Abstract
The yeast exosome is a complex of at least 10 essential 3′–5′ riboexonucleases which is involved in 3′-processing of many RNA species. An exosome-like complex has been found or predicted to exist in other eukaryotes but not in Escherichia coli. The unicellular parasite Trypanosoma brucei diverged very early in eukaryotic evolution. We show here that T.brucei contains at least eight exosome subunit homologs, but only a subset of these associate in a complex. Accordingly, the T.brucei exosome is smaller than that of yeast. Both free and complex-associated homologs are essential for cell viability and are involved in 5.8S rRNA maturation. We suggest that the exosome was present in primitive eukaryotes, and became increasingly complex during subsequent evolution.
Keywords: exoribonucleases/exosome/rRNA processing/Trypanosoma brucei
Introduction
The African trypanosome Trypanosoma brucei causes sleeping sickness in humans and nagana disease in cattle in sub-Saharan Africa. These parasites are transmitted from one mammal to the next by tsetse flies and undergo drastic morphological and biochemical changes in response to the different environments within the insect (procyclic stage) and mammalian (bloodstream stage) hosts (Vickerman, 1985). Trypanosomatid RNA metabolism diverges in several respects from normal eukaryotic paradigms. The two most dramatic examples occur in the editing of mitochondrial RNAs (Estévez and Simpson, 1999), and the processing of cytosolic mRNA. Mature mRNA molecules are generated from long polycistronic precursors via coupled trans splicing and polyadenylation (Ullu et al., 1996) instead of being individually transcribed. The inability to control transcription of individual mRNAs means that regulation of gene expression, which is essential for survival in different environments, has to be exerted almost exclusively at the post-transcriptional level through control of mRNA degradation and translation (Hotz et al., 1997; Di Noia et al., 2000).
Ribosomal RNA processing in T.brucei is also unusual; the large subunit rRNA is cleaved at multiple sites to yield seven stable RNA fragments, two internal 5′ external transcribed spacer region (5′-ETS) cleavages occur and a single 5.8S rRNA species is produced (White et al., 1986; Campbell et al., 1987; Hartshorne and Toyofuku, 1999). Until now, nothing was known about the enzymes involved in either mRNA degradation or rRNA processing in trypanosomes.
The turnover and processing of mRNA and rRNA molecules in prokaryotes and eukaryotes involves 3′–5′ exonucleolytic digestion events. In yeast, a 300–400 kDa complex, the exosome, is responsible for many of these reactions (van Hoof and Parker, 1999; Mitchell and Tollervey, 2000), and is present in both the cytosol and the nucleus. The cytoplasmic complex contains at least 10 different components, Rrp4p, Rrp40p–Rrp46p, Csl4p and Mtr3p, while the nuclear complex has an additional subunit, Rrp6p. All these proteins show 3′–5′ exonucleolytic activity or are predicted to be 3′–5′ exonucleases (Allmang et al., 1999b). Six of them are related to Escherichia coli RNase PH (Rrp41p, Rrp42p, Rrp43p, Rrp45p, Rrp46p and Mtr3p) and three contain an S1 RNA binding domain (Rrp4p, Rrp40p and Csl4p). Rrp44p contains an RNase II-family signature, and Rrp6p is related to RNase D (Allmang et al., 1999b). All these components are essential for cell viability, with the exception of Rrp6p where depletion confers a temperature-lethal phenotype (Allmang et al., 1999b). Human cells also contain cytosolic and nuclear exosome-like complexes, with sizes ranging between 250 and 700 kDa (Mitchell et al., 1997; Brouwer et al., 2001). In Arabidopsis thaliana the Rrp41p homolog AtRrp41p resides in a ∼500 kDa complex (Chekanova et al., 2000). Despite the conserved features and components, the exosomes of different eukaryotes are clearly not identical. Analysis of the Caenorhabditis elegans genome indicates that only three proteins are related to RNase PH, and there is no clear homolog for Rrp4p (van Hoof and Parker, 1999). The six human RNase PH-like genes do not show orthologous pairs with the six yeast ones, and there are no clear homologs for Rrp43p or Mtr3p (Mitchell et al., 1997). Analysis of genome organization in the Archaea suggests that there might be an exosome in these organisms (Koonin et al., 2001), but no exosome-like complex has been detected in E.coli (Deutscher, 1993).
In this paper we present evidence for the existence of a simple exosome complex in T.brucei which is involved in the maturation of 5.8S rRNA.
Results
Identification of homologous proteins of the yeast exosome components in T.brucei
A search for exosome components in the unfinished T.brucei genome sequencing project revealed that there are homologs for the yeast proteins Csl4p, Rrp4p, Rrp6p, Rrp40p, Rrp44p and Rrp45p and two homologs for Rrp41p (Table I). Since in most of the cases the sequences were not complete, we cloned genomic fragments containing the T.brucei genes and then sequenced them to identify the full-length open reading frames (ORFs) (trypanosome genes almost never contain introns). The homologs were defined by several criteria. First, they were the best hits in the trypanosome databases. Secondly, when the complete sequence was re-scanned against DDBJ/EMBL/GenBank and in individual genome databases, the putative homologs again showed the maximal identity and E scores. Thirdly, the typical motifs found in exosome components or putative exosome components in other organisms were identified (Allmang et al., 1999b; van Hoof and Parker, 1999). In the case of TbCSL4, the BLAST scores were far too low to allow definitive identification and we only recognized this protein after we had found it in the purified T.brucei exosome (see below). The low scores for TbCSL4 are due mainly to the fact that the N-terminal domain is not conserved in Csl4p homologs although the functional motifs are present (van Hoof et al., 2000). We found no other RNase PH-like proteins that could be the trypanosome homologs for Rrp42p, Rrp43p, Rrp46p or Mtr3p. It is possible, however, that they are present; the T.brucei genome sequence is not yet complete and partial sequences with low conservation (like TbCSL4) could be unidentifiable.
Table I. Homologs of the yeast exosome components found in the T.brucei partially sequenced genome.
| T.brucei protein (kDa) | DDBJ/EMBL/GenBank accession No. | S.cerevisiae homolog | C.elegans homolog | Motif |
|---|---|---|---|---|
| TbRRP4 (32.7) | AJ308995 | Rrp4p, 39% | none | S1 RNA binding |
| TbRRP6 (78.6) | AJ309000 | Rrp6p, 35% | C14A4.4, 32% | RNase D |
| TbRRP40 (32.0) | AJ308996 | Rrp40p, 33% | F59C6.4, 28% | S1 RNA binding |
| TbRRP41A (27.6) | AJ308997 | Rrp41p, 27% | B0564.1, 28% | RNase PH |
| TbRRP41B (28.0) | AJ309001 | Rrp41p, 25% | C14A4.5, 28% | RNase PH |
| TbRRP44 (109.0) | AJ308998 | Rrp44p, 41% | C04G2.6, 40% | RNase II |
| TbRRP45 (38.9) | AJ308999 | Rrp45p, 30% | F37C12.13, 27% | RNase PH |
| TbCSL4 (32.6) | AJ308994 | Csl4p, 28% | K06A9.1B, 26% | S1 RNA binding |
The T.brucei protein sequences indicated were used to search the S.cerevisiae and C.elegans protein databases using the BLASTP program. The percentage of identity is presented in each case. The molecular masses of the predicted T.brucei peptides are also shown in parentheses.
TbRRP4 and TbRRP45 are present in an 11S complex
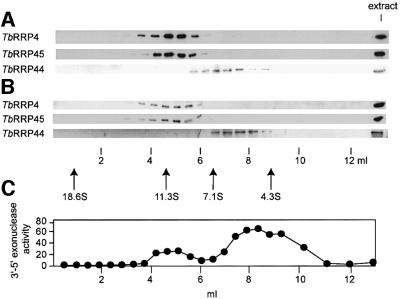
To analyze whether there is an exosome-like complex in T.brucei, polyclonal antibodies were raised in rabbits against TbRRP4, TbRRP44 and TbRRP45. Subcellular fractionation experiments showed that TbRRP4, TbRRP44 and TbRRP45 were in both the cytosol and the nucleus. They were present at similar levels in both bloodstream and procyclic cells so were not developmentally regulated (data not shown). We fractionated cytosolic and nuclear extracts from procyclic T.brucei by glycerol density gradient centrifugation, and individual fractions were tested by immunoblotting analysis and also for 3′–5′ riboexonuclease activity (Figure 1). TbRRP4 and TbRRP45 co-sedimented in a complex with an estimated sedimentation coefficient of ∼11S (240 kDa). In contrast, TbRRP44 sedimented at ∼6S (110 kDa), the expected size of a monomeric form of the protein. No free TbRRP4 or TbRRP45 species was detected. When we measured 3′–5′ riboexonuclease activity in the cytosolic fractions, we observed two peaks, the less active of which sedimented with the complex at ∼11S. The sedimentation behavior of TbRRP4, TbRRP44 and TbRRP45 after fractionation of cytosolic and nuclear extracts from bloodstream trypanosomes resembled that of procyclic cells (data not shown). These results suggest that there is an exosome-like complex in both the cytosol and the nucleus of T.brucei. The T.brucei exosome appeared to be considerably smaller than the yeast exosome, indicating that it might contain fewer components.
Fig. 1. TbRRP4 and TbRRP45, but not TbRRP44, are found in a complex. (A) Cytosolic and (B) nuclear extracts from procyclic T.brucei were fractionated through 10–30% glycerol gradients. Aliquots of each fraction and original extracts were subjected to SDS–PAGE and immunoblotting analysis using antibodies against TbRRP4, TbRRP45 or TbRRP44 (A and B) or assayed for 3′–5′ exonuclease activity (C). The exonuclease activity is expressed as the percentage of substrate digested (see Materials and methods). The sedimentation coefficients of marker proteins processed in parallel are indicated.
TbRRP4 is a processive, 3′–5′ riboexonuclease that does not complement the yeast rrp4-1 allele
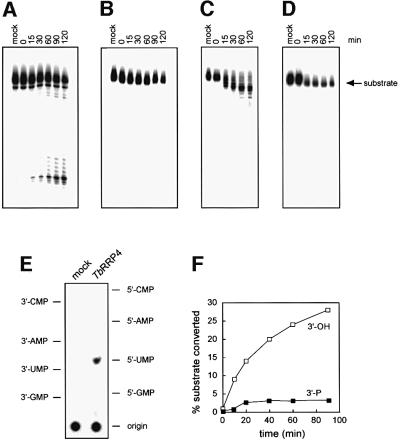
A His6-TbRRP4 fusion protein was expressed in E.coli and purified to homogeneity, in order to test whether it had the exonucleolytic activity expected of a component of the exosome. Incubation of the purified protein with a 5′-labeled substrate resulted in the accumulation of a shorter oligonucleotide product (Figure 2A), whereas incubation with a 3′-labeled substrate led to a loss of signal of the substrate, with no detectable intermediates (Figure 2B). Thin-layer chromatography (TLC) analysis of the reaction products (Figure 2E) revealed that the enzyme generated nucleoside 5′-monophosphates. The exonuclease was unable to digest a substrate blocked at the 3′ end with a phosphate group (Figure 2F) and was inhibited by EDTA (data not shown). These results are indicative of a hydrolytic, processive 3′–5′ exonuclease that degrades RNA molecules with a 3′ hydroxyl group. Yeast Rrp4p is also a hydrolytic 3′–5′ exonuclease but it has a distributive mode of action.
Fig. 2. Recombinant His6-TbRRP4 and the purified T.brucei exosome show 3′–5′ exoribonuclease activity in vitro. Purified His6-TbRRP4 was incubated in the presence of a 5′-labeled (A) or 3′-labeled RNA substrate (B) for the times indicated, and the reactions electrophoresed in PAGE–urea gels. In the mock lane the RNA substrate was incubated for 120 min using the same reaction conditions, without recombinant His6-TbRRP4. Exonuclease activity was assayed for the purified exosome complex under the same conditions using the 5′- (C) or 3′-labeled (D) substrate. (E) Separation of the reaction products by TLC. The RNA substrate was incubated in the absence (mock) or in the presence of His6-TbRRP4. The migration of 5′- and 3′-nucleoside monophosphates is also indicated. (F) Exonuclease activity of His6-TbRRP4 measured in the presence of the RNA substrate without (open squares) or with (filled squares) a phosphate group at the 3′ end.
To test whether TbRRP4 was able to complement an rrp4-1 allele, the gene coding for TbRRP4 was cloned and expressed in yeast. The growth of the yeast strain P58 (Mitchell et al., 1997), transformed with plasmids containing either the TbRRP4 gene, the yeast RRP4 gene or the cloning vector alone (p415GAL) was compared in Sgal–Leu medium plates at 25 or 37°C (see Materials and methods). The yeast RRP4 gene was included as a positive control. Neither the cloning vector alone nor the TbRRP4 gene was able to complement the rrp4-1 allele (data not shown).
Purification of the T.brucei exosome and identification of the components
To analyze the composition of the T.brucei exosome, we purified the complex from procyclic cells using the tandem affinity purification (TAP) method (Rigaut et al., 1999). The TAP tag consists of a calmodulin binding peptide, a tobacco etch virus (TEV) protease cleavage site and two IgG-binding motifs of protein A. We attached the TAP sequence to the C-terminus of TbRRP4, which was known to be present in the 11S complex. TAP-TbRRP4 was expressed in trypanosomes under the control of a tetracycline-inducible EP1 promoter (see Materials and methods). To check that the TAP-tagged protein was functional, we deleted both of the endogenous (unmodified) RRP4 genes from the TAP-TbRRP4-expressing cell line (data not shown). The resulting parasites grew normally in the presence of tetracycline (when the inducible TAP-TbRRP4 was expressed), and stopped growing in the absence of TAP-TbRRP4 expression, showing that TbRRP4 is essential for growth and that the tagged protein was functional.
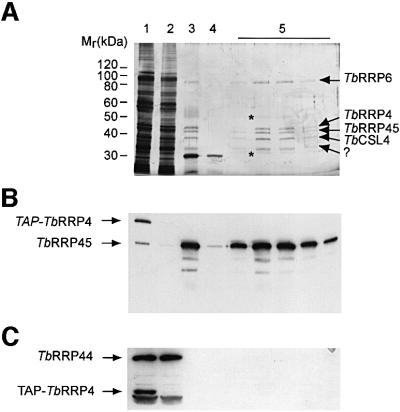
The purification of the T.brucei exosome from a cytosolic extract is shown in Figure 3A. The first step involved binding the protein A component of the tag to IgG–Sepharose. The complex was released from the matrix by addition of the TEV protease, then bound to calmodulin beads in the presence of calcium, via the calmodulin binding peptide. After washing, the complex was eluted with EGTA. The eluted complex was analyzed by SDS–PAGE and silver staining (Figure 3A). Five polypeptides were present in apparently equimolar amounts. Their electrophoretic mobilities corresponded to 90, 45, 42, 38 and 32 kDa (Figure 3A, arrows). In addition, two more, less abundant bands were also seen with apparent molecular weights of 52 and 30 kDa (Figure 3A, asterisks). The 90 kDa band could be resolved as a doublet (90 and 88 kDa) in low-percentage acrylamide gels (data not shown). Both bands correspond to the same protein (see below). Coomassie Blue staining gave similar results except that the 52 kDa band appeared to be more abundant (data not shown). The specificity of the TAP method was monitored by generating a cell line expressing only the TAP tag (see Materials and methods). In this case no bands were detected after purification of the TAP tag peptide (data not shown).
Fig. 3. Purification of the T.brucei exosome. The expression of TAP-tagged TbRRP4 was induced by the addition of tetracycline to the culture medium for 48 h, and the associated proteins purified using the TAP method. (A) Aliquots from each step (or the whole EGTA-eluate fractions) were analyzed by SDS–PAGE and silver staining. Lane 1, S100 extract; lane 2, IgG chromatography flow-through; lane 3, TEV eluate; lane 4, calmodulin chromatography flow-through; lane 5, EGTA eluate (5 fractions). (B and C) Immunoblotting analyses of the TAP fractions using TbRRP45 (B) or TbRRP44 (C) antisera. The protein marker is the 10 kDa protein ladder (Gibco BRL).
The cell line used for the expression of TAP-TbRRP4 and the purification of the exosome still had one TbRRP4 allele (see Materials and methods). From western blotting analysis it became clear that the expression of TAP- TbRRP4 led to depletion of the endogenous TbRRP4 (data not shown), suggesting that perhaps free TbRRP4 is unstable. The same complex composition and stoichiometry were obtained upon expression of TAP-TbRRP4 at a concentration of 1 ng/ml of tetracycline, in which both the endogenous and TAP-TbRRP4 proteins are expressed at similar levels, or at 100 ng/ml tetracycline, when only TAP-TbRRP4 could be detected (data not shown).
Since both TbRRP4 and TbRRP45 co-sediment in an 11S complex (Figure 1), we expected TbRRP45 to copurify with TAP-TbRRP4. Indeed, TbRRP45 was retained in the IgG–Sepharose and calmodulin matrices and was enriched in the EGTA-eluted complex (Figure 3B). TAP-TbRRP4 was also detected with anti-TbRRP45 antibodies (or with anti-TbRRP44 antibodies, see below), because of the presence of the two IgG binding domains within the TAP tag. In contrast, TbRRP44, which is not present in the 11S complex (Figure 1) did not bind to the IgG beads and could not be detected in the purified exosome complex fractions (Figure 3C). When the TAP purification was carried out in the control cell line expressing only the TAP tag, TbRRP45 was detected only in the IgG flow-through, indicating that this protein binds to TbRRP4 and not to the TAP tag (data not shown).
To identify the individual components of the T.brucei exosome, the bands were excised from the gel and subjected to trypsin digestion. The generated peptides were analyzed by MALDI mass spectrometry. The observed peptide masses were compared with those obtained from the virtual tryptic digest of the T.brucei proteins listed in Table I (see Materials and methods). This approach led to the unambiguous identification of TbRRP6, TbRRP4, TbRRP45 and TbCSL4, as indicated in Figure 3A. Both peptides corresponding to the 90 kDa doublet gave a peptide pattern corresponding to TbRRP6. The doublet migration might be indicative of partial post-translational modification. The MALDI pattern obtained from the 32 kDa band (Figure 3A, question mark) did not match any of the proteins listed in Table I, or any known T.brucei protein sequence available in the databases. We are still attempting to identify this and the 53 and 30 kDa bands (Figure 3A, asterisks).
The purified T.brucei exosome was incubated with a 5′-labeled RNA substrate in the same conditions as for recombinant His6-TbRRP4 (Figure 2C). We observed a simultaneous decrease in the lengths of the whole population of substrate molecules, indicative of a distributive riboexonuclease activity. The incubation of the purified exosome with a 3′-labeled substrate led to a loss of signal of the substrate band (Figure 2D). Again, the products of the reaction were 5′-nucleoside monophosphates (data not shown). This indicates that the T.brucei exosome shows a distributive and hydrolytic riboexonuclease activity, as in the case of the yeast exosome (Mitchell et al., 1997). The exonuclease activity of the T.brucei exosome ceased after a 15 min incubation, which may indicate an inactivation of the complex, or that additional factors might be required for proper activity.
The combined molecular weights of the five stoichiometric bands give a value of 217 kDa, which is roughly in agreement with the molecular weight estimated by glycerol gradient analysis, ∼240 kDa. The absence of the 110 kDa TbRRP44 from the purified complex was consistent with the previous results from glycerol gradient centrifugation (Figure 1) and with the small size of the T.brucei exosome compared with that of yeast. None of the peptide masses obtained by MALDI analysis matched TbRRP40, TbRRP41A, TbRRP41B or TbRRP44. It was notable that TbRRP6 was present in stoichiometric amounts in the purified cytosolic exosome complex. In yeast and in human cells, Rrp6p and the human homolog PM-Scl100 are present only in the nuclear complex (Allmang et al., 1999b).
The results so far indicate that the major T.brucei cytosolic exosome species is composed of TbRRP4, TbRRP6, TbRRP45, TbCSL4 and an additional, as yet unidentified, protein. Most TbRRP44 is not associated with the exosome.
Effect on cell growth of the inhibition of the expression of individual exosome components by RNA interference
All components of the yeast exosome, except for Rrp6p, are essential for viability (Allmang et al., 1999b) and are involved in the processing and degradation of many RNAs (reviewed in van Hoof and Parker, 1999). In order to analyze the functions of the proteins listed in Table I we generated conditional mutants. For each gene, we created a trypanosome line that expressed a double-stranded RNA corresponding to the first 500–800 nucleotides, under the control of the tetracycline-inducible promoter (see Materials and methods). Expression of double-stranded RNA in trypanosomes, also known as RNA interference (RNAi), results in depletion of the corresponding mRNA (Shi et al., 2000; Wang et al., 2000). The effect of RNAi was checked by northern blotting for every mRNA, and also by immunoblotting for the proteins for which antisera were available. For every cell line illustrated, the target mRNA virtually disappeared after 48 h in the presence of tetracycline (not shown). The proteins analyzed dropped to ∼10% of the wild-type levels after 24 or 48 h of induction (data not shown). These reductions are in agreement with studies of several other trypanosome genes (Shi et al., 2000; Wang et al., 2000).
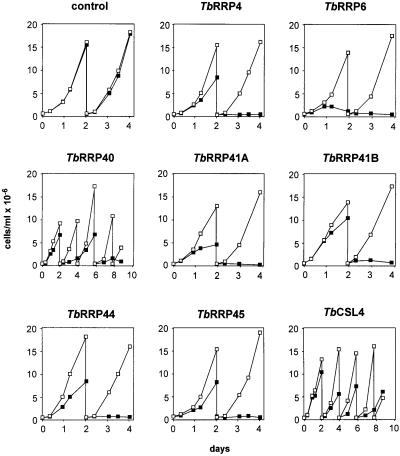
The effect of depletion of each of the homologs on cell growth is shown in Figure 4. After addition of tetracycline to induce RNAi, cell growth was inhibited. Although minor variations in the kinetics were observed (compare TbRRP6 and TbRRP40 growth curves), all the depleted trypanosomes eventually died with the exception of the TbCSL4 line. The variations could indicate that the different exonucleases have a different importance in cell survival, but might also be due to differences in the turnover of the mRNA, the protein product, or both (Wang et al., 2000). These results indicate that all the proteins listed in Table I, with the possible exception of TbCSL4, are essential for the viability of T.brucei.
Fig. 4. Depletion of the exosome protein homologs by inducible RNA interference and effects on growth. Trypanosome lines were created that expressed gene-specific dsRNA in a tetracycline-inducible fashion. Each trypanosome line was grown in the absence (open squares) or presence (filled squares) of 100 ng/ml tetracycline to induce RNAi. Cultures were followed for four to nine days and were diluted to 0.4 × 106 cells/ml every 2 days as required.
Roles of individual exosome component homologs in 5.8S rRNA processing
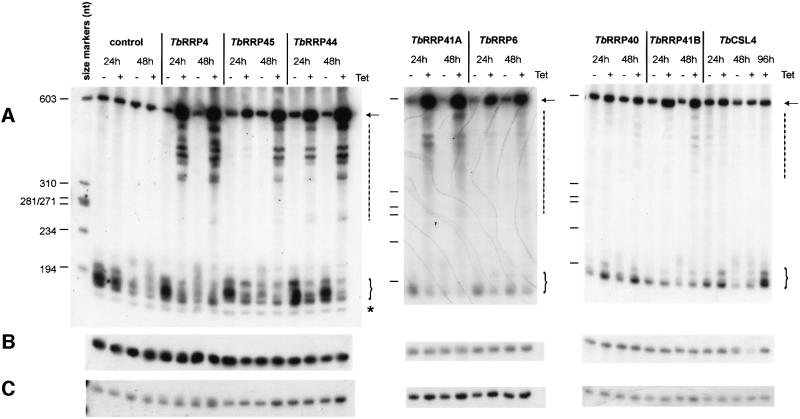
The yeast exosome catalyzes the 3′ trimming of a 7S pre-rRNA precursor to yield mature 5.8S rRNA, and the depletion of any of the yeast exosome components results in the accumulation of both the 7S precursor and incompletely processed pre-rRNA species (Allmang et al., 1999a). A 5.8S rRNA species with short and heterogeneous 3′ extensions, the 6S pre-rRNA, is also detected in wild-type yeast strains; this rRNA precursor is also processed by the exosome (Allmang et al., 1999a). Trypanosoma brucei has a 7S species of 0.6 kb, which contains a 3′ extension of ∼400 nucleotides (Hartshorne and Toyofuku, 1999) and is therefore a likely substrate for the exosome. The effect of RNAi on the maturation of 5.8S rRNA was tested for all the proteins (Figure 5). Total RNA was obtained from uninduced or induced cells, separated on denaturing polyacrylamide gels, and probed with oligonucleotides specific to 3′-extended 5.8S rRNA (Figure 5A), mature 5.8S rRNA (Figure 5B) or (as a loading control) the signal recognition particle RNA (Figure 5C). A 2- to 10-fold accumulation of both full-length and incompletely processed 7S pre-RNA species could be observed upon depletion of TbRRP4, TbRRP45, TbRRP44, TbRRP41A, TbRRP6 and TbRRP41B. Defects were not so evident in the cases of TbRRP40 and TbCSL4, even after prolonged RNAi induction times (Figure 5 and data not shown). Nevertheless, these mutants also showed an accumulation of 6S pre-rRNA and the TbRRP40 mutant also accumulated 7S pre-RNA. These results are summarized in Table II.
Fig. 5. Effect of depletion of each exosome component homolog on 5.8S rRNA maturation in vivo. Total RNA was extracted from parasites grown in the absence (–) or in the presence (+) of tetracycline for 24 or 48 h, and separated in PAGE–urea gels. In the case of TbCSL4, an additional sample taken at 96 h after induction was also included. After electrophoresis the gels were transferred and hybridized to detect (A) extended 5.8S rRNA species, (B) mature 5.8S rRNA or (C) the T.brucei signal recognition particle RNA (loading control). The arrow indicates full-length 7S rRNA and the vertical dashed line shows incompletely processed 7S rRNA species. 6S-like species are indicated with a bracket. The asterisk (A, left panel) indicates the mature 5.8S rRNA, which in this particular blot was not completely stripped from a previous hybridization. The size marker is ØX174 DNA digested with BsuR I (MBI fermentas) that was dephosphorylated and labeled with [γ-32P]ATP and polynucleotide kinase.
Table II. Summary table.
| Protein | Present in the TAP- purified exosome? | Growth phenotype | 5.8S rRNA processing phenotype |
|---|---|---|---|
| TbRRP4 | yes | essential | 1,2 |
| TbRRP6 | yes | essential | 1 |
| TbRRP40 | no | essential | 1,3 |
| TbRRP41A | no | essential | 1,2 |
| TbRRP41B | no | essential | 1,2 |
| TbRRP44 | no | essential | 1,2,4 |
| TbRRP45 | yes | essential | 1,2 |
| TbCSL4 | yes | slow growth | 3 |
The 5.8S rRNA phenotypes observed were: 1, accumulation of full-length 7S pre-rRNA; 2, accumulation of incompletely processed 7S pre-rRNA species; 3, accumulation of 6S pre-rRNA; 4, reduction of 6S pre-rRNA levels.
In yeast mutants, some reduction in the levels of mature 5.8S rRNA was seen after 12–24 h of exosome component depletion (Mitchell et al., 1997; Allmang et al., 1999a,b). No depletion of mature 5.8S rRNA was apparent in T.brucei (Figure 5B). After 48 h of RNAi induction, T.brucei in the presence of tetracycline had divided one to four times, depending on the protein being depleted (Figure 4). RNAi effects on mRNA levels are detected after one cell division (Shi et al., 2000). Around 10% of the normal levels of TbRRP4, TbRRP44 and TbRRP45 persisted after 48 h (data not shown) and this is probably also true of the other exonucleases. This might have been sufficient to maintain normal 5.8S rRNA levels under conditions of no or very slow growth, as insufficient division had occurred to dilute it.
We concluded from these experiments that all the T.brucei exosome component homologs were involved in rRNA processing, although some of them appeared not to be associated with the exosome.
Discussion
We have identified eight T.brucei genes coding for proteins similar to components of the yeast exosome. All but one of the corresponding proteins were essential for cell growth and all were involved in processing of 5.8S rRNA. Our results indicated, however, that the major T.brucei cytosolic exosome species was composed of only five proteins: TbRRP4, TbRRP6, TbRRP45, TbCSL4 and an additional, as yet unknown, protein. TbRRP40, TbRRP41A, TbRRP41B and TbRRP44 were not found in the cytosolic exosome complex.
The simplicity of the trypanosome exosome suggests that this complex has become larger and more elaborate during eukaryotic evolution. Six RNase PH-like proteins are present in the yeast exosome and six human ESTs encode RNase PH homologs. The C.elegans genome, like that of trypanosomes (so far) contains just three RNase PH-like proteins (Allmang et al., 1999b; van Hoof and Parker, 1999). In C.elegans, these could be the homologs of Rrp41p, Rrp45p and Rrp46p, whereas the sequences found in trypanosomes seem to most resemble Rrp45p and Rrp41p (two sequences: TbRRP41A and TbRRP41B) (Table I). The T.brucei Rrp41p homolog TbRRP41B also resembles Rrp46p, but to a lesser extent. A similar situation exists in A.thaliana, which in addition to the genuine Rrp41p homolog AtRrp41p (Chekanova et al., 2000), possesses another RNase PH-like protein, F12M12.180, which gives maximal similarity to Rrp41p when compared with the yeast protein database. Further RNase PH homologs may be discovered as the T.brucei genome is completed; the present level of coverage is probably between 60 and 80%. It is nevertheless striking that, as for C.elegans, no possible homologs have so far been found for Rrp42p, Rrp43p or Mtr3p. In contrast to T.brucei, C.elegans apparently lacks Rrp4p (van Hoof and Parker, 1999; see also Table I).
Using antisera against TbRRP4 and TbRRP45 we found that the cytosolic and nuclear exosomes of T.brucei migrated in glycerol gradients at ∼240 kDa (Figure 1), which was consistent with the small number of subunits seen. TbRRP40, TbRRP41A and TbRRP41B were absent from the cytosolic exosome, but we can not exclude the possibility that they are present in the nuclear exosome in addition to (or instead of) other components, or are involved in exosome assembly. TbRRP44 migrated as a monomer in both cytosolic and nuclear extracts (Figures 1 and 3C), but it is still conceivable that a very small proportion of TbRRP44 is exosome associated.
Overall, these results indicate that the only components that have been conserved and remained exosome associated throughout evolution are RRP6, RRP45 and CSL4, and that exosome association and composition can by no means be predicted on the basis of sequence homology.
In yeast there are additional, non-exosomal RNase II-like proteins which are essential for a whole variety of processes (including cell division, signal transduction, RNA splicing and mitochondrial biogenesis) (Mian, 1997; Luukkonen and Séraphin, 1999). The T.brucei genome also contains at least one other Rnase II-like protein (DDBJ/EMBL/GenBank, No. AJ309002), with a predicted molecular mass of 102 kDa. Trypanosomes expressing RNAi against this protein showed no effects on either cell growth or 5.8 rRNA maturation (data not shown).
Despite the difference in subunit composition, the overall mode of action of the T.brucei and yeast exosomes seems to be similar, i.e. distributive and hydrolytic 3′–5′ riboexonuclease activity. Some of the components of the T.brucei exosome nevertheless exhibit distinct functional properties. The T.brucei Rrp4p homolog, TbRRP4, showed a processive exonucleolytic activity (Figure 2), whereas yeast Rrp4p has a distributive mode of action (Mitchell et al., 1997). The human Rrp4p homolog (hRrp4p) can restore the growth of the rrp4-1 yeast strain at the non-permissive temperature (Mitchell et al., 1997), whereas the TbRRP4 gene could not. This failure could be due to the different exonucleolytic mechanism of TbRRP4 or to an inability of the trypanosome protein to assemble into a functional complex in yeast. All components of the yeast exosome, except Rrp6p, are essential for viability. In T.brucei, in contrast, the Rrp6p homolog TbRRP6 was essential, as were the other exosome component homologs, with the possible exception of TbCSL4 (Figure 4). TbRRP6 was present in stoichiometric amounts in the purified cytosolic exosome, whereas in yeast and in human cells Rrp6p and PM-Scl100 can be detected only in the nuclear complex. We do not yet know whether TbRRP6 has specific cytosolic functions.
The pathways of 5.8S rRNA maturation seem to be quite similar in trypanosomes and yeast; in particular, the presence of the 6S rRNA species may indicate a site within the precursor molecule at which fast processive degradation is replaced by a slow distributive processing (Allmang et al., 1999a). It has been proposed that different components of the yeast exosome play distinct roles in the maturation of 5.8S rRNA (Allmang et al., 1999a). This might also be the case in T.brucei, since different patterns were observed after depletion of different exonucleases. For example, a reduction of 6S pre-rRNA was observed after depletion of TbRRP44, while 6S species accumulated after depletion of TbRRP40 and TbCSL4. Depletion of TbRRP6 resulted in accumulation of 7S pre-rRNA, but incompletely processed 7S species were not readily visible.
It is interesting that similar defects in 5.8S rRNA maturation were observed upon depletion of the proteins examined, irrespective of exosome association. It has been suggested that each exosome subunit in yeast is essential because its absence may cause an exosome assembly failure (van Hoof and Parker, 1999). This is most unlikely to be true for TbRRP44 in T.brucei, and is improbable for TbRRP40, TbRRP41A and TbRRP41B. An alternative scenario is that maturation of 5.8S rRNA is an orchestrated process involving all these proteins, whether or not they are in a complex, and that a defect in any one disrupts the whole pathway. The availability of several of the higher eukaryotic exosome components as separate entities in trypanosomes is an opportunity to determine their functions, independently of effects on exosome complex stability.
Trypanosoma brucei is one of the earliest diverging eukaryotes containing mitochondria (Fernandes et al., 1993). All available data indicate that there is no exosome complex in eubacteria (Deutscher, 1993), but genetic evidence suggests that there may be an exosome precursor in Archaea (Koonin et al., 2001). We suggest that individual exonucleases may have begun to associate in a larger complex at or before the onset of the eukaryotic lineage. As organism complexity grew and RNA processing and its regulation became increasingly more complicated, more subunits were added. The simple exosome of T.brucei provides a glimpse of these early events.
Materials and methods
Cell culture
Bloodstream and procyclic form T.brucei 449 cell lines (Biebinger et al., 1997), stably expressing the tetracycline repressor, were used in all experiments. Cells were grown in the presence of 0.2 µg/ml (bloodstream forms) or 0.5 µg/ml (procyclic forms) phleomycin.
Cloning of T.brucei exosome component homologs
Trypanosoma brucei sequences similar to yeast exosome components were identified by comparing the yeast protein sequences against the unfinished T.brucei genome using the TBLASTN program (Altschul et al., 1990). Oligonucleotides were designed to PCR amplify specific probes, which were used to isolate genomic DNA fragments containing the entire ORFs from partial genomic libraries (Estévez et al., 1997) constructed in pGEM4 or pGEM5 (Promega) vectors. The genomic DNA inserts were sequenced by primer-walking and the ORFs identified using the DNAStar and the Gene Construction Kit programs. Searches for Pfam and Prosite motifs were carried out using the ISREC ProfileScan Server (http://www.isrec.isb-sib.ch/software/PFSCAN_form.html), and by comparison of the T.brucei proteins with their yeast counterparts. The sequence corresponding to the entire TbRRP41B ORF was taken from the unfinished T.brucei chromosome II sequence available at the TIGR Trypanosoma brucei Genome Project.
Overexpression of TbRRP4 in E.coli
A His6-TbRRP4 ORF was PCR amplified from T.brucei genomic DNA, cloned into pET-3a (Stratagene; plasmid pHD1191) and transformed into the E.coli strain BL21(DE3)pLysS (Stratagene). Bacteria were grown at 22°C to an OD600 of 0.2, induced with 1 mM isopropyl-β-d-thiogalactopyranoside and incubated at the same temperature to an OD600 of 1.0. Cell lysis was performed in 10 mM HEPES, 5 mM MgCl2, 0.1% NP-40, 300 mM NaCl, 10% glycerol, 20 mM β-mercaptoethanol, EDTA-free protease inhibitor cocktail (Roche) and 20 mM imidazole pH 7.6. The purification of His6-TbRRP4 using Ni-NTA agarose (Qiagen) was performed according to the manufacturer’s instructions [in the same buffer, lacking the protein inhibitor cocktail but containing 1 mM phenylmethylsulfonyl fluoride (PMSF)], and the protein was eluted at 60 mM imidazol. The enzyme was further purified by AffiGel Blue (Bio-Rad) chromatography and eluted with 1.3 M NaCl. This protein preparation was homogeneous by SDS–PAGE and silver staining. The enzyme was dialyzed against 10 mM HEPES, 5 mM MgCl2, 0.1% NP-40, 10 mM NaCl, 10% glycerol, 1 mM dithiothreitol and 1 mM PMSF pH 7.6, and stored in aliquots at –80°C.
Complementation studies in yeast
The yeast expression vector p415GAL (Mumberg et al., 1994) was used for complementation of the yeast strain P58 (MATα ade2 ade3 leu2 ura3 rrp4-1) (Mitchell et al., 1997). The TbRRP4 and the S.cerevisiae RRP4 ORFs were PCR amplified from genomic DNA and cloned in p415GAL. Complementation of the rrp4-1 allele was assayed in plates containing SGal–Leu medium at 37°C (Mitchell et al., 1997).
Generation of antisera
Polyclonal antibodies against TbRRP4, TbRRP44 and TbRRP45 were raised in rabbits. For TbRRP4, the protein overexpressed in E.coli (see above) was used as an immunogen; ∼750 µg of protein were purified by Coomassie Blue SDS–PAGE gels (Schägger et al., 1988) and processed for immunization (Harlow and Lane, 1988). Antibodies against TbRRP44 and TbRRP45 were raised using the following synthetic peptides as immunogens: NDTGAGGDDHENSGREGIGEESE(C) (TbRRP44), and HHRRPELTVRGSSVIVHPPHERE(C) (TbRRP45). The cysteine residues were added in order to couple the peptides to maleimide-activated keyhole limpet hemocyanin according to the manufacturer’s instructions (Pierce). Immunizations and immunoblotting were performed according to standard procedures (Harlow and Lane, 1988). Antibodies were purified by affinity chromatography using the corresponding peptide coupled to AffiGel-15 (Bio-Rad).
In vitro exoribonuclease assays
The RNA substrate was synthesized in vitro by T7 RNA polymerase transcription (Cunningham and Ofengand, 1990) of plasmid pET-3a (Stratagene) linearized with NdeI, purified in PAGE–urea gels and dephosphorylated using calf-intestine alkaline phosphatase. The purified RNA was labeled either at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase or at the 3′ end using [5′-32P]pCp and RNA ligase. To obtain a 3′-labeled RNA lacking a 3′-phosphate group, an aliquot of [5′-32P]pCp-labeled RNA was dephosphorylated using alkaline phosphatase. Internally-labeled RNA was synthesized in vitro as above, but [α-32P]UTP was included in the reaction and the concentration of non-labeled UTP was lowered to one-fifth. All labeled RNAs were purified in PAGE–urea gels. Exonuclease assays were performed as described (Mitchell et al., 1996) using an excess of substrate over enzyme to ensure that for nearly all substrates, cleavage was the result of the action of only one exonuclease (Bambara et al., 1995). The reactions were incubated at 37°C and stopped by the addition of EDTA. To characterize processivity and direction of decay, the reactions were diluted 1:1 with formamide, heated at 65°C for 5 min and loaded in an 8% PAGE–urea gel. Bands were quantified using a phosphoimager (Fuji). For analysis of the released product, the samples were subjected to TLC in PEI-cellulose plates (Ross, 1999). Commercial 5′- and 3′-nucleoside monophosphates (Sigma and Fluka) were resolved in parallel and used as markers. To analyze the 3′–5′ riboexonuclease activity in glycerol gradient fractions, 5 µl of each sample were tested as above and the enzyme activity was measured as the release of acid-soluble counts from a 3′-labeled and dephosphorylated RNA (Ross, 1999), after an incubation of 15 min at 37°C.
Subcellular fractionation and glycerol gradients
To examine the nuclear exosome, nuclear extracts were obtained from ∼5 × 109 cells (Field and Field, 1996) and centrifuged for 1 h at 100 000 g. The supernatant was allocated and frozen at –80°C. To isolate the cytosolic fraction, the low-speed supernatant obtained after centrifugation of the above cell lysate was further centrifuged for 10 min at 12 000 g. The resulting supernatant was ultracentrifuged and the S100 supernatant was stored as above. The separation on glycerol gradients was done essentially as described (Mitchell et al., 1997), using as markers bovine serum albumin (4.3S), yeast alcohol dehydrogenase (7.4S), bovine catalase (11.3S) and jack bean urease (18.6S).
Purification of the T.brucei exosome using the TAP method
To create a C-terminal, TAP-tagged version of TbRRP4, the TAP tag (Rigaut et al., 1999) was PCR amplified from plasmid pBS1479 (kindly donated by B.Séraphin) and cloned in the T.brucei tetracycline-inducible expression vector pHD678 (Biebinger et al., 1997) to yield pHD918. The T.brucei TbRRP4 ORF was PCR amplified from genomic DNA and cloned into pHD918, yielding pHD924. Trypanosoma brucei procyclic 449 cells in which one TbRRP4 allele was replaced with a blasticidin resistance gene were transfected with either pHD918 or pHD924. Hygromycin-resistant clones were selected and checked for single integration of the plasmid at the ribosomal rDNA intergenic locus by Southern blotting analysis and also for tetracyline inducibility (Biebinger et al., 1997). TAP tag alone (pHD918) or TAP-tagged TbRRP4 (pHD924) expression was induced by adding tetracycline to the medium, and the complex purified as described (Rigaut et al., 1999) from S100 cytosolic extracts obtained from 1 l of cells harvested at a density between 1.0 and 1.5 × 107 cells/ml.
Mass spectrometry and protein identification
The components of the T.brucei exosome complex were resolved by SDS–PAGE and visualized by Coomassie Blue staining. Individual bands were excised, washed repeatedly with H2O and H2O/acetonitrile, shrunk with acetonitrile and digested with 0.25 µg trypsin (sequencing grade modified porcine trypsin from Promega, Madison, WI) in 40 mM ammonium bicarbonate overnight at 37°C. The generated peptides were analyzed by MALDI mass spectrometry. Sample preparation was achieved following the thin film preparation technique (Jensen et al., 1996). MALDI mass spectra were recorded in the positive ion mode with delayed extraction on a reflex II time-of-flight instrument (Bruker-Daltonik GmbH, Bremen, Germany). Proteins were unambiguously identified by comparing the peptide mass fingerprint with the theoretical tryptic digestion of homologous proteins of the exosome found in the T.brucei genome. Theoretical masses of peptides were calculated using the Sherpa software package (http//:128.95.12.16/Development/Sherpa.html). For every protein analyzed, the experimental peptide masses matched the theoretical ones. In addition, mass patterns due to oxidized methionine or oxidized tryptophan were also confirmatory (Simat et al., 1994; Schnölzer and Lehmann, 1997).
RNA techniques
Total RNA was obtained using peqGold Trifast (peqLab, Germany). To study 5.8S rRNA processing, RNA samples were electrophoresed in 5% polyacrylamide–TBE gels, electrotransferred to neutral nylon membranes (1 h at 25 V) and hybridized using standard procedures (Ausubel et al., 1997). The oligonucleotides used were CZ1193 (5′-ACTTTGCTGCGTTCTTCAAC-3′) to detect mature 5.8S rRNA, CZ1427 (5′-GTTTTTATATTCGACACTG-3′) to detect 3′-extended 5.8S rRNA species and CZ1478 (5′-CAACACCGACACGCAACC-3′) for the T.brucei signal recognition particle RNA (Michaeli et al., 1992). Bands were quantified using a phosphoimager (Fuji).
RNA interference
All the constructs used for RNAi were made using the ‘stuffer’ strategy described for T.brucei (Shi et al., 2000). Briefly, PCR fragments (500–800 bp) were amplified from genomic DNA, ligated to a stuffer fragment (Shi et al., 2000) (kindly donated by Elisabetta Ullu) and cloned into the T.brucei expression vector pHD1146 [a pHD678 (Biebinger et al., 1997) derivative lacking the T7 promoter]. The resulting plasmids were linearized with NotI and transfected into procyclic 449 T.brucei cells (Biebinger et al., 1997). Transfectants were selected in 50 µg/ml hygromycin and cloned by limiting dilution. RNAi induction was achieved by adding tetracycline to the medium at a concentration of 100 ng/ml.
More information regarding cloning procedures, and oligonucleotide and plasmid sequences can be obtained from the authors.
Acknowledgments
Acknowledgements
We wish to thank Cristina Guerra for helping in the overexpression of TbRRP4. We are indebted to David Tollervey for providing the yeast strain P58, to Bertrand Séraphin for plasmid pBS1479, to Elisabetta Ullu for the plasmid pSP72 containing the stuffer fragment, to Ralph Jansen for yeast genomic DNA and plasmid p415GAL, and to Toinette Hartshorne, Asunción Delgado, Juan D.Alfonzo and Luis Quijada for valuable comments on the manuscript. All the experiments except the mass spectrometry (T.K.) were planned and carried out by A.M.E., who also wrote the manuscript. C.C. conceived the project, provided occasional technical advice and modified the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft.
References
- Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999a) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Petfalski,E., Podtelejnikov,A., Mann,M., Tollervey,D. and Mitchell,P. (1999b) The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1997) Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York, NY.
- Bambara R.A., Fay,P.J. and Mallaber,L.M. (1995) Methods of analyzing processivity. Methods Enzymol., 262, 270–280. [DOI] [PubMed] [Google Scholar]
- Biebinger S., Wirtz,L.E., Lorenz,P. and Clayton,C. (1997) Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol., 85, 99–112. [DOI] [PubMed] [Google Scholar]
- Brouwer R., Allmang,C., Raijmakers,R., van Aarssen,Y., Vree-Egberts,W., Petfalski,E., van Venrooij,W.J., Tollervey,D. and Pruijn,G.J.M. (2001) Three novel components of the human exosome. J. Biol. Chem., 276, 6177–6184. [DOI] [PubMed] [Google Scholar]
- Campbell D.A., Kubo,K., Clark,C.G. and Boothroyd,J.C. (1987) Precise identification of cleavage sites involved in the unusual processing of trypanosome ribosomal RNA. J. Mol. Biol., 196, 113–124. [DOI] [PubMed] [Google Scholar]
- Chekanova J.A., Shaw,R.J., Wills,M.A. and Belostotsky,D.A. (2000) Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J. Biol. Chem., 275, 33158–33166. [DOI] [PubMed] [Google Scholar]
- Cunningham P.R. and Ofengand,J. (1990) Use of inorganic pyrophos phatase to improve the yield of in vitro transcription catalyzed by T7 RNA polymerase. Biotechniques, 9, 713–714. [PubMed] [Google Scholar]
- Deutscher M.P. (1993) Promiscuous exoribonucleases of Escherichia coli. J. Bacteriol., 175, 4577–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J.M., D́Orso,I., Sánchez,D.O. and Frasch,A.C.C. (2000) AU-rich elements in the 3′-untranslated region of a new mucin-type gene family of Trypanosoma cruzi confers mRNA instability and modulates translation efficiency. J. Biol. Chem., 275, 10218–10227. [DOI] [PubMed] [Google Scholar]
- Estévez A.M. and Simpson,L. (1999) Uridine insertion/deletion RNA editing in trypanosome mitochondria—a review. Gene, 240, 247–260. [DOI] [PubMed] [Google Scholar]
- Estévez A.M., Martínez-Costa,O.H., Sánchez,V. and Aragón,J.J. (1997) Cloning, sequencing and developmental expression of phospho fructokinase from Dictyostelium discoideum. Eur. J. Biochem., 243, 442–451. [DOI] [PubMed] [Google Scholar]
- Fernandes A.P., Nelson,K. and Beverley,S.M. (1993) Evolution of nuclear ribosomal RNAs in kinetoplastid protozoa: perspectives on age and origins of parasitism. Proc. Natl Acad. Sci. USA, 90, 11608–11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H. and Field,M.C. (1996) Leptomonas seymouri, Trypanosoma brucei: a method for isolating trypanosomatid nuclear factors which bind T.brucei single-stranded g-rich telomere sequence. Exp. Parasitol., 83, 155–158. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Hartshorne T. and Toyofuku,W. (1999) Two 5′-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res., 27, 3300–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz H.R., Hartmann,C., Huober,K., Hug,M. and Clayton,C. (1997) Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res., 25, 3017–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen O.N., Podtelejnikov,A. and Mann,M. (1996) Delayed extraction improves specificity in database searches by matrix-assisted laser desorption/ionization peptide maps. Rapid Commun Mass Spectrom., 10, 1371–1378. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Wolf,Y.I. and Aravind,L. (2001) Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res., 11, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen B.G.M. and Séraphin,B. (1999) A conditional U5 snRNA mutation affecting pre-mRNA splicing and nuclear pre-mRNA retention identifies SSD1/SRk1 as a general splicing mutant supressor. Nucleic Acids Res., 27, 3455–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian I.S. (1997) Comparative sequence analysis of ribonucleases HII, III, II, PH and D. Nucleic Acids Res., 25, 3187–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S., Podell,D., Agabian,N. and Ullu,E. (1992) The 7SL RNA homologue of Trypanosoma brucei is closely related to mammalian 7SL RNA. Mol. Biochem. Parasitol., 51, 55–64. [DOI] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2000) Musing on the structural organization of the exosome complex. Nature Struct. Biol., 7, 843–846. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski,E. and Tollervey,D. (1996) The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev., 10, 502–513. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′–5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko,A., Rutz,B., Wilm,M., Mann,M. and Séraphin,B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol., 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Ross J. (1999) Assays for analyzing exonucleases in vitro. Methods, 17, 52–59. [DOI] [PubMed] [Google Scholar]
- Schägger H., Aquila,H. and von Jagow,G. (1988) Coomassie Blue-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for direct visualization of polypeptides during electrophoresis. Anal. Biochem., 173, 201–205. [DOI] [PubMed] [Google Scholar]
- Schnölzer M. and Lehmann,W.D. (1997) Identification of modified peptides by metastable fragmentation in MALDI mass spectrometry. Int. J. Mass Spectrom Ion Processes, 169/170, 263–271. [Google Scholar]
- Shi H., Djikeng,A., Mark,T., Wirtz,E., Tschudi,C. and Ullu,E. (2000) Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA, 6, 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simat T., Meyer,K. and Steinhart,H. (1994) Synthesis and analysis of oxidation, carbonyl condensation compounds of tryptophan. J. Chromatogr. A, 661, 93–99. [Google Scholar]
- Ullu E., Tschudi,C. and Günzl,A. (1996) Trans-splicing in trypanosomatid protozoa. In Smith,D.F. and Parsons,M. (eds), Molecular Biology of Parasitic Protozoa. Oxford University Press, New York, NY, pp. 115–133.
- van Hoof A. and Parker,R. (1999) The exosome: a proteasome for RNA? Cell, 99, 347–350. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Staples,R.R., Baker,R.E. and Parker,R. (2000) Function of the Ski4p (Cs14p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol., 20, 8230–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman K. (1985) Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull., 41, 105–114. [DOI] [PubMed] [Google Scholar]
- Wang Z., Morris,J.C., Drew,M.E. and Englund,P.T. (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem., 275, 40174–40179. [DOI] [PubMed] [Google Scholar]
- White T.C., Rudenko,G. and Borst,P. (1986) Three small RNAs within the 10kb trypanosome rRNA transcription unit are analogous to Domain VII of other eukaryotic 28S rRNAs. Nucleic Acids Res., 14, 9471–9489. [DOI] [PMC free article] [PubMed] [Google Scholar]