Abstract
Heparin-binding epidermal growth factor-like growth factor (HB-EGF), a mitogen and chemotactic factor, binds to two receptor tyrosine kinases, erbB1 and erbB4. Now we demonstrate that HB-EGF also binds to a novel 140 kDa receptor on MDA-MB 453 cells. Purification of this receptor showed it to be identical to N-arginine dibasic convertase (NRDc), a metalloendopeptidase of the M16 family. Binding to cell surface NRDc and NRDc in solution was highly specific for HB-EGF among EGF family members. When overexpressed in cells, NRDc enhanced their migration in response to HB-EGF but not to EGF. Conversely, inhibition of endogenous NRDc expression in cells by antisense morpholino oligonucleotides inhibited HB-EGF-induced cell migration. Anti-erbB1 neutralizing antibodies completely abrogated the ability of NRDc to enhance HB-EGF-dependent migration, demonstrating that this NRDc activity was dependent on erbB1 signaling. Although NRDc is a metalloproteinase, enzymatic activity was not required for HB-EGF binding or enhancement of cell migration; neither did NRDc cleave HB-EGF. Together, these results suggest that NRDc is a novel specific receptor for HB-EGF that modulates HB-EGF-induced cell migration via erbB1.
Keywords: cell motility/EGF family/EGF receptors/metalloendopeptidases
Introduction
Heparin-binding epidermal growth factor-like growth factor (HB-EGF) was first identified in the conditioned medium of macrophages (Higashiyama et al., 1991; reviewed by Raab and Klagsbrun, 1997; Iwamoto and Mekada, 2000). Like other members of the EGF family (Massagué and Pandiella, 1993), HB-EGF is synthesized as a type-1 transmembrane protein that can be shed enzymatically to release a soluble 14–20 kDa growth factor (Raab et al., 1994; Goishi et al., 1995; Gechtman et al., 1999). The transmembrane form of HB-EGF acts in a juxtacrine manner to signal neighboring cells (Higashiyama et al., 1995) and as an adhesion factor, e.g. for blastocysts (Raab et al., 1996). Transmembrane HB-EGF is also the receptor for diphtheria toxin (DT), and mediates DT-induced toxicity (Naglich et al., 1992). The soluble form of HB-EGF is a potent mitogen and chemoattractant for many cell types, including smooth muscle cells (SMC), fibroblasts and keratinocytes (Raab and Klagsbrun, 1997). Ectodomain shedding of transmembrane HB-EGF is required for transactivation of erbB1 by G–protein-coupled receptors (Prenzel et al., 1999), and keratinocyte migration in cutaneous wound healing (Tokumaru et al., 2000). Based on in vitro and in vivo studies, HB-EGF has been implicated as a contributor to wound healing (Marikovsky et al., 1993), blastocyst implantation (Das et al., 1994), SMC hyperplasia as occurs in artherosclerosis (Miyagawa et al., 1995; Nishi et al., 1997) and pulmonary hypertension (Powell et al., 1993), the response to brain injury (Opanashuk et al., 1999; Tanaka et al., 1999), and oncogenic transformation (Fu et al., 1999).
HB-EGF activity is mediated by high-affinity EGF-receptor tyrosine kinases. Four related receptor tyrosine kinases (RTKs) have been identified (reviewed by Hynes and Stern, 1994; Riese and Stern, 1998; Olayioye et al., 2000). These are EGFR/erbB1/HER1, erbB2/HER2/neu, erbB3/HER3 and erbB4/HER4. Some of these receptors exist as isoforms; for example, erbB4 isoforms exist with alterations in the juxtamembrane and PI3-kinase binding domains (Elenius et al., 1997a, 1999; Kainulainen et al., 2000).
The EGF ligands bind to the EGF RTKs with a degree of specificity. EGF, transforming growth factor-α (TGF-α), and amphiregulin (AR), bind exclusively to erbB1. The neuregulins bind to erbB3 and/or to erbB4. HB-EGF, betacellulin (BTC) and epiregulin bind to both erbB1 and erbB4 (Elenius et al., 1997b; Olayioye et al., 2000). No ligand has yet been identified for erbB2, a potent oncogene. The EGF RTKs can heterodimerize, resulting in transactivation of receptors, which expands the signaling potential of the EGF-like ligands (Moghal and Sternberg, 1999; Olayioye et al., 2000).
Our initial observations demonstrated that HB-EGF was a ligand for erbB1 (Higashiyama et al., 1991), that stimulated tyrosine phosphorylation of this RTK (Higashiyama et al., 1992). More recently, we showed that HB-EGF could bind and activate erbB4 (Elenius et al., 1997b). While HB-EGF induced the migration and proliferation of cells expressing erbB1, only migration was induced in cells expressing erbB4. HB-EGF also binds cell surface heparan sulfate proteoglycan (HSPG) via a heparin-binding domain rich in Lys and Arg residues (Higashiyama et al., 1993; Thompson et al., 1994). HSPGs mediate HB-EGF activity, for example, by enhancing the chemotactic response of SMC to HB-EGF (Higashiyama et al., 1993). The adhesion of blastocysts to transmembrane HB-EGF (Raab et al., 1996), and the toxicity of DT (Shishido et al., 1995), are also enhanced by cell surface HSPG.
There is some evidence that possibly novel HB-EGF receptors exist. For example, in analyzing the toxic effects of HB-EGF–Pseudomonas toxin fusion proteins (HB-EGF–PE) on mouse blastocysts, it was observed that HB-EGF–PE, but not TGF-α–PE, was toxic for egfr–/– null (lacking erbB1) blastocysts, suggesting that a receptor other than erbB1 was a target for HB-EGF (Paria et al., 1999). [125I]HB-EGF cross-linking to mouse blastocysts resulted in the formation of 150–160 kDa complexes, which were smaller than the 180–190 kDa complexes formed by HB-EGF with erbB1 and erbB4 (Elenius et al., 1997c). Complexes of 150–160 kDa were also detected in human breast cancer MDA-MB 453, a cell line that expresses little erbB1 or erbB4 (Elenius et al., 1997c).
Given these results, our goal was to purify a possible novel HB-EGF receptor on MDA-MB 453 cells. In this report, we describe the purification and characterization of this HB-EGF receptor, and show it to be identical to an N-terminal-truncated form of N-arginine (R) dibasic convertase (NRDc; nardilysin; EC 3.4.24.61), a member of the M16 family of metalloendopeptidases (Chesneau et al., 1994a,b; Pierotti et al., 1994). Within the EGF family, NRDc is a highly selective receptor for HB-EGF. Furthermore, NRDc expression enhances cell migration induced by HB-EGF via erbB1. We conclude that NRDc is a novel receptor for HB-EGF that modulates HB-EGF-induced migration via erbB1.
Results
Cross-linking of HB-EGF and EGF-family ligands to MDA-MB 453 cells
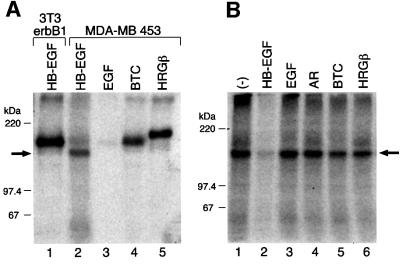
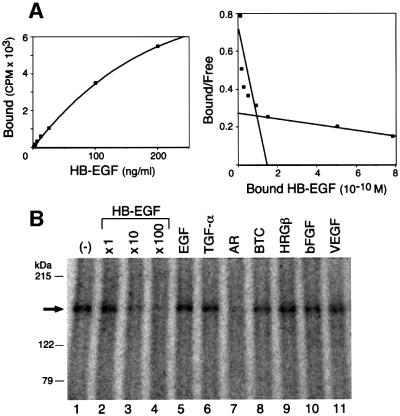
Breast cancer MDA-MB 453 cells (453 cells) express very little erbB1 and were therefore used to analyze HB-EGF binding to non-erbB1 receptors. [125I]HB-EGF cross-linked 453 cells to form a major complex of ∼150 kDa and a minor complex of 180 kDa (Figure 1A, lane 2). The 150 kDa complex was smaller than the 180 kDa complex formed by cross-linking of [125I]HB-EGF to cells expressing only erbB1 (Figure 1A, lane 1) and the 190 kDa complex formed by cross-linking of [125I]HB-EGF to cells expressing only erbB4 (Elenius et al., 1997b). As expected, there was negligible binding of [125I]EGF to 453 cells since EGF binds only to erbB1 (Figure 1A, lane 3). On the other hand, [125I]BTC and [125I]heregulin β (HRGβ) cross-linked 453 cells to form complexes with molecular masses of 190 kDa (Figure 1A, lane 4) and 200 kDa (Figure 1A, lane 5), respectively, consistent with binding to erbB4 and erbB3, respectively.
Fig. 1. Cross-linking of HB-EGF to 453 cells. (A) [125I]HB-EGF (5 ng/ml) was cross-linked to 3T3/erbB1 cells (lane 1) and 453 cells (lane 2). [125I]EGF (lane 3), [125I]BTC (lane 4) and [125I]HRGβ (lane 5) (5 ng/ml each) were bound and cross-linked to 453 cells in 6 cm dishes. The cells were lysed, and proteins were resolved by 6% SDS–PAGE and visualized by autoradiography. (B) [125I]HB-EGF (5 ng/ml) was cross-linked to 453 cells in the presence of 1.25 µg/ml unlabeled HB-EGF, EGF, AR, BTC or HRGβ in 6 cm dishes. The cells were lysed, and proteins were resolved by 6% SDS–PAGE followed by autoradiography as described in (A). The arrows point to the 150 kDa radiolabeled complex containing [125I]HB-EGF and HB-EGF receptor.
The receptor in the 150 kDa HB-EGF complex did not appear to be any of the four known EGF receptor tyrosine kinases. It was not immunoprecipitated with anti-erbB1, anti-erbB2, anti-erbB3 or anti-erbB4 antibodies (data not shown). Neither could the 150 kDa complex formation be inhibited by neutralizing antibodies directed against erbB1, erbB3 or erbB4 (data not shown). Cold excess HB-EGF (Figure 1B, lane 2), but not excess EGF, AR, BTC or HRGβ (Figure 1B, lanes 3–6) competed with binding of [125I]HB-EGF to the 453 cell receptor. Subtracting the molecular weight of recombinant soluble HB-EGF (∼10 kDa), the receptor forming the 150 kDa complex was estimated to have a molecular mass of ∼140 kDa, substantially lower than that of the known erbBs, which are 170–185 kDa. Taken together, it appears that HB-EGF binds to a 140 kDa cell surface-associated receptor on 453 cells in a highly specific manner.
Purification of the specific HB-EGF receptor
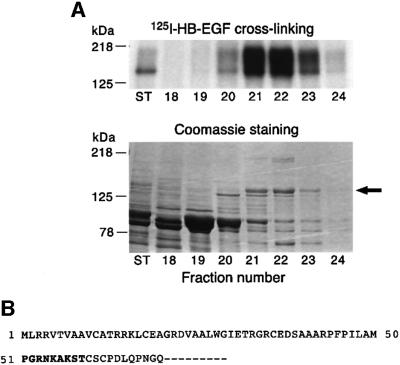
The 140 kDa HB-EGF receptor was purified from 453 total cell lysates in a series of three steps, including SuperQ anion-exchange, hydroxyapatite and MonoQ anion-exchange column chromatography, using an assay in which [125I]HB-EGF was cross-linked in solution to column fractions. After the last step, optimal cross-linking of [125I]HB-EGF yielded a broad complex that appeared mostly in fractions 21 and 22 (Figure 2A, top panel). SDS–PAGE and Coomassie Blue staining of these fractions showed a prominent 140 kDa band, the putative size of the HB-EGF receptor (Figure 2A, bottom panel, arrow). This band was excised and N-terminal amino acid sequencing yielded the sequence PGRNKAKST (Figure 2B). Analysis of the FASTA database indicated a match with amino acids 51–59 of human NRDc. NRDc is a 140 kDa metalloendopeptidase that cleaves selectively at the N-terminus of arginine residues in basic doublets (Chesneau et al., 1994a,b; Pierotti et al., 1994). Thus, the novel 140 kDa HB-EGF receptor appears to be a truncated NRDc missing the predicted 50 N-terminal amino acids.
Fig. 2. Purification of HB-EGF receptor from 453 cells. (A) Upper panel: [125I]HB-EGF was cross-linked to MonoQ column fractions in solution. Lower panel: aliquots of column fractions were analyzed by SDS–PAGE and Coomassie Blue staining. ST is an aliquot of the starting material applied to the MonoQ column, which had been partially purified by SuperQ and hydroxyapatite chromatography. The arrow in the lower panel indicates the band in fraction 22 used for N-terminal protein sequencing. (B) The first 70 amino acids of NRDc are shown. The nine-amino-acid N-terminal amino acid sequence determined by microsequencing is depicted in bold and begins at NRDc amino acid 51.
NRDc binds HB-EGF in solution and on the cell surface
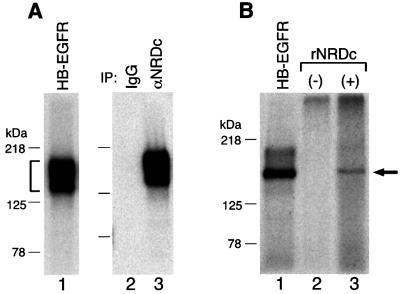
Anti-NRDc antibody (Chesneau et al., 1994a) was used to confirm the identity of the HB-EGF receptor and NRDc (Figure 3A). An aliquot of fraction 21 from the MonoQ column shown in Figure 2A was cross-linked in solution with [125I]HB-EGF to form a complex (Figure 3A, lane 1). This complex was readily immunoprecipitated with anti-NRDc antibody (Figure 3A, lane 3). In addition, cross-linking of [125I]HB-EGF in solution to recombinant rat NRDc, synthesized by in vitro transcription and translation from NRDc cDNA, resulted in the formation of a 150 kDa complex (Figure 3B, lane 3) that co-migrated with a complex of [125I]HB-EGF and HB-EGF receptor partially purified from 453 cells (Figure 3B, lane 1).
Fig. 3. Immunochemical and in vitro translation studies suggest that the HB-EGF specific receptor is NRDc. (A) [125I]HB-EGF was cross-linked in solution to an aliquot of fraction 21 shown in Figure 2A, yielding a 150 kDa complex (lane 1). This complex (bracket) was immuno precipitated with control IgG (lane 2) or anti-NRDc antibody (lane 3). The immunocomplexes were resolved by 6% SDS–PAGE, followed by autoradiography. (B) [125I]HB-EGF was cross-linked in solution in the absence (lane 2) or presence (lane 3) of recombinant NRDc (rNRDc) prepared by in vitro translation of NRDc mRNA. [125I]HB-EGF cross-linking to HB-EGF receptor [the starting material (ST) in Figure 2A, designated here as HB-EGFR] is shown in lane 1 for comparison. The arrow denotes radiolabeled complexes containing [125I]HB-EGF and NRDc.
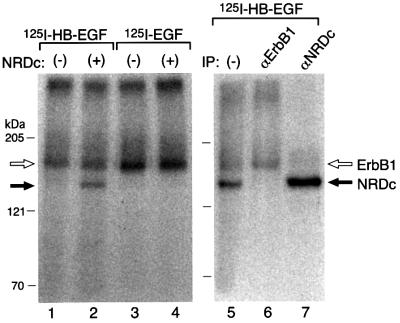
HeLa cells, which express erbB1 but very low levels of NRDc, were transiently transfected with NRDc cDNA. [125I]HB-EGF formed 180 and 150 kDa complexes with HeLa cells transiently expressing NRDc (Figure 4, lane 2), but only a 180 kDa complex in HeLa cells transfected with control vector (Figure 4, lane 1). [125I]EGF formed only 180 kDa complexes in HeLa cells whether they expressed (Figure 4, lane 4) or did not express NRDc (Figure 4, lane 3), evidence that the 180 kDa complex contains erbB1 and that EGF does not bind to cell surface NRDc. Immunoprecipitation with anti-erbB1 antibody (Figure 4, lane 6) or anti-NRDc (Figure 4, lane 7) confirmed that the 180 kDa complex contained erbB1 and the 150 kDa complex contained NRDc, respectively.
Fig. 4. NRDc transiently expressed in HeLa cells binds [125I]HB-EGF. Left: [125I]HB-EGF (5 ng/ml) (lanes 1 and 2) or [125I]EGF (5 ng/ml) (lanes 3 and 4) was cross-linked to HeLa cells transiently transfected with control vector (lanes 1and 3) or NRDc expression vector (lanes 2 and 4). Right: after [125I]HB-EGF cross-linking to HeLa cells transiently transfected with NRDc expression vector, total cell lysates were immunoprecipitated with anti-erbB1 antibody (lane 6) or anti-NRDc antibody (lane 7). Lane 5, total cell lysates prior to immuno precipitation. Immunocomplexes were resolved by 6% SDS–PAGE, followed by autoradiography. Open and solid arrows denote radiolabeled complexes containing [125I]HB-EGF and erbB1, and [125I]HB-EGF and NRDc, respectively.
Characterization of HB-EGF–NRDc interactions
The binding affinity of HB-EGF and rat NRDc in solution was analyzed by Scatchard analysis (Figure 5A). There appeared to be two classes of binding sites: a high affinity site with a Kd value of 0.3–0.6 nM and a low affinity site with a Kd value of 4–6 nM.
Fig. 5. Characterization of HB-EGF–NRDc interactions. (A) Scatchard analysis of HB-EGF binding to NRDc. Left panel: increasing amounts of [125I]HB-EGF (0.78–200 ng/ml) were added to partially purified rat NRDc in solution, followed by immunoprecipitation with anti-NRDc antibody. Immunoprecipitated complex-associated radioactivity was determined by a gamma counter. Non-specific binding was determined by competition with a 200-fold excess of unlabeled HB-EGF. Right panel: the binding data shown in the left panel were analyzed by the method of Scatchard, and best-fit plots were obtained using the LIGAND program. (B) NRDc ligand-binding specificity in solution. [125I]HB-EGF (47 ng/ml) was bound and cross-linked to recombinant rat NRDc in the presence of 47 ng/ml (lane 2), 470 ng/ml (lane 3), 4.7 µg/ml (lane 4) unlabeled HB-EGF, or 4.7 µg/ml unlabeled EGF (lane 5), TGF-α (lane 6), AR (lane 7), BTC (lane 8), HRGβ (lane 9), bFGF (lane 10), or VEGF165 (lane 11). The arrow denotes radiolabeled complexes containing [125I]HB-EGF and NRDc.
To confirm the ligand specificity of NRDc, [125I]HB-EGF was cross-linked in solution to recombinant NRDc in the presence of a 100-fold excess of non-labeled growth factors (Figure 5B). Only excess HB-EGF and AR, but not EGF, TGF–α, BTC, HRGβ or the heparin binding growth factors, basic fibroblast growth factor (bFGF) and vascular endothelial growth factor (VEGF) competed with [125I]HB-EGF binding. These results were similar to those obtained with 453 cells shown previously in Figure 1B, except for AR. Apparently, AR competes with [125I]HB-EGF binding in solution, but not on the cell surface.
NRDc expression enhances HB-EGF-induced migration via erbB1
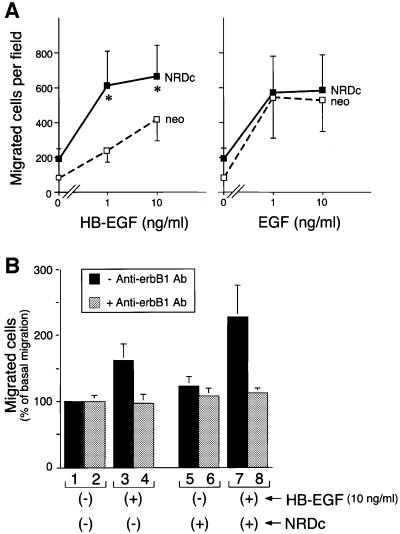
To investigate a possible biological role for HB-EGF–NRDc interactions, we analyzed cell migration, since HB-EGF has been shown to be chemotactic (Higashiyama et al., 1993). HeLa cells were transiently transfected with NRDc and dose-dependent cell migration was analyzed in a Boyden chamber assay. In cells expressing NRDc, migration in response to HB-EGF was enhanced by 2- to 3-fold at 1 ng/ml HB-EGF compared with controls (Figure 6A, left). In contrast, NRDc expression had no significant effect on cell migration induced by EGF (Figure 6A, right).
Fig. 6. Expression of NRDc in HeLa cells enhances HB-EGF-induced migration via erbB1. (A) The migration of HeLa cells transiently transfected with control vector (open squares) or NRDc expression vector (closed squares) in response to HB-EGF (left panel) or EGF (right panel) was measured in a Boyden chamber. Each point represents the mean and standard error of eight (HB-EGF) or six (EGF) independent experiments. Each experiment was carried out in quadruplicate. *P <0.05. (B) Inhibition of NRDc-enhanced cell migration by an anti-erbB1 blocking antibody. HeLa cells transiently transfected with control vector (lanes 1–4) or NRDc expression vector (lanes 5–8) were pre-treated with (gray bar; lanes 2, 4, 6, 8), or without (black bar; lanes 1, 3, 5, 7) 10 µg/ml anti-erbB1 blocking antibody (C225), and then incubated with or without 10 ng/ml HB-EGF in Boyden chamber migration assays. Each bar represents the mean and standard error of four independent experiments, normalized to the value for migrated cell numbers of control vector-transfected cells without C225 treatment, and without HB-EGF stimulation. Each experiment was carried out in quadruplicate.
Anti-erbB1 antibody (C225) inhibits ligand binding to erbB1 and subsequent signaling. C225 treatment totally abolished HB-EGF-induced cell migration (Figure 6B, lane 8 compared with lane 7), including the enhancement that resulted from NRDc overexpression (Figure 6B, lane 8 compared with the differential between lanes 3 and 7). Treatment with AG1478, an erbB1-specific kinase inhibitor, showed similar effects (data not shown). These results demonstrate that NRDc-induced enhancement of migration is mediated via erbB1.
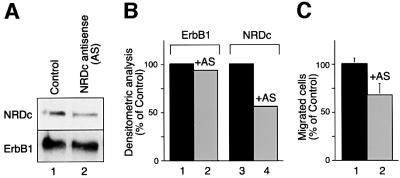
To demonstrate further that NRDc contributes to HB-EGF-induced cell migration, antisense morpholino oligonucleotides (Summerton and Weller, 1997) were used to inhibit NRDc protein synthesis. HaCaT keratinocytes were used for the inhibition studies rather than HeLa cells, since unlike HeLa cells, HaCaT cells express relatively high levels of endogenous NRDc, facilitating analysis of possible inhibition. HB-EGF binding to both HaCaT cell surface erbB1 and NRDc was confirmed by cross-linking (data not shown). The antisense oligonucleotides directed against NRDc inhibited NRDc protein synthesis by ∼45%, but did not inhibit erbB1 synthesis, as demonstrated by western blotting (Figure 7A) and densitometry (Figure 7B). The reduction of NRDc synthesis was accompanied by a 33% decrease in cell migration induced by 10 ng/ml HB-EGF, compared with cells treated with control morpholino oligonucleotides (Figure 7C).
Fig. 7. Antisense morpholino oligonucleotides inhibit NRDc protein synthesis and inhibit HB-EGF-induced migration. (A) HaCaT keratinocytes were treated with a 1.1 µM concentration of either standard morpholino oligonucleotides (control) or NRDc antisense morpholino oligonucleotides (AS) for 48 h. Total cell lysates (140 µg) of oligonucleotide-treated cells were analyzed by western blotting with anti-NRDc antibody. The same membrane was stripped and re-blotted with anti-erbB1 antibody. (B) The intensity of signals in (A) was quantitated by densitometry using the NIH image program. The relative densities of erbB1 (lanes 1 and 2) and NRDc protein levels (lanes 3 and 4) in antisense oligonucleotide-treated cells (lanes 2 and 4) compared to control oligonucleotide-treated cells (lanes 1 and 3) are shown. (C) The migration of HaCaT keratinocytes treated with standard control oligonucleotides (lane 1) or NRDc antisense oligonucleotides (lane 2) in response to HB-EGF (10 ng/ml) was measured in a Boyden chamber. Each value represents the mean and standard error of eight samples from a single experiment, and is representative of the same qualitative effect seen in two such experiments.
NRDc binding to HB-EGF and NRDc-induced migration are independent of NRDc enzymatic activity
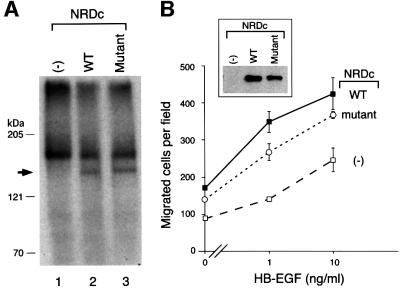
Since NRDc is a metalloproteinase, we considered whether NRDc enzymatic activity was required for HB-EGF binding and/or for mediating HB-EGF-induced responses. An enzymatically inactive NRDc mutant was obtained by replacement of Glu247 in the zinc-binding motif (HXXEH) with Ala. HeLa cells were transfected with wild-type and mutant NRDc cDNA, respectively. As expected, lysates of cells expressing mutant NRDc had low levels of enzymatic activity comparable to that of control vector-transfected cells (data not shown). [125I]HB-EGF cross-linking to cells expressing mutant NRDc resulted in the formation of 180 and 150 kDa complexes (Figure 8A, lane 3), which were identical to complexes formed by cells expressing wild-type NRDc (Figure 8A, lane 2).
Fig. 8. NRDc enzymatic activity is not needed for NRDc binding to HB-EGF or NRDc-induced migration. (A) HB-EGF binds to enzymatically inactive NRDc. [125I]HB-EGF (5 ng/ml) was bound and cross-linked to HeLa cells transiently transfected with control vector (lane 1), a vector expressing wild-type rat NRDc (WT; lane 2) or a vector expressing enzymatically inactive mutant of NRDc (Mutant; lane 3). The arrow denotes radiolabeled complexes containing [125I]HB-EGF and NRDc. (B) Enzymatically inactive NRDc enhances HB-EGF-induced cell migration. Migration of HeLa cells transiently transfected with control vector (open squares), NRDC expression vector (closed squares) or an NRDC expression vector expressing enzymatically inactive (mutant) NRDc (open circles) in response to HB-EGF was measured in a Boyden chamber. Cells were lysed and analyzed by western blotting with anti-NRDc antibody (inset). Each point represents the mean cell number and standard deviations of four different wells from a single experiment, and is representative of the same qualitative effect seen in three such experiments.
Moreover, the enhancement of migration induced by NRDc was not dependent on its enzymatic activity. HeLa cells were transiently transfected with wild-type and mutant NRDc and the two proteins were expressed at comparable levels (Figure 8B, inset). The migration of enzymatically inactive NRDc was well above that of the vector-alone controls, and was comparable to that of cells expressing wild-type enzyme when NRDc protein levels were taken into account (Figure 8B). In three separate experiments, no statistically significant difference was detected in the migration profiles of wild-type or mutant NRDc.
NRDc does not cleave transmembrane HB-EGF or mature HB-EGF
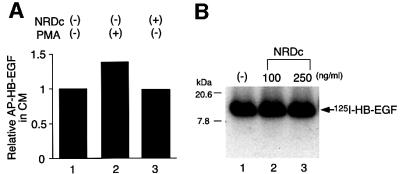
HB-EGF is expressed as a transmembrane protein that can be proteolytically cleaved (shedding) to release soluble HB-EGF, e.g. by phorbol ester (Raab et al., 1994; Goishi et al., 1995). In addition, there are consensus sequences in the heparin-binding domain of HB-EGF for cleavage by NRDc. Accordingly, the ability of NRDc to cleave cell surface transmembrane HB-EGF or HB-EGF in solution was examined. Expression plasmids encoding alkaline phosphatase (AP)–transmembrane HB-EGF fusion protein (Suzuki et al., 1997) were transiently transfected into HeLa cells. Phorbol ester (PMA) induced release of soluble AP–HB-EGF (Figure 9A, lane 2). On the other hand, no increased AP activity was detected in the conditioned medium of cells transiently co-expressing AP–HB-EGF and NRDc (Figure 9A, lane 3), compared with the conditioned medium of cells expressing only AP–HB-EGF (Figure 9A, lane 1). [125I]HB-EGF was incubated with active NRDc in solution. However, no cleavage or degradation of mature HB-EGF was detected (Figure 9B). As a control, the NRDc used was shown to be enzymatically active using dynorphin A as a substrate (data not shown). Taken together, these results indicate that NRDc does not affect the processing of transmembrane HB-EGF or of mature HB-EGF.
Fig. 9. NRDc does not cleave HB-EGF. (A) Cell surface HB-EGF: An expression plasmid encoding AP-transmembrane HB-EGF was co-transfected into HeLa cells with either a control vector (lanes 1 and 2) or an NRDc expression vector (lane 3). After a 24 h incubation, the control vector-transfected cells were treated without (lane 1) or with (lane 2) 100 nM PMA for 30 min. Conditioned medium (CM) of control cells, control cells treated with PMA, or cells expressing NRDc were assayed for relative AP activity compared with control, arbitrarily set as 1. Each bar is the average of triplicate values. (B) HB-EGF in solution. [125I]HB-EGF (60 ng/ml) was incubated with no (lane 1), 100 ng/ml (lane 2) or 250 ng/ml (lane 3) wild-type NRDc in solution for 1 h at 37°C. [125I]HB-EGF size was analyzed on a 15% SDS–PAGE, followed by autoradiography.
Discussion
We have identified a novel and specific 140 kDa HB-EGF receptor that is not one of the known members of the ErbB/HER family of receptor tyrosine kinases. Purification and subsequent N-terminal sequencing revealed it to be an N-terminally truncated form of NRDc, a 140 kDa metalloendopeptidase that has been found to be both cytoplasmic and cell surface associated (Hospital et al., 2000). As evidence, the HB-EGF receptor was immunoprecipitated by anti-NRDc antibody, and [125I]HB-EGF was capable of binding to recombinant NRDc expressed on the surface of HeLa cells and to recombinant NRDc in solution.
The name given to this 140 kDa metalloendopeptidase derives from its in vitro substrate specificity, since it generally cleaves precursor polypeptides at the N-terminus of arginine (R) residues in dibasic sites (Chesneau et al., 1994a,b; Pierotti et al., 1994). It has a zinc-binding motif, HXXEH, and is characterized by a highly acidic stretch of 71 residues (rat; 79% Glu and Asp). NRDc is mainly expressed in developing neural tissue and adult testis, heart and skeletal muscle (Hospital et al., 1997; Fumagalli et al., 1998), tissues that also express HB-EGF (Abraham et al., 1993). Tumor cells generally express high levels of NRDc (Hospital et al., 1997).
Surprisingly, the NRDc sequence does not predict a protein with a transmembrane domain. Depending on the cell type, NRDc is cytoplasmic, secreted, or cell surface associated. In testis, where it was first identified, this enzyme was localized in the cytoplasm of elongated spermatids, but was also secreted (Chesneau et al., 1996). In COS7 and BSC40 cells overexpressing NRDc, it was localized to the cytoplasm and in addition was present at the surface of these cells (Hospital et al., 2000). In our study, 453 and NRDc-transfected HeLa cells expressed NRDc on the cell surface, as demonstrated by cell surface biotinylation and the ability of these cells to bind [125I]HB-EGF. On the other hand, NRDc was secreted by NRDc-transfected CHO cells, but was not cell surface bound (our unpublished data). The nature of the cell surface association is not known. Perhaps NRDc is associated with some as yet unidentified integral cell surface protein, which may be lacking, for example, in CHO cells.
HB-EGF binds to NRDc whether erbB1 is present or not. NRDc can be readily purified as a soluble protein that binds HB-EGF in solution, allowing affinity measurements in the absence of erbB1. Scatchard analysis of [125I]HB-EGF binding to NRDc in solution revealed the existence of two classes of binding sites: a high affinity site with a Kd value of 0.3–0.6 nM and a low affinity site with a Kd value of 4–6 nM. The high affinity Kd is comparable to that of HB-EGF for erbB1, 0.15 nM (Elenius et al., 1997b).
A distinctive property of NRDc is that it is a highly specific receptor for HB-EGF compared with other EGF-family ligands. The cross-linking of [125I]HB-EGF to 453 cell surface was competed by excess unlabeled HB-EGF, but not excess EGF, TGF–α, AR, BTC or HRGβ. The same results occurred in solution, with the exception that AR competed with [125I]HB-EGF binding to NRDc to some degree. The specificity of HB-EGF for NRDc suggests that there is a specific NRDc-binding domain within HB-EGF. HB-EGF has an EGF domain responsible for binding to erbB1, like all other members of the EGF-family (Higashiyama et al., 1991). However, unlike most other EGF-like ligands, HB-EGF also has a highly cationic heparin-binding domain that mediates interactions with cell surface HSPG (Thompson et al., 1994). We have preliminary evidence that HB-EGF binds to NRDc via the HB-EGF heparin-binding domain. A synthetic 21-amino-acid peptide corresponding to the heparin-binding domain of HB-EGF (Higashiyama et al., 1993) specifically inhibited HB-EGF binding to NRDc both on the cell surface and in solution (our unpublished data). AR has a heparin-binding domain similar to the HB-EGF heparin-binding domain (Thorne and Plowman, 1994), which might explain its ability to compete to some extent for HB-EGF binding to NRDc in solution. VEGF and bFGF have heparin-binding domains, but do not interact with NRDc. Together, these results suggest that the specificity of HB-EGF for NRDc might be at least partly due to specific contributions of the HB-EGF heparin-binding domain.
NRDc induces HB-EGF migration, as shown in transient transfection and antisense studies. Transient expression of NRDc in HeLa cells increased the migration of HeLa cells in response to HB-EGF by 2- to 3-fold. On the other hand, transient NRDc expression did not enhance cell migration in response to EGF, consistent with the inability of EGF to bind NRDc. Furthermore, morpholino antisense oligonucleotides (Summerton and Weller, 1997) directed against NRDc inhibited NRDc protein synthesis and HB-EGF-induced migration activity by 45 and 33%, respectively, suggesting a correlation between NRDc levels and HB-EGF-induced migration. The antisense oligonucleotide treatment had no effect on erbB1 expression.
NRDc-induced migration occurs via erbB1, since anti-erbB1 antibodies that inhibit ligand binding and subsequent erbB1 signaling totally abolished HB-EGF-induced cell migration, including the contribution of NRDc overexpression to migration. These results suggest that NRDc expression is a modulator of HB-EGF migration activity via erbB1. The mechanism by which NRDc induces cell migration is not known. NRDc is not a transmembrane protein, has no known kinase motifs and probably does not signal on its own. Preliminary results suggest that NRDc expression significantly inhibits attenuation of erbB1 activation to give a prolonged signal.
NRDc was originally purified and cloned as a zinc metalloendopeptidase (Chesneau et al., 1994a,b; Pierotti et al., 1994). It is possible that the enzymatic activity of NRDc plays a role in mediating HB-EGF–NRDc interactions. However, an enzymatically inactive NRDc mutant that contains an altered zinc-binding site retained the ability to bind HB-EGF and to induce enhanced migration of cells in response to HB-EGF. In addition, NRDc did not cleave the cell-associated transmembrane HB-EGF precursor nor did it cleave soluble HB-EGF in solution. Whether NRDc enzymatic activity is involved in some other aspect of HB-EGF biology, e.g. degradation of extracellular matrix during migration, is not yet known. Insulin degrading enzyme (IDE) is another member of the M16 family. Analogous to NRDc, the enzymatic activity of IDE is not necessary for insulin binding (Gehm et al., 1993). IDE binds not only insulin, but also EGF family members, EGF and TGF-α (Garcia et al., 1989). Thus, there may be several interactions of M16 family metalloendopeptidases with EGF-like growth factors, but independent of enzymatic activity.
In summary, we have demonstrated that NRDc, previously identified as a metalloendopeptidase, is a novel and highly specific receptor for HB-EGF, and that expression of NRDc modulates HB-EGF bioactivity. Our future goal is to elucidate further the mechanisms by which NRDc potentiates HB-EGF bioactivity, and to explore the biological significance of the interaction of HB-EGF and NRDc in vivo.
Materials and methods
Recombinant human HB-EGF was kindly provided by Dr J.Abraham (Scios Nova, Sunnyvale, CA) and Dr S.Higashiyama (Osaka University, Japan). Recombinant human bFGF and VEGF165 were provided by Dr J.Abraham. Recombinant human HRGβ was provided by Dr M.Sliwkowski (Genentech Inc., South San Francisco, CA). Recombinant human EGF, TGF-α, AR and BTC were purchased from R&D (Minneapolis, MN). Recombinant soluble NRDc was partially purified from BSC40 cells infected with recombinant vaccinia virus expressing recombinant NRDc as described previously (Hospital et al., 2000). NRDc-specific rabbit polyclonal antiserum was described previously (Chesneau et al., 1994a). Sheep polyclonal antibody directed against human erbB1 (#06-129) was purchased from UBI (Lake Placid, NY). Mouse monoclonal antibody directed against erbB1 (clone 225) was purchased from NeoMarkers (Fremont, CA).
Cell culture
NIH 3T3 stable cell transfectants expressing erbB1 have been described previously (Elenius et al., 1997b). NIH 3T3/erbB1 and MDA-MB 453 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS), 4.5 g/l glucose and 1% glutamine/penicillin/streptomycin supplement (GPS; Irvine Scientific). HeLa cells were grown in DMEM containing 10% FCS, 1 g/l glucose and 1% GPS. HaCaT keratinocytes (a gift from Michael Detmar) were grown in MEM containing 10% FCS, 1 g/l glucose and 1% GPS.
Radioiodination and chemical cross-linking
HB-EGF, EGF, BTC and HRGβ were iodinated using IODO-BEADS (Pierce) according to the manufacturer’s instructions. Specific activities of 4 × 104–1 × 105 c.p.m/ng were obtained. Cell surface cross-linking was carried out as previously described (Elenius et al., 1997b). Briefly, confluent cells in 6 cm dishes were incubated with 5–25 ng/ml radioiodinated growth factor in binding buffer (DMEM, 20 mM HEPES pH 7.4, 0.1 mg/ml gelatin). After a 2 h incubation on ice, cells were washed with phosphate-buffered saline (PBS) and treated with 200 µM disuccinimidyl suberate (DSS; Pierce) for 15 min at room temperature. To stop the cross-linking reaction, 10× concentrated stop buffer (10 mM Tris–HCl pH 7.4, 250 mM glycine, 2 mM EDTA) was added. After a 1 min incubation on ice, cells were lysed with lysis buffer [10 mM Tris–HCl pH 7.4, 1% NP-40, protease inhibitor cocktail (Roche Molecular Biochemical)]. Aliquots of cell lysates were analyzed by 6% SDS–PAGE. Cross-linked complexes were visualized by autoradiography. For competition of [125I]HB-EGF binding to cell surface, a 250-fold molar excess of unlabeled growth factors was added. For cross-linking of HB-EGF in solution, column fractions or recombinant rat NRDc synthesized by in vitro translation were diluted 1:5–1:10 with HEPES-buffered saline pH 8 and the volume was adjusted to 100 µl. After a 1 h incubation with [125I]HB-EGF (35–50 ng/ml) at room temperature, 1 µl of 20 mM DSS in DMSO was added. After a 15 min incubation at room temperature, 10 µl of 10× concentrated stop buffer were added and 10 µl aliquots of solution were analyzed by 6% SDS–PAGE. For competition of [125I]HB-EGF binding in solution, a 100-fold excess of unlabeled growth factors was added.
Purification and protein micro-sequencing
Approximately 1 × 109 MDA-MB 453 cells were lysed with 75 ml of 20 mM HEPES pH 8, 0.1 M NaCl, 0.5% CHAPS and protease inhibitor cocktail. Total cell lysates were applied to a Toyopearl SuperQ anion-exchange column (Tosohaas) equilibrated with 20 mM HEPES pH 8, 0.1 M NaCl, 0.1% CHAPS. After washing with equilibration buffer, receptor binding activity was eluted with a 60 ml linear gradient of 0.1–1 M NaCl, 20 mM HEPES pH 8 and 0.1% CHAPS at a flow rate of 1 ml/min. Active fractions that cross-linked [125I]HB-EGF were collected and applied to a Macro-Prep Ceramic Hydroxyapatite Type I column (Bio-Rad) equilibrated with 10 mM potassium phosphate buffer pH 7 and 0.1% CHAPS. After washing with equilibration buffer, [125I]HB-EGF binding activity was eluted with a 22.5 ml linear gradient of 10–400 mM potassium phosphate buffer pH 7 and 0.1% CHAPS at a flow rate of 0.5 ml/min. Active [125I]HB-EGF binding fractions eluted at 230–300 mM potassium phosphate buffer were collected and applied to a Mono Q HR 5/5 column (Pharmacia), [125I]HB-EGF binding activity was eluted with a 22.5 ml linear gradient of 0.1–1 M NaCl, 20 mM HEPES pH 8 and 0.1% CHAPS, at a flow rate of 0.5 ml/min. An active HB-EGF binding fraction eluted at ∼0.5 M NaCl (Figure 2, fraction 21) was analyzed by 6% SDS–PAGE. The protein bands were transferred to a Sequi-Blot PVDF membrane (Bio-Rad). The membrane was stained with 0.1% Ponceau S in 5% acetic acid, and a prominent protein around 140 kDa was excised. The protein was N-terminally sequenced using a PE/ABD Procise 494 HT Protein Sequencing System (VGR), a service provided by Dr W.Lane of the Harvard Microchemistry Facility (Cambridge, MA).
Immunoprecipitation
[125I]HB-EGF–receptor complexes were incubated with rabbit anti-NRDc serum (1:200 dilution) or sheep anti-erbB1 (1:200 dilution) antibody for 4 h at 4°C, followed by an incubation with protein G–Sepharose beads for 1 h at 4°C. The protein G-Sepharose beads were washed three times with lysis buffer, and resuspended in SDS–PAGE sample buffer. The samples were boiled for 5 min, and proteins were resolved by 6% SDS–PAGE and visualized by autoradiography.
Western blotting
Cell lysates were separated by 10% SDS–PAGE and transferred to nitrocellulose filters. After blocking with 5% non-fat milk in TBS-T (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.05% Tween-20), filters were incubated with rabbit anti-NRDc antibody (1:5000 dilution) for 16 h at 4°C, followed by a incubation with horseradish peroxidase-conjugated anti-rabbit IgG (Amersham; 1:10 000 dilution) for 1 h at room temperature (RT). The immobilized peroxidase activity was detected by enhanced chemiluminescence (ECL; Amersham).
Transient transfection
The full-length rat cDNA encoding isoform 1 of NRDc was cloned into the pcDNA3 (Invitrogen) mammalian expression vector to generate pcDNA3-rNRD1 as previously described (Hospital et al., 2000). The human and rat proteins share 92% identity (Hospital et al., 1997). HeLa cells were grown in 6 cm dishes and transfected with 0.75 µg of control vector (pcDNA3.1), pcDNA3-rNRD1 or pcDNA3.1-rNRD1-HXXAH using the FuGene 6 transfection reagent (Roche Molecular Biochemical) according to the manufacturer’s instruction. Cells were grown for an additional 40 h and used for cross-linking or cell migration assays.
In vitro transcription and translation
In vitro transcription and translation of rat NRDc were performed using the TNT Coupled Reticulocyte Lysate System (Promega, Madison, WI), according to the manufacturer’s instruction. Briefly, 1 µg of pcDNA3-rNRD1 was incubated with rabbit reticulocyte lysate, RNA polymerase, RNase inhibitor and amino acid mixture at 30°C for 90 min. Ten microliters out of the 50 µl reaction mixture were used for [125I]HB-EGF cross-linking in solution.
Binding in solution and Scatchard analysis
Increasing concentrations (0.78–200 ng/ml) of [125I]HB-EGF were incubated with 40 ng/ml rat NRDc, prepared from BSC-40 cells infected with rNRD1 recombinant vaccinia virus (Hospital et al., 2000), in PBS (total volume: 50 µl). After a 2 h incubation at room temperature, 0.1 µl of anti-NRDc serum were added and incubated for 4 h at 4°C, followed by an incubation with protein G–Sepharose beads for 1 h at 4°C. The protein G–Sepharose beads were washed three times vigorously with PBS, and immunoprecipitated radioactivity was determined with a gamma counter. Non-specific binding was defined as the amount of binding obtained after adding a 200-fold excess of unlabeled HB-EGF together with the labeled HB-EGF. Ligand binding data were used to generate saturation curves and Scatchard plots. Kd values were determined by linear regression analysis using the LIGAND program (Munson and Rodbard, 1980).
Migration assays
Migration assays were performed in a Boyden chamber (Neuro Probe Inc., Gaithersberg, MD) as described previously (Elenius et al., 1997b). Briefly, ∼15 000 cells in DMEM with 0.1% bovine serum albumin were added to wells in the upper chamber and various amounts of HB-EGF or EGF were added to wells in the lower chamber. After a 4 h incubation at 37°C, the membranes were fixed with 4% formaldehyde and the cells were stained with hematoxylin. The non-migrating cells were scraped off the top of the membrane, and the migrated cells at the bottom side of the membrane were counted by phase microscopy. Statistical significance was determined by the paired Student’s t-test.
Enzymatically inactive NRDc
A cDNA encoding an inactive form of rat NRDc was generated by substitution of the 851 bp BglII cassette (376–1126 of the open reading frame) with a mutant cassette obtained by PCR. The Glu247 codon (GAG) of the HXXEH zinc binding motif was replaced by an Ala codon (GCG). In addition, a silent substitution (C→T) was introduced in the following His248 codon to create an NdeI restriction site. Two mutant oligonucleotides [sense: CATTTTTGGCGCATATGGTATTC (O3); antisense: GAATACCATATGCGCCAAAAAGTG (O2)] and two oligonucleotides spanning the 5′ [sense: TCAGATCTAAGTAAT GTGG (O1)] and 3′ [antisense: TATCTGAGAGAAGATCTCC (O4)] BglII sites were synthesized. Two NRDc fragments were amplified with primer pairs of O1 + O2 and O3 + O4, respectively. Both fragments were subsequently mixed and re-amplified with O1 and O4. The resulting 870 bp fragment was cloned into a BglII site of pcDNA3.1/Zeo (Invitrogen) to generate pcDNA3.1-rNRD1-HXXAH. HeLa cells were transiently transfected with pcDNA3.1-rNRD1-HXXAH, and the enzymatic activity of cell lysates was measured using dynorphin A as a substrate, as described previously (Chesneau et al., 1994a). Cells transfected with mutant NRDc cDNA had enzymatic activity comparable to that of cells transfected with control vector and ∼10–20% of the enzymatic activity of cells expressing wild-type NRDc.
Antisense morpholino oligonucleotides
The 25mer morpholino oligonucleotides targeted to a sequence immediately upstream of the AUG translational start site of human NRDc (5′–TCACCACCAAGCTGGAGCGATGGAC-3′) or standard control oligonucleotides (5′–CCTCTTACCTCAGTTACAATTTATA–3′) were introduced into HaCaT keratinocytes according to the manufacturer’s special delivery protocol (GeneTools, LLC, Corvallis, OR). After incubation for 48 h, cells were used for western blotting and migration assays.
HB-EGF ectodomain shedding
Expression plasmids encoding AP–transmembrane HB-EGF fusion protein (Suzuki et al., 1997), were transiently transfected into HeLa cells together with either pcDNA3.1 or pcDNA3-rNRD1. After a 24 h incubation, cells were treated with or without 100 nM phorbol ester (PMA) for another 30 min. The conditioned media of cells were assayed for AP activity as described previously (Suzuki et al., 1997).
Acknowledgments
Acknowledgements
We thank Zeev Gechtman and Shy Soker for helpful discussions and suggestions in protein purification. We also thank Michael Freeman, Katsutoshi Goishi and Gerhard Raab for critically reading the manuscript. We thank Paul Cohen for his constant support and Nabil Seidah for his enthusiasm and helpful discussions. This work was supported by NIH grants GM 47397 (M.K.), by the Japanese Heart Foundation & Bayer Yakuhin Research Grant Abroad (E.N.), by the International Artherosclerosis Society (E.N.) and by a group grant of the Medical Research Council of Canada MRC MGC 11474 (A.P.). V.H. is supported by the Ministère de l’Education du Québec.
References
- Abraham J.A., Damm,D., Bajardi,A., Miller,J., Klagsbrun,M. and Ezekowitz,R.A. (1993) Heparin-binding EGF-like growth factor: characterization of rat and mouse cDNA clones, protein domain conservation across species and transcript expression in tissues. Biochem. Biophys. Res. Commun., 190, 125–133. [DOI] [PubMed] [Google Scholar]
- Chesneau V., Pierotti,A.R., Barre,N., Creminon,C., Tougard,C. and Cohen,P. (1994a) Isolation and characterization of a dibasic selective metalloendopeptidase from rat testes that cleaves at the amino terminus of arginine residues. J. Biol. Chem., 269, 2056–2061. [PubMed] [Google Scholar]
- Chesneau V., Pierotti,A.R., Prat,A., Gaudoux,F., Foulon,T. and Cohen,P. (1994b) N-arginine dibasic convertase (NRD convertase): a newcomer to the family of processing endopeptidases. Biochimie, 76, 234–240. [DOI] [PubMed] [Google Scholar]
- Chesneau V., Prat,A., Segretain,D., Hospital,V., Dupaix,A., Foulon,T., Jegou,B. and Cohen,P. (1996) NRD convertase: a putative processing endoprotease associated with the axoneme and the manchette in late spermatids. J. Cell Sci., 109, 2737–2745. [DOI] [PubMed] [Google Scholar]
- Das S.K., Wang,X.N., Paria,B.C., Damm,D., Abraham,J.A., Klagsbrun, M., Andrews,G.K. and Dey,S.K. (1994) Heparin-binding EGF-like growth factor gene is induced in the mouse uterus temporally by the blastocyst solely at the site of its apposition: a possible ligand for interaction with blastocyst EGF-receptor in implantation. Development, 120, 1071–1083. [DOI] [PubMed] [Google Scholar]
- Elenius K., Corfas,G., Paul,S., Choi,C.J., Rio,C., Plowman,G.D. and Klagsbrun,M. (1997a) A novel juxtamembrane domain isoform of HER4/ErbB4. Isoform-specific tissue distribution and differential processing in response to phorbol ester. J. Biol. Chem., 272, 26761–26768. [DOI] [PubMed] [Google Scholar]
- Elenius K., Paul,S., Allison,G., Sun,J. and Klagsbrun,M. (1997b) Activation of HER4 by heparin-binding EGF-like growth factor stimulates chemotaxis but not proliferation. EMBO J., 16, 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenius K., Allison,G., Das,S.K., Paria,B.C., Dey,S.K. and Klagsbrun,M. (1997c) Interaction of heparin-binding EGF-like growth factor with multiple receptors. In Lichtner,R.B. and Harkins,R.N. (eds), EGF Receptor in Tumor Growth and Progression. Springer-Verlag, Berlin, Germany, pp. 45–64.
- Elenius K., Choi,C., Paul,S., Santiestevan,E., Nishi,E. and Klagsbrun,M. (1999) Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidyl inositol 3-kinase. Oncogene, 18, 2607–2615. [DOI] [PubMed] [Google Scholar]
- Fu S., Bottoli,I., Goller,M. and Vogt,P. (1999) Heparin-binding epidermal growth factor-like growth factor, a v-Jun target gene, induces oncogenic transformation. Proc. Natl Acad. Sci. USA, 96, 5716–5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli P. et al. (1998) Human NRD convertase: a highly conserved metalloendopeptidase expressed at specific sites during development and in adult tissues. Genomics, 47, 238–245. [DOI] [PubMed] [Google Scholar]
- Garcia J.V., Stoppelli,M.P., Decker,S.J. and Rosner,M.R. (1989) An insulin epidermal growth factor-binding protein from Drosophila has insulin-degrading activity. J. Cell Biol., 109, 1301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechtman Z., Alonso,J.L., Raab,G., Ingber,D.E. and Klagsbrun,M. (1999) The shedding of membrane-anchored heparin-binding epidermal-like growth factor is regulated by the Raf/mitogen-activated protein kinase cascade and by cell adhesion and spreading. J. Biol. Chem., 274, 28828–28835. [DOI] [PubMed] [Google Scholar]
- Gehm B.D., Kuo,W.-L., Perlman,R.K. and Rosner,M.R. (1993) Mutations in a zinc-binding domain of human insulin-degrading enzyme eliminate catalytic activity but not insulin binding. J. Biol. Chem., 268, 7943–7948. [PubMed] [Google Scholar]
- Goishi K., Higashiyama,S., Klagsbrun,M., Nakano,N., Umata,T., Ishikawa,M., Mekada,E. and Taniguchi,N. (1995) Phorbol ester induces the rapid processing of cell surface heparin-binding EGF-like growth factor: conversion from juxtacrine to paracrine growth factor activity. Mol. Biol. Cell, 6, 967–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama S., Abraham,J.A., Miller,J., Fiddes,J.C. and Klagsbrun,M. (1991) A heparin-binding growth factor secreted by macrophage-like cells that is related to EGF. Science, 251, 936–939. [DOI] [PubMed] [Google Scholar]
- Higashiyama S., Lau,K., Besner,G.E., Abraham,J.A. and Klagsbrun,M. (1992) Structure of heparin-binding EGF-like growth factor. Multiple forms, primary structure and glycosylation of the mature protein. J. Biol. Chem., 267, 6205–6212. [PubMed] [Google Scholar]
- Higashiyama S., Abraham,J.A. and Klagsbrun,M. (1993) Heparin-binding EGF-like growth factor stimulation of smooth muscle cell migration: dependence on interactions with cell surface heparan sulfate. J. Cell Biol., 122, 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama S., Iwamoto,R., Goishi,K., Raab,G., Taniguchi,N., Klagsbrun,M. and Mekada,E. (1995) The membrane protein CD9/DRAP 27 potentiates the juxtacrine growth factor activity of the membrane-anchored heparin-binding EGF-like growth factor. J. Cell Biol., 128, 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital V., Prat,A., Joulie,C., Cherif,D., Day,R. and Cohen,P. (1997) Human and rat testis express two mRNA species encoding variants of NRD convertase, a metalloendopeptidase of the insulinase family. Biochem. J., 327, 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital V., Chesneau,V., Balogh,A., Joulie,C., Seidah,N.G., Cohen,P. and Prat,A. (2000) N-arginine dibasic convertase (nardilysin) isoforms are soluble dibasic-specific metalloendopeptidases that localize in the cytoplasm and at the cell surface. Biochem. J., 349, 587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes N.E. and Stern,D.F. (1994) The biology of erbB-2/neu/HER-2 and its role in cancer. Biochim. Biophys. Acta, 1198, 165–184. [DOI] [PubMed] [Google Scholar]
- Iwamoto R. and Mekada,E. (2000) Heparin-binding EGF-like growth factor: a juxtacrine growth factor. Cytokine Growth Factor Rev., 11, 335–344. [DOI] [PubMed] [Google Scholar]
- Kainulainen V., Sundvall,M., Maatta,J.A., Santiestevan,E., Klagsbrun, M. and Elenius,K. (2000) A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J. Biol. Chem., 275, 8641–8649. [DOI] [PubMed] [Google Scholar]
- Marikovsky M., Breuing,K., Liu,P.Y., Eriksson,E., Higashiyama,S., Farber,P., Abraham,J. and Klagsbrun,M. (1993) Appearance of heparin-binding EGF-like growth factor in wound fluid as a response to injury. Proc. Natl Acad. Sci. USA, 90, 3889–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. and Pandiella,A. (1993) Membrane-anchored growth factors. Annu. Rev. Biochem., 62, 515–541. [DOI] [PubMed] [Google Scholar]
- Miyagawa J., et al. (1995) Localization of heparin-binding EGF-like growth factor in the smooth muscle cells and macrophages of human atherosclerotic plaques. J. Clin. Invest., 95, 404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghal N. and Sternberg,P. (1999) Multiple positive and negative regulators of signaling by the EGF-receptor. Curr. Opin. Cell Biol., 11, 190–196. [DOI] [PubMed] [Google Scholar]
- Munson P.J. and Rodbard,D. (1980) Ligand: a versatile computerized approach for characterization of ligand-binding system. Anal. Biochem., 107, 220–239. [DOI] [PubMed] [Google Scholar]
- Naglich J.G., Metherall,J.E., Russel,D.W. and Eidels,L. (1992) Expression cloning of a diphtheria toxin receptor: identity with a heparin-binding EGF-like growth factor precursor. Cell, 69, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Nishi E., Kume,N., Ochi,H., Moriwaki,H., Wakatsuki,Y., Higashiyama, S., Taniguchi,N. and Kita,T. (1997) Lysophosphatidylcholine increases expression of heparin-binding epidermal growth factor-like growth factor in human T lymphocytes. Circ. Res., 80, 638–644. [DOI] [PubMed] [Google Scholar]
- Olayioye M., Neve,R., Lane,H. and Hynes,N. (2000) The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J., 19, 3159–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opanashuk L., Mark,R., Porter,J., Damm,D., Mattson,M. and Seroogy,K. (1999) Heparin-binding epidermal growth factor-like growth factor in hippocampus: modulation of expression by seizures and anti-excitotoxic action. J. Neurosci., 19, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paria B.C., Elenius,K., Klagsbrun,M. and Dey,S.K. (1999) Heparin-binding EGF-like growth factor interacts with mouse blastocysts independently of ErbB1: a possible role for heparan sulfate proteoglycans and ErbB4 in blastocyst implantation. Development, 126, 1997–2005. [DOI] [PubMed] [Google Scholar]
- Pierotti A.R., Prat,A., Chesneau,V., Gaudoux,F., Leseney,A.M., Foulon,T. and Cohen,P. (1994) N-arginine dibasic convertase, a metalloendopeptidase as a prototype of a class of processing enzymes. Proc. Natl Acad. Sci. USA, 91, 6078–6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell P.P., Klagsbrun,M., Abraham,J.A. and Jones,R.C. (1993) Eosinophils expressing heparin-binding EGF-like growth factor mRNA localize around lung microvessels in pulmonary hypertension. Am. J. Pathol., 143, 784–793. [PMC free article] [PubMed] [Google Scholar]
- Prenzel N., Zwick,E., Daub,H., Leserer,M., Abraham,R., Wallasch,C. and Ullrich,A. (1999) EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature, 402, 884–888. [DOI] [PubMed] [Google Scholar]
- Raab G. and Klagsbrun,M. (1997) Heparin-binding EGF-like growth factor. Biochim. Biophys. Acta, 1333, F179–F199. [DOI] [PubMed] [Google Scholar]
- Raab G., Higashiyama,S., Hetelekidis,S., Abraham,J.A., Damm,D., Ono,M. and Klagsbrun,M. (1994) Biosynthesis and processing by phorbol ester of the cells surface-associated precursor form of heparin-binding EGF-like growth factor. Biochem. Biophys. Res. Commun., 204, 592–597. [DOI] [PubMed] [Google Scholar]
- Raab G., Kover,K., Paria,B.C., Dey,S.K., Ezzell,R.M. and Klagsbrun,M. (1996) Mouse preimplantation blastocysts adhere to cells expressing the transmembrane form of heparin-binding EGF-like growth factor. Development, 122, 637–645. [DOI] [PubMed] [Google Scholar]
- Riese D.N. and Stern,D. (1998) Specificity within the EGF family/ErbB receptor family signaling network. BioEssays, 20, 41–48. [DOI] [PubMed] [Google Scholar]
- Shishido Y., Sharma,K.D., Higashiyama,S., Klagsbrun,M. and Mekada,E. (1995) Heparin-like molecules on the cell surface potentiate binding of diphtheria toxin to the diphtheria toxin receptor/membrane-anchored heparin-binding epidermal growth factor-like growth factor. J. Biol. Chem., 270, 29578–29585. [DOI] [PubMed] [Google Scholar]
- Summerton J. and Weller,D. (1997) Morpholino antisense oligomer: design, preparation and properties. Antisense Nucleic Acid Drug Dev., 7, 187–195. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Raab,G., Moses,M.A., Fernandez,C.A. and Klagsbrun,M. (1997) Matrix metalloproteinase-3 releases active heparin-binding EGF-like growth factor by cleavage at a specific juxtamembrane site. J. Biol. Chem., 272, 31730–31737. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Sasahara,M., Ohno,M., Higashiyama,S., Hayase,Y. and Shimada,M. (1999) Heparin-binding epidermal growth factor-like growth factor mRNA expression in neonatal rat brain with hypoxic/ischemic injury. Brain Res., 827, 130–138. [DOI] [PubMed] [Google Scholar]
- Thompson S.A. et al. (1994) Characterization of sequences within heparin-binding EGF-like growth factor that mediate interaction with heparin. J. Biol. Chem., 269, 2541–2549. [PubMed] [Google Scholar]
- Thorne B.A. and Plowman,G.D. (1994) The heparin-binding domain of amphiregulin necessitates the precursor pro-region for growth factor secretion. Mol. Cell. Biol., 14, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumaru S. et al. (2000) Ectodomain shedding of epidermal growth factor receptor ligands is required for keratinocyte migration in cutaneous wound healing. J. Cell Biol., 151, 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]