Abstract
Initiation factor IF3 contains two domains separated by a flexible linker. While the isolated N-domain displayed neither affinity for ribosomes nor a detectable function, the isolated C-domain, added in amounts compensating for its reduced affinity for 30S subunits, performed all activities of intact IF3, namely: (i) dissociation of 70S ribosomes; (ii) shift of 30S-bound mRNA from ‘stand-by’ to ‘P-decoding’ site; (iii) dissociation of 30S–poly(U)–NacPhe-tRNA pseudo- initiation complexes; (iv) dissociation of fMet-tRNA from initiation complexes containing mRNA with the non-canonical initiation triplet AUU (AUUmRNA); (v) stimulation of mRNA translation regardless of its start codon and inhibition of AUUmRNA translation at high IF3C/ribosome ratios. These results indicate that while IF3 performs all its functions through a C-domain–30S interaction, the N-domain function is to provide additional binding energy so that its fluctuating interaction with the 30S subunit can modulate the thermodynamic stability of the 30S–IF3 complex and IF3 recycling. The localization of IF3C far away from the decoding site and anticodon stem–loop of P-site-bound tRNA indicates that the IF3 fidelity function does not entail its direct contact with these structures.
Keywords: domain structure/factor recycling/ribosomes/RNA–protein interactions/translation fidelity
Introduction
IF3 is one of the three factors required for initiation of protein synthesis in bacteria (for review see Gualerzi and Pon, 1990; Gualerzi et al., 2000). In Escherichia coli, IF3 is a protein of 180 amino acids encoded by infC, an essential gene (Olsson et al., 1996) mapped at 37.5 min (Sacerdot et al., 1982).
IF3 is an RNA-binding protein displaying, at least in vitro, several possibly interrelated activities. It is known, in particular, that: (i) IF3 antagonizes the association between 30S and 50S subunits, thus supplying the pool of free 30S subunits required for translation initiation (Hershey, 1987 and references therein); (ii) IF3 accelerates the on-rate of codon–anticodon interaction at the P-site, thus stimulating the formation of 30S initiation complexes (Gualerzi et al., 1977; Wintermeyer and Gualerzi 1983); (iii) IF3 promotes the rapid dissociation of fMet-tRNA from initiation complexes formed at the 5′ AUG triplet of leaderless mRNAs (Tedin et al., 1999) as well as the dissociation of pseudo-initiation complexes containing aminoacyl-tRNAs (or tRNAs) other than initiator fMet-tRNA or non-canonical initiation complexes containing fMet-tRNA and triplets other than the initiation triplets AUG, GUG and UUG (Gualerzi et al., 1971; Pon and Gualerzi, 1974; Hartz et al., 1989; Hartz et al., 1990; Haggerty and Lovett, 1993; Sussman et al., 1996); (iv) as a probable consequence of (ii) and (iii), IF3 ensures the efficiency and fidelity of initiation site selection inhibition, if present in a stoichiometric excess over the ribosomes, of translation of mRNAs beginning with non-canonical (e.g. AUU) triplets while stimulating translation of any mRNA regardless of the nature of its start codon if present in correct stoichiometric amounts (La Teana et al., 1993; Pediconi et al., 1995); (v) IF3 induces an adjustment of the mRNA, which is shifted from the ‘stand-by site’ to the ‘P-decoding site’ on the 30S ribosomal subunit (La Teana et al., 1995). The molecular bases of the above activities have so far remained elusive, but it is obvious that they must have a strict correlation with the spatial organization of the IF3 molecule and with its ribosomal binding site.
The three-dimensional (3D) structure of IF3, which has been elucidated by X-ray crystallography in Bacillus stearothermophilus (Biou et al., 1995) and by NMR spectroscopy in E.coli (Fortier et al., 1994; Garcia et al., 1995a,b), consists of two domains separated by a fairly long and flexible linker (Moreau et al., 1997). These two domains interact independently with the 30S ribosomal subunit: the C-domain (IF3C) binds first through some of its loops and α-helices to the 30S subunits, allowing the subsequent interaction of the N-domain (IF3N) via a smaller number of residues (Sette et al., 1999). The two-domain structure of IF3, which is matched by the existence of two binding sites on the 30S ribosomal subunit (McCutcheon et al., 1999; Sette et al., 1999 and references therein), is considered essential for the biological function of the factor. Indeed, models based on the presence of two domains and on their different or fluctuating interactions with the ribosome have been proposed to explain the mechanism of action of IF3 (Pon et al., 1982; de Cock et al., 1999). Furthermore, it was speculated that different IF3 functions might be the prerogative of either one of its two domains (de Cock et al., 1999).
The present findings represent a turning point in our understanding of the relationship between the structure and function of IF3 insofar as they demonstrate that all the above-mentioned functions can be carried out by the isolated C-domain of IF3, and allow us to rationalize that the two-domain structure of IF3 serves the purpose of modulating the thermodynamic stability of the IF3–30S complexes, thereby favouring the cycling of the factor on and off the ribosome.
Results
To study the function(s) of IF3N and IF3C, the infC gene of E.coli was manipulated to produce two vectors each carrying and expressing a different fragment of the gene under the control of an inducible promoter; the first construct encoded IF3N plus almost the entire linker (i.e. from Val7 to Ile91), while the second encoded IF3C preceded by approximately half of the linker (i.e. from Ser80 to the C-terminal Gly180). After induction of the promoters, the two overexpressed domains were purified to electrophoretic homogeneity and tested for their biological activities.
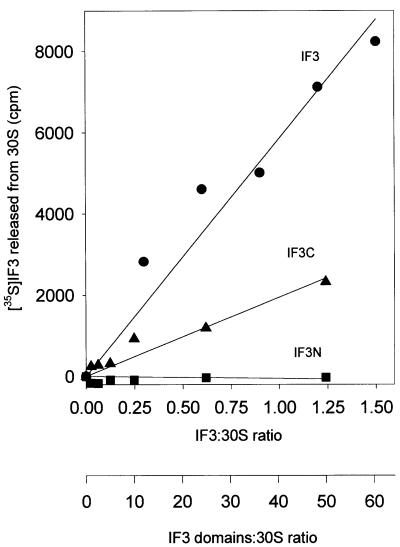
The capacity of isolated IF3N and IF3C to interact with the 30S ribosomal subunits was measured by their ability to compete with intact, 35S-labelled IF3 for binding to the 30S subunits. As seen in Figure 1, increasing amounts of radioactive intact IF3 can be displaced from a fixed amount of 30S ribosomal subunits upon addition of increasing amounts of intact non-radioactive IF3, and the extent of the competition between the two molecules reflected, as expected, their identical affinity for the 30S subunits. In the same experiment, isolated IF3N proved incapable of competing with native IF3, even when present in a 500-fold molar excess over the latter. IF3C, on the other hand, was able to displace intact radioactive IF3 from the 30S subunits, albeit with a lower efficiency than the control non-radioactive IF3, suggesting that native IF3 and its C-terminal domain occupy the same or at least overlapping sites on the 30S subunit. From the extent of the competition between these two molecules, the affinity of the C-domain for the ribosomal subunit can be estimated to be ∼100-fold lower than that displayed by intact IF3, namely Ka ≅ 3 × 105 M–1 instead of ≅ 3 × 107 M–1 (Weiel and Hershey, 1981).
Fig. 1. Interaction between native IF3 and the isolated N-domain (IF3N) and C-domain (IF3C) of IF3 with 30S ribosomal subunits measured through the capacity of increasing amounts of each protein to compete with 35S-labelled native IF3 for a fixed amount of 30S ribosomal subunits. Competition was quantified by incubating a fixed amount of 30S subunits and radioactive IF3 with the indicated amounts of native IF3 (filled circles), IF3C (filled triangles) and IF3N (filled squares), and measuring the amount of radioactive free protein released in the supernatant following centrifugation of the ribosomal subunits by Airfuge (Beckman). Additional experimental details can be found in Materials and methods.
However, that the interaction of IF3C with the 30S ribosomal subunit is strong enough to displace IF3 from its binding site does not ensure per se that the 30S–IF3C interaction is specific and that it could have any functional relevance. Thus, the following experiments were carried out to determine whether, and to what extent, IF3C can replace native IF3 in the various translational functions that are characteristic of this protein. The same activities tested on IF3C were also tested on IF3N, but the results were found to be negative in all cases (not shown), in agreement with the above finding that the isolated N-domain of IF3 does not have a detectable affinity for the ribosomes.
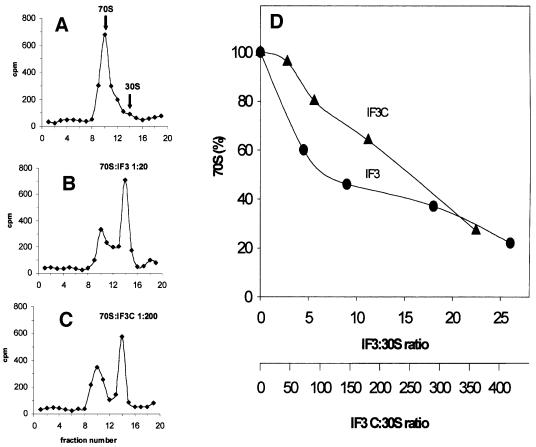
In the first of these experiments, IF3C was tested for the ribosomal subunit anti-association function by which IF3 promotes the dissociation of 70S monomers into 30S and 50S subunits (Figure 2). To allow for a more rigorous and precise quantification of the dissociation, 35S-labelled 30S subunits were mixed with a 1.5-fold stoichiometric excess of non-radioactive 50S subunits to reform 70S monomers, which were first isolated by sucrose density gradient centrifugation and then incubated with increasing amounts of either IF3 or IF3C. Ribosome dissociation was monitored following sucrose density gradient centrifugation and quantification of the radioactivity associated with the monomer (70S) peak and the 30S peak. In Figure 2, we show examples of the sedimentation profiles obtained with ribosomes alone (Figure 2A) or following the addition of 20- and 200-fold stoichiometric excess of IF3 (Figure 2B) and IF3C (Figure 2C); the quantification of the ribosomal dissociation activity induced by increasing amounts of native IF3 or IF3C is shown in Figure 2D. As seen from this graph, IF3C is fully capable of dissociating the ribosomes provided that it is added in amounts ∼50 times higher than those required in the case of intact IF3.
Fig. 2. Dissociation of 70S ribosomes by increasing concentrations of native IF3 and IF3C. The subunit anti-association activity of the protein was measured quantitatively by preparing 70S tight couples containing 35S-labelled 30S subunits and non-labelled 50S subunits. The 70S ribosomes were incubated with the amounts of either native IF3 (filled circles) or IF3C (filled triangles) indicated in the abscissa of (D) and then subjected to sucrose density gradient centrifugation to determine the amount of radioactive 30S sedimenting as 70S or as free 30S. Examples of these analyses are presented for 70S without protein added (A) or following incubation with a 20-fold stoichiometric excess of IF3 (B) or with a 200-fold excess of IF3C (C). The 70S dissociation curve obtained by the addition of increasing amounts of IF3 or IF3C is shown (D).
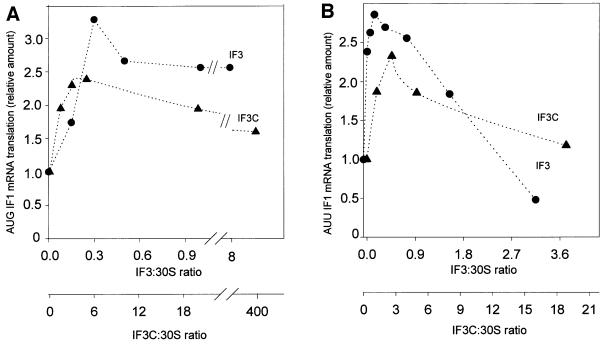
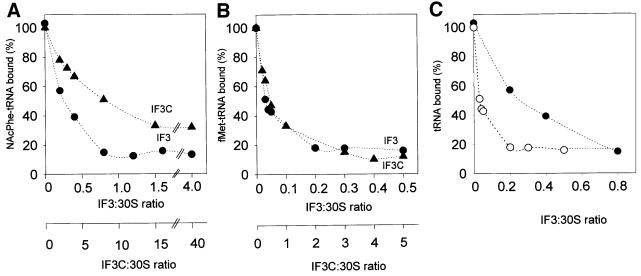
It has been known for a long time that IF3 promotes translation of natural mRNAs; this stimulation occurs regardless of the nature of the initiation triplet (La Teana et al., 1993), a finding that is consistent with the fact that translation of synthetic polynucleotides such as poly(U) and poly(A) can also be stimulated by this factor (Gualerzi et al., 1986). However, an excess of IF3 over the ribosomes inhibits the translation of mRNAs beginning with a non-canonical initiation triplet such as AUU (Brombach and Pon, 1987; La Teana et al., 1993; Pediconi et al., 1995). In the following experiment we have tested whether these activities, typical of intact IF3, are also displayed by IF3C. The results obtained demonstrate that both IF3 and IF3C are able to stimulate translation of infA* mRNA (Calogero et al., 1987) beginning with either a canonical AUG (Figure 3A) or a non-canonical AUU (Figure 3B) triplet and to inhibit translation of the AUU mRNA when present in a stoichiometric excess over the ribosomes (Figure 3B). As pointed out previously, the seemingly paradoxical effect of IF3 (stimulation and inhibition) on the translation of mRNAs beginning with AUU is not a result of the presence of two separate IF3 binding sites on the 30S subunit, but stems instead from an increased rate of fMet-tRNA dissociation from the 30S subunits programmed with AUU-mRNAs (La Teana et al., 1993). This process acquires functional relevance for the regulation of gene expression when excess IF3 slows down the normal flow of 30S initiation complex into the elongation phase of translation by interfering with the joining of 50S subunits to the 30S initiation complex (La Teana et al., 1993). The following experiments demonstrate that like intact IF3, IF3C is also capable of promoting the dissociation of both NacPhe-tRNA from 30S subunits programmed with poly(U) (Figure 4A) and fMet-tRNA from 30S subunits programmed with infA* AUU-mRNA (Figure 4B). Comparison of the activities of IF3 and IF3C in these two tests clearly reveals that the amount of IF3C required to afford the same activity of IF3 is ∼30- and 10-fold higher in the first and second case, respectively (Figure 4A and B). The larger requirement for IF3C compared with IF3 confirms that the C-domain has a lower affinity than the intact molecule for the small ribosomal subunit; on the other hand, the difference in the relative amounts of IF3 and IF3C required to dissociate the pseudo-initiation complex compared with the non-canonical initiation complex indicates that the dissociation mechanism of these complexes might be similar but not identical. This premise is confirmed by the results presented in Figure 4C, which show that the amount of native IF3 required for the dissociation of the two complexes is also different.
Fig. 3. Effect of IF3 and IF3C on the translation of two natural mRNAs as a function of their initiation codon. The reaction mixtures contained IF1 mRNA beginning with either AUG (A) or AUU (B) initiation codon, 70S ribosomes and a 1:1 stoichiometric amount of IF1 and IF2, while the amount of IF3 (filled circles) or IF3C (filled triangles) was varied as indicated in the abscissa. The IF1 product synthesized in each reaction tube was analysed by SDS–PAGE and quantified with a Molecular Imager (Bio-Rad). The results were normalized taking the level of translation obtained in the absence of either IF3 or IF3C as equal to one.
Fig. 4. Dissociation of pseudo-initiation and non-canonical initiation complexes by IF3 and IF3C. The complexes containing 30S subunits and the aminoacyl-tRNAs and templates specified below were prepared as described in Materials and methods, and then subjected to 25-fold dilution with 30 mM Tris–HCl pH 7.1, 50 mM NH4Cl, 15 mM magnesium acetate, 2 mM 2-mercaptoethanol, containing the indicated amounts of either IF3 (filled circles) or IF3C (filled triangles). After nitrocellulose filtration, the IF3-induced dissociation of the complexes was measured through the loss of the corresponding aminoacyl-tRNA radioactivity from the filters. (A) Pseudo-initiation complex of 30S, poly(U) and NAc [14C]Phe-tRNA; (B) non-canonical initiation complex of 30S, AUU IF1 mRNA and f [35S]Met-tRNA; (C) pseudo-initiation complexes of 30S, NAc [14C]Phe-tRNA and poly(U) (filled circles) or non-canonical initiation complexes of 30S, f [35S]Met-tRNA and AUU IF1 mRNA (open circles). Further experimental details are given in Materials and methods.
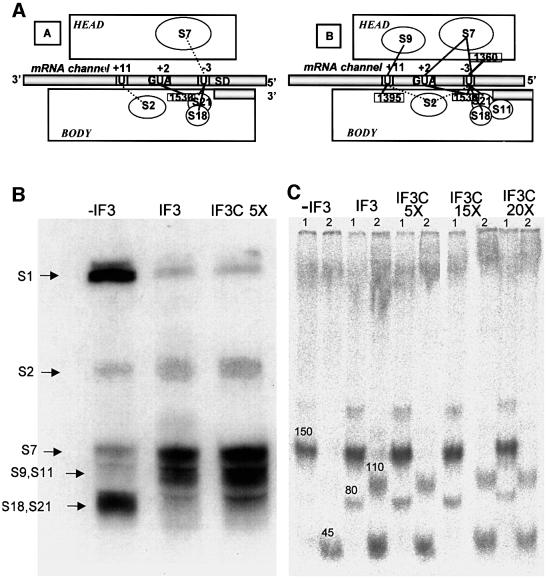
The last of the IF3 functions tested is that of inducing a shift of the 30S-bound mRNA from the ‘stand-by site’ towards the ‘P-decoding site’ (Canonaco et al., 1989; La Teana et al., 1995). The functional significance of this activity is probably two-fold: (i) to favour dissociation of the SD–antiSD interaction while favouring P-site decoding of fMet-tRNA; and (ii) to permit the kinetic selection of the correct initiation site of the mRNA. The mRNA shift is best monitored by site-directed UV-induced cross-linking of the mRNA to specific nucleotides of 16S rRNA and to ribosomal proteins (La Teana et al., 1995). The main changes in the mRNA–30S cross-linking pattern induced by IF3 are shown schematically in Figure 5A, which depicts the identified sites of cross-linking obtained in the absence (i) and presence (ii) of the factor. In the experiments carried out to compare the activity of IF3 with that of IF3C we have monitored the cross-links between the 4-thio Us at positions –3, +2 and +11 of the mRNA and both ribosomal proteins (Figure 5B) and 16S rRNA (Figure 5C). As seen from this figure, IF3 causes the expected changes in the cross-linking of mRNA with S1, S18 and S21(which are decreased), and with S2, S7, S9 and S11 (which are increased), and produces the characteristic cross-link between +11 of mRNA and C1395, and between –3 and A1360 of 16S rRNA. Furthermore, it can be seen from the same figure that the mRNA–30S cross-linking pattern obtained in the presence of a <5-fold excess of IF3C is very similar to that obtained with the native factor, indicating that the same or a very similar mRNA-shifting function of IF3 can also be performed very efficiently by its isolated C-domain.
Fig. 5. Effect of IF3 and IF3C on the mRNA shift on the 30S ribosomal subunit. (A) To illustrate the IF3-induced repositioning of the mRNA on the 30S subunit described previously (La Teana et al., 1995), we show the main site-directed mRNA–30S cross-linking sites obtained in the absence (left) and presence (right) of IF3. (B) Autoradiography of the SDS–PAGE separation of the ribosomal proteins (indicated by the arrows on the left side) cross-linked to the mRNA in initiation complexes formed, as indicated above the corresponding lanes, in the absence or presence of either IF3 or IF3C (at a 5-fold higher concentration than IF3). (C) Autoradiography of RNaseH digestion products of 16S rRNA UV cross-linked to 32P-labelled mRNA within 30S–mRNA complexes prepared in the presence of IF1, IF2 and, where indicated, IF3 or increasing amounts of IF3C. The RNaseH digestion was carried out in the presence of oligonucleotides complementary to three regions of 16S rRNA, namely (A) 1298–1307, (B) 1382–1391 and (C) 1491–1507. Two separate digestions were performed with two different pairs of oligonucleotides, A/B (lanes marked 1) and B/C (lanes marked 2), respectively. The bands marked 45 and 150, which are obtained with all types of complexes, correspond to cross-links between –3 and +2 of mRNA and a single site (1530) of 16S rRNA, while the two additional bands marked 80 and 110, obtained exclusively from the complexes containing IF3 or IF3C, correspond to cross-links between –3 of the mRNA and 1360 of 16S rRNA, and between +11 of the mRNA and 1395 of 16S rRNA, respectively. The experimental details are identical to those described previously and it should be noted that although the electrophoretic analysis does not allow the separation of S9/S11 and S18/S21, all these proteins were demonstrated to be cross-linked to the mRNA by immunological analysis (Dontsova et al., 1991; La Teana et al., 1995).
Discussion
Translation initiation factor IF3 performs several functions, at least one of which is essential for cell viability (Olsson et al., 1996). In particular, IF3 stimulates on and off rates of ‘30S initiation complex’ formation (Gualerzi et al., 1977), ensuring translational efficiency and, at the same time, fidelity. In fact, IF3 promotes the dissociation of coded or non-coded, charged or uncharged elongator tRNAs from 30S subunits (Gualerzi et al., 1971, 1979; Hartz et al., 1989) and the dissociation of fMet-tRNA bound to 30S subunits in response to non-canonical triplets (e.g. AUU) or to a canonical start codon AUG when this is at the 5′-terminus of a leaderless mRNA (La Teana et al., 1993; Tedin et al., 1999). In addition, IF3 stimulates translation of natural mRNAs (regardless of their start codon) and of synthetic polynucleotide templates (Gualerzi et al., 1986; La Teana et al., 1993), but can also act as a selective translational repressor of mRNAs beginning with non-canonical triplets (e.g. AUU and GUA) by inhibiting their translation when present in a stoichiometric excess over the ribosomes (La Teana et al., 1993; Sussman et al., 1996; Haggerty and Lovett, 1997). Finally, IF3 promotes a re-positioning of the mRNA from the ‘stand-by site’ to the ‘P-decoding site’ on the 30S subunit (Canonaco et al., 1989; La Teana et al., 1995) and ensures a pool of free 30S subunits causing the dissociation of 70S ribosomes (Subramanian and Davis, 1970).
To explain the existence of such a large number of functions with a single mechanism, it was suggested that the ‘translation fidelity’ and the other functions of IF3 are a result of an IF3-induced conformational change of the 30S subunit (Pon and Gualerzi, 1974; Pon et al., 1982). Thus, based on the topographical data available and on the functional and structural properties of IF3 known at the beginning of the 1980s, it had been suggested that IF3 contains two active sites: one interacting with the head and the other with the platform of the 30S subunit. A fluctuating interaction of IF3 involving its two binding sites and the two 30S binding sites was proposed to affect the conformational dynamics of the ribosomal subunit, causing a ‘nodding’ of the head, which influences P-site decoding and other 30S functions (Pon et al., 1982).
The subsequent elucidation of the 3D structure of IF3 led to the discovery of the rather striking structural organization of this protein, which was indeed found to contain two domains (Fortier et al., 1994; Biou et al., 1995; Garcia et al., 1995a,b) separated by a long and flexible linker (Moreau et al., 1997; Hua and Raleigh, 1998). The two domains move (Moreau et al., 1997) and bind to the 30S subunit (Sette et al., 1999) independently. As a result of these and other findings, de Cock et al. (1999) have interpreted their recent NMR and genetic data to suggest that IF3 binds to the 30S with both N- and C-domain, and that the linker between these two domains acts as a strap, which triggers a conformational change of the 30S subunit, essentially re-proposing the same model of Pon et al. (1982). Furthermore, it was suggested that different functions might be carried out independently by either one of the two domains of IF3. In particular, ribosomal binding and ribosome dissociation activity are attributed to the C-domain, while the ‘initiation fidelity function’ is suggested to reside in the N-domain. In fact, whereas biochemical, genetic and spectroscopic evidence indicates that most of the residues involved in IF3–30S interaction are localized in the C-terminal domain of IF3 (Sette et al., 1999 and references therein), some chemical modifications and mutations in the N-proximal portion of the linker can affect the fidelity function of IF3 without affecting its 30S binding capacity (Bruhns and Gualerzi, 1980; De Bellis et al., 1992; Sacerdot et al., 1996; Sussman et al., 1996).
Contrary to the above predictions, in this paper we demonstrate that isolated IF3C not only binds to the 30S ribosomal subunits, as suggested by previous data (Garcia et al., 1995b), but can also perform all known functions of the intact molecule, provided that its concentration is sufficiently high to compensate for its lower affinity for the 30S subunit. In contrast, no function was detected that could be carried out by isolated IF3N and under no circumstance was the addition of isolated IF3N found to improve the activity of isolated IF3C (not shown). Thus, our results demonstrate that the two-domain organization of IF3 has no immediate bearing on the mechanism by which this factor carries out the five functions that can be measured in vitro. However, if isolated IF3C is capable of carrying out all functions of the intact factor, what then is the function of the N-domain and what kind of information is transferred by the linker from one side of the molecule to the other and from one side of the ribosome to the other?
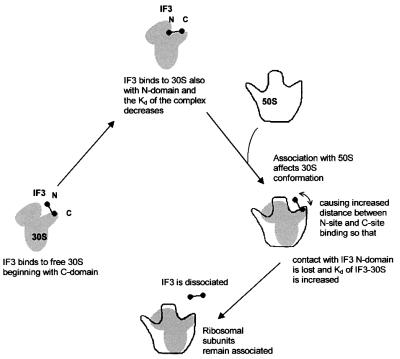
Considering that the affinity of IF3C for the ribosome was shown to be approximately two orders of magnitude lower than that of the intact molecule (Figure 1), and taking into account the finding that IF3 interacts with the 30S subunits initially through its C-domain and then with the N-domain as well as the evidence for the existence of two physically separated binding sites for IF3 on the 30S subunit (Sette et al., 1999 and references therein), we suggest that the two-domain structure is required to modulate binding and release of IF3 to and from the 30S subunit. In the present model (Figure 6), like in that proposed almost two decades ago (Pon et al., 1982), IF3 establishes two main contacts with the 30S subunit: a thermodynamically more stable interaction between its C-domain and the platform of the free 30S subunit, and a much weaker interaction involving the N-domain and a second site located on the neck or head of the 30S subunit. The postulated function of this second interaction is that of reducing the dissociation rate of IF3, thereby stabilizing the 30S–IF3 complex. However, when the 50S ribosomal subunit interacts with this complex, it induces a conformational change whereby the platform and body of the 30S subunit move with respect to each other (Lata et al., 1996; Gabashvili et al., 1999), widening the gap between the two IF3 binding sites. Having lost the stabilizing interaction through its N-domain, IF3 rapidly falls off the 30S subunit, yielding to the process of subunit association. This model is consistent with a large number of both current and old findings and, in particular, with the apparently puzzling properties of the linker, to which our model does not attribute a particularly sophisticated role except that of ensuring a physical connection between IF3C and its N-terminal ‘anchor’. In fact, the physical integrity of the linker is the only condition for functional cooperation between the two IF3 domains; furthermore, the linker, which remains in an extended conformation and accessible to trypsin even when IF3 is ribosome bound (de Cock et al., 1999), does not display particularly relevant sequence conservation. In addition, the length and the hydrophilic and positive character of the linker, which appear to be somewhat conserved, can be altered, at least to some extent, without detectable effect on IF3 function in vivo. Only very drastic changes of its length or complete charge inversion (from +5 to –5) can lead to the loss of IF3 activity (Yu and Spremulli, 1997; de Cock et al., 1999).
Fig. 6. Involvement of the two IF3 domains in the modulation of IF3 binding to and release from 30S ribosomal subunits. The model rationalizes the existence of two domains interacting with two separate sites on the 30S ribosomal subunit. IF3 binds initially through its C-domain (main contact), and this allows the interaction of the N-domain with the second site (minor contact), which enhances the thermodynamic stability of the complex. The interaction with the 50S subunit induces a conformational change of the 30S subunit, which increases the distance between the two IF3 sites and causes the consequent loss of the minor contact between the subunit and the N-domain of IF3. This results in the reversal of the thermodynamic stabilization that had been provided by the 30S–N-domain interaction and favours the dissociation of IF3 from the small subunit.
Two alternative mechanisms have been suggested to explain the function of IF3 in determining initiation fidelity. According to the ‘direct inspection’ model, IF3 accomplishes its task by establishing a physical contact with at least some of the template and tRNA bases involved in codon–anticodon interaction (Meinnel et al., 1999) and/or with the anticodon stem–loop of initiator tRNA (Hartz et al., 1990). The alternative model, originally proposed by Pon and Gualerzi (1974), postulates an indirect mechanism in which IF3 affects the conformational dynamics of the 30S ribosomal subunits, whereby the dissociation rates of canonical, non-canonical and pseudo-30S initiation complexes are differentially increased, resulting in a kinetic discrimination against the latter two. The present finding that IF3C is fully competent in performing the IF3 fidelity function offers, in light of its localization on the 30S subunit, a definite proof in favour of the indirect mechanism. In fact, the location of IF3 and of IF3C on the 30S ribosomal subunit has been determined by cryo-electron microscopy (McCutcheon et al., 1999) and very recently by crystallography (Pioletti et al., 2001). The crystallographic localization is on the solvent side of the upper tip of the platform where it makes direct contacts with proteins S18, S2, S7 and S11 as well as with RNA helices 23, 26 and 45 (Pioletti et al., 2001), while that surmised by cryo-electron microscopy is on the opposite side of the platform facing the 50S subunit (McCutcheon et al., 1999). Although in sharp contrast with one another, both localizations place IF3C too far away from P-site decoding as well as from the anticodon stem–loop of P-site-bound tRNA (Cate et al., 1999; Schluenzen et al., 2000; Wimberly et al., 2000) to allow the C-domain to establish a physical contact with either structure.
Additional data clash with the ‘direct inspection’ model. In fact, the codon–anticodon interaction and anticodon stem–loop of the initiator tRNA, which are the specific features ‘inspected’ by IF3 (Hartz et al., 1990; Meinnel et al., 1999), are buried within the P-site, where they are inaccessible to chemical probing (Hüttenhofer and Noller, 1992), and surrounded and held by six molecular clamps (Cate et al., 1999). Thus, it is difficult to envisage how 30S-bound IF3 might contact the codon and the anticodon stem–loop in a way that ensures the required discrimination, which, as mentioned above, is not restricted to non-initiator tRNA or to non-best-fit codon–anticodon interactions, but also includes fMet-tRNA when bound in response to a 5′ AUG start codon or to an internal 5′ (CUG) or 3′ (AUU) wobbling triplet.
If the mechanism sustaining the fidelity function of IF3 is that based on the IF3-induced conformational change of the 30S subunit (Pon and Gualerzi, 1974), the present data obviously imply that this change does not require the two-domain structure of IF3 as isolated IF3C can perform the same job. Several clues exist that an IF3C-induced con formational change of the 30S subunit can indeed occur. First, IF1, a protein smaller than IF3C and constituted by a single domain, was shown to cause distinct, long-range conformational changes upon binding to the 30S subunit, a finding clearly indicating that domains of the 30S subunit are endowed with structural flexibility and that their conformation can be sensitive to interaction with relatively small protein ligands (Carter et al., 2001). Secondly, IF3 induces changes in three intra-rRNA cross-links (U793/G1517, C967/C1400 and C1402/C1501) within the decoding region of the ribosomal P-site (Shapkina et al., 2000), and preliminary data indicate that isolated IF3C promotes the same alterations of the cross-linking pattern induced by native IF3 (P.Wollenzien, personal communication). Thirdly, we have shown here (Figure 5B and C) that isolated IF3C can induce the same changes of mRNA– rRNA and mRNA–r-protein cross-linking patterns caused by native IF3.
Finally, the finding that all functions of the entire molecule can be performed by IF3C poses the question of whether they could be attributed to a single or to more than one mechanism, and if different roles might be played by different parts of IF3C. Vis-à-vis its 100- to 120-fold lower affinity for the 30S ribosomal subunits, IF3C induces the same effects produced by native IF3 when offered in amounts exceeding those of IF3 by ∼45- to 55-fold (for dissociation of 70S), 20- to 30-fold (for stimulation of translation), 30-fold (for dissociation of pseudo-initiation complexes), 10-fold (for dissociation of non-canonical initiation complexes) and 5-fold (for mRNA shift). These differences in the relative efficiency by which IF3C can replace native IF3 in its various functions could be a result of the fact that in some tests IF3C must compete directly with other ligands (native IF3, the 50S subunit) or could reflect the different rates of the various events examined in relation to the on and off rates which characterize the ribosomal interactions of either IF3 or IF3C.
However, these differences could also indicate the existence of more than one mechanism, especially in the case of the dissociation of the pseudo- and non-canonical initiation complexes, two activities detected by essentially the same test. Direct comparison of the two activities (Figure 4A and B) demonstrates not only that IF3C is comparatively more efficient in dissociating the non-canonical complexes than pseudo-initiation complexes, but also that the dissociation of the pseudo-initiation complexes requires a fairly larger amount of native IF3 than that of the non-canonical initiation complexes (Figure 4C). A single mechanism could still be at the basis of these two IF3 activities if the two types of aminoacyl-tRNAs (initiator versus non-initiator), possibly because of the structural differences of their anticodon stem–loops, occupy somewhat different positions on the ribosome, so as to sense somewhat differently the same IF3-induced conformational change of the 30S. A final answer as to whether all IF3 functions are caused by a single mechanism can probably come from a detailed genetic characterization of the active sites of IF3C implicated in the various IF3 functions.
Materials and methods
General preparations
Escherichia coli MRE600 30S ribosomal subunits and 70S ribosomes, fMet-tRNAfMet and purified initiation factors IF1, IF2 and IF3 were prepared as described previously (Pawlik et al., 1981; Ohsawa and Gualerzi, 1983). NAc-Phe-tRNA was prepared as described (Haenni and Chapeville, 1966). In vitro transcribed mRNAs were produced and purified as described (La Teana et al., 1993).
35S-labelling of IF3 and 70S ribosomes
Escherichia coli strain UT5600 (Elish et al., 1988) carrying pcI857 and pIM302 (Brombach and Pon, 1987) was grown at 30°C in 1.5 l of M9 minimal medium supplemented with 0.05% casaminoacids, 0.005% vitamin B1, 2 mM MgSO4, 0.08 mg/ml amino acid mixture containing proline, tryptophan and leucine, kanamycin (25 µg/ml) and ampicillin (60 µg/ml). When the A590 reached 1.4, the overproduction of labelled IF3 was achieved by adding 480 µl of [35S]Met/[35S]Cys Promix (14.3 mCi/ml; Amersham) to the culture and shifting the temperature from 30 to 42°C. Incubation continued for 20 min at 42°C, followed by a further incubation at 37°C for 30 min. Radioactive IF3 and 70S ribosomes were purified according to the standard procedures mentioned above.
Genetic manipulations
To overproduce IF3N and IF3C, the desired DNA regions of E.coli infC were amplified by PCR using pIM302 as template and two oligonucleotide primers for IF3N (5′-dCGGGATCCGTTCAAACGGCGCG and 5′-dGGAATTCTTAGATAACTTTTTGC) and two for IF3C (5′-dCGGGATCCTCTAAGGAACAGAAG and 5′-dGGAATTCACAGACAGTGCTA). Both pairs of primers introduce a BamHI and an EcoRI site at the proximal and distal terminus of the amplified fragments, so that upon digestion with BamHI and EcoRI, the resulting DNA fragments encoding 85 (IF3N) and 101 (IF3C) amino acids were ligated into the corresponding sites of pGEX-2T (Pharmacia). These vectors were introduced into E.coli BL21 (DE3)-pLysS (Studier, 1991) and the resulting strains used for overexpression of the two IF3 domains.
Expression and purification of IF3N and IF3C
Overexpression of glutathione S-transferase (GST)–IF3N or GST–IF3C was obtained by isopropyl-β-d-thiogalactopyranoside induction (0.8 mM) of the appropriate cell cultures grown at 30°C in Luria–Bertani medium containing ampicillin (60 µg/ml) to an A600 of 1 essentially as described by the supplier of pGEX-2T (Pharmacia). After 3 h of further incubation, the cells were harvested by centrifugation, resuspended in buffer I (10 mM Tris–HCl pH 7.7, 60 mM NH4Cl, 10 mM magnesium acetate, 5 mM β-mercaptoethanol) and rapidly frozen at –80°C.
Essentially the same procedure was used to purify IF3N and IF3C. The cells were disrupted by sonication and subjected to centrifugation for 1 h at 14 000 r.p.m. (Sorvall SA600 rotor). NH4Cl was added to the supernatant to a final concentration of 1 M and ribosomes were sedimented by ultracentrifugation at 30 000 r.p.m. for 15 h (Beckman 70Ti rotor). The supernatant was diluted with buffer II (20 mM Tris–HCl pH 7.1, 0.1 mM EDTA, 10% glycerol, 5 mM 2-mercaptoethanol, 0.2 mM phenylmethylsulfonyl fluoride, 0.2 mM benzamidine) to reduce the NH4Cl to 100 mM, and loaded onto a phosphocellulose column (75 ml bed volume) equilibrated with buffer II containing 100 mM NH4Cl. GST–IF3N or GST–IF3C was eluted with a 1.5 l linear NH4Cl gradient from 200 to 900 mM NH4Cl in buffer II. Following SDS–PAGE analysis, the fractions containing GST–IF3N or GST–IF3C were pooled, dialysed against buffer III pH 7.3 (10 mM Na2HPO4, 1.8 mM KH2PO4, 140 mM NaCl, 2.7 mM KCl) and subjected to affinity chromatography on a glutathione–Sepharose column prior to cleavage with thrombin according to the supplier’s instructions (Pharmacia). After cleavage, the pool was loaded onto a DE52 column equilibrated with buffer II pH 8 containing 25 mM NH4Cl. IF3N or IF3C was recovered in the flow through, loaded onto a phosphocellulose column (2 ml bed volume) and eluted with buffer II containing 1 M NH4Cl. Fractions containing IF3N or IF3C, as determined by A280 measurement, were pooled, dialysed against buffer II containing 200 mM NH4Cl and stored at –80°C.
Airfuge binding experiments
Each reaction tube contained, in 175 µl of buffer IV (20 mM Tris–HCl pH 7.7, 5 mM magnesium acetate, 200 mM NH4Cl), 5% sucrose, 100 pmol of 30S ribosomal subunits, 250 pmol of 35S-labelled IF3 (105 c.p.m./pmol) and increasing amounts of non-radioactive IF3, IF3N or IF3C. The tubes were then subjected to 1 h centrifugation at 30 p.s.i. in a Beckman Airfuge at 4°C (actual running T = 11°C). At the end of the centrifugation, the unbound radioactive IF3 present in 75 µl of the supernatant was quantified by scintillation counting.
In vitro translation
The infA* gene encoding IF1 (Calogero et al., 1987) was modified by site-directed mutagenesis to create a degenerate initiation codon. AUG was converted to AUU using the oligonucleotide 5′-dGGTATACTATTGCGAAAG and the ‘Altered site in vitro mutagenesis’ protocol (Promega). The resulting mRNA, obtained by in vitro transcription with T7 RNA polymerase, was used for translation assays carried out essentially as described (La Teana et al., 1993; Brandi et al., 1996). Each reaction mixture contained 30 pmol of high-salt-washed 70S ribosomes, 15 pmol of infA* mRNA, 0.3 µmol of [35S]methionine (1000 Ci/mmol; Amersham), 30 pmol each of IF1 and IF2, and increasing amounts of IF3, IF3N or IF3C. After 15 min incubation at 37°C, 15 µl were withdrawn for determination of the radioactivity insoluble in trichloroacetic acid.
Dissociation of ternary complexes
IF3-dependent release of NAc[14C]Phe-tRNA from pseudo-initiation complexes containing poly(U) and 30S subunits was measured as described (Pon and Gualerzi, 1974) in the presence of increasing amounts of IF3 (up to 120 pmol) or IF3C (up to 1200 pmol). IF3-dependent release of fMet-tRNA from non-canonical initiation complexes containing AUU mRNA and 30S subunits was measured as described (La Teana et al., 1993) in the presence of increasing amounts of IF3 (up to 15 pmol) or IF3C (up to 150 pmol).
Ribosome dissociation activity
[35S]30S ribosomal subunits, obtained from dissociation of labeled 70S particles through sucrose density gradient centrifugation in the presence of 1 mM magnesium acetate, were re-associated with non-radioactive 50S ribosomal subunits (1.5-fold excess) and the ‘hybrid’ 70S purified on a sucrose gradient in the presence of 6 mM magnesium acetate.
Dissociation experiments were carried out as follows: 5 pmol of the ‘hybrid’ 70S were incubated in 100 µl of 10 mM Tris–HCl pH 7.7 containing 100 mM NH4Cl, 6 mM magnesium acetate and 6 mM 2-mercaptoethanol with increasing amounts of IF3 or IF3C for 5 min at 37°C. The reaction mixtures were then loaded onto 10–30% sucrose density gradients in a SW60 rotor and run at 40 000 r.p.m. for 2 h. Fractions of 200 µl were collected and the radioactivity determined by scintillation counting.
mRNA–30S complex formation and cross-linking reaction
The 32P-labelled mRNA analogue 4N/-3, containing 4-thio-U at three different positions (La Teana et al., 1995), was transcribed from synthetic DNA templates using T7 RNA polymerase and purified by gel electrophoresis as described (Stade et al., 1989). Binding reactions were carried out in 100 µl of 20 mM Tris–HCl pH 7.8 containing 100 mM NH4Cl, 7 mM MgCl2, 1 mM dithiothreitol, 200 pmol of 30S ribosomal subunits (pre-incubated for 5 min at 50°C), 150 pmol of IF1, 150 pmol of 32P-labelled mRNA and 200 pmol of IF3 or increasing amounts of IF3C. After incubation at 37°C for 15 min, the complexes were subjected to UV irradiation (>300 nm) for 3 min at 4°C as described (Tate et al., 1990), and the cross-linked products were separated by two successive sucrose gradient centrifugations (Dontsova et al., 1991).
Analysis of the cross-linking sites
Identification of the cross-linking sites on 16S RNA was carried out essentially as described (La Teana et al., 1995). RNase H digestion of the 16S rRNA was performed in the presence of two pairs of oligonucleotides: the first two were complementary to regions 1298–1307 and 1382–1391, and the second two to regions 1382–1391 and 1491–1507 of 16S rRNA. Ribosomal proteins cross-linked to mRNA were identified essentially as described (La Teana et al., 1995).
Acknowledgments
Acknowledgements
The financial support of grants from the Italian CNR (Progetto Strategico ST74), CNR-MURST(95/95) to C.O.G. and Italian MURST (PRIN 2000) to C.L.P., A.L.T. and C.O.G. is gratefully acknowledged.
References
- Biou V., Shu,F. and Ramakrishnan,V. (1995) X-ray crystallography shows that translational initiation factor IF3 consists of two compact α/β domains linked by an α-helix. EMBO J., 14, 4056–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi A., Pietroni,P., Gualerzi,C.O. and Pon,C.L. (1996) Post-transcriptional regulation of CspA expression in Escherichia coli. Mol. Microbiol., 19, 231–240. [DOI] [PubMed] [Google Scholar]
- Brombach M. and Pon,C. (1987) The unusual translational initiation codon AUU limits the expression of the infC (initiation factor IF3) gene of Escherichia coli.Mol. Gen. Genet., 208, 94–100. [DOI] [PubMed] [Google Scholar]
- Bruhns J. and Gualerzi,C. (1980) Structure–function relationship in Escherichia coli initiation factors: role of tyrosine residues in ribosomal binding and functional activity of IF-3. Biochemistry, 19, 1670–1676. [DOI] [PubMed] [Google Scholar]
- Calogero R.A., Pon,C.L. and Gualerzi,C.O. (1987) Chemical synthesis and in vivo hyperexpression of a modular gene coding for Escherichia coli translational initiation factor IF1. Mol. Gen. Genet., 208, 63–69. [DOI] [PubMed] [Google Scholar]
- Canonaco M.A., Gualerzi,C.O. and Pon,C.L. (1989) Alternative occupancy of a dual ribosomal binding site by mRNA affected by translation initiation factors. Eur. J. Biochem., 182, 501–506. [DOI] [PubMed] [Google Scholar]
- Carter A.P., Clemons,W.M.,Jr, Brodersen,D.E., Morgan-Warren,R.J., Hartsch,T., Wimberly,B.T. and Ramakrishnan,V. (2001) Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science, 291, 498–501. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Yusupov,M.M., Yusopova,G.Z., Earnest,T.N. and Noller,H.F. (1999) X-ray crystal structures of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- De Bellis D., Liveris,D., Goss,D., Ringquist,S. and Schwartz,I. (1992) Structure–function analysis of Escherichia coli translation initiation factor IF3: tyrosine 107 and lysine 110 are required for ribosome binding. Biochemistry, 31, 11984–11990. [DOI] [PubMed] [Google Scholar]
- De Cock E., Springer,M. and Dardel,F. (1999) The inter-domain linker of Escherichia coli initiation factor IF3: a possible trigger of translation initiation specificity. Mol. Microbiol., 32, 193–202. [DOI] [PubMed] [Google Scholar]
- Dontsova O., Kopylov,A. and Brimacombe,R. (1991) The location of mRNA in the ribosomal 30S initiation complex; site-directed cross-linking of mRNA analogues carrying several photo-reactive labels simultaneously on either side of the AUG start codon. EMBO J., 10, 2613–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elish M.E., Pierce,J.R. and Earhart,C.F. (1988) Biochemical analysis of spontaneous fepA mutants of Escherichia coli. J. Gen. Microbiol., 134, 1355–1364. [DOI] [PubMed] [Google Scholar]
- Fortier P.L., Schmitter,J.M., Garcia,C. and Dardel,F. (1994) The N-terminal half of initiation factor IF3 is folded as a stable independent domain. Biochimie, 76, 376–383. [DOI] [PubMed] [Google Scholar]
- Gabashvili I.S., Agrawal,R.K., Grassucci,R., Squires,C.L., Dahlberg,A.E. and Frank,J. (1999) Major rearrangements in the 70S ribosomal 3D structure caused by a conformational switch in 16S ribosomal RNA. EMBO J., 18, 6501–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia C., Fortier,P.L., Blanquet,S., Lallemand,J.Y. and Dardel,F. (1995a) 1H and 15N resonance assignments and structure of the N-terminal domain of Escherichia coli initiation factor 3. Eur. J. Biochem., 228, 395–402. [PubMed] [Google Scholar]
- Garcia C., Fortier,P.L., Blanquet,S., Lallemand,J.Y. and Dardel,F. (1995b) Solution structure of the ribosome-binding domain of E.coli translation initiation factor 3. Homology with the U1 A protein of the eukaryotic spliceosome. J. Mol. Biol., 254, 247–259. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon,C.L. (1990) Initiation of mRNA translation in prokaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Gualerzi C., Pon,C.L. and Kaji,A. (1971) Initiation factor dependent release of aminoacyl-tRNAs from complexes of 30S ribosomal subunits, synthetic polynucleotide and aminoacyl tRNA. Biochem. Biophys. Res. Commun., 45, 1312–1319. [DOI] [PubMed] [Google Scholar]
- Gualerzi C., Risuleo,G. and Pon,C. (1977) Initial rate kinetic analysis of the mechanism of initiation complex formation and the role of initiation factor IF-3. Biochemistry, 16, 1684–1689. [DOI] [PubMed] [Google Scholar]
- Gualerzi C., Risuleo,G. and Pon,C. (1979) Mechanism of the spontaneous and initiation factor 3-induced dissociation of 30S aminoacyl-tRNA polynucleotide ternary complexes. J. Biol. Chem., 254, 44–49. [PubMed] [Google Scholar]
- Gualerzi C.O., Pon,C.L., Pawlik,R.T., Canonaco,M.A., Paci,M. and Wintermeyer,W. (1986) Role of the initiation factors in Escherichia coli translational initiation. In Hardesty,B. and Kramer,G. (eds), Structure, Function and Genetics of Ribosomes. Springer-Verlag, New York, NY, pp. 621–641.
- Gualerzi C.O., Brandi,L., Caserta,E., La Teana,A., Spurio,R., Tomšic,J. and Pon,C.L. (2000) Translation initiation in bacteria. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 477–494.
- Haenni A.L. and Chapeville,F. (1966) The behaviour of acetyl phenylalanyl soluble ribonucleic acid in polyphenylalanine synthesis. Biochim. Biophys. Acta, 114, 135–148. [DOI] [PubMed] [Google Scholar]
- Haggerty T.J. and Lovett,S.T. (1993) Suppression of recJ mutations of E.coli by mutations in translation initiation factor IF3. J. Bacteriol., 175, 6118–6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty T.J. and Lovett,S.T. (1997) IF3-mediated suppression of a GUA initiation codon mutation in the recJ gene of Escherichia coli. J. Bacteriol., 179, 6705–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D., McPheeters,D.S. and Gold,L. (1989) Selection of the initiator tRNA by E.coli initiation factors. Genes Dev., 3, 1899–1912. [DOI] [PubMed] [Google Scholar]
- Hartz D., Binkley,J., Hollingsworth,T. and Gold,L. (1990) Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev., 4, 1790–1800. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B. (1987) Protein synthesis. In Neidhardt,F.C., Ingraham,J.L., Low,K.B., Magasanik,B., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 613–647.
- Hua Y. and Raleigh,D.P. (1998) On the global architecture of initiation factor IF3: a comparative study of the linker regions from the Escherichia coli protein and the Bacillus stearothermophilus protein. J. Mol. Biol., 278, 871–878. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A. and Noller,H.F. (1992) Hydroxyl radical cleavage of tRNA in the ribosomal P site. Proc. Natl Acad. Sci. USA, 89, 7851–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata K.R., Agrawal,R.K., Penczek,P., Grassucci,R., Zhu,J. and Frank,J. (1996) Three-dimensional reconstruction of the Escherichia coli 30S ribosomal subunit in ice. J. Mol. Biol., 262, 43–52. [DOI] [PubMed] [Google Scholar]
- La Teana A., Pon,C.L. and Gualerzi,C.O. (1993) Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc. Natl Acad. Sci. USA, 90, 4161–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A., Gualerzi,C.O. and Brimacombe,R. (1995) From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA, 1, 772–782. [PMC free article] [PubMed] [Google Scholar]
- McCutcheon J.P., Agrawal,R.K., Philips,S.M., Grassucci,R.A., Gerchman, S.E., Clemons,W.M.,Jr, Ramakrishnan,V. and Frank,J. (1999) Location of translational initiation factor IF3 on the small ribosomal subunit. Proc. Natl Acad. Sci. USA, 96, 4301–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Sacerdot,C., Graffe,M., Blanquet,S. and Springer,M. (1999) Discrimination by Escherichia coli initiation factor IF3 against initiation on non-canonical codons relies on complementarity rules. J. Mol. Biol., 290, 825–837. [DOI] [PubMed] [Google Scholar]
- Moreau M., de Cock,E., Fortier,P.L., Garcia,C., Albaret,C., Blanquet,S., Lallemand,J.Y. and Dardel,F. (1997) Heteronuclear NMR studies of E.coli translation initiation factor IF3. Evidence that the inter-domain region is disordered in solution. J. Mol. Biol., 266, 15–22. [DOI] [PubMed] [Google Scholar]
- Ohsawa H. and Gualerzi,C. (1983) Chemical modification in situ of Escherichia coli 30S ribosomal proteins by the site-specific reagent pyridoxal phosphate. Inactivation of the aminoacyl-tRNA and mRNA binding sites. J. Biol. Chem., 258, 150–156. [PubMed] [Google Scholar]
- Olsson C.L., Graffe,M., Springer,M. and Hershey,J.W.B. (1996) Physiological effects of translation initiation factor IF3 and ribosomal protein L20 limitation in Escherichia coli.Mol. Gen. Genet., 250, 705–714. [DOI] [PubMed] [Google Scholar]
- Pawlik R.T., Littlechild,J., Pon,C.L. and Gualerzi,C. (1981) Purification and properties of Escherichia coli translational initiation factors. Biochem. Int., 2, 421–428. [Google Scholar]
- Pediconi D., Spurio,R., La Teana,A., Jemiolo,D., Gualerzi,C.O. and Pon,C.L. (1995) Translational regulation of infC operon in Bacillus stearothermophilus. Biochem. Cell Biol., 73, 1071–1078. [DOI] [PubMed] [Google Scholar]
- Pioletti M. et al. (2001) Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J., 20, 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon C.L. and Gualerzi,C. (1974) Effect of initiation factor 3 binding on the 30S ribosomal subunits of Escherichia coli. Proc. Natl Acad. Sci. USA, 71, 4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon C.L., Pawlik,R.T. and Gualerzi,C. (1982) The topographical localization of IF3 on Escherichia coli 30S ribosomal subunits as a clue to its way of functioning. FEBS Lett., 137, 163–167. [DOI] [PubMed] [Google Scholar]
- Sacerdot C., Fayat,G., Dessen,P., Springer,M., Plumbridge,J.A., Grunberg-Manago,M. and Blanquet,S. (1982) Sequence of a 1.26-kb DNA fragment containing the structural gene for E.coli initiation factor IF3: presence of an AUU initiator codon. EMBO J., 1, 311–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdot C., Chiaruttini,C., Engst,K., Graffe,M., Milet,M., Mathy,N., Dondon,J. and Springer,M. (1996) The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol., 21, 331–346. [DOI] [PubMed] [Google Scholar]
- Schluenzen F. et al. (2000) Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell, 102, 615–623. [DOI] [PubMed] [Google Scholar]
- Sette M., Spurio,R., van Tilborg,P., Gualerzi,C.O. and Boelens,R. (1999) Identification of the ribosome binding sites of translation initiation factor IF3 by multidimensional heteronuclear NMR spectroscopy. RNA, 5, 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapkina T.G., Dolan,M.A., Babin,P. and Wollenzien,P. (2000) Initiation factor 3-induced structural changes in the 30S ribosomal subunit and in complexes containing tRNA(f)(Met) and mRNA. J. Mol. Biol., 299, 615–628. [DOI] [PubMed] [Google Scholar]
- Stade K., Rinke-Appel,J. and Brimacombe,R. (1989) Site-directed cross-linking of mRNA analogues to the Escherichia coli ribosome; identification of 30S ribosomal components that can be cross-linked to the mRNA at various points 5′ with respect to the decoding site. Nucleic Acids Res., 17, 9889–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W. (1991) Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol., 219, 37–44. [DOI] [PubMed] [Google Scholar]
- Subramanian A.R. and Davis,B.D. (1970) Activity of initiation factor F3 in dissociating Escherichia coli ribosomes. Nature, 228, 1273–1275. [DOI] [PubMed] [Google Scholar]
- Sussman J.K., Simons,E. and Simons,R.W. (1996) Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol., 21, 347–360. [DOI] [PubMed] [Google Scholar]
- Tate W., Greuer,B. and Brimacombe,R. (1990) Codon recognition in polypeptide chain termination: site-directed crosslinking of termination codon to Escherichia coli release factor 2. Nucleic Acids Res., 18, 6537–6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedin K., Moll,I., Grill,S., Resch,A., Graschopf,A., Gualerzi,C.O. and Bläsi,U. (1999) Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol., 31, 67–77. [DOI] [PubMed] [Google Scholar]
- Weiel J. and Hershey,J.W.B. (1981) Fluorescence polarization studies of the interaction of Escherichia coli protein synthesis initiation factor 3 with 30S ribosomal subunits. Biochemistry, 20, 5859–5865. [DOI] [PubMed] [Google Scholar]
- Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,W. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W. and Gualerzi,C. (1983) Effect of Escherichia coli initiation factors on the kinetics of N-AcPhe-tRNAPhe binding to 30S ribosomal subunits. A fluorescence stopped-flow study. Biochemistry, 22, 690–694. [DOI] [PubMed] [Google Scholar]
- Yu N.J. and Spremulli,L.L. (1997) Structural and mechanistic studies on chloroplast translational initiation factor 3 from Euglena gracilis. Biochemistry, 36, 14827–14835. [DOI] [PubMed] [Google Scholar]