Abstract
Ribosomal precursor particles are assembled in the nucleolus before export into the cytoplasm. Using a visual assay for nuclear accumulation of 60S subunits, we have isolated several conditional-lethal strains with defects in ribosomal export (rix mutants). Here we report the characterization of a mutation in an essential gene, RIX7, which encodes a novel member of the AAA ATPase superfamily. The rix7-1 temperature-sensitive allele carries a point mutation that causes defects in pre-rRNA processing, biogenesis of 60S ribosomal subunits, and their subsequent export into the cytoplasm. Rix7p, which associates with 60S ribosomal precursor particles, localizes throughout the nucleus in exponentially growing cells, but concentrates in the nucleolus in stationary phase cells. When cells resume growth upon shift to fresh medium, Rix7p–green fluorescent protein exhibits a transient perinuclear location. We propose that a nuclear AAA ATPase is required for restructuring nucleoplasmic 60S pre-ribosomal particles to make them competent for nuclear export.
Keywords: AAA ATPase/nuclear transport/ribosomal biogenesis/ribosomal export
Introduction
The nucleolus is a specialized subnuclear compartment in which most steps of ribosome biogenesis take place. This process starts with the synthesis of two pre-rRNA transcripts, pre-5S rRNA and a large RNA polymerase I transcript (35S pre-rRNA in yeast), which is processed to the mature 25S/28S, 18S and 5.8S rRNAs (Kressler et al., 1999). The 5S RNA is transcribed by RNA polymerase III and recruited separately to the assembling ribosome (Dechampesme et al., 1999). Ribosomal proteins are synthesized in the cytoplasm and imported into the nucleus, where they associate with newly transcribed pre-rRNAs to generate pre-ribosomal particles (Woolford, 1991). In contrast to pre-rRNA processing and modification, very little is known about the assembly pathway for eukaryotic ribosomal subunits (Venema and Tollervey, 1999). However, it is clear that the assembly of pre-ribosomes is closely coordinated with rRNA maturation and transport. A succession of steps leads to the formation of particles competent for release from the nucleolus and export from the nucleus (for a recent review see Kressler et al., 1999).
The mechanism of ribosome biogenesis appears to be well conserved throughout eukaryotes. Many components involved in this complex process were identified and characterized in Saccharomyces cerevisiae, where >60 trans-acting factors are required to synthesize the mature 60S and 40S subunits. These include rRNA-modifying enzymes, endonucleases, exonucleases, RNA helicases and proteins associated with the small nucleolar RNAs (snoRNAs) (Kressler et al., 1999). The majority of these factors are enriched in the nucleolus. However, some are detected in the cytoplasm (Kressler et al., 1999) or associated with the nuclear pore complex (NPC) (Rout et al., 2000).
Saccharomyces cerevisiae has proven to be useful for the analysis of the NPC and the nuclear export of proteins, tRNA or mRNA (Amberg et al., 1992; Kadowaki et al., 1992; Doye and Hurt, 1997; Segref et al., 1997; Stade et al., 1997; Hellmuth et al., 1998; Sarkar et al., 1999). The analysis of nuclear export of ribosomes has been aided by the development of functional green fluorescent protein (GFP)-tagged ribosomal protein reporters. We have previously reported an in vivo export assay that uses the large subunit reporter Rpl25p–eGFP (Hurt et al., 1999; Gadal et al., 2001). Rpl25p is imported into the nucleus and assembles with ribosomes by direct binding to the rRNA inside the nucleolus (Van Beekvelt et al., 2000). This assay showed that a subset of nucleoporins and the Ran system are required for nuclear 60S subunit export. Analysis using a different ribosomal protein–GFP fusion gave similar results (Stage-Zimmermann et al., 2000). We have exploited the Rpl25p–eGFP assay to identify new factors required for nuclear 60S subunit export (Gadal et al., 2001; Milkereit et al., 2001).
Here we report the characterization of the rix7-1 mutant, which is complemented by YLL034c encoding a previously uncharacterized member of the AAA family of ATPases. AAA proteins act in a variety of cellular functions, including cell cycle regulation, protein degradation, organelle biogenesis and vesicular transport; hence, the origin of their name ‘ATPases associated with different activities’. The AAA domain is highly conserved among this family and a common mechanism of action has been proposed. A nucleotide-dependent conformational switch is proposed to apply tension to bound proteins, allowing AAA proteins to unfold polypeptides, dissociate protein– protein interactions or generate unidirectional movement (Rouiller et al., 2000; Vale, 2000). We show that Rix7p is located in the nucleus, and is required for 60S biogenesis and subsequent nuclear export.
Results
Isolation of the rix7-1 mutant, which is impaired in the export of ribosomes from the nucleolus to the cytoplasm
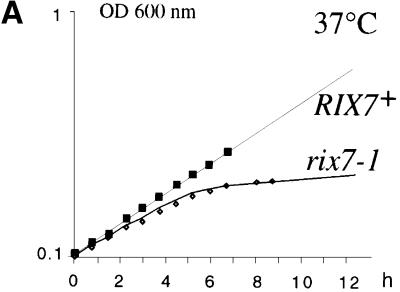
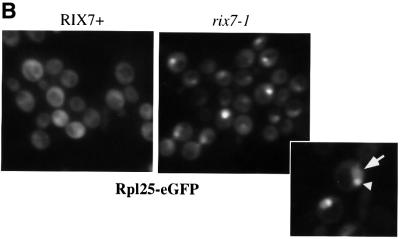
The screen for nuclear accumulation of the large subunit reporter Rpl25p–eGFP (Gadal et al., 2001) identified rix7-1, which shows slow growth at permissive temperatures (doubling times of 2.8 h for rix7-1 strains at 23°C and of 2 h for the wild-type strain) and ceases growth 5 h after shift to the restrictive temperature of 37°C (Figure 1A). At 23°C, Rpl25p–eGFP was cytoplasmic with nuclear exclusion in the rix7-1 strain, but strong nuclear accumulation was observed 2–3 h after transfer to 37°C (Figure 1B). The rix phenotype was, therefore, visible before a distinct growth defect was observed at 37°C. The pattern of accumulation of Rpl25p–eGFP inside the nucleus was found to be different in the various rix mutants (Gadal et al., 2001). In some rix mutants, Rpl25p–eGFP accumulates throughout the nucleoplasm, whereas in other mutants, including rix7-1, Rpl25p–eGFP accumulates predominantly in the nucleolus (Figure 1B, insert). This suggests that following transfer of the rix7-1 cells to 37°C, release of pre-ribosomal particles from the nucleolus to the nucleoplasm is inhibited.
Fig. 1. Nuclear accumulation of Rpl25p–eGFP in rix7-1 cells. (A) Growth curve of rix7-1 and isogenic wild-type cells at 37°C. (B) rix7-1 and isogenic wild-type cells expressing Rpl25p–eGFP were grown at 23°C, before shift for 5 h to 35°C and inspection in the fluorescence microscope. The arrowhead points to nuclear staining and the triangle to nucleolar staining.
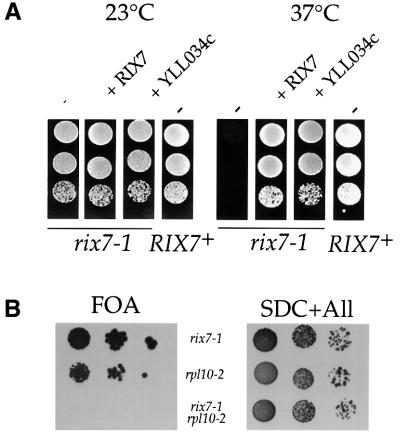
The wild-type RIX7 gene was cloned by complementation of the rix7-1 temperature-sensitive (ts) phenotype, and corresponds to locus YLL034c, which encodes an essential and novel protein of 92 kDa (see below). Cloned YLL034c complemented the growth defect of rix7-1 (Figure 2A) and rescued the nuclear accumulation of Rpl25p–eGFP. However, a few percent of the complemented cells still exhibited nuclear accumulation of Rpl25p–eGFP (data not shown). Similarly, the pre-rRNA processing defect was not completely rescued by expression of YLL034c (see below), indicating that the rix7-1 mutation is semi-dominant. To show that the rix7-1 mutation lies in the YLL034c gene locus and to isolate the mutant allele, DNA from the chromosomal locus was recovered by PCR from rix7-1 and isogenic wild-type strains. A single nucleotide substitution was found in the YLL034c open reading frame of rix7-1, changing proline (224) to leucine. The recovered rix7 allele was inserted into an ARS/CEN plasmid. When expressed in a rix7-Δ strain, the cloned allele conferred ts growth and nuclear accumulation of Rpl25p–eGFP similar to the original rix7-1 strain. Mild nuclear accumulation of Rpl25p–eGFP was also observed when the recovered rix7 allele was expressed in a wild-type strain (data not shown), consistent with the semi-dominant phenotype of rix7-1. We conclude that the Pro224→Leu mutation in YLL034c is responsible for the rix7-1 phenotype.
Fig. 2. rix7-1 is complemented by YLL034c, which is genetically linked to RPL10. (A) rix7-1 is complemented by YLL034c. Growth of wild-type yeast (RIX7+) and the rix7-1 ts strain harbouring an empty plasmid (-), a plasmid with a 10 kb genomic insert containing YLL034c (pRIX7) or a plasmid containing only YLL034c. YPD plates were incubated for 4 days at 23 or 37°C. (B) Synthetic lethal relationship between rix7-1 and rpl10-2. Synthetic lethality was analysed by the ability to lose wild-type RPL10 present on a URA3-containing plasmid in the haploid double mutant rix7-1/rpl10-2 (see Table I). For comparison, the growth of haploid single mutants rix7-1and rpl10-2 is shown. Growth was analysed after 4 days at 23°C on 5-fluoroorotic acid (FOA)-containing plates (left panel) or SDC+All (minimal glucose medium plus all nutrients) plates.
Next, we wanted to know whether the RIX7 gene is functionally linked to other components involved in ribosomal export. Mutations in the ribosomal protein Rpl10p, a late nuclear assembling component of 60S ribosomal subunits, can give rise to a rix phenotype (Gadal et al., 2001). Rpl10p acts, at least in part, by recruiting Nmd3p to the 60S ribosomal subunit. Nmd3p, in turn, is an adapter for the exportin Xpo1p, which exports NES-containing proteins from the nucleus to the cytoplasm (Ho et al., 2000; Gadal et al., 2001). As shown in Figure 2B, the rix7-1 mutation is synthetically lethal with the rpl10-2 allele, indicating that Rix7p and Rpl10p also functionally interact.
Rix7p is a conserved nuclear member of the AAA ATPase family
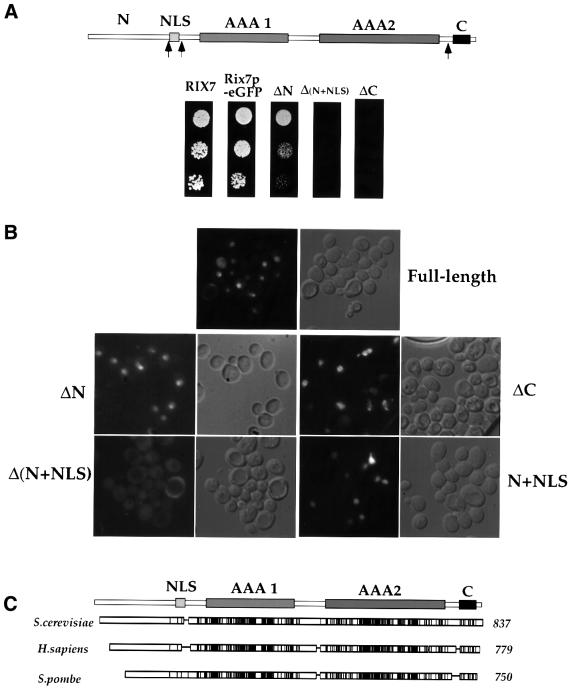
A search for conserved motifs in Rix7p revealed the presence of two AAA ATPase domains (residues 241–430 and 569–737; Neuwald et al., 1999) and a putative bipartite nuclear localization signal (NLS) in the N-terminal region (residues 175–195; Figure 3A, upper panel). To identify functionally important regions within Rix7p, a set of N- and C-terminal deletions was generated. A construct deleted of the N-terminal amino acids 1–174 (ΔN) was functional, whereas a longer N-terminal deletion of 1–202, including the putative NLS (ΔN+NLS), no longer supported growth (Figure 3A). A short C-terminal deletion of 776–837 (ΔC) also leads to loss of function (Figure 3A). The short C-terminal region of Rix7p that lies outside of the AAA domains is, therefore, essential for cell growth.
Fig. 3. Domain organization of Rix7p. (A) Upper panel: schematic presentation of the Rix7p sequence consisting of an N-domain (N), an NLS (residues 175–195), two consecutive AAA domains (residues 241–430 and 569–737) and a short C-terminal domain (residues 789–824). Arrows indicate the position of N- or C-terminal deletions. Lower panel: functional analysis of the Rix7p deletion constructs ΔN, Δ(N+NLS) and ΔC. The complementing activity of Rix7p–GFP, ΔN, ΔN+NLS and ΔC was tested in a RIX7 shuffle strain by plating cells on 5-FOA-containing plates (4 days at 23°C). (B) Intracellular location of Rix7p–GFP and the derived N- and C-terminal deletions. Strains expressing either Rix7p–eGFP or the deletion constructs ΔN, Δ(N+NLS), ΔC and N+NLS were analysed at 23°C during the exponential growth phase by fluorescence microscopy and Nomarski imaging. (C) Schematic presentation of the conserved amino acids within Rix7p homologues. Black or grey rectangles denote identical and homologous residues, respectively. NCB.GenPep accession Nos: S.cerevisiae Rix7p (S64785), and S.pombe (T39584) and Homo sapiens (U68140) homologues.
Fluorescence microscopy revealed that full-length Rix7p–GFP is nuclear in exponentially growing cells (Figure 3B). The functional Rix7pΔN–GFP shows a similar localization, but removal of the putative NLS (ΔN+NLS) causes a cytoplasmic location (Figure 3B). Furthermore, the corresponding (N+NLS)–GFP construct is nuclear (Figure 3B). Thus, Rix7p contains a single functional NLS in its N-terminal region (Figure 3A). However, a construct with the short C-terminal deletion (ΔC), although still nuclear, is not functional, demonstrating that this part of Rix7p plays a different essential role.
Rix7p shows high homology to all members of the AAA ATPase superfamily (data not shown). However, human NVL (Germain-Lee et al., 1997) and Schizo saccharomyces pombe SPBC16E9.10c may be the orthologues of Rix7p since additional conserved motifs can be identified in these two proteins, including the essential C-terminal domain, in addition to the two AAA domains (Figure 3C). No clear homology was detected within the non-essential N-terminal domain of Rix7p, but a putative NLS is present just before the first AAA domain of the human and S.pombe counterparts (data not shown).
Rix7p is involved in 60S ribosomal subunit biogenesis
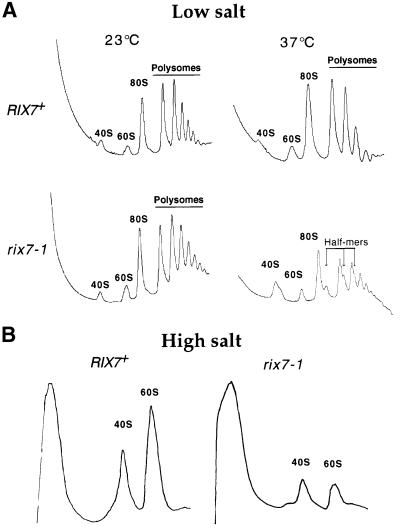
The nucleolar/nuclear accumulation of Rpl25p–eGFP observed in the rix7-1 ts mutant (see Figure 1B) and the localization of Rix7p (see Figure 3B) suggest a role of Rix7p in the biogenesis of large ribosomal subunits. Defects in 60S subunit biogenesis generally result in decreased free 60S subunits and the appearance of ‘half-mer’ polysomes (polysomes containing an unjoined 40S subunit at the initiation site). The sucrose density gradient profile of polysomes isolated from rix7-1 cells grown at the permissive temperature was similar to that of the isogenic wild type (Figure 4A). Following transfer of the rix7-1 strain to 37°C, the free 60S subunit peak was reduced, while the 40S peak increased (Figure 3A). The polysomal peaks were also reduced and half-mers were readily detected.
Fig. 4. rix7-1 is impaired in 60S subunit biogenesis. Polysomal profiles (OD260 nm) after sucrose density gradient centrifugation derived from the rix7-1 mutant and wild-type strain grown at permissive temperature (23°C) and 4 h after shift to restrictive temperature (37°C) under low salt (100 mM KCl) (A) and high salt (0.8 M KCl) conditions (B). The positions of 40S, 60S and 80S ribosomal particles, polysomes and ‘half-mers’ are indicated.
To observe the decrease in 60S ribosomal subunits in rix7-1 cells more directly, 40S and 60S subunits were dissociated with high salt (0.8 M KCl) before sucrose density gradient centrifugation (Figure 4B). The 60S:40S ratio measured by UV absorption is ∼2 in wild-type strains, but decreased to 0.85 in the rix7-1 strain after shift to 37°C (Figure 4B). This strong depletion of 60S subunits suggests that Rix7p functions in large subunit biogenesis.
Pre-rRNA processing is defective in rix7-1 mutant strains
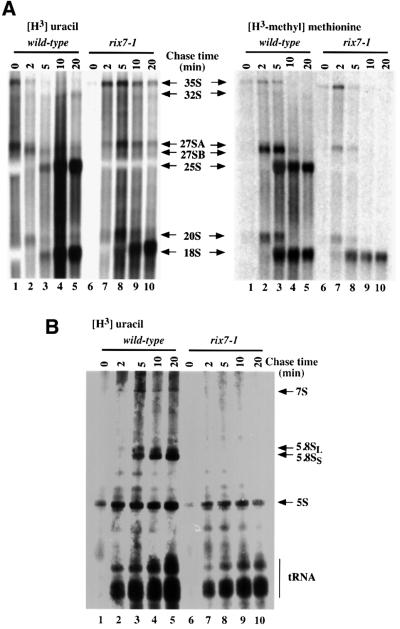
To test whether the depletion of 60S ribosomal subunits observed in the rix7-1 strain is a consequence of defects in pre-rRNA processing, this was analysed by northern hybridization (Figure 5B and C), primer extension (Figure 5D) and pulse–chase analysis (Figure 6). Following transfer of the rix7-1 strain to 37°C for 2 h, the precursors on the pathway of 25S and 5.8S rRNA synthesis, the 27SB, 7S and 6S pre-rRNAs, are very strongly reduced (Figure 5). By 4 h after transfer to 37°C, the mature 5.8S and 25S rRNAs are seen to be depleted, consistent with the reduced amount of 60S subunit in the rix7-1 strain at 37°C. In addition, the 35S pre-rRNA was accumulated, accompanied by some reduction in the levels of the 27SA2 and 20S pre-rRNAs. A low level of the 23S pre-rRNA, which is generated from the 35S by cleavage at site A3 in the absence of prior cleavage at sites A0, A1 and A2, was also detected. These phenotypes reveal a delay in the three early cleavages at sites A0, A1 and A2 in the rix7-1 strain.
Fig. 5. Northern analysis of pre-rRNA processing in rix7-1 cells. (A) Schematic diagram of the pre-rRNA processing pathway. The 35S pre-rRNA precursor contains the sequences for the mature 18S, 5.8S and 25S rRNAs separated by the two internal transcripted spacers, and flanked by the 5′-ETS and 3′-ETS. The locations of the known processing sites and rRNA precursor molecules are indicated. (B) Wild type (lanes 1 and 2) and rix7-1 (lanes 3–5) were analysed following growth at 25°C (0 h samples), and 2 and 6 h after transfer to 37°C. Pre-rRNA species of high molecular weight were detected as indicated, as are the oligonucleotides used. (C) Pre-rRNA species of low molecular weight from wild type (lanes 1–3) and rix7-1 (lanes 4–6) were analysed in the same way. (D) Primer extension analysis of wild-type (lanes 1 and 2) and rix7-1 (lanes 3–5) cells.
Fig. 6. Pulse–chase analysis of pre-rRNA processing in the rix7-1 strain. Wild-type (lanes 1 –5) and rix7-1 (lanes 6–10) strains were labelled with [3H]uracil for 1 min or with [3H]methyl methionine for 2 min, and then chased with unlabelled uracil or methionine for the times indicated. (A) High molecular weight RNA species. (B) Low molecular weight RNA species.
Primer extension analysis indicated that cleavage at sites A2 (Figure 5D), A0 and A3 (data not shown) continued. Primer extension stops at sites B1L and B1S were detected at low levels, but with no clear change in their relative efficiencies (Figure 5D). All cleavages were accurate at the nucleotide level. We conclude that processing of the pre-rRNA at sites A3, B1L and B1S continues in the rix7-1 strain, followed by rapid degradation of the 27SB pre-rRNAs. In wild-type cells, a low level of a pre-rRNA that extends from site A2 to site E (the 3′ end of the 5.8S rRNA) is observed (Figure 5C). In the rix7-1 mutant, this is lost and replaced by a species that extends from A2 to C2 (the 3′ end of the 7S pre-rRNA). We conclude that some pre-rRNA cleavage at site C2 also continues in the mutant strains. Together with the continued processing at B1, this indicates that some level of the 7S pre-rRNA continues to be synthesized and rapidly degraded. Notably, no intermediates on the pathway of 25S/5.8S synthesis were accumulated in the rix7-1 strains, making it unlikely that loss of the mature rRNAs is primarily due to a defect in pre-rRNA cleavage.
Pulse–chase labelling of the rix7-1 strain 2 h after transfer to 37°C, with either [3H]uracil or [3H]methyl methionine, showed a dramatic reduction in the synthesis of both the 25S and 5.8S rRNAs (Figure 6). The 35S pre-rRNA was synthesized and showed some retardation in its processing. The 27SA pre-rRNA was also synthesized with some delay in the rix7-1 strain, but the 27SB and 7S pre-rRNAs were not detected. Synthesis of the 20S pre-rRNA and 18S rRNA continued, with a delay consistent with that in 35S processing. Some reduction in the amount of 18S rRNA was seen, probably due to the reduced growth rate of the mutant. Synthesis of tRNAs and of the 5S rRNA continued in the rix7-1 strain, with a reduction in accumulation consistent with the reduced growth.
Together, these data indicate that strains defective in Rix7p are not primarily defective in pre-rRNA cleavage, but rapidly degrade the 27SB, and possibly 7S, pre-rRNA. This suggests that Rix7p is involved in a structural rearrangement required for correct assembly, and therefore stability, of the 60S ribosomal subunit. The kinetic delay in processing of the early sites on the pathway of 18S rRNA is probably indirect, since this has been seen for many other mutants defective in 60S synthesis (Kressler et al., 1999; Venema and Tollervey, 1999).
Rix7p associates with pre-ribosomes
The observed defects in 60S ribosomal biogenesis suggest that Rix7p functionally interacts with pre-ribosomes inside the nucleus. Three major ribosomal precursor particles are reported from yeast (Trapman et al., 1975). An early 90S particle, which contains the 35S pre-rRNA associated with many early assembling ribosomal proteins, is subsequently cleaved into 66S and 43S pre-ribosomes, the precursors to the mature 60S and 40S subunits, respectively. The 66S pre-ribosomal particle contains the 27S and 7S pre-rRNA species, whereas the 43S particle contains the 20S pre-rRNA. Protein complexes associated with the pre-ribosomal particles, the Noc1p–Noc2p and Noc2p– Noc3p complexes, have been shown to co-fractionate with the 35S and the 27S/7S pre-rRNA, respectively, under the extraction conditions used here (Milkereit et al., 2001).
The association of protein A (ProtA)-tagged Rix7p with pre-ribosomal particles was assessed by sucrose gradient centrifugation and western blotting (Figure 7). The ribosomal protein Rpl10p served as a marker for 60S subunits, 80S ribosomes, and polysomes, and the Noc3p protein as a marker for 66S pre-ribosomes (see also Milkereit et al., 2001). When whole-cell lysates were prepared under low salt conditions, most Rix7p was found on the top of the sucrose gradient. However, a pool of Rix7p–ProtA is also seen in denser fractions of the sucrose gradient, with a peak in a zone where 60S subunits are found (Figure 7A). A more careful analysis of the fractions around 60S subunits revealed that Rix7p–ProtA co-peaks with 60S subunits, but also tails into deeper fractions of the gradient, in which pre-ribosomal particles such as 66S and 90S particles can be found (Figure 7B). In contrast, when the yeast cell extract was treated with high salt prior to centrifugation, the pool of Rix7p–ProtA peaking around 60S subunits disappeared, but 60S and 40S subunits were readily detectable (data not shown). We conclude that under steady-state conditions, a fraction of Rix7p is associated with intranuclear precursor particles to 60S large subunits. Rix7p is strongly predicted to be an enzyme and we expect only transient interactions with the pre-ribosomal particles.
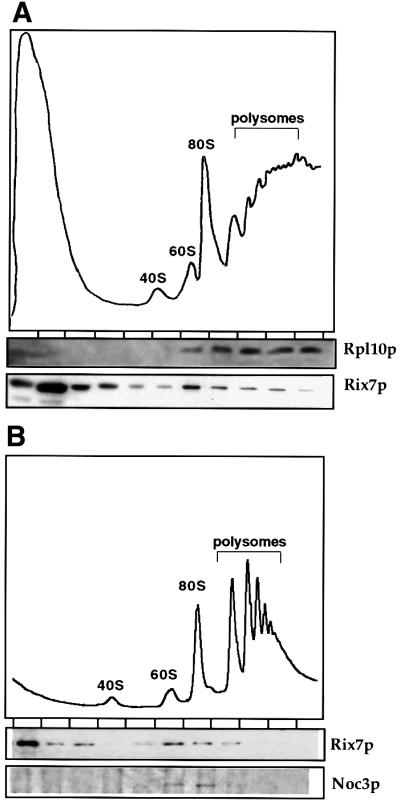
Fig. 7. A pool of Rix7p co-fractionates with pre-ribosomal particles to 60S large subunits. (A) Analysis of polysomal ribosome fractions derived from cells expressing Rix7p–ProtA. Fractions were analysed by SDS–PAGE and western blotting using α-Rpl10p and α-ProtA antibodies, respectively. (B) Comparison of Rix7p–ProtA and the 66S pre-ribosomal subunit marker Noc3p (Milkereit et al., 2001) on sucrose gradients. Fractions were analysed by SDS–PAGE and western blotting using α-Noc3p and α-ProtA antibodies, respectively.
Rix7p changes its intranuclear distribution depending on the growth condition
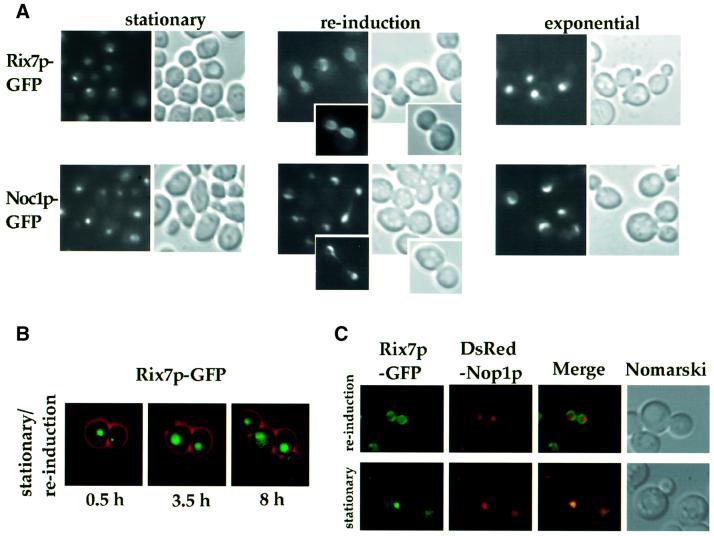
As shown above, Rix7p tagged with GFP and expressed from an ARS/CEN plasmid exhibits a generalized nuclear localization with no clear sub-nuclear concentration (see Figure 3B). We expressed Rix7p–GFP as a construct integrated at the RIX7 gene locus to avoid overexpression. When cells were grown to stationary phase (OD600 between 5 and 10), the Rix7p–GFP signal became weaker, but was concentrated in a distinct nuclear region, apparently corresponding to the nucleolus (Figure 8A, stationary; see below). When cells in stationary phase were transferred to fresh medium for 2 h, the Rix7p–GFP signal increased and exhibited a transient perinuclear location (Figure 8A, re-induction). After prolonged growth in fresh medium, Rix7p–GFP finally regained an overall nuclear distribution (Figure 8A, exponential). As a control we used a bona fide nucleolar protein tagged with GFP, Noc1p–GFP (Milkereit et al., 2001), which is found in a similar distinct nuclear spot (i.e. the nucleolus) in starved cells, but remains nucleolar and does not exhibit a perinuclear location upon re-induction of growth (Figure 8A, re-induction).
Fig. 8. Location of Rix7p–GFP in living cells grown under different growth conditions. (A) The Rix7–GFP (genomic integration) strain was grown at 23°C to stationary phase (OD600 = 5), growth re-induced in fresh YPD medium for 3 h at 23°C, and cells grown further to exponential phase (OD600 = 0.5). The GFP signal was monitored by fluorescence microscopy. Cells were also visualized by Nomarski imaging. The nucleolar protein Noc1p–GFP (Milkereit et al., 2001), which does not exhibit a transient perinuclear location during growth induction, served as a control. (B) Time-lapse microscopy of Rix7p–GFP from stationary cells and after re-induction in fresh YPD medium. (C) Rix7–GFP cells also expressing DsRed–Nop1p (pUN100-DsRed-NOP1) were analysed by fluorescence microscopy under stationary phase conditions and after growth in fresh YPD medium. The Rix7p–GFP and DsRed–Nop1p images were merged to detect co-localization.
This change in the intranuclear localization of Rix7p points to a dynamic localization under different growth conditions. This was also followed by time-lapse fluorescence microscopy. Similar to the above observations, Rix7p–GFP was concentrated in a distinct area of the nucleus in starved cells (Figure 8B, 0.5 h). After re-induction of growth through transfer to fresh medium (Figure 8B, 3.5 h), the nucleolar concentration of Rix7p–GFP changed to a distribution throughout the entire nucleoplasm (Figure 8B, 3.5 and 8 h). No specific changes in the pattern of Rix7p–GFP distribution were seen during mitosis.
To show that Rix7p–GFP concentrates in the nucleolus at stationary phase, it was co-expressed with the nucleolar marker DsRed–Nop1p (Gadal et al., 2001). The green fluorescent signal of Rix7p–GFP and the red fluorescent signal of DsRed–Nop1p largely overlapped in starved cells (Figure 8C, stationary), indicating that Rix7p–GFP indeed concentrates in the nucleolus. The peripheral nuclear location of Rix7p–GFP seen after re-induction of growth is not due to an extended nucleolus. Under these conditions, the nucleolus remains distinct and small, and does not overlap significantly with the Rix7p–GFP signal (Figure 8C, re-induction).
We conclude from these studies that Rix7p can shuttle between the nucleolus, nucleoplasm and nuclear periphery. Alterations in the distribution of Rix7p may correlate with changes in ribosome biogenesis and transport in response to nutrient availability.
Discussion
Here, we report that a member of the superfamily of AAA ATPases (Rix7p) associates with precursor particles to 60S ribosomal subunits, and plays a role in large subunit assembly and subsequent export to the cytoplasm.
While many members of the ‘DEAD-box’ family of ATPases (helicases) have been implicated in ribosome assembly, this is the first report of a role for an AAA-type ATPase in ribosome biogenesis. AAA proteins are found in eukaryotes, prokaryotes and Archaea, and each organism has many different AAA proteins, which function in diverse cellular processes. The genome of S.cerevisiae encodes ∼50 different AAA ATPases. The unifying feature of the AAA superfamily is an ATPase domain of 220 amino acids, whose structural fold has been solved recently (for reviews see Vale, 2000; Zhang et al., 2000). While the biological functions of the various members of this superfamily seem unrelated, a common mechanism is emerging from structural studies. AAA proteins assemble into oligomers, in most cases forming hexameric rings. A nucleotide-dependent conformational switch applies tension to bound substrates, allowing AAA proteins to unfold or dissociate attached proteins (Vale, 2000). We have no direct evidence for the oligomerization of Rix7p, but rix7-1 exhibits a semi-dominant phenotype consistent with the incorporation of both intact and mutated subunits into an oligomeric complex. Similar observations have been made for NSF, ClpA and regulatory subunits of the proteasome (Vale, 2000).
It seems reasonable to assume that activities that can disassemble protein complexes will be required during the various stages of ribosome synthesis and during transport of pre-ribosomes from the nucleolus to the cytoplasm. A vast number of trans-acting factors participate in ribosome synthesis. More than 50 protein factors and >100 snoRNAs are predicted to associate with and subsequently dissociate from each pre-rRNA molecule during its intranuclear lifespan. Moreover, pre-rRNA processing in yeast is very fast, implying a rapid binding and release of many factors. This presumably entails a considerable free energy change and is, therefore, likely to require a substantial input of energy. This need may, in part, be met by ATP hydrolysis, and Rix7p could be required for these steps. Growing yeast cells have enough copies of the U3 snoRNP to support ribosome synthesis for only ∼1 min in the absence of recycling (at 2000 ribosomes/min), and other pre-rRNA processing factors appear to have roughly comparable abundances, underlining the potential importance of such an activity.
The very large Balbiani ring pre-mRNAs have been extensively studied in Chironomus tentans. For these, substantial structural rearrangement can be seen to precede nuclear export, accompanied by the loss of some but not all hnRNP proteins (Sun et al., 1998). It appears very probable that the export of ribosomal subunits will involve energy-dependent structural rearrangements. Close homologues of Rix7p exist in eukaryotes, but not in Archaea and bacteria, suggesting a function in ribosome synthesis that is conserved only among eukaryotes. Preparation of the ribosome subunits for nuclear transport could be such an activity. We have reported previously that a late structural rearrangement appears to convert salt-labile pre-60S particles to the mature subunits (Tollervey et al., 1993). Rix7p would be an obvious candidate to participate in this rearrangement.
The pre-rRNA processing phenotype observed in the rix7-1 mutant strain is typical of defects seen in many other mutants that are defective in 60S subunit biogenesis. In almost all cases, the early pre-rRNA cleavages at sites A0–A2 show at least some inhibition, even when there is no clear or direct relationship between the mutant product and these cleavages (see also Milkereit et al., 2001; reviewed in Kressler et al., 1999; Venema and Tollervey, 1999). Recycling of processing and assembly factors may be inhibited in rix7-1 cells by either a direct defect in disassembly or the nucleoplasmic accumulation of inappropriately structured 60S ribosome precursors. In either case, depletion of the pool of free factors available is envisaged to inhibit the assembly and processing of newly synthesized pre-rRNAs.
In summary, we have shown that a conserved member of the AAA-type ATPase superfamily, Rix7p, is involved in 60S subunit biogenesis. Rix7p is associated with precursor particles of the 60S subunits and apparently accompanies them from the nucleolus to the nucleoplasm. Our data suggest a model in which Rix7p acts to restructure the 60S subunits within the nucleus prior to their transport through the NPCs. In the absence of this restructuring, the subunits may not be competent for export, and are retained and rapidly degraded in the nucleus. The lack of Rix7p activity may also slow recycling of other processing factors, potentially explaining the delay in the early pre-rRNA processing steps seen in the mutant strains.
Materials and methods
Yeast strains, DNA recombinant work, and microbiological techniques
Yeast strains used in this study are listed in Table I. Microbiological techniques, plasmid transformation and recovery, mating, sporulation of diploids and tetrad analysis were carried out essentially as described (Santos-Rosa et al., 1998). DNA recombinant work was performed according to Maniatis et al. (1982).
Table I. Yeast strains.
| Name | Genotype | Origin |
|---|---|---|
| RS453a | MATa, ade2, leu2, ura3, his3, trp1 | Hurt et al. (1999) |
| FY23 | MATa, ura3, trp1, leu2 | derived from S288C |
| FY86 | MATa, ura3, his3, leu2 | derived from S288C |
| rix7-1 original | MATa, ura3, his3, leu2, rix7-1 | isolated from ts collection (Amberg et al., 1992) |
| rix7-1 a | MATa, ura3, his3, leu2, trp1, rix7-1 | offspring of rix7-1 original × FY86 |
| rix7-1 α | MATα, ura3, his3, leu2, trp1, rix7-1 | offspring of rix7-1 original × FY86 |
| RIX7 Shuffle | MATa, leu2, ura3, his3, met15, lys2, rix7::KANMX4 +pRS316-RIX7(ARS/CEN, RPL25, URA3) | derived from Euroscarf strain Y21522 |
| DEHQ1-1 | MATα, ade2, his3, trp1, leu2, ura3, can1, qsr1Δ1::HIS3 +pHFF22 (ARS/CEN, URA3, QSR1) | Eisinger et al. (1997) |
| RPL10Δ rix7-1 | MATα, ade2, his3, trp1, leu2, ura3, qsr1Δ1::HIS3, rix7-1 +pHFF22 (ARS/CEN, URA3, QSR1) | offspring of rix7-1 a × DEHQ1-1 |
| RIX7–eGFP | MATα, leu2, ura3, his3, met15, lys2, rix7::KANMX4 +pRS315-RIX7-eGFP(ARS/CEN, LEU2, Rix7-eGFP) | derived from RIX7 Shuffle |
| RIX7–GFP | MATa, ura3, trp1, his3, leu2, RIX7-GFP::KANMX4 | offspring of FY23 × FY86 |
| RIX7–ProtA | MATa, ura3, trp1, his3, leu2, RIX7-ProtA::TRP1 | offspring of FY23 × FY86 |
Plasmid constructions
Plasmids pUN100-DsRed-Nop1, pRS316-Rpl25-eGFP, pNOPPA1L, pNOPGFP1L and pFA6a-GFP(S65T)-KANMX6 were described previously (Hellmuth et al., 1998; Longtine et al., 1998; Gadal et al., 2001). pFA6a-(2*ProtA-TEV)-TRP1 was derived from pFA6a-GFP(S65T)-TRP1 by cloning a PCR-generated fragment coding for 2×ProtA-TEV from pNOPPA1L in place of the GFP coding sequence.
RIX7 including its 5′-UTR was amplified by PCR from yeast genomic DNA. The derived fragment was cut with SacI–BamHI and fused in-frame to the eGFP variant present in pRS315-RPL25-eGFP, generating pRS315-RIX7-eGFP. The pRS315-rix7-1-eGFP plasmid was constructed in the same way, but using yeast genomic DNA extracted from the rix7-1 strain.
Three PCR-generated fragments containing the 5′-UTR of RIX7 and different C-terminal deletions of the RIX7 coding sequence were generated from yeast genomic DNA: N (corresponding to residues 1–174), N+NLS (corresponding to residues 1–202) and ΔC (corresponding to residues 1–776). These three PCR-generated fragments were cut with SacI–BamHI and fused in-frame to the eGFP variant present in pRS315-RPL25-eGFP. N-terminal deletions of RIX7 were amplified in two steps. The RIX7 5′-UTR was amplified by PCR from yeast genomic DNA, generating an NdeI site at the ATG codon of RIX7. This fragment was then cloned as a SacI–BamHI fragment into pRS315-RPL25-eGFP to generate pRS315-RIX7::eGFP. Two fragments corresponding to the C-terminal part of RIX7 coding sequence were generated by PCR: ΔN (residues 174–837) and ΔN+NLS (residues 202–837). These fragments were cut with NdeI–BamHI and cloned into pRS315- RIX7::eGFP. These deletion constructs were expressed under the control of the RIX7 promoter using the ATG start codon at the NdeI site.
Strain constructions
Genomic integration of GFP in-frame with RIX7 was obtained as described previously (Longtine et al., 1998). Genomic integration of ProtA was carried out in the same way, but using the pFA6a-(2*ProtA-TEV)-TRP1 vector.
Cloning of RIX7/YLL034c
A yeast genomic library in a LEU2-containing ARS/CEN plasmid (Gautier et al., 1997) was transformed into the rix7-1 strain. From colonies growing at the restrictive temperature (37°C) was isolated plasmid pRIX7 with a genomic insert. The complementing plasmid contained the YLL034c gene. pYLL034c harbouring only the RIX7 gene was cloned and shown to complement the ts growth defect of the rix7-1 mutant. pRS316-RIX7 was generated by subcloning of YLL034c into pRS316.
Pulse–chase and northern analysis of rRNA
Pulse–chase labelling of rRNA and analysis of rRNA processing by northern hybridization were performed as described (Tollervey, 1987; Tollervey et al., 1993).
Oligonucleotides used were: 003: TGTTACCTCTGGGCCC; 004: CGGTTTTAATTGTCCTA; 007: CTCCGCTTATTGATATGC; 008: CATGGCTTAATCTTTGAGAC; 013: GGCCAGCAATTTCAAGTTA; 017: GCGTTGTTCATCGATGC; 020: TGAGAAGGAAATGACGCT; 219: GAAGCGCCATCTAGATG; 5′ A0L: GGTCTCTCTGCTGCCGG.
Fluorescence microscopy
pRS315-RPL25-eGFP or pRS316-RPL25-eGFP was introduced into yeast cells by transformation, and selected on SDC-leu medium or SDC-ura medium, respectively. Individual transformants were grown in liquid SDC-leu medium at 23°C to an OD600 nm of ∼1, before shift to 37°C in liquid YPD medium. After centrifugation, cells were resuspended in water, mounted on a slide and viewed in the fluorescence microscope. In vivo, the GFP signal was examined in the fluorescent channel; the DsRed used in fusion with Nop1p was examined in the rhodamine channel of a Zeiss Axioskop fluorescence microscope and pictures were obtained with a Xillix Microimager CCD camera. Digital pictures were processed by the software program Improvision (Open lab) and Photoshop 4.0.1 (Adobe).
Time-lapse microscopy
Time-lapse microscopy was performed using the RIX7–GFP strain. Exponential growing cells (interval: 120 s; exposure: 0.2 s; transmission: 25%; Z-Planes: 3; Optovar: 1×; objective: 100× 1.3 PH3 oil) or stationary culture (interval: 180 s; exposure: 0.3 s; transmission: 50%; Z-Planes: 3; Optovar: 1×; objective: 100× 1.3 PH3 oil) were followed by confocal microscopy.
Miscellaneous
The search for functional domains was performed using the pfscan package (www.isrec.isb-sib.ch/software/PFSCAN). SDS–PAGE and western blot analysis were performed according to Siniossoglou et al. (1996), and isolation of ribosomes under low and high salt conditions by sucrose gradient centrifugation as described in Tollervey et al. (1993). Whole-cell lysates and fractions from the sucrose gradient were separated by SDS–PAGE and analysed by western blotting using the indicated antibodies. Polyclonal antiserum against Rpl10p was a kind gift of Dr Trumpower (Hanover, NH). Affinity-purified α-Noc3p antibodies were kindly obtained from Dr Tschochner (BZH, Heidelberg, Germany).
Acknowledgments
Acknowledgements
We are grateful to Dr Trumpower for providing antibodies against ribosomal proteins. We thank Dr K.Sträßer for critical reading of the manuscript. E.H. was a recipient of grants from the Deutsche Forschungs gemeinschaft (Schwerpunktprogramm ‘Funktionelle Architektur des Zellkerns’), E.P. and D.T. are funded by the Wellcome Trust, and O.G. is a holder of a HFSP fellowship.
References
- Amberg D.C., Goldstein,A.L. and Cole,C.N. (1992) Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev., 6, 1173–1189. [DOI] [PubMed] [Google Scholar]
- Dechampesme A.M., Koroleva,O., Leger-Silvestre,I., Gas,N. and Camier,S. (1999) Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J. Cell Biol., 145, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V. and Hurt,E.C. (1997) From nucleoporins to nuclear pore complexes. Curr. Opin. Cell Biol., 9, 401–411. [DOI] [PubMed] [Google Scholar]
- Eisinger D.P., Dick,F.A. and Trumpower,B.L. (1997) Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol. Cell. Biol., 17, 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauβ,D., Kessl,J., Trumpower,B., Tollervey,D. and Hurt,E. (2001) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a NES-containing factor Nmd3p that associates with the large subunit protein Rpl10p. Mol. Cell. Biol., 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier T., Bergès,T., Tollervey,D. and Hurt,E. (1997) Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol., 17, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain-Lee E.L., Obie.,C. and Valle,D. (1997) NVL: a new member of the AAA family of ATPases localized to the nucleus. Genomics, 44, 22–34. [DOI] [PubMed] [Google Scholar]
- Hellmuth K., Lau,D.M., Bischoff,F.R., Künzler,M., Hurt,E.C. and Simos,G. (1998) Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol., 18, 6374–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H., Kallstrom,G. and Johnson,A.W. (2000) Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol., 151, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E., Hannus,S., Schmelzl,B., Lau,D., Tollervey,D. and Simos,G. (1999) A novel in vivo assay reveals inhibition of ribosomal nuclear export in Ran-cycle and nucleoporin mutants. J. Cell Biol., 144, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Zhao,Y. and Tartakoff,A.M. (1992) A conditional yeast mutant deficient in mRNA transport from nucleus to cytoplasm. Proc. Natl Acad. Sci. USA, 89, 2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Linder,P. and De La Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A., Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch,E.T. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Milkereit P. et al. (2001) Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell, 105, 499–509. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind,L., Spouge,J.L. and Koonin,E.V. (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and dissassembly of proteins complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- Rouiller I., Butel,V.M., Latterich,M., Milligan,R.A. and Wilson-Kubalek,E.M. (2000) A major conformational change in p97 AAA ATPase upon ATP binding. Mol. Cell, 6, 1485–1490. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Aitchison,J.D., Suprapto,A., Hjertaas,K., Zhao,Y. and Chait,B.T. (2000). The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol., 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno,H., Simos,G., Segref,A., Fahrenkrog,B., Panté,N. and Hurt,E. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol., 18, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Azad,A.K. and Hopper,A.K. (1999) Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 96, 14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma,K., Doye,V., Hellwig,A., Huber,J., Lührmann,R. and Hurt,E.C. (1997) Mex67p which is an essential factor for nuclear mRNA export binds to both poly(A)+ RNA and nuclear pores. EMBO J., 16, 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer,C., Rieger,M., Doye,V., Tekotte,H., Weise,C., Emig,S., Segref,A. and Hurt,E.C. (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell, 84, 265–275. [DOI] [PubMed] [Google Scholar]
- Stade K., Ford,C.S., Guthrie,C. and Weis,K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T., Schmidt,U. and Silver,P.A. (2000) Factors affecting nucler export of the 60S subunits in vivo. Mol. Biol. Cell, 11, 3777–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Alzhanova-Ericsson,A.T., Visa,N., Aissouni,Y., Zhao,J. and Daneholt,B. (1998) The hrp23 protein in the Balbiani ring pre-mRNP particles is released just before or at the binding of the particles to the nuclear pore complex. J. Cell Biol., 142, 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. (1987) A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J., 6, 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen,H., Jansen,R.P., Kern,H. and Hurt,E.C. (1993) Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation and ribosome assembly. Cell, 72, 443–457. [DOI] [PubMed] [Google Scholar]
- Trapman J., Retel,J. and Planta,R.J. (1975) Ribosomal precursor particles from yeast. Exp. Cell Res., 90, 95–104. [DOI] [PubMed] [Google Scholar]
- Vale R.D. (2000) AAA proteins: lords of the ring. J. Cell Biol., 150, 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beekvelt C.A., Kooi,E.A., de Graaff-Vincent,M., Riet J., Venema,J. and Raue,H.A. (2000) Domain III of Saccharomyces cerevisiae 25 S ribosomal RNA: its role in binding of ribosomal protein L25 and 60 S subunit formation. J. Mol. Biol., 296, 7–17. [DOI] [PubMed] [Google Scholar]
- Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- Woolford J.L. Jr (1991) The structure and biogenesis of yeast ribosomes. Adv. Genet., 29, 63–118. [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. (2000) Structure of the AAA ATPase p97. Mol. Cell, 6, 1473–1484. [DOI] [PubMed] [Google Scholar]