Abstract
Phosphorylation on serines or threonines preceding proline (Ser/Thr-Pro) is a major signaling mechanism. The conformation of a subset of phosphorylated Ser/Thr-Pro motifs is regulated by the prolyl isomerase Pin1. Inhibition of Pin1 induces apoptosis and may also contribute to neuronal death in Alzheimer’s disease. However, little is known about the role of Pin1 in cancer or in modulating transcription factor activity. Here we report that Pin1 is strikingly overexpressed in human breast cancers, and that its levels correlate with cyclin D1 levels in tumors. Overexpression of Pin1 increases cellular cyclin D1 protein and activates its promoter. Furthermore, Pin1 binds c-Jun that is phosphorylated on Ser63/73-Pro motifs by activated JNK or oncogenic Ras. Moreover, Pin1 cooperates with either activated Ras or JNK to increase transcriptional activity of c-Jun towards the cyclin D1 promoter. Thus, Pin1 is up-regulated in human tumors and cooperates with Ras signaling in increasing c-Jun transcriptional activity towards cyclin D1. Given the crucial roles of Ras signaling and cyclin D1 overexpression in oncogenesis, our results suggest that overexpression of Pin1 may promote tumor growth.
Keywords: cancer/c-Jun/cyclin D1/Pin1/Ras signaling
Introduction
The reversible phosphorylation of proteins on serine/threonine residues preceding proline (pSer/Thr-Pro) is a key regulatory mechanism for the control of various cellular processes, including cell division and transcription (reviewed by Hunter and Karin, 1992; Nurse, 1994; Nigg, 1995; Treisman, 1996; Whitmarsh and Davis, 1996; Karin et al., 1997). For example, various growth factors and oncoproteins, such as oncogenic Ras, trigger a signaling cascade leading to the activation of c-Jun N-terminal kinases (JNKs), which phosphorylate c-Jun on Ser63/73-Pro and enhance its transcriptional activity towards c-Jun target genes, including cyclin D1 (Binetruy et al., 1991; Smeal et al., 1991; Derijard et al., 1994; Hinds et al., 1994; Albanese et al., 1995, 1999; Fantl et al., 1995; Sicinski et al., 1995; Robles et al., 1998; Bakiri et al., 2000). Overexpression of cyclin D1 often occurs in a variety of human cancers (Hunter and Pines, 1994), including ∼50% of human breast tumors (Bartkova et al., 1994; Gillett et al., 1994; Lin et al., 2000). Importantly, cyclin D1 can act as an oncogene that contributes to cell transformation by complementing a defective E1A oncogene (Hinds et al., 1994). Conversely, inhibition of cyclin D1 expression causes growth arrest in tumor cells (Schrump et al., 1996; Arber et al., 1997; Driscoll et al., 1997; Kornmann et al., 1998). Moreover, knockout of cyclin D1 in mice blocks the proliferation of breast epithelial cells and retina, and inhibits tumor development in response to Ha-Ras (Fantl et al., 1995; Sicinski et al., 1995; Robles et al., 1998; Rodriguez-Puebla et al., 1999). These results indicate that cyclin D1 plays an important role during oncogenesis, acting as a downstream mediator of Ras activity during tumor development, and that phosphorylation of c-Jun on Ser63/73-Pro motifs is an important mechanism for the Ras-dependent up-regulation of cyclin D1. However, it is not clear whether the c-Jun activity is further regulated after Pro-directed phosphorylation.
Compelling evidence supports an additional and crucial signaling mechanism, which affects the state of Pro-directed phosphorylation sites, namely the conformational change induced by phosphorylation-specific prolyl isomerization. Such conformational change can regulate protein function (Zhou et al., 1999). The phosphorylated Ser/Thr-Pro moiety exists in two distinct, slowly interconverting conformations: cis and trans. This conformational change introduces kinks into a peptide chain, thereby determining protein structure and function (Fischer, 1994; Galat and Metcalfe, 1995; Schmid, 1995; Hunter, 1998; Zhou et al., 1999). Significantly, phosphorylation on Ser/Thr-Pro motifs further restrains the already slow cis/trans prolyl isomerization of peptide bonds (Yaffe et al., 1997; Schutkowski et al., 1998), and also renders them resistant to the catalytic action of conventional peptidyl-prolyl cis/trans isomerases (PPIases), including cyclophilins and FK506-binding proteins (Yaffe et al., 1997). In contrast, Pin1 represents a new subfamily of highly conserved and phosphorylation-specific PPIases that isomerize only the phosphorylated Ser/Thr-Pro bonds, and not their non-phosphorylated counterparts (Yaffe et al., 1997). Pin1 contains an N-terminal WW domain and a C-terminal PPIase domain (Lu et al., 1996; Ranganathan et al., 1997). The WW domain functions as a pSer/Thr-binding module, interacting with specific pSer/Thr-Pro motifs present in a defined subset of phosphoprotein substrates, including Cdc25C, tau, Myt1, S6 kinase, Rab4 and the C-terminal domain of RNA polymerase II (Lu et al., 1999b). At the substrate, the PPIase domain of Pin1 isomerizes specific pSer/Thr-Pro bonds, and regulates protein function and dephosphorylation (Yaffe et al., 1997; Shen et al., 1998; Lu et al., 1999b; Zhou et al., 1999, 2000). For example, in the case of Cdc25C, Pin1 binds phosphorylated Cdc25C, and inhibits its activity to dephosphorylate and activate Cdc2 (Shen et al., 1998; Zhou et al., 2000). However, in the case of tau, Pin1 binds Alzheimer’s disease-associated phosphorylated tau and restores its biological function to promote microtubule assembly (Lu et al., 1999a; Zhou et al., 2000). These results indicate that Pin1 plays an important role in the regulation of a defined subset of phosphorylated proteins.
Functionally, Pin1 is critical for cell proliferation in vivo. Temperature-sensitive mutations or deletion of the Ess1 gene (the Pin1 homologue in budding yeast) result in mitotic arrest and nuclear fragmentation (Hanes et al., 1989; Hani et al., 1995, 1999; Lu et al., 1996). These arrested cells have defective 3′ end formation of pre-mRNA, and decreased levels of some mRNAs (Hani et al., 1999; Wu et al., 2000). However, it remains to be determined whether these defects are primarily due to the effect of Ess1 on the general transcription machinery, as suggested, or secondarily due to the fact that these cells are arrested in mitosis with fragmented nuclei, or both. Inhibition of the Pin1 function in human tumor cells using expression of the Pin1 antisense RNA or dominant-negative mutants induces mitotic arrest and apoptosis (Lu et al., 1996; Rippmann et al., 2000; P.J.Lu, X.Z.Zhou, Y.-C.Liou, J.P.Noel and K.P.Lu, submitted). Similarly, depletion of Pin1 in Alzheimer’s disease brain may also contribute to neuronal death (Lu et al., 1999a). Furthermore, depletion of Pin1 in Xenopus extracts induces premature mitotic entry and disrupts a DNA replication checkpoint (Winkler et al., 2000). These results together suggest that the level and function of Pin1 are pivotal for cell proliferation. However, the level and role of Pin1 in human cancer have not yet been described.
Here we show that Pin1 is overexpressed in most human breast cancer cell lines and many human breast cancer tissues. Furthermore, the Pin1 levels correlate significantly with the grade of the tumors, according to Bloom and Richardson’s classification system (Bloom and Richardson, 1957), and with the level of cyclin D1 in the tumors. Moreover, Pin1 increases levels of cellular cyclin D1 mRNA and protein, and activates its promoter through the AP-1 site. Importantly, Pin1 binds to phosphorylated c-Jun and increases its transcriptional activity towards the cyclin D1 promoter, in cooperation either with activated JNK or oncogenic Ras. The effects of Pin1 on the c-Jun transcriptional activity depend on both the isomerase activity of Pin1 and phosphorylation of c-Jun on Ser63/73. In contrast, inhibition of endogenous Pin1 reduces the transcriptional activity of phosphorylated c-Jun. These results demonstrate that Pin1 is up-regulated in human tumor samples and cooperates with Ras signaling in increasing c-Jun transcriptional activity towards cyclin D1. Given the crucial roles of the activated Ras signaling and cyclin D1 overexpression in the development of cancer, our results suggest that overexpression of Pin1 may promote tumor growth.
Results
Pin1 is overexpressed in human breast tumors and its levels correlate with the tumor grade
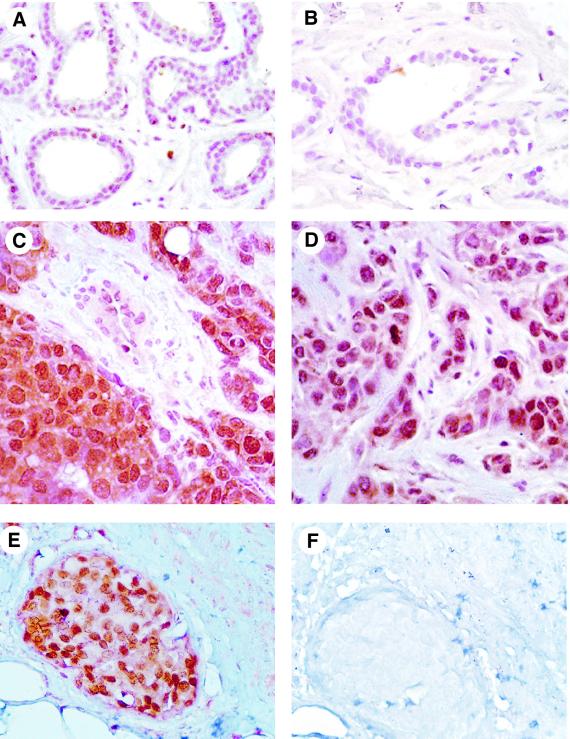
To examine the role of Pin1 in cancer, we examined the expression of Pin1 in normal human breast tissues and breast tumors by immunohistochemistry and immunoblotting with affinity-purified anti-Pin1 antibodies, as described earlier (Lu et al., 1999a). Normal breast epithelial cells showed weak but detectable Pin1 staining primarily in the nucleus (Figure 1A and B). In contrast, carcinoma cells were strongly positive for the Pin1 staining (Figure 1C–E), while surrounding normal connective tissue, blood vessels, adipose and stromal cells stained only weakly for Pin1 (Figure 1E). In these tumor cells, Pin1 staining was detected at high levels in the cytoplasm, in addition to intensive staining in the nucleus (Figure 1C–E). To ensure that these signals indeed represent Pin1, the Pin1-specific antibodies were depleted using glutathione S-transferase (GST)–Pin1 beads prior to immunostaining. Figure 1F shows that the Pin1-depleted antibodies showed no immunoreactivity, confirming the specificity of the antibodies, as described (Lu et al., 1999a). Immunohistochemistry in other cancer types revealed high Pin1 levels in some tumors, including colon cancer, lymphomas, melanoma, prostate and brain tumors, but rarely in others, such as sarcoma (data not shown). Since we had access to a large number of breast cancer samples, we focused this study on breast cancer.
Fig. 1. Immunostaining of Pin1 in human breast cancer. Sections from paraffin-embedded tissues were subjected to an antigen retrieval treatment, followed by immunostaining with anti-Pin1 antibodies. Non-cancerous tissues (A and B; normal breast with mild fibrocystic changes) show weak, but detectable, Pin1 staining, while invasive ductal carcinomas (C and D) or ductal carcinoma in situ (E) show intense Pin1 staining. To show the specificity of Pin1 antibodies, Pin1-specific antibodies were first depleted using GST–Pin1 beads and then used to stain the breast sections (F).
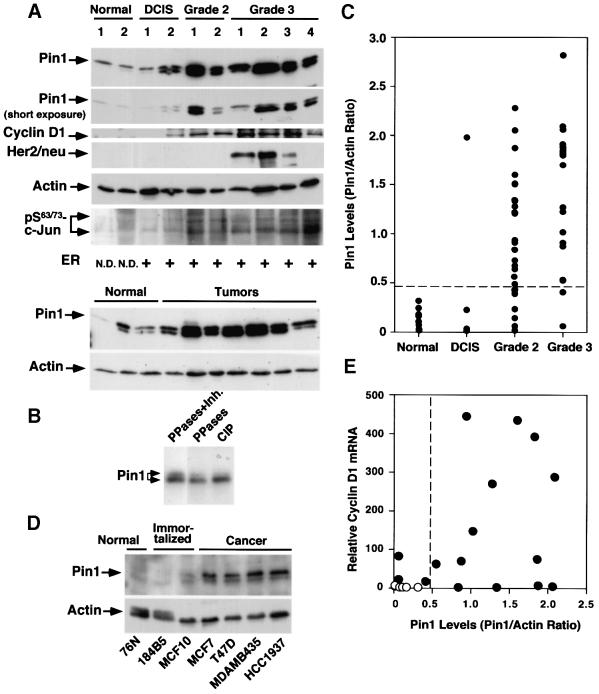
To evaluate Pin1 expression in breast cancer quantitatively, we ground fresh, normal or tumor breast tissues in liquid nitrogen and subjected the lysates directly to immunoblotting analysis with various antibodies, followed by semi-quantification of protein levels using Imagequant, as described (Lu et al., 1999a). Pin1 was generally detected as a doublet in immunoblots, especially in tumor tissues where Pin1 was overexpressed. Upon dephosphorylation with protein phosphatases PP2A and PP1, or calf intestine phosphatase (CIP), the intensity of the upper band decreased, while the lower one increased (Figure 2B). In addition, Pin1 displays a mitosis- and phosphorylation-specific mobility shift during the cell cycle (P.J.Lu, X.Z.Zhou, Y.-C.Liou, J.P.Noel and K.P.Lu, submitted). These results indicate that the Pin1 doublet is likely to be due to the electrophoretic mobility difference of phosphorylated and dephosphorylated Pin1. Interestingly, the upper phosphorylated band appeared to be predominant in the normal tissues, whereas the lower dephosphorylated band was more abundant in the cancerous tissues where Pin1 was overexpressed (Figure 2A), suggesting that there are more mitotic cells and/or the kinase(s) responsible for the Pin1 phosphorylation might be limited in tumor cells.
Fig. 2. Pin1 overexpression in human breast cancer cell lines and patient tissues, and its correlation with the Bloom and Richardson grade of tumors. (A) Comparison of Pin1 levels and known breast tumor markers in normal and cancerous human breast tissues. Normal breast and cancer tissues were pulverized in liquid nitrogen, and equal amounts of total protein were separated on SDS-containing gels and transferred to membranes. The membranes were cut into five pieces and subjected to immunoblotting analysis using antibodies to Pin1, cyclin D1, HER2/neu, phosphorylated Ser63/73-c-Jun and actin, respectively. The estrogen receptor status was determined by radioimmunoassay and defined as positive when its levels were >10 fmol/l. The estrogen receptor status in normal controls was not determined (N.D.). Note that Pin1 was detected in immunoblots as a doublet due to phosphorylation. (B) Phosphatase treatment abolishes the double-band pattern of Pin1 in immunoblots. Tumor cell lysates were treated either with a mixture of PP1 and PP2A (PPases) in the presence (lane 1) or absence (lane 2) of the phosphatase inhibitor okadaic acid (Inh.), or CIP (lane 3). (C) Pin1 levels in 10 normal breast tissues and different stages of 51 human breast cancer samples. Pin1 levels were determined by immunoblotting analysis, as in (A), and semi-quantified using Imagequant. Actin was used as an internal control, and the Pin1 level in each sample was expressed as the Pin1:actin ratio. (D) Comparison of Pin1 levels in mammary epithelial cell lines. The same amounts of total lysates prepared from normal human mammary epithelial cell lines (Normal), spontaneously immortalized normal human mammary epithelial cell lines (Immortalized) and human breast carcinoma-derived cell lines (Cancer) were subjected to immunoblotting analysis with Pin1 or actin antibodies. (E) Correlation of Pin1 protein levels with cyclin D1 mRNA. RNA was isolated from six normal and 16 cancerous tissues, cDNA synthesized and subjected to real-time PCR for the quantitative analysis of cyclin D1 mRNA expression. The Pearson correlation coefficient was 0.47 (p <0.05).
To compare the levels of Pin1 in different human tissues, we used actin as an internal control, and expressed the Pin1 level in each sample as a Pin1:actin ratio. We defined Pin1 overexpression as higher than the mean plus three times the standard deviation (x̄ + 3 SD) of normal controls (Figure 2C; Table I). In 10 normal and 51 primary human breast cancer tissues examined, we observed striking differences in the levels of Pin1 protein expression (Figure 2A and C). One out of four DCIS tumors, 20 out of 28 (71.4%) grade II tumors and 17 out of 19 (89.5%) grade III tumors, according to Bloom and Richardson’s classification system, overexpressed Pin1 (Figure 2C). Although we observed considerable inter-individual variations, especially in grade II and III tumors (Figure 2C), the mean expression level of Pin1 was ∼10 times higher in cancer samples than in the normal controls (Table I). Furthermore, Pin1 levels positively correlated with the Bloom and Richardson grade in invasive breast cancer, as analyzed by the Kruskal–Wallis test (Glantz, 1997) (Figure 2B; Table II). Similar results were also obtained using a monoclonal antibody against Pin1 for immunostaining and immunoblotting analyses (data not shown). The levels of Pin1 in four cell lines derived from human breast cancers were considerably higher than those in either normal mammary epithelial cells or two cell lines established from normal mammary epithelial cells (Figure 2D). Together, these results indicate that Pin1 is overexpressed in many human breast cancer tissues and cell lines, and its levels are correlated with the tumor grade.
Table I. Clinical and pathological characteristics of breast tissues.
| Normal | Carcinoma |
||||
|---|---|---|---|---|---|
| Total | In situ | Grade 2 | Grade 3 | ||
| Pin1 positive | 0/10a | 38/51 (75%) | 1/4 (25%) | 20/28 (71%) | 17/19 (89%) |
| x̄ ± SD | 0.114 ± 0.106 | 1.072 ± 0.719 | 0.564 ± 0.948 | 0.924 ± 0.609 | 1.399 ± 0.717 |
| Cyclin D1 | 0/10 | 24/51 (47%) | 2/4 (50%) | 10/28 (36%) | 12/19 (63%) |
| HER2/neu | 0/10 | 8/51 (16%) | 0/4 (0%) | 4/28 (14%) | 4/19 (21%) |
| Estrogen receptor | N.D.b | 34/50c (68%) | 3/4 (75%) | 20/28 (71%) | 11/18 (61%) |
| Age median (range) | 57 (22–91) | 65 (28–90) | 72 (43–80) | 65 (31–90) | 60 (28–78) |
Tumors were pathologically classified into ductal carcinoma in situ (in situ) and invasive grade 2 and 3 carcinoma, according to the criteria of Bloom and Richardson. Levels of Pin1 in tissues were determined by immunoblotting analysis and semi-quantified using Imagequant, with the results being expressed as Pin1/actin ratio. Pin1 was defined positive when the Pin1/actin ratio was higher than the mean plus three times the standard deviation (x̄ ± 3 SD) of normal controls. Cyclin D1 and HER2/neu were determined by immunoblotting and categorized as either positive or negative by the presence or absence of the respective proteins. Estrogen receptor was defined positive when its levels were >10 fmol/l, as determined by radioimmunoassay.
aNumber of cases examined.
bEstrogen receptors in controls not determined.
cEstrogen receptor determination on one patient not available.
Table II. Correlation of the Pin1 level with clinical and pathological characteristics.
| No. of cases | Pin1 level (x̄ ± SD) | p | |
|---|---|---|---|
| Normal | 10 | 0.114 ± 0.106 | <0.0001b |
| Tumor | 51 | 1.072 ± 0.716 | |
| Tumor grade | |||
| grade 2 | 28 | 0.924 ± 0.609 | 0.02b |
| grade 3 | 19 | 1.399 ± 0.717 | |
| Cyclin D1a | |||
| positive | 24 | 1.364 ± 0.715 | 0.01b |
| negative | 27 | 0.824 ± 0.631 | |
| HER2/neua | |||
| positive | 8 | 1.317 ± 0.732 | 0.10 |
| negative | 43 | 1.027 ± 0.713 | |
| Estrogen receptora | |||
| positive | 34 | 1.011 ± 0.718 | 0.32 |
| negative | 16 | 1.238 ± 0.720 |
The significance of the differences in Pin1 levels between various clinical and pathological categories was analyzed by the Kruskal–Wallis test.
aAnalyses performed only on tumors.
bThe differences are statistically significant when p ≤0.05 and highly significant when p ≤0.01.
Up-regulation of Pin1 correlates with cyclin D1 levels in breast tumor tissues and elevates cellular cyclin D1 expression in breast cell lines
Amongst other breast cancer tumor markers, Pin1 levels did not appear to correlate with either estrogen receptor or HER2/neu expression, but did correlate significantly with cyclin D1 overexpression (Tables I and II). As shown previously (Bartkova et al., 1994; Gillett et al., 1994), cyclin D1 was overexpressed in ∼50% of the patient samples (24 out of 51 cases). Importantly, Pin1 was overexpressed in 20 out of 24 cyclin D1-overexpressing tumors, and Pin1 levels in cyclin D1-overexpressing tumors were on average about twice as high as those in cyclin D1-negative tumors (Figure 2A; Table II). In order to establish a link between Pin1 overexpression and cyclin D1 transcription, we performed quantitative real-time PCR to detect cyclin D1 mRNA expression in 6 out of the 10 normal tissues and 16 out of the 51 breast cancer tissues, from which we were able to isolate total RNA. Figure 2E shows relative cyclin D1 mRNA levels as a function of Pin1 protein levels. While a few patients had high Pin1 but low cyclin D1 mRNA levels, all but one patient with high cyclin D1 mRNA levels also displayed high Pin1 levels, which is consistent with the results on cyclin D1 protein levels (Table II). Statistical analysis revealed that there was again a positive correlation between Pin1 protein levels and cyclin D1 mRNA expression (r = 0.47, p <0.05).
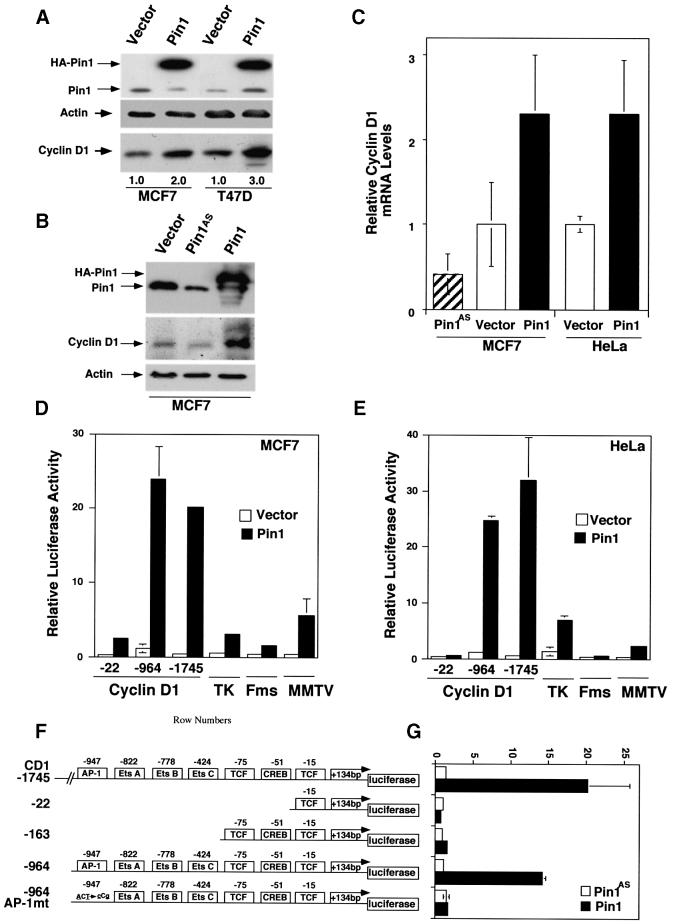
The correlation between Pin1 and cyclin D1 expression suggested that overexpression of Pin1 might increase the expression of endogenous cyclin D1. To examine this possibility, we transiently transfected a Pin1 expression construct into two breast cancer-derived cell lines, MCF7 and T47D cells, and then examined the effects on endogenous cyclin D1 levels. Pin1 overexpression led to 2- to 3-fold increases in cyclin D1 protein levels in both cell lines, while the expression of actin remained constant (Figure 3A). To examine whether the depletion of Pin1 affected cyclin D1 expression, we used MCF7 and HeLa cells because their Pin1 levels can be increased or decreased by expressing a sense or antisense Pin1 construct, respectively (Figure 3B), as described previously (Lu et al., 1996). Overexpression of Pin1 significantly increased the levels of cyclin D1 protein and mRNA in both cells (Figure 3B and C and data not shown). In contrast, depletion of Pin1 significantly reduced the levels of cyclin D1 protein and mRNA in MCF7 cells (Figure 3B and C). Since these experiments were performed between 24 and 36 h after transfection, and since manipulation of Pin1 levels affects the cell cycle only after 48–72 h post-transfection (Lu et al., 1996), the observed effects of Pin1 on cyclin D1 are unlikely to be related to cell cycle arrest. These results indicate that high levels of Pin1 correlate with the overexpression of cyclin D1 on both RNA and protein levels in human breast cancer tissues, and that overexpression of Pin1 increases cellular cyclin D1 mRNA and protein levels in cell lines.
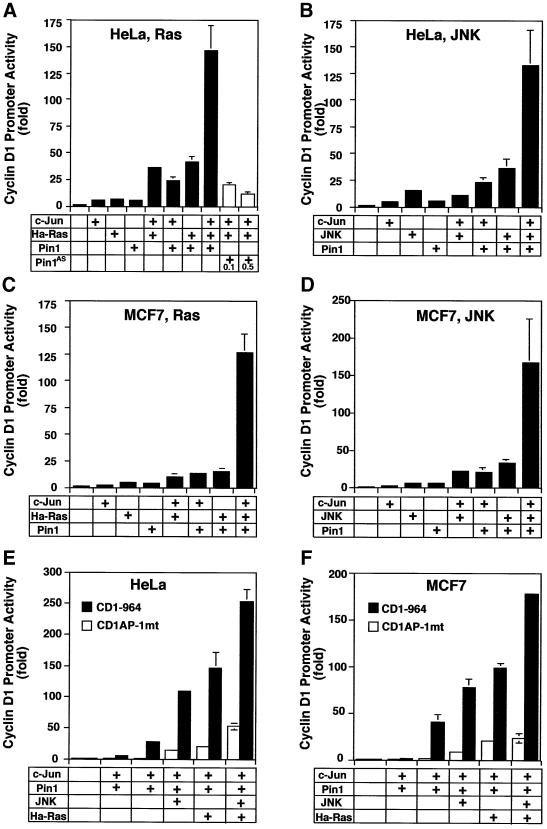
Fig. 3. Pin1 elevates cyclin D1 protein and activates the cyclin D1 promoter via the AP-1 site. (A) Increase in cellular cyclin D1 protein by Pin1. MCF7 or T47D cells were transfected with Pin1 or control vector, followed by immunoblotting analysis of the cell lysates with antibodies against Pin1 and cyclin D1, with actin as a control. Cyclin D1 levels were semi-quantified using Imagequant and are presented below the image; the level in the vector control was defined as 1. (B) Manipulation of Pin1 levels in cells causes changes in cyclin D1 levels. MCF7 cells were transiently transfected with the control vector or a construct expressing HA-Pin1 or antisense Pin1 (Pin1AS), followed by immunoblotting analysis with anti-cyclin D1, -Pin1 or -actin antibodies. (C) Overexpression and depletion of Pin1 increase and decrease levels of cyclin D1 mRNA. MCF7 or HeLa cells were transfected with constructs encoding for Pin1 sense, antisense or vector control as indicated in the figure. After 24 h, mRNA was isolated, cDNA synthesized and subjected to real-time PCR to obtain relative cyclin D1 mRNA levels. (D and E) Activation of cyclin D1, but not TK, c-fms or MMTV promoter by Pin1. MCF7 (D) or HeLa (E) cells were transiently transfected with Pin1 or the vector and various reporter constructs, followed by assaying the luciferase activity. pRL-TK Renilla luciferase reporter construct was co-transfected in each sample to normalize for transfection efficiency. The activity of the reporter luciferase was expressed relative to that in control vector-transfected cells, which is defined as 1. All results are expressed as x̄ ± SD of independent duplicate cultures. Note that to detect the maximal effect of Pin1 on various promoters, we used 0.5 µg of Pin1 cDNA per transfection in this experiment, which was higher than in other experiments described here. (F) Schematic representation of cyclin D1 (CD1) pA3LUC basic reporter constructs and its mutants. Possible transcription factor-binding sites are indicated. –964CD1AP-1mt was same as the wild-type –964CD1construct except for two base-pair substitutions at the consensus AP-1 site. (G) Activation of the cyclin D1 promoter by Pin1 via the AP-1 site. HeLa cells were co-transfected with various cyclin D1 reporter constructs as indicated in (F) and Pin1 sense or antisense (Pin1AS) construct, followed by assaying the luciferase activity. Note that for this experiment 200 ng Pin1 sense or antisense cDNA were used, while in subsequent co-transfection experiments only 50 ng/assay were used.
Pin1 activates the cyclin D1 promoter
Although cyclin D1 overexpression is found in ∼50% of breast cancer patients (Bartkova et al., 1994; Gillett et al., 1994), gene amplification accounts for only 10% of these cases (Fantl et al., 1993). Therefore, other mechanisms, such as up-regulation of gene transcription, must play a substantial role in the overexpression of cyclin D1. To determine whether Pin1 regulates the transcription of cyclin D1, we measured the effects of Pin1 on the cyclin D1 promoter using cyclin D1–luciferase reporter constructs. Two cyclin D1–reporter constructs were tested: one (–1745CD1) corresponds to the original fragment of cyclin D1 5′ sequence cloned from the PRAD1 breakpoint (Motokura and Arnold, 1993), and the other (–964CD1) is the minimum 5′ sequence that retains the responsiveness to activated Ras (Albanese et al., 1995). Both –1745CD1 and –964CD1 reporters were strongly activated in response to expression of Pin1 both in MCF7 and HeLa cells (Figure 3D and E). These results indicate that Pin1 activates the cyclin D1 promoter and that the –964CD1 promoter fragment retains the complete responsiveness to Pin1.
It has recently been shown that Pin1/Ess1p binds the phosphorylated C-terminal domain of RNA polymerase II and may regulate the general transcription machinery in yeast (Wu et al., 2000). To determine whether activation of the cyclin D1 promoter by Pin1 is due to its effect on the general transcription machinery, we examined the effect of Pin1 on several other unrelated promoters. To detect the maximal effect of Pin1 on various promoters, we used 500 ng of Pin1 cDNA per transfection. In contrast, out of many other promoters examined, including thymidine kinase (TK), c-fms (M-CSF receptor) and MMTV promoters, Pin1 either had no effect or had minor transactivating effects (Figure 3D and E), indicating that activation of the general transcription machinery by Pin1 is very low, which is consistent with a recent report (Chao et al., 2001). Therefore, the above results indicate that Pin1 specifically activates the cyclin D1 promoter.
To further confirm the specificity of the Pin1 action on the cyclin D1 promoter, we identified the element in the cyclin D1 promoter that is responsible for Pin1 activation. The –964CD1 promoter fragment contains binding sites for various transcription factors, including a CREB site, four TCF sites, three Ets sites and one AP-1 site (Albanese et al., 1995; Tetsu and McCormick, 1999) (Figure 3F). To determine which promoter is necessary for Pin1 responsiveness, we used two deletion constructs containing either 22 bp (–22CD1) or 163 bp (–163CD1) of the cyclin D1 promoter as reporters. Low concentrations (50–200 ng) of Pin1 did not have any significant transactivating effect either on the –22CD1 or the –163CD1 reporter (Figure 3F and G), while at high concentrations (>200 ng per transfection) Pin1 could also transactivate the –163CD1 promoter containing the TCF sites (Ryo et al., 2001). At low concentrations, i.e. ≤200 ng, Pin1 significantly transactivated both the –1745CD1 and –964CD1 promoters (Figure 3F and G). These results confirm that Pin1 does not affect the cyclin D1 promoter activity via the general transcriptional machinery but through specific sequences such as the AP-1 and/or Ets sites. To examine the importance of the AP-1 site, we used a mutant promoter, –964CD1AP-1mt, containing only two base-pair substitutions at the consensus AP-1 site, as described (Albanese et al., 1995). Elimination of the AP-1 site almost completely abolished the ability of Pin1 to activate the cyclin D1 promoter (Figure 3F and G). These results indicate that the AP-1 site is essential for activation of the cyclin D1 promoter by Pin1.
Pin1 binds c-Jun phosphorylated on Ser63/73-Pro motifs
The AP-1 site mutation in the cyclin D1 promoter that disrupts the Pin1 transactivating activity also abolishes cyclin D1 expression induced by the activation of Ras or c-Jun (Albanese et al., 1995), suggesting that Pin1 might affect the same pathway as that regulated by Ras or c-Jun. Activation of Ras triggers a signaling cascade, leading to activation of the c-Jun N-terminal kinase JNK, which phosphorylates c-Jun on Ser63/73-Pro to increase its transcriptional activity towards its target genes, including cyclin D1 (Binetruy et al., 1991; Smeal et al., 1991; Derijard et al., 1994; Hinds et al., 1994; Albanese et al., 1995, 1999; Fantl et al., 1995; Sicinski et al., 1995; Robles et al., 1998; Bakiri et al., 2000). In fact, Ras-mediated tumorigenesis depends on signaling pathways with cyclin D1 as an important intermediary protein (Robles et al., 1998). Since Pin1 binds and regulates the function of a defined subset of proteins phosphorylated on certain Ser/Thr-Pro motifs (Shen et al., 1998; Lu et al., 1999a), it is possible that Pin1 might activate the cyclin D1 promoter via modulation of the the activity of phosphorylated c-Jun.
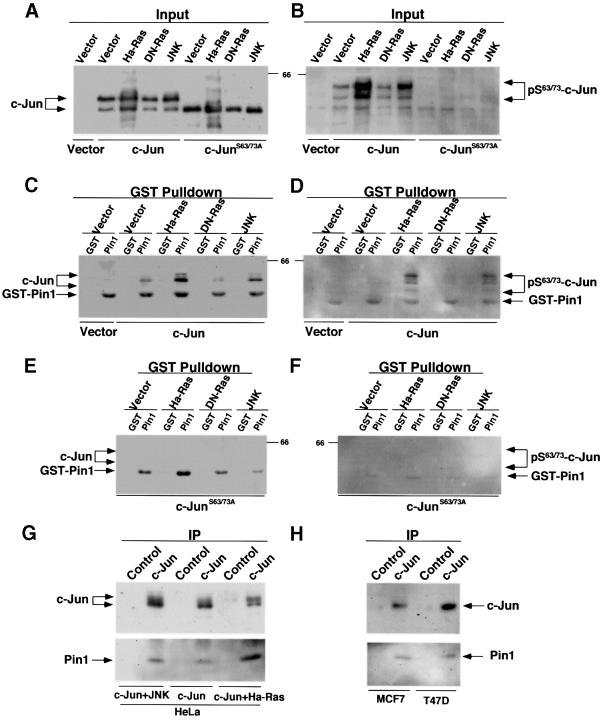
A well established and successful procedure to identify Pin1 substrates has been the use of GST–Pin1 pulldown experiments to determine whether Pin1 binds to c-Jun, and whether the binding depends on phosphorylation of c-Jun on specific Ser-Pro motifs, as demonstrated for many other Pin1 substrates (Yaffe et al., 1997; Crenshaw et al., 1998; Shen et al., 1998; Lu et al., 1999b). To increase phosphorylation of c-Jun on Ser63/73-Pro, we co-transfected c-Jun with a constitutively activated form of JNK (Derijard et al., 1994). Alternatively, we co-transfected c-Jun with a further upstream activator, the oncogenic Harvey-Ras (Ha-Ras or RasL61), which activates a MAK kinase pathway, leading to activation of JNK (Smeal et al., 1991; Derijard et al., 1994). To reduce phosphorylation of c-Jun on Ser63/73-Pro, we co-transfected c-Jun with the dominant-negative Ras (DN-Ras or RasN17) (Smeal et al., 1991; Derijard et al., 1994). As expected, phosphorylation of c-Jun on Ser63/73-Pro was increased to similar extents by either activated JNK or Ha-Ras, but significantly decreased by DN-Ras, as detected by antibodies specifically recognizing phosphorylated Ser63/73 in c-Jun (Figure 4A and B). Notably, following activation of JNKs by UV radiation or serum stimulation, c-Jun has been shown to be phosphorylated on several Ser-Thr sites, which resulted in a considerable shift in electrophoretic mobility of the protein, migrating as multiple bands in SDS gels (Ui et al., 1998). Furthermore, mutation of c-Jun on Ser63 and Ser73 abolishes the mobility shift (Ui et al., 1998). We observed a similar mobility shift for wild-type c-Jun, but not c-JunS63/73A, after co-transfection either with Ha-Ras or activated JNK (Figure 4A–D). Importantly, although there was no binding between GST and c-Jun, weak binding between GST–Pin1 and c-Jun was detec ted when only c-Jun was transfected (Figure 4C). Furthermore, the binding was significantly increased by co-transfection either with activated JNK or oncogenic Ha-Ras, but not with DN-Ras (Figure 4C). Moreover, c-Jun bound by Pin1 was also phosphorylated on Ser63/73-Pro, as indicated by phosphorylated Ser63/73-specific antibodies (Figure 4D). To examine further the importance of phosphorylation on Ser63/73 for Pin1 binding, we used a c-Jun mutant, c-JunS63/73A, which contains double Ala substitutions at Ser63 and Ser73 (Smeal et al., 1991). In contrast to wild-type c-Jun, the mutant protein did not display a significant mobility shift and was not recognized by phosphorylated Ser63/73-specific antibodies (Figure 4A and B), as shown previously (Ui et al., 1998). Importantly, little, if any, mutant protein was precipitated by Pin1 (Figure 4E and F). These results indicate that phosphorylation of c-Jun on Ser63/73-Pro is important for the Pin1 binding. Thus, Pin1 binds to c-Jun via phosphorylated Ser63/73-Pro motifs.
Fig. 4. Pin1 binds to c-Jun phosphorylated on Ser63/73-Pro. (A and B) Modulation of c-Jun phosphorylation by Ras or JNK. HeLa cells were co-transfected with c-Jun or c-JunS63/73A and Ha-Ras, DN-Ras, activated JNK or control vector. Cells were harvested and cellular proteins were subjected to immunoblotting analysis with antibodies against c-Jun (A) or phosphorylated Ser63/73-c-Jun (B). (C and D) Interaction between Pin1 and c-Jun phosphorylated on Ser63/73-Pro. The same cellular proteins as those described in (A) were incubated with GST–agarose beads that had been pre-incubated with either GST alone or GST–Pin1. Proteins associated with the beads were subjected to immunoblotting analysis with antibodies against c-Jun (C) or phosphorylated Ser63/73-c-Jun (D). Note that GST–Pin1 was non-specifically recognized by monoclonal antibodies, as shown previously (Yaffe et al., 1997; Lu et al., 1999b). (E and F) No interaction between Pin1 and c-JunS63/73A. The same cellular proteins as those described in the (A) were incubated with GST–agarose beads containing GST or GST–Pin1, and bound proteins were subjected to immunoblotting analysis with antibodies against c-Jun (E) or phosphorylated Ser63/73-c-Jun (F). (G and H) Co-immunoprecipitation of transfected (G) or endogenous (H) c-Jun with endogenous Pin1. HeLa cells were co-transfected with c-Jun and Ha-Ras or JNK. c-Jun was immunoprecipitated from transfected HeLa cells (G) or non-transfected breast cancer cell lines (H) with polyclonal c-Jun antibodies or non-related antibodies (Control), and then subjected to immunoblotting using monoclonal anti-c-Jun antibodies (upper panel) or anti-Pin1 antibodies (lower panel).
To confirm these GST–Pin1 protein pulldown results, we performed co-immunoprecipitation experiments between endogenous Pin1 and transfected c-Jun in the presence or absence of activated JNK or Ha-Ras, as well as co-immunoprecipitations between endogenous Pin1 and c-Jun in breast cancer cell lines expressing high levels of both proteins. Endogenous Pin1 was detected in anti-c-Jun immunoprecipitates from transfected (Figure 4G) and non-transfected cells (Figure 4H). Furthermore, more Pin1 was co-immunoprecipitated by anti-c-Jun antibodies if c-Jun was co-transfected with activated JNK or Ha-Ras (Figure 4G). These results indicate that Pin1 binds c-Jun in vivo in breast cancer cell lines, and that the binding is increased when c-Jun is phosphorylated on Ser63/73- Pro motifs by activated JNK or Ha-Ras. These results demonstrate that Pin1 binds phosphorylated c-Jun both in vitro and in vivo.
Pin1 cooperates with either oncogenic Ha-Ras or activated JNK to increase transcriptional activity of c-Jun towards the cyclin D1 promoter
Given that Pin1 binds phosphorylated c-Jun, we asked whether Pin1 also modulates the activity of c-Jun. To address this question, we examined the effect of Pin1 on the transcriptional activity of c-Jun towards the cyclin D1 promoter in the presence or absence of Ha-Ras or activated JNK. When Pin1 cDNA was co-transfected with c-Jun, Pin1 cooperated moderately with c-Jun in activating the cyclin D1 promoter in both MCF7 and HeLa cells (Figure 5). The activity of the cyclin D1 promoter in cells co-transfected with Pin1 and c-Jun was 3- to 5-fold higher than that in cells transfected with either Pin1 or c-Jun alone (Figure 5A–D). The most dramatic potentiation of cyclin D1 reporter gene activity was observed when c-Jun was activated by JNK or Ha-Ras in the presence of Pin1; cyclin D1 promoter activity was increased up to 150-fold, or higher, in both cell lines (Figure 5A–D). The combination of JNK, Ras, c-Jun and Pin1 resulted in a further small increase in transactivation (Figure 5E and F, last bars), consistent with the idea that Ras and JNK act on the same target c-Jun. However, when the AP-1 site mutant cyclin D1 promoter was used in the same assay, only ≤10% of the transactivation measured for the wild-type promoter was observed (Figure 5E and F), indicating that transactivation of the cyclin D1 promoter by c-Jun, activated by Pin1, JNK or Ras, is dependent on the intact AP-1-binding site. These results indicate that Pin1 cooperates either with activated JNK or oncogenic Ras to dramatically activate the cyclin D1 promoter. These cooperative effects are expected because Pin1 can regulate the transcriptional activity of c-Jun only after it has been phosphorylated by them.
Fig. 5. Pin1 cooperates either with Ha-Ras or activated JNK in enhancing the activity of c-Jun to activate the cyclin D1 promoter. HeLa cells (A, B and E) or MCF7 (C, D and F) were co-transfected with vector, c-Jun, or c-Jun with or without Ha-Ras (A and C) or activated JNK (B and D) and then subjected to the luciferase assay with the –964 cyclin D1 Luc promoter construct as reporter gene. In the same system, a reporter gene construct with an AP-1 site mutant fails to respond to Pin1 in combination with c-Jun, JNK or Ha-Ras (E and F).
To examine whether endogenous Pin1 is important for Ha-Ras to increase the transcriptional activity of c-Jun, we used Pin1AS to reduce cellular Pin1 levels (Figure 3B). When c-Jun and Ha-Ras were co-transfected with different concentrations of the Pin1AS construct, the transcriptional activity of c-Jun decreased significantly in a concentration-dependent manner (Figure 5A), indicating that inhibiting endogenous Pin1 decreases the ability of phosphorylated c-Jun to activate the cyclin D1 promoter. These results indicate that Pin1 cooperates with Ha-Ras or activated JNK to increase the activity of c-Jun toward the cyclin D1 promoter.
Pin1 contains a WW domain and a PPIase domain, which bind and isomerize specific pSer/Thr-Pro motifs, respectively, and both these activities are normally required for Pin1 to modulate the function of its phosphoprotein substrates, such as Cdc25C and tau (Ranganathan et al., 1997; Yaffe et al., 1997; Shen et al., 1998; Lu et al., 1999a,b). To examine whether only one, or both, of these activities is required for Pin1 to modulate the activity of c-Jun we used Pin1 mutants, Pin1R68,69A, Pin1W34A and Pin1S16E, which contain mutations at the key residues either in the PPIase domain (R68, R69) or the WW domain (W34 or S16), and fail to isomerize pSer/Thr-Pro bonds or to bind phosphoproteins (including c-Jun; data not shown), respectively (Shen et al., 1998; Lu et al., 1999b; Zhou et al., 2000). In contrast to wild-type protein, these Pin1 mutants neither increased the transcriptional activity of c-Jun towards the cyclin D1 promoter nor cooperated with Ha-Ras to activate c-Jun (Figure 6A and B). Neither did the mutants affect the levels of c-Jun phosphorylation (data not shown). These results indicate that both phosphoprotein-binding and phosphorylation-specific isomerase activities of Pin1 are required for its ability to modulate the activity of c-Jun.
Fig. 6. The effects on the transcriptional activity of c-Jun depend on the phosphoprotein-binding and PPIase activity of Pin1 as well as phosphorylation of c-Jun on Ser63/73. (A) Abolishing the Pin1 effect by inactivating its PPIase activity. HeLa cells were co-transfected with vectors, c-Jun, or c-Jun + Ha-Ras, and Pin1 or its PPIase-negative mutant Pin1R68,69A, and then subjected to luciferase assay. Pin1R68,69A fails to isomerize phosphorylated Ser/Thr-Pro bonds (Yaffe et al., 1997). (B) Abolishing the Pin1 effect by inactivating its phosphoprotein-binding activity. HeLa cells were co-transfected with vectors, c-Jun, or c-Jun + Ha-Ras and green fluorescent protein (GFP)–Pin1 or its WW domain point mutants, and then subjected to luciferase assay. GFP–Pin1W34A and GFP–Pin1S16E did not bind phosphoproteins, as shown (Lu et al., 1999b). Note that GFP fusion proteins were used because the WW domain Pin1 mutants were not stable in cells, but they were stable as GFP fusion proteins, although expressed at reduced levels (data not shown). Although the absolute maximal luciferase activity was not as high as in other experiments, which is likely to be due to lower levels of GFP fusion proteins being expressed, the overall trends were the same. (C) Inhibiting the ability of Pin1 to increase the c-Jun activity by DN-Ras. Cells were co-transfected with c-Jun or c-Jun + Pin1, and increasing amounts of DN-Ras, and then subjected to the luciferase assay. (D and E) Abolishing the cooperative effect between Pin1 and Ha-Ras or activated JNK by mutating c-Jun phosphorylation sites Ser63/73. HeLa cells were co-transfected with various amounts of Pin1, c-Jun or c-JunS63/73A construct, and Ha-Ras (D) or activated JNK (E) and then subjected to the luciferase assay.
The above results suggest that Pin1 may increase the activity of c-Jun by binding and isomerizing its pSer/Thr-Pro motifs, as it does to Cdc25C and tau (Shen et al., 1998; Lu et al., 1999a; Zhou et al., 2000). In this case, down-regulation of the Ras-dependent phosphorylation of c-Jun should reduce the effect of Pin1 on c-Jun, and mutations of the c-Jun phosphorylation sites that Pin1 binds to should abolish the Pin1 effect. To examine the first assumption, we co-transfected cells with Pin1, c-Jun and DN-Ras to examine the effect of DN-Ras on the ability of Pin1 to activate the cyclin D1 promoter. DN-Ras reduced both phosphorylation of c-Jun on Ser63/73 and the ability of Pin1 to bind c-Jun (Figure 4A–D). Indeed, DN-Ras not only inhibited the ability of c-Jun to activate the cyclin D1 promoter, as shown previously (Albanese et al., 1995), but also inhibited the ability of Pin1 to enhance the activity of c-Jun 5- to 7-fold (Figure 6C). These results suggest that the Ras-dependent phosphorylation of c-Jun is important for the Pin1 function on c-Jun. To examine the second assumption, we used the mutant c-JunS63/73A, which failed to bind Pin1 (Figure 4E and F). Pin1 almost completely failed to cooperate either with activated JNK or oncogenic Ha-Ras to increase the ability of c-JunS63/73A to induce the cyclin D1 promoter (Figure 6D and E), indicating that phosphorylation of c-Jun on Ser63/73 is essential for Pin1 to induce the cyclin D1 promoter. These results indicate that phosphorylation of c-Jun on Ser63/73, induced by the Ras-dependent signaling pathway, is essential for Pin1 to increase transcription of the cyclin D1 promoter. Thus, Pin1 binds phosphorylated c-Jun and potentiates its transcriptional activity towards cyclin D1 in response to activation of Ras or JNK.
Discussion
Previous studies have demonstrated that depletion of Pin1 induces apoptosis and is also observed in neuronal cell death in Alzheimer’s disease (Lu et al., 1996, 1999a). We show here the striking overexpression of Pin1 in a large fraction of breast cancers. Furthermore, Pin1 levels correlate significantly with the grade of the breast tumors according, to Bloom and Richardson’s classification system, although the relationship between Pin1 levels and the prognosis of cancer patients remains to be determined. Consistent with our findings is the observation that Pin1 is one of the genes that are most drastically suppressed by up-regulation of Brca1, as detected in cDNA array screening and northern analysis (MacLachlan et al., 2000). In addition, the level of Pin1 in breast cancer cell lines is much higher than that in either normal or non-transformed mammary epithelial cells. Although further studies are needed to elucidate the mechanisms leading to overexpression of Pin1, these results demonstrate for the first time that Pin1 is up-regulated markedly in many human tumor samples.
The significance of Pin1 overexpression in cancer is further substantiated by our findings that Pin1 cooperates with activated JNK or Ha-Ras in increasing the transcriptional activity of phosphorylated c-Jun to activate the cyclin D1 promoter. Overexpression of cyclin D1 is found in 50% of patients with breast cancer (Bartkova et al., 1994; Gillett et al., 1994). Furthermore, overexpression of cyclin D1 contributes to cell transformation (Hinds et al., 1994), whereas inhibition of cyclin D1 expression by antisense expression causes growth arrest of tumor cells (Schrump et al., 1996; Arber et al., 1997; Driscoll et al., 1997; Kornmann et al., 1998). Disruption of the cyclin D1 gene in mice blocks the proliferation of breast epithelial cells and reduces tumor development in response to Ha-Ras (Fantl et al., 1995; Sicinski et al., 1995; Robles et al., 1998). These results indicate that cyclin D1 plays an important role during oncogenesis, especially during Ras-mediated tumorigenesis (Rodriguez-Puebla et al., 1999). Oncogenic Ras induces the cyclin D1 promoter via its AP-1 site (Albanese et al., 1995). Although the AP-1 complex is composed of the c-Jun and c-Fos proteins, c-Jun is the most potent transactivator in the complex (Angel et al., 1989; Chiu et al., 1989; Abate et al., 1991) and is elevated in Ha-Ras-transformed cells, in which c-Fos is down-regulated (Thomson et al., 1990; Binetruy et al., 1991). In addition to the regulation of protein levels, the activity of c-Jun is enhanced by phosphorylation induced by growth factors, oncogenic proteins, or stress conditions. Although different pathways may be involved, they eventually lead to activation of JNKs, which phosphorylate c-Jun on two critical N-terminal Ser-Pro motifs (S63/73–P) and enhance its transcriptional activity (Binetruy et al., 1991; Smeal et al., 1991; Hunter and Karin, 1992; Derijard et al., 1994; Hinds et al., 1994; Albanese et al., 1995, 1999; Fantl et al., 1995; Sicinski et al., 1995; Whitmarsh and Davis, 1996; Karin et al., 1997; Robles et al., 1998; Bakiri et al., 2000). Thus, phosphorylation of c-Jun on Ser63/73-Pro is a key regulatory mechanism that converts inputs from various signaling pathways into changes in gene expression. However, it has not been described previously whether the activity of phosphorylated c-Jun is further regulated after phosphorylation.
We have found that Pin1 not only binds phosphorylated c-Jun, but also dramatically increases its ability to activate the cyclin D1 promoter in cooperation either with activated JNK or oncogenic Ha-Ras. In contrast, inhibition of endogenous Pin1 reduces the transcriptional activity of phosphorylated c-Jun, indicating that endogenous Pin1 is also required for the optimal activation of c-Jun. The significance of this Pin1-dependent regulation is further substantiated by our findings that up-regulation of Pin1 not only correlates with cyclin D1 overexpression in breast cancer tissues, but also induces cyclin D1 expression in breast cancer cell lines. Thus, Pin1 is a potent modulator of phosphorylated c-Jun in inducing cyclin D1 expression, presumably by regulating the conformation of the phosphorylated Ser-Pro motifs in c-Jun (Figure 7). The importance of Pin1 in the regulation of cyclin D1 expression has been further supported by our recent identification of cyclin D1 as one of the Pin1-induced genes in breast cancer cells in the differential display screen (Ryo et al., 2001), and by our phenotypic analysis of Pin1-deficient mice (Y.-C.Liou, A.Ryo, H.K.Huang, P.J.Lu, F.Fujimori, T.Uchida, R.Bronson, T.Hunter and K.P.Lu, submitted). Although Pin1–/– mice have previously been shown to develop normally (Fujimori et al., 1999), we have uncovered that they display a range of cell proliferative abnormalities, including decreased body size, retinal degeneration and neurological abnormalities. Moreover, in Pin1-deficient adult females, the breast epithelial compartment failed to undergo the massive proliferative changes caused by pregnancy (Y.-C.Liou, A.Ryo, H.K.Huang, P.J.Lu, F.Fujimori, T.Uchida, R.Bronson, T.Hunter and K.P.Lu, submitted). Significantly, many features of these Pin1-deficient mice, such as retinal degeneration and mammary gland impairment, are also characteristic of cyclin D1-deficient mice (Fantl et al., 1995; Sicinski et al., 1995). Moreover, cyclin D1 levels were significantly reduced in Pin1-deficient retina and breast epithelial cells from pregnant mice (Liou et al., submitted). These results provide the genetic evidence for an essential role of Pin1 in maintaining cell proliferation and cyclin D1 expression, and further support a role of Pin1 in oncogenesis. Abnormal activation of the Ras-dependent signaling pathway and cyclin D1 overexpression are a common and critical mechanism during the development of many malignancies, such as breast, skin and colon cancer (Fantl et al., 1995; Sicinski et al., 1995; Robles et al., 1998; Rodriguez-Puebla et al., 1999). Indeed, Pin1 is significantly overexpressed in many of these human tumors (G.M.Wulf and K.P.Lu, unpublished data), suggesting that it plays a positive role for cell proliferation during oncogenesis (Figure 7).
Fig. 7. Role of Pin1 in regulating the transcriptional activity of phosphorylated c-Jun towards the cyclin D1 promoter. Oncogenic Ha-Ras activates JNKs, which phosphorylate c-Jun on two critical amino terminal Ser-Pro motifs, enhancing its transcriptional activity. Pin1 is up-regulated in breast cancer and functions as a potent regulator of phosphorylated c-Jun to induce cyclin D1 expression, presumably by altering the conformation of the phosphorylated Ser-Pro motifs (insert). Double arrows, up-regulation; the asterisk indicated the activated form of proteins.
In summary, our results show that Pin1 is strikingly overexpressed in human breast cancer tissues, and cooperates with activated Ras signaling in increasing c-Jun transcriptional activity towards the cyclin D1 gene. Given the well established role of activated Ras signaling and cyclin D1 overexpression during oncogenesis, our study suggests that overexpression of Pin1 may promote tumor growth. In addition, since inhibition of the Pin1 enzymatic activity triggers tumor cells to enter apoptosis, overexpressed Pin1 may act as a novel anti-cancer target.
Materials and methods
Analysis of protein and mRNA levels in patient samples
Fifty-one cancerous and 10 normal breast tissue specimens were randomly selected. The malignancy of infiltrating carcinomas was scored according to Bloom and Richardson’s classification system (Bloom and Richardson, 1957). Tissue from the core of the tumor was snap frozen in liquid nitrogen and pulverized using a Microdismembrator (Braun). About 10 µg of the pulverized tissues were resuspended in 100 µl of SDS sample buffer. Immunoblotting with anti-Pin1, anti-cyclin D1, anti-Her2/neu and anti-actin antibodies was performed as described (Shen et al., 1998; Lu et al., 1999a), as was immunohistochemistry using anti-Pin1 polyclonal or monoclonal antibodies (Lu et al., 1999a). Levels of Pin1 and actin were semi-quantified using Imagequant, as described (Lu et al., 1999a). mRNA was isolated using the Trizol reagent (Gibco) and cDNA was synthesized using Superscript (Gibco). Twenty-five nanograms of cDNA were used per real-time PCR run with primers specific for cyclin D1, and GAPDH as an internal control. All real-time PCR runs were performed in duplicate and analyzed according to the manufacturer’s instructions (Applied Biosystems). The significance of the differences in Pin1 levels between clinical and pathological categories was analyzed using the Kruskal–Wallis test (Glantz, 1997). The Pearson correlation coefficients were obtained using the SAS software (Release 6.12; SAS Institute Inc., Cary, NC).
Determination of Pin1 levels and the effects of Pin1 on cyclin D1 expression in cell lines
The levels of Pin1 in normal (76N), spontaneously immortalized but not transformed (184B5 and MCF10), and transformed (MCF7, T47D, MDAMB435 and HCC1937) mammary epithelial cell lines were determined by subjecting total cellular proteins to immunoblotting analysis with anti-Pin1 polyclonal antibodies. To examine the nature of the double band, a tumor lysate was incubated at 30°C for 60 min in the presence of 100 nM okadaic acid (Sigma), PP1 and PP2A (Upstate Biotechnology) or CIP. To examine the effects of Pin1 on cyclin D1 expression, Pin1 cDNA was subcloned into pcDNA3 vector (Invitrogen) and transfected into MCF7, T47D or HeLa cells for 36 h, followed by determining the level of Pin1 and cyclin D1 by immunoblotting analysis with anti-Pin1 and anti-cyclin D1 antibodies, respectively, as described (Lu et al., 1996; Shen et al., 1998), and cyclin D1 mRNA by real-time PCR, as described above.
Determination of the Pin1-c–Jun interaction
To examine the interaction between Pin1 and phosphorylated c-Jun, HeLa cells were co-transfected with c-Jun or c-JunS63/73A and the oncogenic Ha-Ras, consititutively active JNK, DN-Ras or the control vector for 24 h. The cells were lysed in a lysis buffer containing 1% Triton X-100, and the supernatants incubated with 10 µl of agarose beads containing various GST–Pin1 proteins or control GST for 2 h at 4°C. The precipitated proteins were washed five times in the buffer containing 1% Triton X-100 before being subjected to immunoblotting analysis using antibodies against c-Jun or c-Jun phosphorylated on Ser63/73 (New England Biolabs), as described (Yaffe et al., 1997; Shen et al., 1998; Lu et al., 1999a,b). For co-immunoprecipitation, we used anti-c-Jun polyclonal antibodies (Santa Cruz) and unrelated polyclonal antibodies (Pericentrin antibodies) as a control. The pre-cleared lysates were incubated for 2 h with the respective antibodies, and the immune complexes were collected with protein A beads (Sigma) and subjected to immunoblotting with anti-Pin1 or anti-c-Jun antibodies. The ability of the Pin1 WW domain and PPIase domain mutants to bind phosphoproteins (MPM-2 or c-Jun) and to isomerize pSer/Thr-Pro bonds were determined, as described (Yaffe et al., 1997; Lu et al., 1999a,b).
Promoter reporter assays
Various cyclin D1–luciferase reporter constructs, c-Jun and Ras constructs were gifts from R.Pestell (Albert Einstein College of Medicine), M.Karin (University of California at San Diego) and L.Feig (Tufts University), respectively, and have been confirmed by DNA sequencing. Luciferase reporter constructs for TK, c-fms and MMTV were purchased. Superfect (Qiagen) was used for transfections. Reporter gene assays were performed with the Dual-luciferase reporter assay system (Promega) at 24–36 h after transfection. One nanogram of pRL-TK (Promega) Renilla luciferase was co-transfected in each sample as an internal control for transfection efficiency. Expression of all transfected genes was confirmed by immunoblotting analysis with the respective antibodies. The amounts of DNA used in transfection were carefully titrated for each construct; typically, only ∼50 ng of each DNA were used, with exceptions indicated in the text. The activity of the reporter luciferase was expressed relative to the activity in control vector-transfected cells, which was defined as 1. Similar results were obtained in at least three different experiments. All results are expressed as x̄ ± SD of independent duplicate cultures. Since Pin1AS induces mitotic arrest and apoptosis at 48–72 h after transfection (Lu et al., 1996), all experiments with Pin1AS were performed before 36 h, when no significant apoptotic cells were observed, as described previously (Lu et al., 1996).
Acknowledgments
Acknowledgements
We are grateful to L.Schnipper for his support and advice, R.Davis, B.Neel, L.Cantley, S.Korsmeyer, D.Medina, T.Hunter, X.D.Fu and S.Sands for constructive discussions, to M.Hueffner for supplying the tumor samples, to M.Karin, R.Pestell, L.Feig, J.Blenis, A.Toker and D.Tanien for reagents, and to the members of the Lu laboratory, especially X.Zhou and P.Lu, for their important contributions. G.M.W. and A.R. are fellows of the DOD Breast Cancer Research Program and Japan Society for the Promotion of Science, respectively. K.P.L. is a Pew Scholar and a Leukemia and Lymphoma Society Scholar. This study was supported by NIH grants R01GM56230 and GM58556 to K.P.L.
References
- Abate C., Luk,D. and Curran,T. (1991) Transcriptional regulation by Fos and Jun in vitro: interaction among multiple activator and regulatory domains. Mol. Cell. Biol., 11, 3624–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese C. et al. (1999) Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem., 274, 34186–34195. [DOI] [PubMed] [Google Scholar]
- Albanese C., Johnson,J., Watanabe,G., Eklund,N., Vu,D., Arnold,A. and Pestell,R.G. (1995) Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem., 270, 23589–23597. [DOI] [PubMed] [Google Scholar]
- Angel P., Smeal,T., Meek,J. and Karin,M. (1989) Jun and v-jun contain multiple regions that participate in transcriptional activation in an interdependent manner. New Biol., 1, 35–43. [PubMed] [Google Scholar]
- Arber N. et al. (1997) Antisense to cyclin D1 inhibits the growth and tumorigenicity of human colon cancer cells. Cancer Res., 57, 1569–1574. [PubMed] [Google Scholar]
- Bakiri L., Lallemand,D., Bossy-Wetzel,E. and Yaniv,M. (2000) Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J., 19, 2056–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., Lukas,J., Muller,H., Lutzhoft,D., Strauss,M. and Bartek,J. (1994) Cyclin D1 protein expression and function in human breast cancer. Int. J. Cancer, 57, 353–361. [DOI] [PubMed] [Google Scholar]
- Binetruy B., Smeal,T. and Karin,M. (1991) Ha-Ras augments c-Jun activity and stimulates phosphorylation of its activation domain. Nature, 351, 122–127. [DOI] [PubMed] [Google Scholar]
- Bloom H.J.G. and Richardson,W.W. (1957) Histological grading and prognosis in breast cancer: a study of 1049 cases, of which 359 have been followed for 15 years. Br. J. Cancer, 11, 359–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, S.H., Greenleaf,A.L. and Price,D.H. (2001) Juglone, an inhibitor of the peptidyl-prolyl isomerase Pin1, also directly blocks transcription. Nucleic Acids Res., 29, 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R., Angel,P. and Karin,M. (1989) Jun-B differs in its biological properties from, and is a negative regulator of, c-Jun. Cell, 59, 979–986. [DOI] [PubMed] [Google Scholar]
- Crenshaw D.G., Yang,J., Means,A.R. and Kornbluth,S. (1998) The mitotic peptidyl-prolyl isomerase, Pin1, interacts with Cdc25 and Plx1. EMBO J., 17, 1315–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B., Hibi,M., Wu,I.H., Barrett,T., Su,B., Deng,T., Karin,M. and Davis,R.J. (1994) JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell, 76, 1025–1037. [DOI] [PubMed] [Google Scholar]
- Driscoll B., Wu,L., Buckley,S., Hall,F.L. Anderson,K.D. and Warburton,D. (1997) Cyclin D1 antisense RNA destabilizes pRb and retards lung cancer cell growth. Am. J. Physiol., 273, L941–949. [DOI] [PubMed] [Google Scholar]
- Fantl V., Smith,R., Brookes,S., Dickson,C. and Peters,G. (1993) Chromosome 11q13 abnormalities in human breast cancer. Cancer Surv., 18, 77–94. [PubMed] [Google Scholar]
- Fantl V., Stamp,G., Andrews,A., Rosewell,I. and Dickson,C. (1995) Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev., 9, 2364–2372. [DOI] [PubMed] [Google Scholar]
- Fischer G. (1994) Peptidyl-prolyl cis/trans isomerases. Angew. Chem. Int. Ed. Engl., 33, 1415–1436. [Google Scholar]
- Fujimori F., Takahashi,K., Uchida,C. and Uchida,T. (1999) Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G0 arrest. Biochem. Biophys. Res. Commun., 265, 658–663. [DOI] [PubMed] [Google Scholar]
- Galat A. and Metcalfe,S.M. (1995) Peptidylproline cis/trans isomerases. Prog. Biophys. Mol. Biol., 63, 67–118. [DOI] [PubMed] [Google Scholar]
- Gillett C., Fantl,V., Smith,R., Fisher,C., Bartek,J., Dickson,C., Barnes,D. and Peters,G. (1994) Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res., 54, 1812–1817. [PubMed] [Google Scholar]
- Glantz S.A. (1997) Primer of Biostatistics. McGraw Hill Health Professions Division, New York, NY.
- Hanes S.D., Shank,P.R. and Bostian,K.A. (1989) Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast, 5, 55–72. [DOI] [PubMed] [Google Scholar]
- Hani J., Schelbert,B., Bernhardt,A., Domdey,H., Fischer,G., Wiebauer,K. and Rahfeld,J.U. (1999) Mutations in a peptidylprolyl-cis/trans-isomerase gene lead to a defect in 3′-end formation of a pre-mRNA in Saccharomyces cerevisiae. J. Biol. Chem., 274, 108–116. [DOI] [PubMed] [Google Scholar]
- Hani J., Stumpf,G. and Domdey,H. (1995) PTF1 encodes an essential protein in Saccharomyces cerevisiae, which shows strong homology with a new putative family of PPIases. FEBS Lett., 365, 198–202. [DOI] [PubMed] [Google Scholar]
- Hinds P.W., Dowdy,S.F., Eaton,E.N., Arnold,A. and Weinberg,R.A. (1994) Function of a human cyclin gene as an oncogene. Proc. Natl Acad. Sci. USA, 91, 709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (1998) Prolyl isomerase and nuclear function. Cell, 92, 141–143. [DOI] [PubMed] [Google Scholar]
- Hunter T. and Karin,M. (1992) The regulation of transcription by phosphorylation. Cell, 70, 375–387. [DOI] [PubMed] [Google Scholar]
- Hunter T. and Pines,J. (1994) Cyclins and cancer II: cyclin D and CDK inhibitors come of age. Cell, 79, 573–582. [DOI] [PubMed] [Google Scholar]
- Karin M., Liu,Z. and Zandi,E. (1997) AP-1 function and regulation. Curr. Opin. Cell Biol., 9, 240–246. [DOI] [PubMed] [Google Scholar]
- Kornmann M., Arber,N. and Korc,M. (1998) Inhibition of basal and mitogen-stimulated pancreatic cancer cell growth by cyclin D1 antisense is associated with loss of tumorigenicity and potentiation of cytotoxicity to cisplatinum. J. Clin. Invest., 101, 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Xia,W., Wang,J.C., Kwong,K.Y., Spohn,B., Wen,Y., Pestell,R.G. and Hung,M.C. (2000) β-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc. Natl Acad. Sci. USA, 97, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.P., Hanes,S.D. and Hunter,T. (1996) A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature, 380, 544–547. [DOI] [PubMed] [Google Scholar]
- Lu P.J., Wulf,G., Zhou,X.Z., Davies,P. and Lu,K.P. (1999a) The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature, 399, 784–788. [DOI] [PubMed] [Google Scholar]
- Lu P.J., Zhou,X.Z., Shen,M. and Lu,K.P. (1999b) A function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science, 283, 1325–1328. [DOI] [PubMed] [Google Scholar]
- MacLachlan T.K., Somasundaram,K., Sgagias,M., Shifman,Y., Muschel,R.J., Cowan,K.H. and El-Deiry,W.S. (2000) BRCA1 effects on the cell cycle and the DNA damage response are linked to altered gene expression. J. Biol. Chem., 275, 2777–2785. [DOI] [PubMed] [Google Scholar]
- Motokura T. and Arnold,A. (1993) PRAD1/cyclin D1 proto-oncogene: genomic organization, 5′ DNA sequence and sequence of a tumor-specific rearrangement breakpoint. Genes Chromosomes Cancer, 7, 89–95. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. (1995) Cyclin-dependent protein kinases: key regulators of the eukaryote cell cycle. BioEssays, 17, 471–480. [DOI] [PubMed] [Google Scholar]
- Nurse P. (1994) Ordering S phase and M phase. Cell, 79, 547–550. [DOI] [PubMed] [Google Scholar]
- Ranganathan R., Lu,K.P., Hunter,T. and Noel,J.P. (1997) Structural and functional analysis of the mitotic peptidyl-prolyl isomerase Pin1 suggests that substrate recognition is phosphorylation dependent. Cell, 89, 875–886. [DOI] [PubMed] [Google Scholar]
- Rippmann J.F. et al. (2000) Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell Growth Differ., 11, 409–416. [PubMed] [Google Scholar]
- Robles A.I. et al. (1998) Reduced skin tumor development in cyclin D1-deficient mice highlights the oncogenic ras pathway in vivo. Genes Dev., 12, 2469–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Puebla M.L., Robles,A.I. and Conti,C.J. (1999) ras activity and cyclin D1 expression: an essential mechanism of mouse skin tumor development. Mol. Carcinog., 24, 1–6. [PubMed] [Google Scholar]
- Ryo A., Nakamura,M., Wulf,G., Liou,Y.-C. and Lu,K.P. (2001) Prolyl isomerase Pin1 regulates turnover and subcellular localization of β-catenin by inhibiting its interacition with APC. Nature Cell Biol., 3, in press. [DOI] [PubMed] [Google Scholar]
- Schmid F.X. (1995) Prolyl isomerases join the fold. Curr. Biol., 5, 993–994. [DOI] [PubMed] [Google Scholar]
- Schrump D.S., Chen,A. and Consoli,U. (1996) Inhibition of lung cancer proliferation by antisense cyclin D. Cancer Gene Ther., 3, 131–135. [PubMed] [Google Scholar]
- Schutkowski M., Bernhardt,A., Zhou,X.Z., Shen,M., Reimer,U., Rahfeld,J.U., Lu,K.P. and Fischer,G. (1998) Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry, 37, 5566–5575. [DOI] [PubMed] [Google Scholar]
- Shen M., Stukenberg,P.T., Kirschner,M.W. and Lu,K.P. (1998) The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev., 12, 706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicinski P. et al. (1995) Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell, 82, 621–630. [DOI] [PubMed] [Google Scholar]
- Smeal T., Binetruy,B., Mercola,D.A., Birrer,M. and Karin,M. (1991) Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature, 354, 494–496. [DOI] [PubMed] [Google Scholar]
- Tetsu O. and McCormick,F. (1999) β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature, 398, 422–426. [DOI] [PubMed] [Google Scholar]
- Thomson T.M., Green,S.H., Trotta,R.J., Burstein,D.E. and Pellicer,A. (1990) Oncogene N-ras mediates selective inhibition of c-fos induction by nerve growth factor and basic fibroblast growth factor in a PC12 cell line. Mol. Cell. Biol., 10, 1556–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. (1996) Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol., 8, 205–215. [DOI] [PubMed] [Google Scholar]
- Ui M., Sonobe,M.H., Ito,T., Murakami,M., Okazaki,S., Takada,M., Sato,T. and Iba,H. (1998) Biochemical and functional analysis of highly phosphorylated forms of c-Jun protein. FEBS Lett., 429, 289–294 [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J. and Davis,R.J. (1996) Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med., 74, 589–607. [DOI] [PubMed] [Google Scholar]
- Winkler K.E., Swenson,K.I., Kornbluth,S. and Means,A.R. (2000) Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science, 287, 1644–1647. [DOI] [PubMed] [Google Scholar]
- Wu X., Wilcox,C.B., Devasahayam,G., Hackett,R.L., Arevalo-Rodriguez,M., Cardenas,M.E., Heitman,J. and Hanes,S.D. (2000) The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J., 19, 3727–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M.B. et al. (1997) Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science, 278, 1957–1960. [DOI] [PubMed] [Google Scholar]
- Zhou X.Z., Lu,P.J., Wulf,G. and Lu,K.P. (1999) Phosphorylation-dependent prolyl isomerization: a novel signaling regulatory mechanism. Cell. Mol. Life Sci., 56, 788–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.Z. et al. (2000) Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol. Cell, 6, 873–883. [DOI] [PubMed] [Google Scholar]