Abstract
Meiotic homologous pairing is crucial to proper homologous recombination, which secures subsequent reductional chromosome segregation. We have identified a novel meiosis-specific protein of fission yeast Schizosaccharomyces pombe, Meu13p, to be a molecule that is required for proper homologous pairing and recombination. Rec12p (homologue of Saccharomyces cerevisiae Spo11p), which is essential for the initiation of meiotic recombination, is also shown for the first time to participate in the pairing process of S.pombe. Meu13p, however, contributes to pairing through a recombination-independent mechanism, as disruption of the meu13+ gene reduces pairing whether the rec12+ gene is deleted or not. We also demonstrate a dynamic nature of homologous pairing in living meiotic cells, which is markedly affected by meu13 deletion. Meu13p is not required for telomere clustering and the nuclear movement process, which are well known requirements for efficient pairing in S.pombe. Based on these results, together with the localization of Meu13p on meiotic chromatin, we propose that Meu13p directly promotes proper homologous pairing and recombination.
Keywords: chromosome dynamics/Hop2/synapsis/telomere/TBPIP
Introduction
Meiosis is a process that produces genetically recombined gametes and thus increases genetic diversity. Genetic recombination occurs during the meiotic prophase when homologous chromosomes become paired and form crossovers and non-crossovers (Paques and Haber, 1999). Some of the crossovers persist until metaphase I as physical links called chiasmata, and these direct chromosomal segregation (reviewed in Hawley, 1988). Defects in pairing lead to defective recombination, which in turn impairs the faithful segregation of homologous chromosomes.
While pairing is being established, homologous chromo somes undergo a complex series of reorganization events that increase their compaction (Scherthan et al., 1992), change their orientation in the nucleus (reviewed in Hiraoka, 1998), and resolve spatial and topological problems among and within themselves (von Wettstein et al., 1984; Kleckner and Weiner, 1993; Zickler and Kleckner, 1998). In many eukaryotes, pairing culminates in synapsis, wherein homologous chromosomes are physically connected along their entire lengths by a proteinaceous structure called the synaptonemal complex (SC) (reviewed in von Wettstein et al., 1984). However, observations of presynaptic alignment and the presence of a significant level of homologous pairing in asynaptic organisms suggest that SC formation is involved in maintaining rather than initiating pairing (reviewed in Roeder, 1997). Chromosomes also interact with each other via DNA-strand exchange during the meiotic homologous recombination process. Again, these interactions are not prerequisites for pairing; rather, they seem to contribute to the maintenance of pairing. Supporting this is that numerous yeast mutants deficient in homologous recombination do not completely lack pairing, although the amount of pairing is decreased and synapsis is defective in these mutants (Loidl et al., 1994; Weiner and Kleckner, 1994; Nag et al., 1995). In Caenorhabditis elegans and Drosophila melanogaster, recombination is also not required for pairing to occur, nor for synapsis, as shown by spo11 mutants that are incapable of initiating homologous recombination (Dernburg et al., 1998; McKim and Hayashi-Hagihara, 1998; McKim et al., 1998). However, whether homologous recombination is not a prerequisite for pairing in all organisms is at present unclear, as spo11 mutants of Saccharomyces cerevisiae, Coprinus cinereus, Arabidopsis thaliana and Mus musculus do show defects in homologous pairing and/or synapsis (Weiner and Kleckner, 1994; Baudat et al., 2000; Celerin et al., 2000; Kneitz et al., 2000; Romanienko and Camerini-Otero, 2000; Grelon et al., 2001). Schizosaccharomyces pombe, an asynaptic yeast, is proposed to utilize homologous recombination instead of SC to maintain pairing (Walker and Hawley, 2000), although supportive evidence of this notion is lacking to date.
In S.pombe, meiosis is easily induced by simply removing the nitrogen source from the culture medium, whereupon h+/– diploid cells enter directly into meiosis (azygotic meiosis) and haploid cells of the opposite mating type, h+ and h–, conjugate and then enter meiosis (zygotic meiosis) (Egel, 1989). After meiosis has been induced, the telomeres cluster at the spindle pole body (SPB), after which the nucleus begins to oscillate between the cell poles and quickly attains an elongated shape called the ‘horse-tail’. The nuclear movement continues until shortly before meiotic division (Chikashige et al., 1994, 1997; Hiraoka et al., 2000). The horse-tail period includes the meiotic prophase that is defined in higher eukaryotes (Robinow, 1977). Analysis of mutants defective in sister chromatid cohesion, telomere clustering or nuclear movement suggest that these events are necessary for proper homologous pairing and/or recombination (Molnar et al., 1995; Shimanuki et al., 1997; Cooper et al., 1998; Nimmo et al., 1998; Yamamoto et al., 1999; Niwa et al., 2000). Here we describe a meiosis-specific protein, Meu13p, which facilitates homologous pairing and recombination, and whose identification sheds light on the requirements that allow these processes to occur efficiently.
Results
Isolation and characterization of meu13+
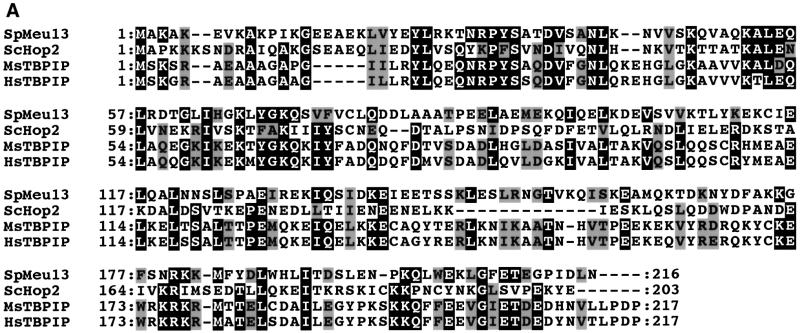
We isolated the meu13+ cDNA clone during the screening of a subtracted cDNA library that was constructed to identify meiosis-specific genes in S.pombe (meu: meiotic expression upregulated). Cloning and sequencing of the corresponding meu13+ genomic clone revealed that it spans five exons and four introns bearing canonical splicing sequences. The predicted Meu13 protein is 217 amino acids long. As shown in Figure 1A, it displays significant sequence similarity to proteins from budding yeast (Hop2p) (Leu et al., 1998), mice (TBPIP; Tanaka et al., 1997) and humans (TBPIP; Ijichi et al., 2000), as amino acid sequence identity among these proteins reaches 40% in their N-termini.
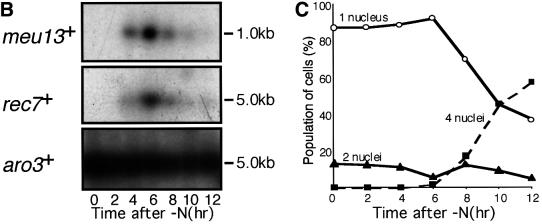
Fig. 1. Sequence comparison of Meu13p and meiosis-specific expression of meu13+. (A) The meu13+ gene encodes a 25 kDa protein similar to Hop2p of S.cerevisiae and TBPIP of mice and humans. Identical amino acids are highlighted in black. Grey boxes indicate the similar amino acids. (B) meu13+ expression as analysed by northern blotting. Total RNA was extracted from h+/– diploids (CD16-5) at the indicated times after meiosis was induced by nitrogen starvation. The RNA was blotted and probed with cDNA of meu13+, rec7+ or aro3+. The last one was used as a loading control (Nakanishi and Yamamoto, 1984). (C) The meiotic profile of the diploids used for RNA extraction. The population of cells stained with Hoechst 33342 that have one nucleus (before nuclear division), two nuclei [after a trace of mitotic nuclear division (0–6 h) or after meiosis I (after 8 h)] and four nuclei (after meiosis II) are shown.
The meu13+ gene is specifically expressed in the early phase of meiosis
To analyse the expression of the meu13+ gene in detail, we performed northern hybridization using a meu13+ cDNA probe on total RNA extracted from diploid cells that were synchronously undergoing meiosis. As shown in Figure 1B, the expression of meu13+ began to increase after meiosis was induced by nitrogen starvation and it peaked 6 h after meiotic induction. The expression of meu13+ was not detected in vegetatively growing cells (Figure 1B, 0 h time point), nor in G1-arrested cells (data not shown). Considering that the second meiotic division occurred 2 h after the peak of meu13+ expression (Figure 1C), meu13+ transcripts thus appear to be expressed specifically in the early meiotic phase. The expression pattern of meu13+ consistently resembled that of the rec7+ gene (Figure 1B, middle panel), which is transiently expressed early in meiosis and is believed to be required only in the early steps of meiotic recombination (Lin et al., 1992).
Meu13p localizes on chromosomes and forms foci during the mid- to late-horse-tail period
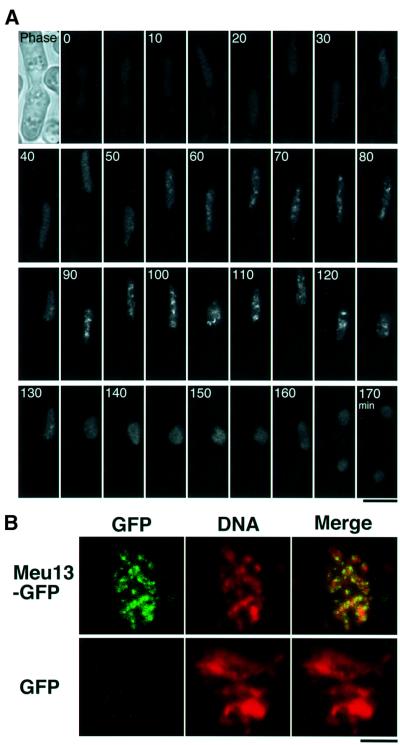
To investigate the expression of the Meu13 protein and its cellular localization, the gene that encodes the green fluorescing Meu13–GFP (green fluorescent protein) fusion protein was integrated into the S.pombe genome to replace the chromosomal meu13+ gene. Thus, this fusion gene is under control of the native promoter and the timing of its expression is expected to be the same as that of the native meu13+ gene. Indeed, its expression pattern is quite similar to that observed by northern blotting analysis (see below). In addition, the normal spore formation in the Meu13–GFP integrant strain suggests that Meu13–GFP is fully functional (data not shown). After meiosis was induced, a faint nuclear GFP signal appeared at about the time karyogamy occurs, after which the GFP signal intensity increased during the horse-tail period. Thus, GFP signal foci were seen from 68 ± 11 to 164 ± 19 min after karyogamy (in eight independent observations), and disappeared after the nucleus stopped moving at the end of the horse-tail period. Prior to the onset of meiotic nuclear division, the GFP signal faded. During meiosis I a faint nuclear GFP signal was seen (Figure 2A). These signals are specific to Meu13p and not derived from GFP alone or background fluorescence because this pattern of GFP fluorescence was not detected when GFP alone was expressed (data not shown). We also studied the localization of Meu13–GFP on surface-spread chromatin (Figure 2B) and observed GFP foci on the chromatin of the zygote expressing Meu13–GFP, whereas no GFP signal was seen in the control strain that expressed GFP alone. These observations strongly suggest that Meu13p functions on chromosomes during the mid- to late-horse-tail period.
Fig. 2. Meu13–GFP foci during the mid- to late-horse-tail period and localization on surface-spread chromatin. (A) Time-lapse observation of Meu13–GFP in living zygote. Observations were made at 5 min intervals. Projections of sectioned images are shown. The number in each panel is the time in minutes after the first image. Phase, the phase contrast image of a zygote. The scale bar represents 5 µm. (B) Meu13–GFP signals on surface-spread chromatin. Zygotes expressing Meu13–GFP or GFP alone were spread on the surface of a glass slide and observed under a fluorescent microscope. DNA, chromatin stained with Hoechst 33342. GFP, GFP signals that were only detected for zygotes expressing Meu13–GFP. Merge, Meu13–GFP (green) clearly seen on a spread chromatin (red). The scale bar represents 5 µm.
Meu13p is required for efficient crossover recombination
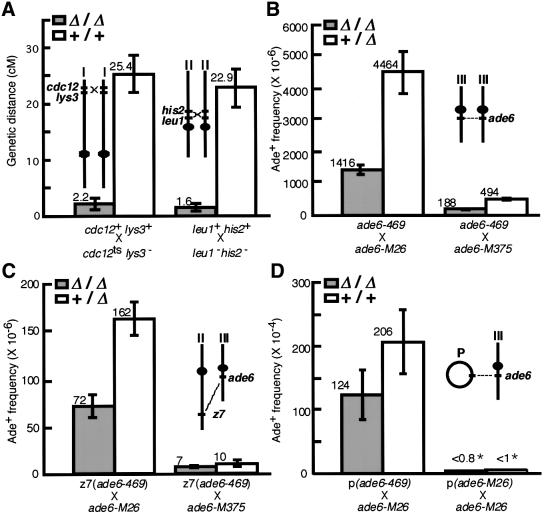
To investigate the loss-of-function phenotype of Meu13p, a null mutant (Δmeu13) was created. The results above prompted us to examine meiotic recombination activity in this strain because it is one of the major chromosomal events occurring during the mid-to-late horse-tail period. We first investigated crossover recombination of zygotic meiosis by tetrad analysis, which allowed us to measure the genetic distances between two pairs of loci (Figure 3A): one near the end of chromosome I (cdc12-lys3) and the other near the centromere of chromosome II (leu1-his2). In crosses between Δmeu13 strains (Δ/Δ), the genetic distance between cdc12 and lys3, and between leu1 and his2, respectively, decreased to 8.6 and 7.0% of the distance in crosses between wild-type strains (+/+). This phenotype is recessive as no significant decrease in genetic distance was observed in crosses between Δmeu13 and wild-type strains (data not shown). Thus, Meu13p is required for efficient crossover recombination in meiosis.
Fig. 3. Meiotic recombination is decreased in Δmeu13. (A) Genetic distance was measured during zygotic meiosis. The chromosomal positions of the loci and centromeres are sketched in insets. Crosses indicated as Δ/Δ and +/+ correspond to the cross between Δmeu13 strains (NP32-2C and -2D) and between wild-type strains (NP32-2A and -2B), respectively. Of these two cross types, 210 and 292 tetrads were analysed, respectively, of which 157 and 162 tetrads could each produce four viable spores (spore viability is 90.1 and 55.4%, respectively). Only the tetrads that could generate four viable spores were analysed to calculate genetic distance. Error bars indicate the standard errors. (B–D) The average frequencies of Ade+ recombinant spores generated by intragenic recombination in four independent experiments are shown with standard errors. The loci locations used in each recombination assay are shown in each inset. (B) Allelic intragenic recombination between ade6 alleles in a cross of Δmeu13 ade6-469 (NP57-100) with Δmeu13 ade6-M26 (NP55-26) or Δmeu13 ade6-M375 (NP54-62), or in a cross of Δmeu13 ade6-469 (NP57-100) with ade6-M26 (GP-1540) or ade6-M375 (GP936). (C) Ectopic intragenic recombination between ade6 alleles in a cross of Δmeu13 ade6-469 (NP57-100) with Δmeu13 ade6-M26 (NP55-26) or Δmeu13 ade6-M375 (NP54-62), or in a cross of Δmeu13 ade6-469 (NP57-100) with ade6-M26 (GP-1540) or ade6-M375 (GP936). The level of recombination between M375 and 469 was too low (∼10–6) to be analysed for statistical significance. (D) Ectopic intragenic recombination between ade6 alleles on a high-copy-number plasmid and a chromosomal locus occurring in the self-conjugated homothallic strain Δmeu13 ade6-M26 (NP36-24A) or ade6-M26 (NP36-39D) bearing plasmids carrying the ade6-469 gene or ade6-M26 gene. *p >0.99.
Meu13p is required for efficient intragenic recombination
To further characterize the meiotic recombination in Δmeu13 cells, we next examined allelic intragenic recombination between two different mutant alleles of ade6. We used three ade6 mutant alleles, namely, M26, M375 and 469. Of these, M26 displays hot spot activity, i.e. when crossed with other alleles the frequency of Ade+ recombinants increases 10- to 15-fold compared with the other mutants (Gutz, 1971). In crosses between Δmeu13 strains (Δ/Δ, Figure 3B), the frequency of Ade+ recombinant spores decreased to 31% in M26 × 469 crosses and to 38% in M375 × 469 crosses relative to the frequencies in crosses of Δmeu13 to the wild type (Δ/+, Figure 3B).
We also examined ectopic intragenic recombination in two different situations, namely, chromosome-by-chromosome and plasmid-by-chromosome recombination. For the first, we used a pair of ade6 alleles that were located at different chromosomal loci, one at the natural position of ade6 on chromosome III and the other at the pac1 locus on chromosome II (z7, Figure 3C) (Virgin and Bailey, 1998). The frequency of Ade+ recombinant spores between these two ectopic loci decreased to 44% (in the M26 and 469 pair) in a cross between Δmeu13 strains compared with a cross of Δmeu13 with the wild type (Figure 3C). This decrease was smaller than that for allelic recombination but still statistically significant (t-test; P <0.006). Thus, Meu13p function appears to be required for both ectopic and allelic intragenic recombination.
To examine chromosome-by-plasmid recombination, we used the M26 allele at the natural position of ade6 on chromosome III together with the 469 allele on a high-copy-number plasmid. We employed the same assay used in the screening for recombination-deficient rec– mutants (Ponticelli and Smith, 1989). Although the average frequency of Ade+ recombinant spores from Δmeu13 decreased to 59% of that from the wild type (Figure 3D, left), this decrease was statistically less significant (t-test, P <0.06) and smaller than that for chromosome-by-chromosome recombination. It may be that spontaneous reversion is increased in Δmeu13, which would have the effect of reversing the decrease in recombination. However, we found that the level of spontaneous reversion was negligible (Figure 3D, right). Thus, Δmeu13 is not as defective in plasmid-by-chromosome recombination as it is in chromosome-by-chromosome recombination. This suggests that Meu13p is not involved in a process that is generally required for recombination but rather that it participates in a process that facilitates homologous recombination, particularly between chromosomal loci.
Homologous pairing is decreased in Δmeu13
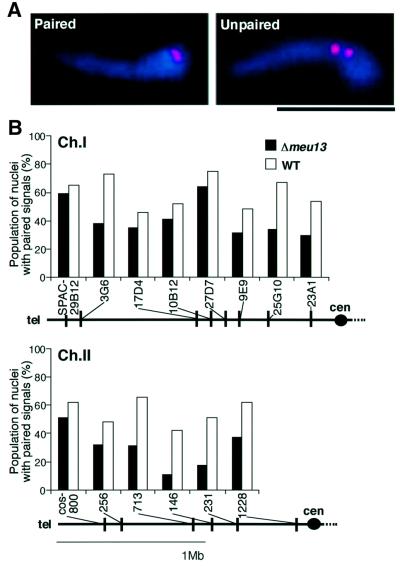
The pairing between homologous chromosomal loci is believed to be a prerequisite for efficient homologous recombination and therefore we examined the homologous pairing in Δmeu13 cells. We performed fluorescent in situ hybridization (FISH) analysis on a random sample of horse-tail nuclei from asynchronous meiotic cells with 14 probes mapping to the arm region of chromosomes. A single signal in a nucleus was interpreted as indicating paired homologous loci, while two distinct signals were assumed to be unpaired homologous loci (Figure 4A). These assumptions should be valid because the nuclei with unpaired homologous loci that were observed as a single signal were estimated to comprise 2–3% of the total horse-tail nuclei, which is much less frequent than the nuclei displaying a single signal (see Materials and methods). We found that, in general, the population of nuclei with paired signals in Δmeu13 was lower than that in the wild type for any of the loci tested (Figure 4B).
Fig. 4. Homologous pairing is decreased in Δmeu13. (A) Typical merged images of the chromosomal loci probed with a Cy3-labelled cos146 probe (red) in FISH and DNA stained with Hoechst 33342 (blue) in a zygote; the loci are paired (left) or unpaired (right). The scale bar represents 5 µm. (B) The population (%) of the nuclei that have paired FISH signals in a random sample of 80–100 horse-tail nuclei from asynchronous zygotes is shown as a histogram. Solid boxes, Δmeu13 cells (NP40-1B); open boxes, wild-type cells (NP40-1C). The chromosomal positions of the observed loci are sketched under the histograms. A black dot indicates the position of the centromere (cen).
In S.pombe, sister chromatid cohesion, telomere clustering and the subsequent nuclear movement correlate with the efficient homologous pairing (Molnar et al., 1995; Yamamoto et al., 1999; Niwa et al., 2000). Sister chromatid cohesion seemed to be normal in Δmeu13 as no obvious difference was detected between wild type and Δmeu13 in a population of cells with split FISH signals. Moreover, we did not detect any difference between Δmeu13 and wild type in telomere clustering and nuclear movement. Telomeres and SPB were visualized by simultaneous FISH and indirect immunofluorescence, in which all the Δmeu13 cells showed clustered telomeres at SPB (Figure 5A). Nuclear movement in a living cell was investigated with an ectopically expressed nuclear GFP marker (TB19; Ding et al., 2000). Movement of the nucleus of Δmeu13 was indistinguishable from that of wild type (Figure 5B). Thus, Meu13p appears to be required for the efficient pairing of homologous loci not through these well characterized events.
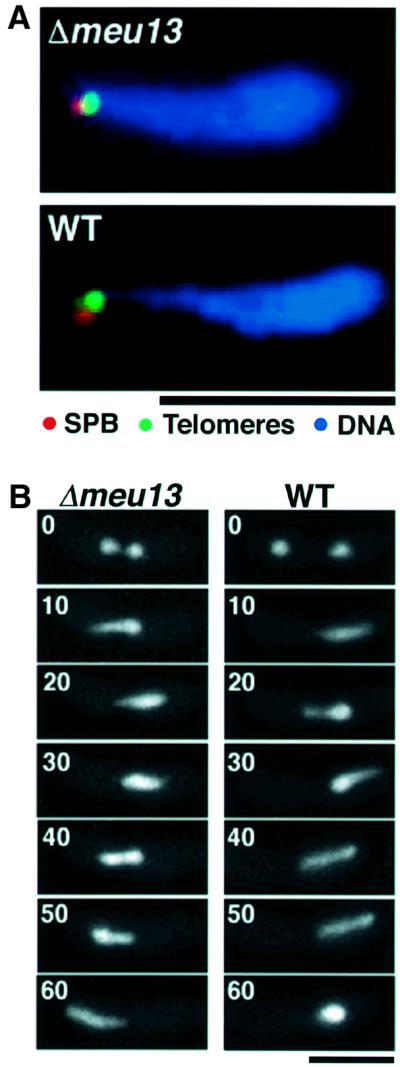
Fig. 5. Intact telomere clustering and nuclear movement in Δmeu13 cells. (A) Merged images of fluorescent immunolocalization and in situ hybridization detecting the spindle pole body (SPB, red), telomeric regions probed by cos212 (green) and DNA stained with Hoechst 33342 (blue) in representative horse-tail nuclei are shown. In both Δmeu13 (NP40-1B, upper panel) and wild-type (NP40-1C, lower panel) zygotes, telomeres clustered and associated with the SPB. The scale bar represents 5 µm. (B) Time-lapsed images of nuclear GFP marker (TB19) in living Δmeu13 (NP40-1B, left column) and wild-type (NP40-1C, right column) zygotes. In the first panels two nuclei before karyogamy are shown, and the number in each panel is the time in minutes after the first panel. The scale bar represents 10 µm.
Homologous pairing is also decreased in Δrec12 but less severely than in Δmeu13
The reduced level of homologous pairing in Δmeu13 suggests that Meu13p directly participates in the pairing process. However, it could also merely be a consequence of a decrease in recombination as homologous recombination may contribute to pairing by searching for homology, or by producing physical links between chromosomal DNA. We therefore investigated whether recombination participates in the pairing in S.pombe by using the rec12 deletion mutant, which completely lacks meiotic recombination (De Veaux et al., 1992). The gene product of S.pombe rec12+ is homologous to the S.cerevisiae Spo11p, which introduces double strand breaks (DSBs) during an initial step in meiotic recombination (Keeney et al., 1997). Rec12p is also required for DSB formation during S.pombe meiosis (Cervantes et al., 2000).
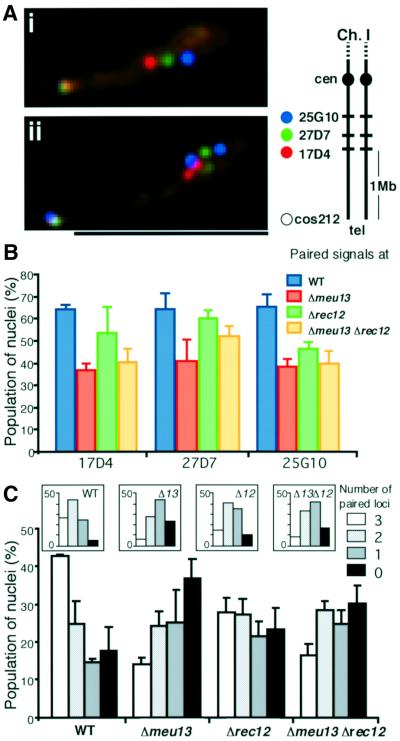
Homologous pairing was investigated by multi-locus FISH, in which three loci on the same chromosomal arm were simultaneously visualized with FISH, and the pairing of these loci scored (Figure 6A). In wild-type cells, the population of nuclei with paired signals was ∼65% of the randomly sampled horse-tail nuclei for each locus, whereas it was 55% in Δrec12 and 40% in Δmeu13 cells (Figure 6B). Thus, homologous pairing is not abolished but it is decreased when recombination is eliminated in S.pombe, although the decrease was smaller than that seen in Δmeu13. Sister chromatid cohesion seemed to be normal, and telomere clustering and nuclear movement occurred properly in Δrec12 (data not shown), indicating that Rec12p is not necessary for these events, as was also observed for Meu13p.
Fig. 6. Homologous pairing is reduced but is still higher in recombin ation-deficient mutants than in Δmeu13. (A) Typical samples of merged images of the hybridization signals of the SPAC-17D4 (red), -27D7 (green), -2510 (blue) and cos212 (greenish white) probes in a multi-locus FISH assay and Hoechst 33342 staining of DNA (orange). (i) All three loci probed are paired as only a single dot of each colour is seen. (ii) All three loci are unpaired as two dots for each colour are seen. A schematic representation of the cosmid positions used as probes is shown in an inset. The scale bar represents 5 µm. (B) The average population (%) of nuclei with paired signals in random samples of horse-tail nuclei, shown with standard deviations as error bars. Three to four independent FISH experiments, each of which includes a scoring of 80–100 nuclei, were performed for each strain. (C) The average population (%) of the samples classified according to the number of paired loci (0–3). Standard deviations as error bars are indicated. The inset shows the distribution that is expected when each locus pairs independently from other loci.
Relative to the wild type, the difference in the pairing of each locus in the Δrec12 and Δmeu13 mutants was at most 25%. However, the combination of paired loci appeared to differ markedly among these strains. As shown in Figure 6C, the population of horse-tail nuclei with paired or unpaired signals for all three loci was greater than would be expected if each locus behaved independently (Figure 6C, insets). This bias toward all-or-none pairing indicates that the neighbouring chromosomal loci have a tendency to associate or dissociate concurrently. Due to this tendency, the population of the nuclei showing no pairing in Δmeu13 was ∼2-fold greater than that observed in the wild type (Figure 6C).
While pairing was reduced in Δrec12, the reduction is less severe than that observed for Δmeu13. We found that pairing could be decreased further by deleting meu13+ in the Δrec12 mutant (Figure 6B). Moreover, as demonstrated by time-lapse observation, the initial increase in pairing was affected in Δmeu13 but not in Δrec12 (see below). These results clearly demonstrate that the decrease in pairing in Δmeu13 is not merely a consequence of the decrease in recombination, which in turn strongly suggests that Meu13p directly participates in the pairing process.
Pairing occurs in a dynamic fashion
To further investigate the requirement of Meu13p and Rec12p in the pairing process, we observed homologous chromosomal regions in living zygotes over time. A tandem repeat of the Escherichia coli lac operator sequence was integrated into the S.pombe genome at the cut3+ locus, which is located at the centre of the short arm of chromosome II. The fusion gene encoding GFP–LacI-NLS, which binds to the lac operator, was then expressed in this strain. This allowed us to visualize two homologous loci on the chromosomal arm regions by GFP fluorescence. In the wild-type background we found that these homologous loci repeatedly associated and dissociated in the horse-tail nucleus that was oscillating back and forth between the cell poles (Figure 7A). Strains with the Δmeu13 or Δrec12 genetic background also showed such dynamic pairing (data not shown), but the paired GFP signals were less frequent (see below).
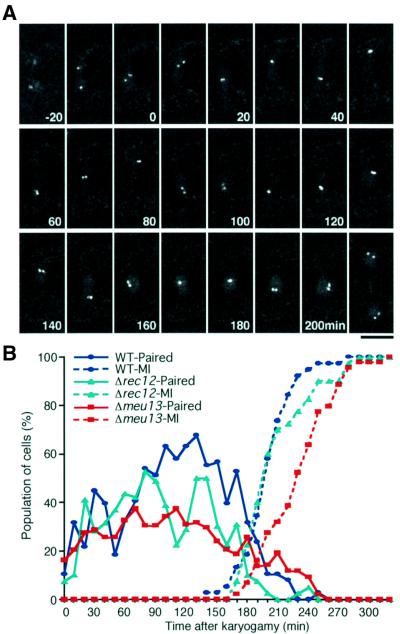
Fig. 7. Homologous pairing is a dynamic process. (A) Time-lapse observation, taken at 10 min intervals, of a single wild-type zygote expressing GFP–LacI, which is located in the nucleus (diffuse signal) and binds to lacO target sequences in the arm region of chromosome II (cut3 locus, bright dots). Projections of sectioned images are assembled in a time course. The number in each panel indicates the time in minutes after the first image. Before nuclear fusion, two separated GFP dots are seen (from –20 to 0 min). After nuclear fusion (occurring between –10 and 0 min), the GFP dots move back and forth between the zygote poles that are at the top and the bottom of each panel: this oscillatory motion reflects horse-tail nuclear movement. Repeated association (e.g. at 30 min) and dissociation of GFP dots (e.g. at 70 min) occurs. During the period between cessation of the horse-tail nuclear movement and commencement of meiosis I (from 160 to 200 min), the GFP dots stay at the centre of the zygote. After the onset of meiosis I (between 200 and 210 min), a pair of GFP dots can be seen in each half of the zygote. This indicates separation of sister chromatid arms at meiosis I. (B) The changes of the population of cells that have paired GFP signals, as measured by multiple live observations. Approximately forty cells were observed independently for each strain. The blue line indicates the wild-type pairing profile. The population of cells with paired signals is low (∼30%) for the initial 60 min (time 0 set to the time of karyogamy). From 70 to 160 min, it increases and remains at a high level (∼60%). There is a similar initial increase in Δrec12 (green line), whereas no such increase is observed for Δmeu13 (red line). The dotted line represents the accumulation of cells that passed the first meiotic division; this indicates that meiotic progression occurs highly synchronously between individual cells of the same strain.
In order to quantify the pairing activity over time, we scored the occurrence of paired signals every 10 min from karyogamy to the first meiotic nuclear division in ∼40 live individuals of each strain. As shown in Figure 7B, the duration from karyogamy to the first meiotic division was nearly constant among the individual cells of the same strain. This enabled us to examine the pairing in a group of cells that were undergoing meiotic progression relatively synchronously. For Δmeu13 and Δrec12 mutant cells, meiosis I was defined to occur when more than two chromatin masses had emerged, because unequal partitioning of the chromatin mass was frequently observed in these strains (data not shown). In the wild-type strain, during the initial 70 min the population of cells with paired signals was ∼30%, after which they increased to ∼60%. This frequency was maintained until 170 min, after which it decreased prior to the onset of meiosis I (Figure 7B, blue line). In Δmeu13, there was no significant difference from the wild type in the initial 70 min but the subsequent increase in pairing observed in the wild type was completely absent (Figure 7B, red line). In Δrec12, the initial increase in pairing also occurred similarly to the wild type (Figure 7B, green line). In the middle of the meiotic prophase, transient decrease of pairing was observed (90–120 min), and in later phase the level of pairing was intermediate between those of wild type and Δmeu13. This difference in pairing dynamics between Δmeu13 and Δrec12 is consistent with the multi-locus FISH results that demonstrated that Meu13p promotes homologous pairing independently of Rec12p activity.
Discussion
We report here the isolation and characterization of the S.pombe meiosis-specific gene, meu13+, whose product is localized on chromosomes at the meiotic prophase, and facilitates homologous recombination and pairing. Meu13p defines a novel class of factors that promote homologous pairing because Meu13p promotes pairing independently of recombination and any other already known requirements for homologous pairing.
Homology to Hop2 and TBPIP
Schizosaccharomyces pombe Meu13p, S.cerevisiae Hop2p, and mouse and human TBPIP display a high level of conservation in their amino acid sequences. Hop2p is meiosis specific and localizes on meiotic chromosomes (Leu et al., 1998). The mouse TBPIP gene is highly expressed in the testis, and its product is most abundantly detected in the nuclei of primary and secondary spermatocytes (Tanaka et al., 1997); furthermore, its expression is not detected in premature testes, nor in testes that are defective in forming sperm (K.Nabeshima and H.Nojima, unpublished). Human TBPIP is also expressed in the testis (Ijichi et al., 2000). These observations suggest that these proteins may share a conserved molecular function in meiosis.
There are, however, important differences in the meiotic phenotype between the meu13 and hop2 null mutants. First, Δhop2 arrests at the pachytene stage whereas Δmeu13 completes meiosis. Secondly, the frequency of ectopic recombination is increased in Δhop2 but decreased in Δmeu13. In S.cerevisiae, the hop2 mutant arrests by the ‘pachytene checkpoint’ (Leu and Roeder, 1999), which causes arrest at the pachytene stage in a group of mutants that are defective in recombination and/or SC formation (reviewed in Roeder, 1997). In S.pombe, none of the meiotic recombination-deficient mutants identified so far shows a meiotic arrest phenotype. Thus, S.pombe presumably lacks an arrest mechanism in response to a deficiency in recombination. This feature enabled us to examine the ectopic recombination of Δmeu13 spores, while in Δhop2 it could be examined only in diploid cells obtained from return-to-growth experiments. Thus, the discrepancy in the ectopic recombination phenotype between Δmeu13 and Δhop2 could either be due to differences in the regulation of meiotic progression or it could simply reflect differences between the assay methods used.
Another major difference between the meu13 and hop2 null mutants is that S.cerevisiae Δhop2 displays inappropriate SC formation between heterologous chromosomes (Leu and Roeder, 1999). Heterologous synapsis cannot be detected in Δmeu13 because S.pombe does not form an SC (Olson et al., 1978; Hirata and Tanaka, 1982; Bähler et al., 1993) and thus this phenotypic difference does not necessarily mean that there is functional difference between Hop2p and Meu13p. In fact, Leu et al. (1998) pointed out the possibility that Hop2p may have a role in the pairing process that affects subsequent SC formation.
Dynamic nature of homologous pairing
The observation of the pairing dynamics with the GFP reporter construct provides the first direct evidence for unstable homologous pairing in the meiotic prophase, as has been suggested by Scherthan et al. (1994) and proposed in a ‘kissing’ model by Kleckner and Weiner (1993). In the meiotic prophase of other eukaryotes, homologous association may also be unstable but this may be difficult to measure in synaptic organisms because unstable homologous associations would only last for a limited period of time before SC terminate them. In this respect, S.pombe, which is an asynaptic organism, has an advantage in studying the nature of these unstable associations.
The observation of pairing in living cells also enabled us to assess pairing in a precise time course. This approach appears to be useful to know when a gene product works for pairing. Meu13p appears to be required for increasing pairing at the beginning of the pairing process, whereas Rec12p seems not to be required for it. We observed a transient drop in pairing in Δrec12 in the middle of prophase (90–120 min after karyogamy). This drop is not a fluctuation but probably meaningful because the sample size is large enough (∼40 independent samples) and the drop is seen over four contiguous timepoints. This result suggests that Rec12p is required for pairing only in a short period of time in the middle of prophase, which is consistent with the result that Δrec12 shows only a minor pairing defect in multi-locus FISH experiments. Rec12p may have a role in maintaining pairing probably through a recombination process.
In S.pombe, the association of homologous chromosomes seems to rely primarily on the chance contact between homologous loci, which appears to be facilitated by the oscillatory movement of telomere-bundled chromosomes as previously proposed (Hiraoka, 1998; Yamamoto et al., 1999). It does not follow, however, that homologous chromosomes become associated unidirectionally from the telomeres. Instead, the association of homologous chromosomes occurs at multiple interstitial sites, as was shown by the variety of paired and unpaired loci combinations in the multi-locus FISH assay (data not shown). A similar pattern of pairing at multiple sites along chromosomes has also been observed in other organisms (reviewed in Roeder, 1997). However, the pairing is not completely random, as neighbouring loci have a tendency to pair concurrently. This is probably due to spatial constraints within the chromosome.
Homologous pairing and recombination
Our results reported here clearly demonstrate that maximizing homologous pairing in S.pombe requires Meu13p and Rec12p. However, that the homologous pairing is reduced but still significant in the Δrec12 mutant indicates that homologous recombination is dispensable for the initiation of homologous pairing. Furthermore, Meu13p promotes pairing in a Δrec12 mutant as indicated in the disruption of the meu13 gene in the Δrec12 mutant. These findings suggest a mechanism that promotes homologous pairing independently of recombination. Meu13p is certainly involved in such a mechanism.
In addition to homologous pairing, Meu13p may also take part in homologous recombination. If homologous pairing is perturbed, it is expected that the chance of contact between ectopic loci increases, which leads to ectopic recombination when these loci have homologous sequences. An increase in ectopic recombination in meiosis is indeed observed for some of the mutants with a pairing defect of S.cerevisiae (Goldman and Lichten, 2000) and S.pombe (Niwa et al., 2000). In the Δmeu13 mutant, despite its perturbed pairing, ectopic recombination is not increased but rather decreased. Therefore, Meu13p may be directly involved in a part of homologous recombination.
Localization of Meu13p on spread nuclei strongly suggests that Meu13p acts directly on chromatin to exert its function. The linear arrangement of nuclear foci of Meu13p resembles a shape of linear element (LE), which is probably equivalent to the unsynapsed axial cores found in other eukaryotes and proposed to provide structural support for homologous interaction (Bähler et al., 1993). It should be interesting to investigate whether Meu13p is a component of LE. We favour the possibility that Meu13p may help establish a fundamental chromosomal structure that serves as a basis of meiotic chromosomal events including homologous pairing and recombination. Further understanding of the molecular function of Meu13p should provide new insights into the mechanisms that underlie meiotic homologous pairing and recombination.
Materials and methods
Strains, media and genetic manipulations
Complete medium YPD or YE containing 75 mg/l adenine sulfate, minimal medium EMM2, sporulation medium ME or EMM2-N, and germination medium YEade or EMMG was used (Alfa et al., 1993). Standard S.pombe genetic techniques (Gutz et al., 1974; Moreno et al., 1991) were employed throughout. Strains used are listed in Table I.
Isolation of meu13+
An S.pombe cDNA library was constructed from meiosis-specific mRNA by subtracting the mRNA of G1-arrested diploid cells from the mRNA of synchronous meiotic diploid cells (Watanabe et al., 2001). A total of 400 randomly selected clones were examined for meiosis specificity by northern blot analysis using total RNA extracted from a diploid strain (CD16-5) undergoing synchronous meiosis. Using the clone k508, which contains a sequence involved in early meiosis, as a probe, a genomic DNA clone was isolated by hybridization screening of an S.pombe genomic DNA library (H.Nojima, unpublished). The 4.2 kb genomic insert was completely sequenced and recorded in the DDBJ/EMBL/GenBank database (accession No. AB017038).
Gene disruption and GFP tagging of meu13+
The S.pombe ura4+ gene or the S.cerevisiae LEU2 gene was inserted between two MunI sites (at positions –11 and +913), which replaced the entire meu13+ open reading frame (ORF) except for the last six amino acids. Subsequently, the fragment between two HindIII sites (at original positions –1210 and +2030) was used to replace the genomic copy of meu13+ in a diploid strain by homologous recombination (as determined by Southern blotting). A fragment between two HindIII sites (at original positions –1210 and +2030) containing additional GFP-coding sequences and the nmt1 poly-adenylation signal sequence at the 3′ end of the meu13+ ORF was used to replace the disrupted meu13 genomic copy in the meu13 deletion mutant NP40-1B (as verified by Southern blotting). The Meu13–GFP replacements (NP70) were selected with 5-fluoroorotic acid.
Surface spreading of chromatin
Chromatin spreading was performed according to Bähler et al. (1993). Late-log phase cultures of NP70 and NP117-1 (in which only the GFP-coding sequence was integrated) in EMM2 liquid medium were transferred onto EMM2-N agar plates and incubated for 10 h, then collected and subjected to chromatin spreading.
Plasmid construction
For the assay of plasmid-by-chromosome recombination, the PvuII–SmaI fragment containing an ade6 mutant gene, obtained from the plasmids pade6-M26 (for pKN92) or -469 (for pKN94) (Szankasi et al., 1988), was inserted into a plasmid containing the S.cerevisiae LEU2 gene and 2µDNA. For the integration of the E.coli lacO repeat into the cut3+ locus, a derivative of pMK2A (Nabeshima et al., 1998) that contains the lacO repeat from the plasmid pAFS59 (Straight et al., 1997) and the carboxyl half of the cut3+ gene from pYS335 that rescues the cut3-477 mutant upon integration into the cut3 locus (Saka et al., 1994) were used.
Determination of the frequency of meiotic recombination events
Crossover recombination. Parental strains were mated for 2 days at 26°C. Ascospores were dissected with a micro-manipulator (MS series200; Singer instrument, UK) and germinated at 26°C for 4 days. The resultant colonies were replicated to examine their auxotrophy at 26°C or their temperature sensitivity at 36°C. Genetic distance was calculated according to the formula 50(TT+6NPD)/(PD+TT+NPD) (Perkins, 1949), where TT, NPD and PD indicate the number of tetratypes, non-parental ditypes and parental ditypes, respectively.
Intragenic recombination. Strains were grown in YE with uracil and crossed. Spores were treated with 0.5% glusulase (NEN Life Science Products, Inc.) for 16 h at 36°C and checked microscopically for complete digestion of contaminating vegetative cells in the spore suspension. Spores were plated on EMMGA containing supplements and germinated at 30°C for 4–5 days. The frequency of Ade+ spores was calculated to be the number of colonies on plates without adenine divided by that on plates with adenine.
Plasmid-by-chromosome recombination. The procedure in Ponticelli and Smith (1989) was followed with minor modifications. The homothallic strain carrying the ade6-M26 mutation and the pKN92 plasmid (or the pKN94 plasmid, which was used as a control) were grown in EMM2 with adenine. Before meiotic induction, a small portion was plated on EMM2 containing supplements to determine the frequency of Ade+ cells (fcell) or leu– cells (k). Cells were mated at 28°C for 3–4 days and spores were treated as described above. The frequency of Ade+ spores yielded by meiosis was calculated according to the formula (fspore – fcell)/(1 – k2), where fspore is the frequency of recombinant Ade+ spores.
Indirect immunofluorescence and FISH
Simultaneous FISH to telomeres and indirect immunofluorescence to a component of SPB (Sad1) were carried out as described in Chikashige et al. (1997). FISH was carried out as described in Yamamoto et al. (1999) with some modifications: cells were fixed in EMM2-N with 3.7% formaldehyde and 1% glutaraldehyde for 15 min. Cell wall digestion was performed in PEMS containing 0.5 mg of Zymolyase-100T and 0.1 mg/ml Novozyme-234 for 1 h at 36°C. Quenching was performed twice with PEM containing 0.1 M glycine for 20 min. RNase treatment was performed with PEMBL containing 0.1 mg/ml RNase A for 6–8 h at 36°C. The cosmid cos212 was used to probe the ends of chromosomes I and II (Funabiki et al., 1993). Cosmids SPAC-29B12, 3G6, 17D4, 10B12, 27D7, 9E9, 25G10 and 23A1 (from the Sanger Centre, Cambridge, UK) and cos800, 256, 713, 146, 231 and 1228 (kindly donated by Dr Yanagida, Kyoto Univerity, Japan) from the ordered cosmid libraries (Hoheisel et al., 1993; Mizukami et al., 1993) were used to probe the arm regions. Cosmid probes were labelled with Cy3-dUTP (Amersham Pharmacia Biotech) using terminal transferase reactions (Boehringer Mannheim). In multi-locus FISH, SPAC17D4, 27D7 and 25G10 were labelled with Cy3-dUTP, DIG-dUTP (Boehringer Mannheim) and Cy5-dUTP (Amersham Pharmacia Biotech), respectively, while cos212 was independently labelled with each of these dyes. A FITC-conjugated anti-DIG antibody (Boehringer Mannheim) was used to detect DIG-labelled probes. Multicolour images were obtained on a computerized CCD microscope using Resolve3D software on a Silicon Graphics workstation (Hiraoka et al., 1991). The three-dimensional (3D)-sectioning images (15 sections at 0.2 µm focus intervals) obtained for each sample were processed to remove out-of-focus information and analysed using SoftWolks™ software (Applied Precision).
If two dots were <0.3 µm apart, they were considered as a single signal, because the diameter of each signal is >0.3 µm. A single signal can contain coincidentally associated signals of unpaired loci. The probability of this should be similar to the association between loci on heterologous chromosomes that are mapped to similar physical distances from their telomeres, if the telomeres are clustered. The frequency of loci association was probed with SPAC27D7 (on chromosome I) and cos256 (on chromosome II), both of which map to ∼1 Mb from their telomeres, and was found to be 2% in wild-type cells and 3% in Δmeu13 cells. These frequencies were regarded to be equivalent to the frequency of false pairing. All values of pairing shown in this work contain this false pairing.
Construction of cells with a GFP-marked chromosome
NP35, which carries the integrated GFP–LacI-NLS gene at the his7 locus and the lacO array at the cut3 locus (verified by Southern blotting), was used to generate the prototroph strains NP43-20 (wild type), NP44-58 (Δmeu13) and NP204-26 (Δrec12), which were used in live observations.
Live observation of GFP fusion-expressing cells
Cells incubated on an EMM2-N plate at 26°C for 8–12 h were suspended in EMM2-N medium and mounted on a glass-bottomed culture dish for observation under a computerized microscope as described (Ding et al., 1998). During the observation of the GFP-marked chromosome, 3D time-lapse images were taken with a 0.3 s exposure at 10 min intervals, with 10 optical sections made at 0.3 µm intervals for each time point. For observation of nuclear movement, Δmeu13 (NP40-1B) or wild type (NP40-1C) was transformed with TB19–GFP expression plasmid (Ding et al., 2000). For Meu13–GFP observation, images were taken with a 0.3 s exposure at 5 min intervals, with six optical sections made at 0.5 µm intervals for each time point. Projected images obtained with DeltaVision™ software were analysed.
Table I. Strains used in this study.
| Strain | Genotype |
|---|---|
| CD16–1a | h+/h– ade6-M210/ade6-M216 cyh1+/cyh1 lys5+/lys5-391 |
| GP1594b | h+ ade6-469 ura4-D18 leu1-32 |
| GP936b | h– ade6-M375 ura4-D18 leu1-32 |
| GP1540b | h– ade6-M26 ura4-D18 leu1-32 |
| GP1123b | h+ ade6-D1 ura4-D18 leu1-32 z7::(ade6-469-ura4+) |
| NP1-6A | h– ade6-M210 leu1-32 ura4-D18 |
| NP1-6B | h– ade6-M210 leu1-32 ura4-D18 meu13::ura4+ |
| NP1-6C | h+ ade6-M216 leu1-32 ura4-D18 his2 meu13::ura4+ |
| NP1-6D | h+ ade6-M216 leu1-32 ura4-D18 his2 |
| NP32-2A | h+ ura4-D18 leu1-32 his2 |
| NP32-2B | h– ura4-D18 cdc12 lys3 |
| NP32-2C | h+ ura4-D18 leu1-32 his2 meu13::ura4+ |
| NP32-2D | h– ura4-D18 cdc12 lys3 meu13::ura4+ |
| NP35 | h+ ade6-M210 ura4-D18 leu1-32 cut3-477::cut3+::(lacOr) his7::(GFP-LacI-NLS) |
| NP36-39D | h90 ade6-M26 ura4-D18 leu1-32 |
| NP36-24A | h90 ade6-M26 ura4-D18 leu1-32 meu13::ura4+ |
| NP40-1B | h90 ade6-M216 ura4-D18 leu1-32 meu13::ura4+ |
| NP40-1C | h90 ade6-M216 ura4-D18 leu1-32 |
| NP43-20 | h90 his7::(GFP-LacI-NLS) cut3-477::cut3+::(lacOr) |
| NP44-58 | h90 his7::(GFP-LacI-NLS) cut3-477::cut3+::(lacOr) ura4-D18 meu13::ura4+ |
| NP53 | h90 leu1-32 ade6-L52 rec12-152::LEU2 |
| NP54-1 | h90 leu1-32 ura4-D18 ade6-L52 meu13::ura4+ rec12–152::LEU2 |
| NP54-62 | h– ade6-M375 ura4-D18 leu1-32 meu13::LEU2 |
| NP55-26 | h– ade6-M26 ura4-D18 leu1-32 meu13::LEU2 |
| NP56-46 | h+ ade6-D1 ura4-D18 leu1-32 z7::(ade6-469-ura4+) meu13::LEU2 |
| NP57-100 | h+ ade6-469 ura4-D18 leu1-32 meu13::LEU2 |
| NP70 | h90 ade6-M216 ura4-D18 leu1-32 meu13::meu13-GFP |
| NP117-1 | h90 ade6-M216 ura4-D18 leu1-32 meu13::GFP |
| NP204-26 | h90 his7::(GFP-LacI-NLS) cut3-477::cut3+::(lacOr) leu1-32 rec12::LEU2 |
Provided by aChikashi Shimoda and bGerald Smith.
Acknowledgments
Acknowledgements
We would like to thank Drs Mitsuhiro Yanagida, Chikashi Shimoda, Ayumu Yamamoto, Kenichi Mizuno, Kunihumi Ohta, Yoshinori Watanabe and Gerry Smith for providing strains and plasmids; Dr Mitsuhiro Yanagida and the Sanger Centre for providing cosmid DNA; Drs Yoshimi Tanaka, Ayumu Yamamoto, Da-Qiao Ding, Yuji Chikashige, Monica Molnar, Jürg Kohli, Peter Munz, Jeffery Virgin and Gerry Smith for technical suggestions and critical discussion; and Rumi Kurokawa for technical assistance with FISH. We are also grateful to Drs David Alexander and Hubert Renauld for critically reading the manuscript. This work was supported by a grant from the Ministry of Education, Science and Culture of Japan to H.N. and K.N. (scientific research on priority areas and promoted scientific research A, respectively), grants from the Osaka Cancer Society, The Naito Foundation and The Uehara Foundation to H.N., and a grant from the Human Frontier Science Program and Japan Science and Technology Corporation to Y.H.
References
- Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) Experiments With Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bähler J., Wyler,T., Loidl,J. and Kohli,J. (1993) Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol., 121, 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Manova,K., Yuen,J.P., Jasin,M. and Keeney,S. (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking spo11. Mol. Cell, 6, 989–998. [DOI] [PubMed] [Google Scholar]
- Celerin M., Merino,S.T., Stone,J.E., Menzie,A.M. and Zolan,M.E. (2000) Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J., 19, 2739–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes M.D., Farah,J.A. and Smith,G.R. (2000) Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell, 5, 883–888. [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Ding,D.Q., Funabiki,H., Haraguchi,T., Mashiko,S., Yanagida,M. and Hiraoka,Y. (1994) Telomere-led premeiotic chromosome movement in fission yeast. Science, 264, 270–273. [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Ding,D.Q., Imai,Y., Yamamoto,M., Haraguchi,T. and Hiraoka,Y. (1997) Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J., 16, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.P., Watanabe,Y. and Nurse,P. (1998) Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature, 392, 828–831. [DOI] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald,K., Moulder,G., Barstead,R., Dresser,M. and Villeneuve,A.M. (1998) Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell, 94, 387–398. [DOI] [PubMed] [Google Scholar]
- De Veaux L.C., Hoagland,N.A. and Smith,G.R. (1992) Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics, 130, 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D.-Q., Chikashige,Y., Haraguchi,T. and Hiraoka,Y. (1998) Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci., 111, 701–712. [DOI] [PubMed] [Google Scholar]
- Ding D.-Q., Tomita,Y., Yamamoto,A., Chikashige,Y., Haraguchi,T. and Hiraoka,Y. (2000) Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells, 5, 169–190. [DOI] [PubMed] [Google Scholar]
- Egel R. (1989) Mating-type genes, meiosis, and sporulation. In Nasim,A., Young,P. and Johnson,B. (eds), Molecular Biology of the Fission Yeast. Academic Press, New York, NY, pp. 31–73.
- Funabiki H., Hagan,I., Uzawa,S. and Yanagida,M. (1993) Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol., 121, 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman A.S. and Lichten,M. (2000) Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc. Natl Acad. Sci. USA, 97, 9537–9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M., Vezon,D., Gendrot,G. and Pelletier,G. (2001) AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J., 20, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H. (1971) Site specific induction of gene conversion in Shizosaccharomyces pombe. Genetics, 69, 317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H., Heslot,U., Leupold,U. and Loprineo,N. (1974) Shizosaccharo myces pombe. In King,R.C. (ed.), Handbook of Genetics. Plenum Press, New York, NY, pp. 395–446.
- Hawley S. (1988) Exchange and chromosomal segregation in eukaryotes. In Kucherlapati,R. and Smith,G.R. (eds), Genetic Recombination. American Society for Microbiology, Washington, DC, pp. 497–527.
- Hiraoka Y. (1998) Meiotic telomeres: a matchmaker for homologous chromosomes. Genes Cells, 3, 405–413. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Swedlow,J.R., Paddy,M.R., Agard,D.A. and Sedat,J.W. (1991) Three-dimensional multiple-wavelength fluorescence micro scopy for the structural analysis of biological phenomena. Semin. Cell Biol., 2, 153–165. [PubMed] [Google Scholar]
- Hiraoka Y., Ding,D.Q., Yamamoto,A., Tsutsumi,C. and Chikashige,Y. (2000) Characterisation of fission yeast meiotic mutants based on live observation of meiotic prophase nuclear movement. Chromosoma, 109, 103–109. [DOI] [PubMed] [Google Scholar]
- Hirata A. and Tanaka,K. (1982) Nuclear behavior during conjugation and meiosis in the fission yeast Schizosaccharomyces pombe. J. Gen. Appl. Microbiol., 28, 263. [Google Scholar]
- Hoheisel J.D., Maier,E., Mott,R., McCarthy,L., Grigoriev,A.V., Schalkwyk,L.C., Nizetic,D., Francis,F. and Lehrach,H. (1993) High resolution cosmid and P1 maps spanning the 14 Mb genome of the fission yeast S. pombe. Cell, 73, 109–120. [DOI] [PubMed] [Google Scholar]
- Ijichi H., Tanaka,T., Nakamura,T., Yagi,H., Hakuba,A. and Sato,M. (2000) Molecular cloning and characterisation of a human homologue of TBPIP, a BRCA1 locus-related gene. Gene, 248, 99–107. [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux,C.N. and Kleckner,N. (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell, 88, 375–384. [DOI] [PubMed] [Google Scholar]
- Kleckner N. and Weiner,B.M. (1993) Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harb. Symp. Quant. Biol., 58, 553–565. [DOI] [PubMed] [Google Scholar]
- Kneitz B. et al. (2000) MutS homolog 4 localisation to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev., 14, 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Leu J.Y. and Roeder,G.S. (1999) The pachytene checkpoint in S.cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell, 4, 805–814. [DOI] [PubMed] [Google Scholar]
- Leu J.Y., Chua,P.R. and Roeder,G.S. (1998) The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell, 94, 375–386. [DOI] [PubMed] [Google Scholar]
- Lin Y., Larson,K.L., Dorer,R. and Smith,G.R. (1992) Meiotically induced rec7 and rec8 genes of Schizosaccharomyces pombe. Genetics, 132, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Klein,F. and Scherthan,H. (1994) Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J. Cell Biol., 125, 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S. and Hayashi-Hagihara,A. (1998) mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev., 12, 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S., Green-Marroquin,B.L., Sekelsky,J.J., Chin,G., Steinberg,C., Khodosh,R. and Hawley,R.S. (1998) Meiotic synapsis in the absence of recombination. Science, 279, 876–878. [DOI] [PubMed] [Google Scholar]
- Mizukami T. et al. (1993) A 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell, 73, 121–132. [DOI] [PubMed] [Google Scholar]
- Molnar M., Bähler,J., Sipiczki,M. and Kohli,J. (1995) The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics, 141, 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa,T., Straight,A.F., Murray,A., Chikashige,Y., Yamashita,Y.M., Hiraoka,Y. and Yanagida,M. (1998) Dynamics of centromeres during metaphase–anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell, 9, 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D.K., Scherthan,H., Rockmill,B., Bhargava,J. and Roeder,G.S. (1995) Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics, 141, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N. and Yamamoto,M. (1984) Analysis of the structure and transcription of the aro3 cluster gene in Schizosaccharomyces pombe. Mol. Gen. Genet., 195, 164–169. [DOI] [PubMed] [Google Scholar]
- Nimmo E.R., Pidoux,A.L., Perry,P.E. and Allshire,R.C. (1998) Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature, 392, 825–828. [DOI] [PubMed] [Google Scholar]
- Niwa O., Shimanuki,M. and Miki,F. (2000) Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J., 19, 3831–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson L.W., Eden,U., Egel-Mitani,M. and Egel,R. (1978) Asynaptic meiosis in fission yeast? Hereditis, 89, 189–199. [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D.D. (1949) Biochemical mutants in the smut fungus Ustilago maydis. Genetics, 34, 607–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli A.S. and Smith,G.R. (1989) Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics, 123, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow C.F. (1977) The number of chromosomes in S. pombe: light microscopy of stained preparations. Genetics, 87, 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G.S. (1997) Meiotic chromosomes: it takes two to tango. Genes Dev., 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Romanienko P.J. and Camerini-Otero,R.D. (2000) The mouse spo11 gene is required for meiotic chromosome synapsis. Mol. Cell, 6, 975–987. [DOI] [PubMed] [Google Scholar]
- Saka Y., Sutani,T., Yamashita,Y., Saitoh,S., Takeuchi,M., Nakaseko,Y. and Yanagida,M. (1994) Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J., 13, 4938–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Loidl,J., Schuster,T. and Schweizer,D. (1992) Meiotic chromosome condensation and pairing in Saccharomyces cerevisiae studied by chromosome painting. Chromosoma, 101, 590–595. [DOI] [PubMed] [Google Scholar]
- Scherthan H., Bähler,J. and Kohli,J. (1994) Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J. Cell Biol., 127, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki M., Miki,F., Ding,D.Q., Chikashige,Y., Hiraoka,Y., Horio,T. and Niwa,O. (1997) A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol. Gen. Genet., 254, 238–249. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Marshall,W.F., Sedat,J.W. and Murray,A.W. (1997) Mitosis in living budding yeast: anaphase A but no metaphase plate. Science, 277, 574–578. [DOI] [PubMed] [Google Scholar]
- Szankasi P., Heyer,W.D., Schuchert,P. and Kohli,J. (1988) DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination host spot allele ade6-M26. J. Mol. Biol., 204, 917–925. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Nakamura,T., Takagi,H. and Sato,M. (1997) Molecular cloning and characterisation of a novel TBP-1 interacting protein (TBPIP): enhancement of TBP-1 action on Tat by TBPIP. Biochem. Biophys. Res. Commun., 239, 176–181. [DOI] [PubMed] [Google Scholar]
- Virgin J.B. and Bailey,J.P. (1998) The M26 hotspot of Schizosaccharo myces pombe stimulates meiotic ectopic recombination and chromosomal rearrangements. Genetics, 149, 1191–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D., Rasmussen,S.W. and Holm,P.B. (1984) The synaptonemal complex in genetic segregation. Annu. Rev. Genet., 18, 331–413. [DOI] [PubMed] [Google Scholar]
- Walker M.Y. and Hawley,R.S. (2000) Hanging on to your homolog: the roles of pairing, synapsis and recombination in the maintenance of homolog adhesion. Chromosoma, 109, 3–9. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Miyashita,K., Saito,T.T., Yoneki,T., Kakihara,Y., Nabeshima,K., Kishi,Y.A., Shimoda,C. and Nojima,H. (2001) Comprehensive isolation of meiosis specific genes identifies many unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res., 29, 2327–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner B.M. and Kleckner,N. (1994) Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell, 77, 977–991. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., West,R.R., McIntosh,J.R. and Hiraoka,Y. (1999) A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol., 145, 1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. and Kleckner,N. (1998) The leptotene–zygotene transition of meiosis. Annu. Rev. Genet., 32, 619–697. [DOI] [PubMed] [Google Scholar]