORC and Cdc6 are no longer required for DNA replication once licensing is complete (Hua and Newport, 1998; Rowles et al., 1999), this weakened binding may help to prevent re-licensing of replicated DNA.
Fig. 2. Events occurring at a replication origin in the Xenopus early embryo from late mitosis to late G1. (A) Assembly of ORC onto origin DNA. (B) Binding of Cdc6 and RLF-B/Cdt1 onto ORC. (C) Multiple Mcm(2–7) heterohexamers are assembled onto each origin to license it. (D) Once licensing is complete Cdc6 is removed, whilst ORC binds less tightly (unshaded ORC denotes weak binding to DNA). (E) Cdc7 is recruited to the licensed origin and phosphorylates Mcm(2–7) complexes (purple circles denote phosphorylation). (F) Following nuclear assembly, CDK activity induces the assembly of Cdc45 onto the origin.
Inactivation of the licensing system prior to the start of S phase is likely to be mediated by several redundant mechanisms. Early results showed that in order for replicated (G2) nuclei to re-replicate when transferred to fresh egg extract, the nuclear envelope had to be permeabilized prior to transfer (Blow and Laskey, 1988). This suggests that one or more components of the licensing system are excluded from G2 nuclei. One promising candidate for this nuclear exclusion is Cdc6, because in Xenopus extracts (and mammalian cells) Cdc6 is exported from the nucleus as a consequence of CDK activation in S phase (Saha et al., 1998; Pelizon et al., 2000). Although removal of CDK phosphorylation sites from Xenopus Cdc6 prevented it from being excluded from nuclei, this did not cause re-replication of DNA (Pelizon et al., 2000), suggesting that other mechanisms for preventing re-licensing of replicated DNA must exist. One general mechanism that has been identified in yeast is the inactivation of various licensing components by CDKs active during S, G2 and M phases of the cell cycle (reviewed in Diffley, 1996). However, although CDKs are capable of directly inhibiting origin licensing in Xenopus (Hua et al., 1997; Mahbubani et al., 1997), CDK inhibition in G2 does not induce re-licensing (Sun et al., 2000). This may be due to the presence in Xenopus of geminin, a specific RLF-B/Cdt1 inhibitor (McGarry and Kirschner, 1998; Wohlschlegel et al., 2000; Tada et al., 2001). Geminin accumulates during S phase and G2, but is abruptly degraded at the metaphase–anaphase transition, just as the licensing system is activated (McGarry and Kirschner, 1998). Indeed, geminin has been shown to be the major inhibitor of origin licensing present in metaphase-arrested egg extracts (Tada et al., 2001). Further experimentation is needed, however, to disentangle the different roles of nuclear exclusion, CDKs and geminin in preventing re-licensing of replicated DNA.
S phase-promoting factor
In order for licensed origins to initiate replication, they must be acted upon by the Cdc7 and CDK protein kinases that constitute SPF (Figure 1). The major physiological substrate of the Cdc7 kinase appears to be Mcm(2–7) (reviewed in Johnston et al., 1999), whilst the key CDK substrates for the initiation of replication have yet to be identified. Xenopus Cdc7 is recruited to chromatin from late mitosis through to the end of S phase (Jares and Blow, 2000). This recruitment occurs only on licensed chromatin, but does not require the continued presence of ORC or Cdc6 once they have fulfilled their essential role in licensing. Since Mcm(2–7) are very good substrates for the Cdc7 kinase both in vivo and in vitro, the recruitment of Cdc7 to chromatin is likely to be mediated by direct binding to Mcm(2–7) (Figure 2E). Only once Xenopus Cdc7 has phosphorylated Mcm(2–7) can CDKs execute their essential function for the initiation of DNA replication (Jares and Blow, 2000; Walter, 2000).
Three different types of CDK are known to be present in the early Xenopus embryo: Cdk1–cyclin B, Cdk1–cyclin A and Cdk2–cyclin E. Cyclins A and B are periodically degraded at the end of mitosis, whilst cyclin E levels remain approximately constant throughout the early embryonic cell cycle. Both Cdk1–cyclin A and Cdk2–cyclin E (but not Cdk1–cyclin B) can promote DNA replication in the Xenopus system (Strausfeld et al., 1996). However, Cdk2–cyclin E seems competent to support the normal programme of DNA replication on its own, as the rate of DNA replication is not reduced when re-synthesis of cyclin A is blocked by protein synthesis inhibitors. As a consequence of both Cdc7 and CDK function, the Cdc45 protein is recruited to the chromatin, possibly by binding to Mcm(2–7) (Figure 2F) (Mimura and Takisawa, 1998; Jares and Blow, 2000; Mimura et al., 2000; Walter, 2000). Cdc45 recruitment is thought to be one of the final steps before replication forks are initiated.
For chromosomal DNA to be precisely duplicated, it is essential that the licensing system and SPF act sequentially on the DNA. Above, I have discussed how the licensing system is inactivated after S phase has started; but how is SPF activity suppressed during late mitosis and G1 whilst origins are being licensed (Figure 1)? In somatic cell cycles this is achieved by restricting the abundance of cyclins during G1. This cannot occur in the Xenopus early embryo, however, since cyclin E remains stable and active throughout the cell cycle (Rempel et al., 1995). Instead, SPF is unable to act early in the cell cycle before nuclear assembly is complete, because CDKs can only induce initiation within an intact nucleus. Once nuclear assembly is complete, initiation rapidly ensues. It is unclear why S phase-inducing CDK activity is dependent on nuclear assembly. It seems to be more than just a mechanism for concentrating CDK activity, since attempts to induce initiation in the cytoplasm by inducing high CDK activity have not been successful (P.Jares and J.J.Blow, unpublished data). It cannot reflect a requirement for nuclear ultrastructure since licensed chromatin efficiently replicates in soluble nuclear extracts (Walter et al., 1998). One possibility is that the full activation of Cdk2–cyclin E may depend on Cdc34-mediated proteolysis that is only active within the nucleoplasm (Yew and Kirschner, 1997). Biochemical analysis of soluble nuclear extracts that support replication initiation (Walter et al., 1998) appears to be a promising means of answering these questions.
Although the activities regulating chromosome replication in the early Xenopus embryo appear to be the same as those involved in somatic cell cycles, they are controlled in slightly different ways. Instead of their abundance being controlled, as occurs in somatic cells, the activities of key regulatory proteins in the Xenopus early embryo are controlled via their subcellular localization. On exit from mitosis, whilst the chromatin is freely accessible to cytoplasmic proteins, origins are assembled and licensed, but once nuclear assembly is complete, key parts of the licensing system are inactivated. CDKs are activated only within nuclei to induce the initiation of replication. This exploitation of differences in subcellular localization is an elegant way of ensuring precise chromosome replication in the short embryonic cell cycles where extensive proteolysis would be wasteful.
Acknowledgments
Acknowledgements
Thanks to Dave Gilbert, Ben Hodgson, Neil Perkins and Anna Woodward for comments on this manuscript. This work was supported by the Cancer Research Campaign (CRC; grant SP2385/0101).
References
- Blow J.J. and Laskey,R.A. (1986) Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell, 47, 577–587. [DOI] [PubMed] [Google Scholar]
- Blow J.J. and Laskey,R.A. (1988) A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature, 332, 546–548. [DOI] [PubMed] [Google Scholar]
- Blow J.J., Gillespie,P.J., Francis,D. and Jackson,D.A. (2001) Replication origins in Xenopus egg extract are 5–15 kb apart and are activated in clusters that fire at different times. J. Cell Biol., 152, 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal A.B., Kriegstein,H.J. and Hogness,D.S. (1974) The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb. Symp. Quant. Biol., 38, 205–223. [DOI] [PubMed] [Google Scholar]
- Carpenter P.B., Mueller,P.R. and Dunphy,W.G. (1996) Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature, 379, 357–360. [DOI] [PubMed] [Google Scholar]
- Chong J.P.J., Mahbubani,M.H., Khoo,C.-Y. and Blow,J.J. (1995) Purification of an Mcm-containing complex as a component of the DNA replication licensing system. Nature, 375, 418–421. [DOI] [PubMed] [Google Scholar]
- Coleman T.R., Carpenter,P.B. and Dunphy,W.G. (1996) The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell, 87, 53–63. [DOI] [PubMed] [Google Scholar]
- DePamphilis M.L. (1999) Replication origins in metazoan chromosomes: fact or fiction? BioEssays, 21, 5–16. [DOI] [PubMed] [Google Scholar]
- Diffley J.F. (1996) Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev., 10, 2819–2830. [DOI] [PubMed] [Google Scholar]
- Donovan S., Harwood,J., Drury,L.S. and Diffley,J.F. (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA, 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R.M. and Laskey,R.A. (1980) Regulated DNA replication of DNA microinjected into eggs of Xenopus laevis. Cell, 21, 761–771. [DOI] [PubMed] [Google Scholar]
- Hua X.H. and Newport,J. (1998) Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol., 140, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X.H., Yan,H. and Newport,J. (1997) A role for Cdk2 kinase in negatively regulating DNA replication during S phase of the cell cycle. J. Cell Biol., 137, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. and Méchali,M. (1992) Plasmid replication in Xenopus eggs and egg extracts: a 2D gel electrophoretic analysis. Nucleic Acids Res., 20, 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. and Méchali,M. (1993) Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J., 12, 4511–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Maric,C. and Méchali,M. (1995) Transition in specification of embryonic metazoan DNA replication origins. Science, 270, 994–997. [DOI] [PubMed] [Google Scholar]
- Jares P. and Blow,J.J. (2000) Xenopus Cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev., 14, 1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Johnston L.H., Masai,H. and Sugino,A. (1999) First the CDKs, now the DDKs. Trends Cell Biol., 9, 249–252. [DOI] [PubMed] [Google Scholar]
- Kearsey S.E. and Labib,K. (1998) MCM proteins: evolution, properties and role in DNA replication. Biochim. Biophys. Acta, 1398, 113–136. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Mimura,S., Nishimoto,S., Takisawa,H. and Nojima,H. (1995) Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell, 81, 601–609. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Mimura,S., Nishimoto,S., Masuda,T., Nojima,H. and Takisawa,H. (1997) Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J., 16, 3320–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. and Diffley,J.F. (2001) Is the MCM2–7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev., 11, 64–70. [DOI] [PubMed] [Google Scholar]
- Lohka M.J. and Masui,Y. (1983) Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science, 220, 719–721. [DOI] [PubMed] [Google Scholar]
- Mahbubani H.M., Paull,T., Elder,J.K. and Blow,J.J. (1992) DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res., 20, 1457–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbubani H.M., Chong,J.P., Chevalier,S., Thömmes,P. and Blow,J.J. (1997) Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J. Cell Biol., 136, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D., Moreau,J. and Mechali,M. (2000) XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature, 404, 622–625. [DOI] [PubMed] [Google Scholar]
- McGarry T.J. and Kirschner,M.W. (1998) Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell, 93, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Mimura S. and Takisawa,H. (1998) Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase cdk. EMBO J., 17, 5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S., Masuda,T., Matsui,T. and Takisawa,H. (2000) Central role for Cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells, 5, 439–452. [DOI] [PubMed] [Google Scholar]
- Murray A.W. and Kirschner,M.W. (1989) Cyclin synthesis drives the early embryonic cell cycle. Nature, 339, 275–280. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Lygerou,Z., Nishimoto,T. and Nurse,P. (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature, 404, 625–628. [DOI] [PubMed] [Google Scholar]
- Pelizon C., Madine,M.A., Romanowski,P. and Laskey,R.A. (2000) Unphosphorylatable mutants of Cdc6 disrupt its nuclear export but still support DNA replication once per cell cycle. Genes Dev., 14, 2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova T.A. and Blow,J.J. (2000) Sequential MCM/P1 subcomplex assembly is required to form a heterohexamer with replication licensing activity. J. Biol. Chem., 275, 2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel R.E., Sleight,S.B. and Maller,J.L. (1995) Maternal Xenopus Cdk2–cyclin E complexes function during meiotic and early embryonic cell cycles that lack a G1 phase. J. Biol. Chem., 270, 6843–6855. [DOI] [PubMed] [Google Scholar]
- Romanowski P., Madine,M.A., Rowles,A., Blow,J.J. and Laskey,R.A. (1996) The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol., 6, 1416–1425. [DOI] [PubMed] [Google Scholar]
- Rowles A., Chong,J.P., Brown,L., Howell,M., Evan,G.I. and Blow,J.J. (1996) Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell, 87, 287–296. [DOI] [PubMed] [Google Scholar]
- Rowles A., Tada,S. and Blow,J.J. (1999) Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci., 112, 2011–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P., Chen,J.J., Thome,K.C., Lawlis,S.J., Hou,Z.H., Hendricks,M., Parvin,J.D. and Dutta,A. (1998) Human CDC6/Cdc18 associates with Orc1 and cyclin–cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol., 18, 2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld U.P., Howell,M., Descombes,P., Chevalier,S., Rempel,R.E., Adamczewski,J., Maller,J.L., Hunt,T. and Blow,J.J. (1996) Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopus egg extracts. J. Cell Sci., 109, 1555–1563. [DOI] [PubMed] [Google Scholar]
- Sun W., Hola,M., Pedley,K., Tada,S., Blow,J.J., Todorov,I.T., Kearsey,S.E. and Brooks,R.F. (2000) The replication capacity of intact mammalian nuclei in Xenopus egg extracts declines with quiescence, but the residual DNA synthesis is independent of Xenopus MCM proteins. J. Cell Sci., 113, 683–695. [DOI] [PubMed] [Google Scholar]
- Tada S., Li,A., Maiorano,D., Méchali,M. and Blow,J.J. (2001) Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nature Cell Biol., 3, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thömmes P., Kubota,Y., Takisawa,H. and Blow,J.J. (1997) The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J., 16, 3312–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J.C. (2000) Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem., 275, 39773–39778. [DOI] [PubMed] [Google Scholar]
- Walter J., Sun,L. and Newport,J. (1998) Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell, 1, 519–529. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Dwyer,B.T., Dhar,S.K., Cvetic,C., Walter,J.C. and Dutta,A. (2000) Inhibition of eukaryotic replication by geminin binding to Cdt1. Science, 290, 2309–2312. [DOI] [PubMed] [Google Scholar]
- Yew P.R. and Kirschner,M.W. (1997) Proteolysis and DNA replication: the CDC34 requirement in the Xenopus egg cell cycle. Science, 277, 1672–1676. [DOI] [PubMed] [Google Scholar]