Abstract
Oxanine (Oxa, O), one of the major damaged bases from guanine generated by NO- or HNO2-induced nitrosative deamination, has been considered as a mutagen-potent lesion. For exploring more detailed properties of Oxa, large-scale preparation of Oxa-containing oligodeoxynucleotide (Oxa-ODN) with the desired base sequence is a prerequisite. In the present study, we have developed a chemical synthesis procedure of Oxa-ODNs and characterized thermodynamic properties of Oxa in DNA strands. First, 2′-deoxynucleoside of Oxa (dOxo) obtained from 2′-deoxyguanosine by HNO2-nitrosation was subjected to 5′-O-selective tritylation to give 5′-O-(4,4′-dimethoxytrityl)-dOxo (DMT-dOxo) with a maximum yield of 70%. Subsequently, DMT-dOxo was treated with conventional phosphoramidation, which resulted in DMT-dOxo-amidite monomer with a maximum yield of 72.5%. The amidite obtained was used for synthesizing Oxa-ODNs: the coupling yields for Oxa incorporation were over 93%. The prepared Oxa-ODNs were employed for analyzing the thermodynamic properties of DNA duplexes containing base-matches of O:N [N; C (cytosine), T (thymine), G (guanine) or A (adenine)]. Melting temperatures (Tm) and thermodynamic stability () were found to be lower by 6.83∼13.41°C and 2.643∼6.047 kcal mol−1, respectively, compared with those of oligodeoxynucleotides, which had the same base sequence except that O:N was replaced by G:C (wild type). It has also been found that Oxa-pairing with cytosine shows relatively high stability in DNA duplex compared with other base combinations. The orders of were O:C > O:T > O:A > O:G. The chemical synthesis procedure and thermodynamic characteristics of Oxa-ODNs established here will be helpful for elucidating the biological significance of Oxa in relation to genotoxic and repair mechanisms.
INTRODUCTION
In 1996, we demonstrated for the first time that 2′-deoxyoxanosine (dOxo) forms as one of the main deaminated products from 2′-deoxyguanosine (dGuo) through nitric oxide (NO)- or nitrous acid (HNO2)-induced nitrosative oxidation (1). It was well known that DNA bases with exocyclic amino groups are converted to deaminated forms through nitrosative stress or high temperature (2–5), i.e. 2′-deoxyuridine (dUrd), 2′-deoxythymidine (dThd), 2′-deoxyinosine (dIno) and 2′-deoxyxanthosine (dXao) are resulted by the oxidative deamination from 2′-deoxycytidine (dCyd), 5-methyl-2′-deoxycytidine (5medCyd), 2′-deoxyadenosine (dAdo) and dGuo, respectively (4). Compared with dXao and other deaminated nucleosides (dIno, dUrd and dThd), in which exocyclic nitrogen is oxidized, dOxo is a unique deaminated form in which an endocyclic nitrogen atom of guanine (Gua) is substituted by an oxygen atom (1,6,7).
Since then, theoretical and experimental efforts have been made to elucidate the chemical mechanism of dOxo formation, together with dXao, from dGuo by nitrosative deamination (7–11). Glaser and co-workers (7,10,11) presented that the ring-opened intermediate of Gua pyrimidine, after loss of dinitrogen in Gua diazonium ions, leads to the formation of oxanine (Oxa, base moiety of dOxo) as well as xanthine (Xan, base moiety of dXao). Furthermore, we have shown that the Oxa formation mechanism involves in such a ring-opened intermediate of Gua by isolating and characterizing the Gua-diazoate intermediate (10).
Since dOxo, which is a DNA lesion of dGuo, could be produced in the cellular system by NO, HNO2 or other nitrosating agent (1,6,12), its genotoxic properties including deglycosylation susceptibility, base-pairing stability and base-incorporation patterns have been analyzed (13,14). If Oxa is formed in DNA strands, Oxa can exist for a sufficient time since dOxo moiety is not easily hydrolyzed due to its stable N-glycosidic bond between base and sugar moieties. Also, it was suggested that Oxa could form two hydrogen bondings with either cytosine (Cyt) or thymine (Thy). Therefore, the Oxa generated in cellular genomes may induce the misincorporation of incorrect nucleotides causing G:C to A:T transition (13). In addition, our recent work has revealed that Oxa mediates a novel genotoxic mechanism related to the formation of DNA–protein cross-link (DPC) (15). Since Oxa has an O-acylisourea structure, it is likely that Oxa reacts with intracellular nucleophiles under physiological conditions and would lead to novel mutagenic or lethal events (15–17). In the case of the base-excision repair (BER) system, we observed that bacterial AlkA (3-methyladenine DNA glycosylase II) and endonuclease VIII possess repair activities on Oxa in oligodeoxynucleotides (ODNs) (18). Recently, Cao and co-workers (19,20) reported that bacterial endonuclease V and human AAG (alkyladenine glycosylase) shows BER activity on Oxa. However, another recent report by our group demonstrated that in the case of Oxa-related genotoxicity (Oxa-mediated DPC formation and its relevant events), the nucleotide excision and recombination repair (NER) system would play a more efficient role than BER system (21).
Oxa-containing oligodeoxynucleotide (Oxa-ODN) needs to be prepared to elucidate the biological significance of Oxa from biophysical properties to genotoxic threats in detail. Up to now, enzymatic methods using DNA polymerases, such as T4 polymerase (exo−) or Pol I klenow fragment, have been applied for the preparation of Oxa-ODNs [in this method, 2′-deoxyoxanosine triphosphate (dOTP) was used as a monomer and incorporated to pair with Cyt in DNA strands by DNA polymerase] (14,19–21). However, since the enzymatic method is not appropriate for the large-scale preparation and desired-sequence design, the chemical synthesis method has been required for extending biological and biotechnological research using Oxa-ODNs. Especially, to reveal the Oxa-induced cellular mechanisms, a large amount of Oxa-ODNs should be used as bait molecules for trapping Oxa-response enzymes or DPC-relevant proteins and as reporter molecules for identifying the Oxa-related genotoxic events in cellular system. Also, various kinds of Oxa-ODNs should be prepared for analyzing more detailed biophysical and biochemical properties of Oxa, such as thermodynamic characterization and structural analysis (NMR or X-ray crystallography). In addition, since Oxa has a unique property of reacting with several chemical functional groups such as amine or amine-containing molecules, Oxa-ODNs could be subjected to the recent hot topics of DNA-based nano-biotechnology.
In the present study, we report, for the first time, on the chemical synthesis procedures for preparing dOxo-amidite monomer and Oxa-ODNs. Also, we show the detailed characterization of the synthesized Oxa-ODNs in terms of thermodynamic stabilities of DNA duplexes containing Oxa base-pairings.
MATERIALS AND METHODS
Materials
2-Cyanoethyl-N,N,N′,N′-tetraisopropylphosphoramidite was purchased from Aldrich Chemical Co. (Milwaukee, WI) and all other chemicals for chemical synthesis were obtained from Wako Pure Chemicals (Osaka, Japan). Solvents were acquired from Nacalai Tesques (Osaka, Japan) and anhydrous solvent was used for organic reactions. The reagents for oligonucleotide synthesis (including CPG column and appropriately protected normal nucleosides) were obtained from Glen Researches Co. (Sterling, VA). The enzymes such as nuclease P1 and alkaline phosphatase required for ODN digestion were purchased from Roche Diagnostics (Mannheim, Germany).
Instrument systems
The reversed phase (RP-) HPLC system used for collection of dOxo was composed of a Tosho PX-8010 (controller), a CCPM (pump) and a UV-8010 (UV detector) (Tosho Co, Tokyo, Japan) with a preparative COSMOSIL C18-PAQ column [250 × 28 mm, 5 µm; Nacalai Tesques, (Osaka, Japan)]. For analysis and preparation of DMT-dOxo-amidite, the normal phase (NP-) HPLC system consisted of a Hitachi L-6200 (controller + pump) and an L4000 (UV detector) (Hitachi Co, Tokyo, Japan) with an Ultron VX-SIL column [150 × 4.6 mm (for analysis) or 250 × 20 mm (for preparation), 5 µm; Shinwa Co. (Kyoto, Japan)]. For purification of synthesized ODNs, an RP-HPLC system constructed with a Tosho PX-8020 (controller), a DP-8020 (pump), a CO-8020 (temperature controller) and a PD-8020 (diode detector) with an Ultron VX-ODS column [150 × 4.6 mm (for analysis) or 250 × 10 mm (for purification), 5 µm; Shinwa Co.] was used. NMR spectra were measured on a JEOL ECA-600 NMR spectrometer (1H-NMR; 600 MHz, 13C-NMR; 150 MHz) at 30°C. The chemical shift was referenced to tetramethylsilane (TMS) in DMSO-d6 or CD3CN. The high resolution (HR-) EI mass spectrum was recorded on a JEOL DX-300 at 30 eV. Synthesis of ODNs was carried out with an Applied Biosystems 3400 DNA synthesizer [Applied Biosystems (Foster, LA)]. UV spectra of ODNs were measured on a Shimadzu UV-260 UV-Vis spectrophotometer equipped with an SPR-5 temperature controller. Melting temperature analysis was performed with Shimadzu TMSPC-8 Tm analysis system [Shimadzu Co (Kyoto, Japan)]. CD spectra of DNA complex were obtained on a Jasco spectropolarimeter J-720 [Japan Spectrocopic Co. (Tokyo, Japan)] interfaced with a microcomputer and equipped with temperature controller.
Chemical synthesis of dimethoxytritylated dOxo-phosphoramidite
2′-Deoxyoxanosine (dOxo):dGuo (4 mmol) was incubated with sodium nitrite (NaNO2; 100 mM) in 400 ml of sodium acetate buffer (3.0 M, pH 3.7) at 45°C for 4 h. After neutralization with sodium hydroxide (NaOH; 5 M), reaction mixtures were reduced to half of the volume by rotary evaporator. The solution was filtrated by Millex GS [0.22 µm; Millipore (Bradford, MA)] and applied to a preparative RP-HPLC system [isocratic elution, sodium phosphate buffer (400 µM, pH 7.4) containing 10% (v/v) acetonitrile (CH3CN); flow rate, 6 ml/min]. Further purification and freeze drying of the appropriate fraction gave a white product of dOxo (0.6 mmol, 15% yield from dGuo) [1H NMR (600 MHz, D2O at 30°C): δ = 7.98 (s, 1H, H-2), 6.26 (dd, 1H, H-1′), 4.61 (ddd, 1H, H-3′), 4.12 (ddd, 1H, H-4′), 3.79 (m, 2H, H-5′,5′), 2.77 (ddd, 1H, H-2′), 2.52 (ddd, 1H, H-2′); UV: λmax = 245, 286 (pH 7); MS(FAB) m/z 269 (M+H+)].
5′-O-(4,4′-dimethoxytrityl)-2′-deoxyoxanosine (DMT-dOxo):4-4′-Dimethoxytritylchloride (DMT-Cl, 0.6∼0.8 mmol) was added to a suspension of dOxo (0.4 mmol) in dry dimethylformamide (DMF; 3 ml) and then, imidazole (0.8 mmol) and diisopropylethylammonium mesylate (DIEA-Mes, 0.8 mmol) were added to the solution. DIEA-Mes was prepared as reported by Kataoka and Hayakawa (22). The reaction mixture was allowed to stir at room temperature for 3 h. During this period, the mixture became homogeneous. The reaction mixture was poured into water (900 ml) and the resulting precipitate was collected by suction filtration and dried at room temperature for 24 h. The crude product was re-dissolved in dichloromethane (CH2Cl2) and dried using sodium sulfate (Na2SO4). The re-dissolved solution was concentrated and re-crystallized in hexane to give a white product of DMT-dOxo (0.28 mmol, 70% yield from dOxo). [1H NMR (600 MHz, DMSO-d6 at 30°C): δ = 7.88 (br, 2H, NH2), 7.80 (s, 1H, H-2), 7.31-6.74 (m, 13H, Ar-H), 6.04 (dd, 1H, H-1′), 5.26 (s, 1H, 3′-OH), 4.39 (m, 1H, H-3′), 3.85 (m, 1H, H-4′), 3.75 (s, 6H, Ar-OCH3), 3.06 (m, 2H, H-5′,5′), 2.54 (ddd, 1H, H-2′), 2.22 (ddd, 1H, H-2′); MS(FAB) m/z 571 (M+H+)].
5′-O-(4,4′-dimethoxytrityl)-2′-deoxyoxanosine-3′-O-(2-cynoethyl)-N,N-diisopropyl phosphoramiditethe (DMT-dOxo-amidite):DMT-dOxo (0.35 mmol) and tetrazole (0.35 mmol) was dissolved in anhydrous CH2Cl2 (3 ml) and then, 2-cyanoethyl-N,N,N′,N′-tetraisopropylphosphoramidite (0.525 mmol) was added to this solution. The homogeneous mixture was allowed to stir at ambient temperature for 3 h. The resulting solution was quenched by the addition of methanol (MeOH; 1 ml) followed by diisopropylethylamine (DIEA; 0.15 ml) and ethyl acetate (EtOAc; 3 ml), and washed with 10% sodium hydrogen carbonate (NaHCO3; 3 ml) three times. The organic layer was dried by using Na2SO4 and concentrated to give a viscous oil. The crude product was re-dissolved in CH2Cl2 (2 ml) and subjected to a preparative NP-HPLC system [isocratic elution, EtOAc:CH2Cl2:MeOH =65:35:0.33 (by vol); flow rate, 6 ml/min]. Reduced-pressure evaporation of the appropriate fraction gave white product of DMT-dOxo-amidite (0.29 mmol, 72.5% yield from DMT-dOxo). [1H NMR (600 MHz, CD3CN at 30°C) and 13C NMR (150 MHz, CD3CN at 30°C) were arranged in Table 1; 31P NMR (320 MHz, CD3CN at 30°C): δ = 151.094, 151.278 ppm (diastereoisomer); MS(FAB) m/z 772 (M+H+)].
Table 1.
13C, 1H and 31P NMR data of dimethoxytrityl-protected phoshoramidite of dOxo (DMT-dOxo-amidite)
| Amidite A | Amidite B | |||
|---|---|---|---|---|
| 31P | 151.094 | 31P | 151.278 | |
| 13C | 1H | 13C | 1H | |
| 1 | ||||
| 2 | 137.644 | 7.618 s | 137.572 | 7.625 s |
| 3 | ||||
| 3a | 153.355 | 153.443 | ||
| 4 | ||||
| 5 | 155.102 | 155.110 | ||
| 5-NH2 | 5.971 s | 5.976 s | ||
| 6 | ||||
| 7 | 160.550 | 160.590 | ||
| 7a | 113.744 | 113.696 | ||
| R1 | 84.806 | 6.103 dd | 84.661 | 6.106 t |
| R2 | 39.474 | 2.765 ddd | 39.510 | 2.781 ddd |
| 2.484 ddd | 2.544 ddd | |||
| R3 | 74.238 | 4.716 dddd | 74.635 | 4.688 dddd |
| R4 | 86.660 | 4.160 ddd | 86.404 | 4.124 ddd |
| R5 | 64.656 | 3.312 dd | 64.648 | 3.278 dd |
| 3.286 dd | 3.241 dd | |||
| A | 129.095 | 7.401 t | 129.031 | 7.387 d |
| B | 128.855 | 7.268 t | 128.855 | 7.258 t |
| C | 127.893 | 7.213 t | 127.885 | 7.206 t |
| D | 146.136 | 146.128 | ||
| A′ | 131.130 | 7.277 t | 131.090 | 7.261 d |
| B′ | 114.065 | 6.827 d | 114.073 | 6.818d |
| C′ | 159.748 | 159.748 | ||
| C′-OCH3 | 55.955 | 3.759 s | 55.947 | 3.754 s |
| D′ | 136.899 | 136.827 | ||
| A″ | 131.066 | 7.277 d | 131.050 | 7.261 d |
| B″ | 114.065 | 6.815 d | 114.073 | 6.808 d |
| C″ | 159.732 | 159.732 | ||
| C″-OCH3 | 55.955 | 3.757 s | 55.947 | 3.753 s |
| D″ | 136.833 | 136.827 | ||
| E | 87.177 | 87.169 | ||
| iPr-CH3 | 24.941 | 1.170 d | 24.977 | 1.155 d |
| iPr-CH | 44.097 | 3.599 dq | 44.129 | 3.577 dq |
| iPr′-CH3 | 24.941 | 1.157 d | 24.893 | 1.078 d |
| iPr′-CH | 44.097 | 3.599 dq | 44.129 | 3.577 dq |
| -OCH2 | 59.384 | 3.693 dddd | 59.400 | 3.805 ddt |
| 3.639 dddd | 3.730 ddt | |||
| -CH2CN | 21.043 | 2.534 ddd | 21.111 | 2.683 t |
| 2.486 ddd | ||||
| -CN | 119.505 | 119.657 | ||
Note: for naming and numbering of each atom, please see Figure 2.
Synthesis and purification of oxanine-containing ODNs
ODNs were synthesized by phosphoramidite chemistry beginning with 5′-DMT-DNA base bound CPG support (0.2 or 1 µmol scale CPG). For coupling of deoxynucleoside phosphoramidites, the standard protocol was used with a DNA synthesizer (ABI 3400 DNA synthesizer). The 5′-terminal DMT group was not removed by the synthesizer. The synthesized DMT-on oligomers were cleaved from the CPG support and deprotected by treatment with a solution of the conc. NaOH (0.5 M) at 37°C for 24 h. After treatment with triethylamine-acetate buffer (TEAA; 2 M, pH 7.0), the mixture solution was subjected to a Poly-Pak cartridge [Glen Researches Co. (Sterling, VA)] to give a purified DMT-off ODN. Detritylated ODN was further purified by an RP-HPLC system using a gradient of CH3CN using Eluent A [5% CH3CN in 100 mM TEAA (pH 7.0)] and Eluent B [15% CH3CN in 100 mM TEAA (pH 7.0)]; 26% (0 min)–90% (40 min) of Eluent B (flow rate : 2 ml/min). The appropriate sample was subjected to the final confirmation by an RP-HPLC system with the same gradient condition mentioned above (flow rate: 1 ml/min).
The purified ODNs were subjected to enzymatic digestion to analyze the component nucleosides. The ODN (10 µg) was dissolved in 50 µl of sodium acetate buffer (50 mM, pH 5.8) containing zinc chloride (5 mM). The DNA was digested to deoxynucleoside monophosphates by the addition of nuclease P1 (8 U) and incubation at 37°C for 3 h. Following addition of 50 µl sodium acetate buffer (50 mM, pH 7.4), phosphate groups were removed with alkaline phosphatase (15 U) and phosphodiesterase I (0.01 U; optional treatment) by incubation at 37°C for 6 h. The reaction mixture was directly analyzed by an RP-HPLC system with a linear gradient of 0% (0 min)–20% (20 min) of CH3CN in 100 mM TEAA at a flow rate of 1 ml/min.
Biophysical characterization of oxanine-containing ODNs
For melting temperature analysis, all DNA solutions were prepared in a phosphate buffer composed of 1 M NaCl, 10 mM Na2HPO4 and 1 mM Na2EDTA adjusted to pH 7.0. ODN concentrations were determined on the absorbance measured at 260 nm using ɛ values of nucleosides. Melting curves of DNA duplexes were obtained for the solutions containing a 1:1 strand ratio of ODNs with an increase in temperature from 20 to 90°C at a rate of 0.2°C/min. The melting temperatures were measured at various total concentrations (Ct) of ODN (between 1 and 50 µM). The thermodynamic parameters (enthalpy change, ΔH0; entropy change, ΔS0) for each DNA duplex formation were estimated by the linear van't Hoff equation, versus ln(Ct/4) (23). For analysis of the whole DNA structure, CD spectra of DNA duplex were measured. All CD spectra were recorded from 350 to 210 nm at 25°C with a scan speed 100 nm/min in a jacketed cylindrical cuvette with a path length of 10 mm. The cuvette-holding chamber was flushed with a constant stream of dry N2 gas to avoid moisture condensation on the cuvette exterior. The total strand concentration of the samples was 16 µM in the same buffer solutions as used for the melting temperature studies. All the CD data were accumulated 10 times and processed using a noise reduction program.
RESULTS AND DISCUSSION
Chemical preparation of DMT-dOxo-amidite
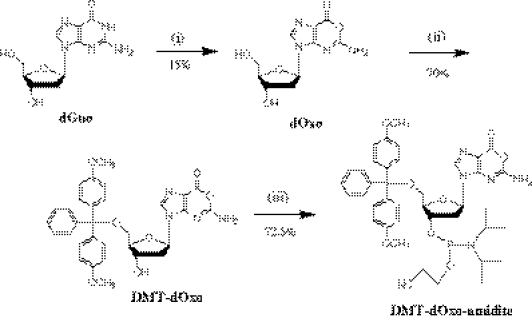
As summarized in Figure 1, a chemical preparation procedure of dimethoxytrityl-protected amidite of dOxo (DMT-dOxo-amidite) was developed in this study.
Figure 1.
Chemical synthesis of dimethoxytrityl-protected phoshoramidite of dOxo (DMT-dOxo-amidite): (i) dOxo from dGuo in NaNO2/sodium acetate buffer (3.0 M, pH 3.7), 4 h. (ii) Dimethoxytritylation of dOxo using dimethoxytritylchloride, imidazole, and diisopropylethylammonium mesylate in dry DMF, 3 h. (iii) Phosphoramidation of DMT-dOxo using 2-cyanoethyl- N,N,N′,N′-tetraisopropylphosphoramidite and tetrazole in anhydrous CH3CN, 3 h.
In the first step (preparation of dOxo), as reported previously (1), dGuo was incubated in a weakly acidic HNO2 solution, which was mediated by NaNO2 (100 mM) in a sodium acetate buffer (3.0 M, pH 3.7), at 45°C. Even though dGuo was not completely converted to deaminated products such as dXao and dOxo, the above nitrosative reaction was terminated in 4 h by neutralization of NaOH because the products including dOxo began to precipitate after 4 h incubation: It should be noted that the precipitates are difficult to re-dissolve in aqueous solution, making the recovering process of dOxo complex. HNO2-treated dGuo solution was subjected to large-scale isocratic RP-HPLC separation to purify dOxo. The mobile phase employed in the preparative RP-HPLC was sodium phosphate buffer (400 µM, pH 7.4) containing 10% CH3CN: low concentrated phosphate salt was found to be efficient for successful separation of dOxo from the reaction mixture (Supplementary Figure S1). Finally, sodium phosphate salts were removed from the collected dOxo fraction by another RP-HPLC system (mobile phase condition; 8% CH3CN aqueous solution). The final dOxo yield from dGuo was ∼15%: the conversion from the reacted dGuo was similar to that of our previous report (22∼25%). In other paper, it was reported that dOxo could be chemically synthesized from dIno, through photochemical transformation by UV-irradiation of 1-hydoxy derivatives of dIno (24). This chemical procedure is composed of several steps and the yield of dOxo was not higher than that achieved by the nirosation conversion from dGuo (the yield of dOxo from dIno is 26%). Moreover, for obtaining highly purified dOxo, RP-HPLC separation is also required. Therefore, our method described above is more effective so far.
In the second step (preparation of DMT-dOxo), a 5′-O selective dimethoxytritylation method was employed on the basis of another previous report (22). DIEA-Mes (2.0 equiv. to dOxo) was applied to acquire the 5′-O selectivity of tritylation (22). dOxo, DMT-Cl, DIEA-Mes and imidazole were incubated and their mole ratios were optimized. It was found that the amount of DMT-dOxo was dependent on the amount of DMT-Cl. However, when the ratio of DMT-Cl to dOxo is over 2:1, a large amount of by-product was observed to be formed (in the case that the ratio was over 3:1, the portion of by-product was ∼45%). Ratios of DIEA-Mes and imidazole were not determinant factors to the yield of DMT-dOxo. The best ratio of dOxo, DMT-Cl, DIEA-Mes and imidazole was determined as 1:2:2:2. This 5′-O selective dimethoxytritylation method is efficient to yield 80% conversion of dOxo to DMT-dOxo (Supplementary Figure S2). By-product (<10%) and unreacted dOxo (<10%) were easily removed by the combination of large-scale precipitation in water and re-crystallization in hexane. The final yield for the conversion of dOxo to DMT-dOxo was ∼70%, which is comparable with that of DMT-dGuo synthesis (74%) obtained by a similar method (22).
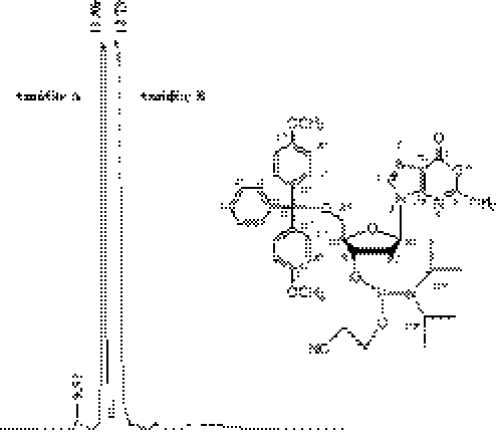
In the third step (preparation of DMT-dOxo-amidite), DMT-dOxo was subjected to the normal phosphoramidation method, in which tetrazole and 2-cyanoethyl- N,N,N′,N′-tetraisopropylphosphoramidite were used. To increase the purity and the yield of DMT-dOxo-amidite, the product was purified by preparative NP-HPLC. It should be noted that TEA, generally used as an additive modifier for increasing the resolution of NP-HPLC, was not appropriate for separation of DMT-dOxo-amidite. Instead, a small amount of MeOH (0.33%) was added to the NP-HPLC mobile phase composed of EtOAc and CH2Cl2 (65:35). As shown in Figure 2, two peaks of DMT-dOxo-amidite (diastereoisomer) were obtained. In this preparation step, DMT-dOxo was subjected to the phosphoramidite modification without protection of exo-NH2. Therefore, this step using unprotected nucleoside substrate could be expected to generate a large quantity of by-product. However, the final yield from DMT-dOxo was as high as 72.5% (max.), which is comparable with the phosphoramidation yields obtained by the previous methods composed of several steps without base protection (25). Although the yield is less than that in the cases of protected normal deoxynucleotides (∼90%), this high conversion without large by-product would be practically sufficient since DMT-dOxo-amidite is obtained from dGuo in only three steps. The structural assignment performed using 1H, 13C and 31P NMR evidenced the purified sample as DMT-dOxo-amidite (Supplementary Figure S-3). NMR data for each diastereoisomer of DMT-dOxo-amidite are summarized in Table 1.
Figure 2.
NP-HPLC chromatogram of dimethoxytrityl-protected phoshoramidite of dOxo (DMT-dOxo-amidite). NP-HPLC condition: elution; isocratic with EtOAc:CH2Cl2:MeOH = 65:35:0.33, flow rate; 1 ml/min, column; Ultron VX-SIL (150 × 4.6 mm, 5 µm), temperature; ambient, detection; 11.86 and 12.79 min (diastereoisomer).
Chemical incorporation of Oxa base into ODNs
Using DMT-dOxo-amidite monomer prepared above, several kinds of oxanine-containing oligodeoxynucleotides (Oxa-ODNs) were synthesized as listed in Table 2.
Table 2.
List of ODNs synthesized in this study
| ID | Sequence | cf |
|---|---|---|
| Sequence for spectroscopic or enzymatic analysis | ||
| OT | 5′-OT-3′ | O:Oxanine base |
| 5mer Oxa | 5′-GCOAT-3′ | O:Oxanine base |
| Sequence for coupling test | ||
| COG-18 | 5′-TCT CTT CCO GCC TAC GAT-3′ | O:Oxanine base |
| TOA-18 | 5′-TCT CTT CTO ACC TAC GAT-3′ | O:Oxanine base |
| GOC-18 | 5′-TCT CTT CGO CCC TAC GAT-3′ | O:Oxanine base |
| AOT-18 | 5′-TCT CTT CAO TCC TAC GAT-3′ | O:Oxanine base |
| Complementary sequence for Tm analysis | ||
| N_COG-18 | 5′-ATC GTA GGC NGG AAG AGA-3′ | N = A,G,T,C |
| N_TOA-18 | 5′-ATC GTA GGT NAG AAG AGA-3′ | N = A,G,T,C |
| Control ODN for Tm analysis | ||
| CGG-18 | 5′-TCT CTT CCG GCC TAC GAT-3′ | |
| TGA-18 | 5′-TCT CTT CTG ACC TAC GAT-3′ | |
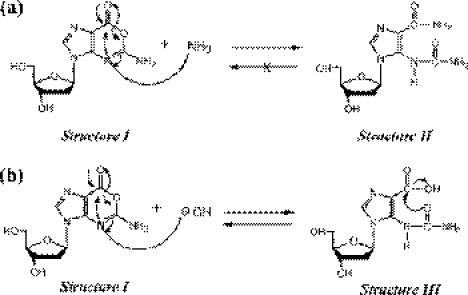
In the conventional method, ammonium hydroxide solution (28% NH4OH) is used for the CPG-cleavage and base-protection removal after the oligonucleotide synthesis. However, conc. NH4OH-treatment is not applicable for Oxa-ODN synthesis since Oxa base has high reactivity with primary amine as reported previously (16,17), and therefore, as suggested in Figure 3a, it reacts with NH3 to form irreversible ring-opened compound: after 1 h incubation of dOxo in conc. NH4OH solution, irreversible compound was observed by RP-HPLC and found to be a ring-opened form of dOxo-NH3 adduct (Structure II of Figure 3a) (HPLC data not shown). On the other hand, Oxa base was reported to exist in the ring opening/closure equilibrium in NaOH solution (13,26). As shown in Figure 3b, since Structure III, which is formed at high pH, is reversible to Structure I in neutral conditions, the Oxa base can be left intact. After 24 h incubation of dOxo in NaOH solution, no other compound derived from dOxo was observed in RP-HPLC data. Therefore, in the present study, conc. NaOH solution (0.5 M) was applied to CPG-cleavage and base-protection removal after the synthesis of Oxa-ODNs. For efficient base-protection removal by NaOH, Ac-dC amidite (acetyl protected dC amidite), Pac-dC amidite (phenoxyacetyl protected dA amidite) and iPr-Pac-dG amidite (4-isopropyl-pheoxyacetyl protected dG amidite) were used instead of benzoyl protected amidite (dC and dA) or isobutyl protected amidite (dG).
Figure 3.
Proposed mechanism of Oxa base response to NH3OH (a) and NaOH (b), which shows ring-opened form of dOxo-NH3 adduct (a: Structure II is irreversible with Structure I) and ring opening/closure forms of dOxo in the presence of hydroxide (b: Structure III is reversible with Structure I).
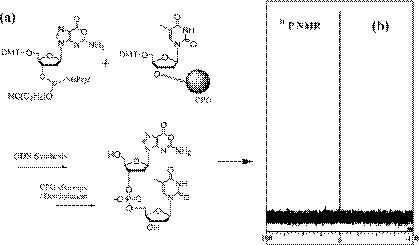
Next, we explored whether any change of Oxa takes place in ODN synthesis by analysis of OT-ODN (sequence 5′-OT-3′; O = Oxa and T = Thy). After DMT-dOxo-amidite was incorporated into the dT-CPG column (1 µmol scale), as depicted in Figure 4a, the DMT-off sample of OT-ODN was analyzed. When amine-unprotected phosphoramidite was employed in ODN synthesis, some by-products may be generated, i.e. free exo-NH2 of amidite might react with another activated phosphoramidite monomer. If this is the case, two or several peaks would appear in 31P NMR spectra. Another 31P NMR analysis reported that the amine-unprotected dAdo and dCyd induce some amount of by-product, whereas amine-free dGuo amidite present no by-product (25). In the case of OT-ODN sample, only one signal was observed at 0 p.p.m. in 31P NMR, as shown in Figure 4b, indicating that no by-product was produced in amine-free Oxa incorporation. Moreover, enzymatic digestion of the obtained OT-ODN sample with nuclease P1 and alkaline phosphatase showed only two peaks of dOxo and dThd (data not shown). These results demonstrate that the synthesized Oxa-ODN possesses intact form of Oxa.
Figure 4.
31P NMR analysis of the synthesized Oxa-ODN, which shows no change of Oxa base during the DNA synthesis using DMT-dOxo-amidite (a: schematic procedure of OT-ODN (5′-OT-3′) preparation. b: 31P NMR (320 MHz) data of the synthesized OT-ODN in CD3CN at 30°C).
Then, as summarized in Table 2, several kinds of Oxa-ODNs and their complementary ODNs were synthesized. In cases of synthesis of Oxa-ODNs, DMT cation released from detritylation of each step was monitored to evaluate the coupling efficiency before and after incorporation of DMT-dOxo-amidite into ODN. For each sequence, 1 µmol CPG-scale ODN synthesis was performed three times to get the average values of the coupling yield. As shown in Table 3, all the coupling yields of DMT-dOxo-amidite were observed to be over 93%. The coupling yields of Oxa after Gua (G), Ade (A), Cyt (C) and T were 93.39, 96.28, 98.06 and 99.09%, respectively. After Oxa incorporation, the coupling yield of the next amidite monomer was not influenced (c∼d column in Table 3; the efficiencies of C, T, G and A were 97.82, 96.60, 97.28 and 97.60%, respectively). Moreover, before and after DMT-dOxo-amidite incorporation, the average of step-wise coupling yield was unchanged (a∼b column versus d∼e column in Table 3). In all these cases, the average values of step-wise coupling yield during the Oxa-ODN synthesis were around 97% (a∼e column in Table 3). These results imply that the incorporated Oxa, amine-free form, does not mediate any severe influence to ODN chemical synthesis.
Table 3.
Coupling efficiency in chemical synthesis of several Oxa-ODNs
| Sequence | Position | Coupling yield | ||||
|---|---|---|---|---|---|---|
| e d c b a | a∼ba | b∼c (%) | c∼d (%) | d∼ea | a∼ea | |
| COG-18 | 5′-TCT CTT CCO GCC TAC GAT-3′ | 96.96% ± 2.44% | 93.39 | 97.82 | 96.92 ± 2.59% | 96.70 ± 2.52% |
| TOA-18 | 5′-TCT CTT CTO ACC TAC GAT-3′ | 96.58% ± 2.23% | 96.28 | 96.60 | 97.64 ± 1.19% | 97.02 ± 1.79% |
| GOC-18 | 5′-TCT CTT CGO CCC TAC GAT-3′ | 97.09% ± 2.30% | 98.06 | 97.28 | 96.59 ± 1.26% | 97.16 ± 2.23% |
| AOT-18 | 5′-TCT CTT CAO TCC TAC GAT-3′ | 97.28% ± 1.86% | 99.09 | 97.60 | 97.42 ± 1.32% | 97.40 ± 1.86% |
aAverage step-wise coupling yield.
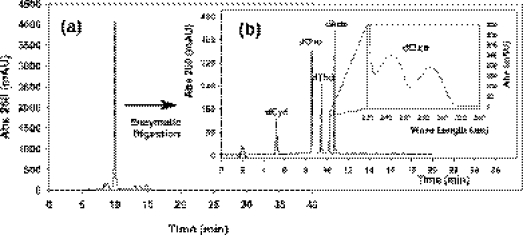
As given in Figure 5, more detailed status of Oxa-ODN was presented by exemplifying 5mer Oxa-ODN (sequence 5′-GCOAT-3′). After conc. NaOH (0.5 M) was used for CPG-cleavage and deprotection of the synthesized ODN sample, alkali-tolerant Poly-Pak cartridge was employed for desalting NaOH and purifying Oxa-ODN. As shown in Figure 5a, the detritylated 5mer Oxa-ODN was obtained with over 90% purity. The appropriate fraction of Oxa-ODN was collected by further RP-HPLC purification and was subjected to enzymatic digestion with nuclease P1 and alkaline phosphatase. As presented in Figure 5b, the corresponding peaks of all the expected deoxynucleosides including dOxo were observed. Mass spectroscopy analysis (Supplementary Figure S-4) also supported that 5mer Oxa-ODN is the normal oligomers composed of dOxo and other four deoxynucleosides. All the data demonstrate that the Oxa incorporated into ODN can survive the various steps during and post DNA synthesis, i.e. the chemical preparation method established here produce Oxa-ODN successfully.
Figure 5.
RP-HPLC separation of detritylated 5mer Oxa-ODN (5′-GCOAT-3′) purified by Poly-Pak cartridge (a), and DNA monomer, dCyd, dGuo, dThd, dOxo and dAdo formed by the digestion of with nuclease P1 and alkaline phosphatase (b). HPLC condition: mobile phase; 100 mM TEAA solution (pH 7.0) with a gradient of CH3CN [for (a), 7.6% (0 min)∼ 14% (40 min) (please see Materials and Methods) and for (b), 0% (0 min)∼20% (20 min)], with flow rate; 1 ml/min, column; Ultron VX-ODS columns [150 × 4.6 mm, 5 µm; Shinwa Co. (Kyoto, Japan)].
Thermodynamic characterization of Oxa-ODNs
To see whether the base-paring between Oxa and normal bases affects DNA duplex conformation or not, CD analysis has been performed for all the DNA complementation types (Supplementary Figure S-5). Similarly to a previous report (13), all CD data were almost identical to each other, indicating that such Oxa-pairing with normal bases does not evoke any severe influence on the whole conformation of DNA duplex. However, the local structure around Oxa in DNA duplex might be affected by the base-pair type correlated to thermodynamic stability of Oxa-containing DNA duplex (Oxa-DNA duplex).
For analyzing the thermodynamic stability of Oxa-DNA duplex, melting temperature and thermodynamic parameters were obtained using the prepared Oxa-ODNs as summarized in Table 4. In a previous report (13), we have already shown the melting temperatures (Tm) for Oxa-containing oligothymidine, d(T5OT6), which was prepared by the HPLC purification of HNO2-treated d(T5GT6). The Tm for the duplexes of d(T5OT6)/d(A6NA5) (N = C, G, A and T) were 14.1∼19.3°C, which were much lower than that of d(T5GT6)/d(A6CA5) (Tm = 32.8°C). Although these were the Tm values reported for Oxa-DNA duplex for the first time, the thermodynamic properties of Oxa-DNA duplex have not been elucidated in detail because of the low Tm values.
Table 4.
Melting temperatures and thermodynamic parameters of DNA duplexes containing O:N and G:C base-pairing
| Type | Sequence | X | N | Tma (°C) | ΔTmb(°C) | −ΔH0 (kcal mol−1) | −ΔS0 (cal mol−1 K−1) | c (kcal mol−1) | d (kcal mol−1) |
|---|---|---|---|---|---|---|---|---|---|
| O | C | 64.49 | −6.83 | 130.396 | 371 | 15.293 | 2.643 | ||
| -CXG- | 5′-TCT CTT CCX GCC TAC GAT-3′ | O | T | 61.15 | −10.17 | 126.664 | 365 | 13.494 | 4.442 |
| -GNC- | 3′-AGA GAA GGN CGG ATG CTA-5′ | O | G | 62.57 | −8.75 | 105.994 | 303 | 11.889 | 6.047 |
| O | A | 62.68 | −8.64 | 113.296 | 323 | 13.073 | 4.863 | ||
| G | C | 71.32 | 0 | 135.595 | 379 | 17.936 | 0 | ||
| O | C | 56.80 | −9.92 | 130.574 | 381 | 12.332 | 3.648 | ||
| -TXA- | 5′-TCT CTT CTX ACC TAC GAT-3′ | O | T | 55.37 | −11.35 | 124.935 | 367 | 11.118 | 4.862 |
| -ANT- | 3′-AGA GAA GAN TGG ATG CTA-5′ | O | G | 53.31 | −13.41 | 115.422 | 339 | 10.191 | 5.789 |
| O | A | 54.51 | −12.21 | 117.079 | 343 | 10.563 | 5.417 | ||
| G | C | 66.72 | 0 | 132.679 | 376 | 15.980 | 0 |
aTm was measured in case that the total concentrations of ODNs is 4 µM.
bΔTm was calculated by Tm−Tm(G:C).
c was calculated by ΔH0 − (310.15 × ΔS0).
d was calculated by (G:C).
As represented in Table 4, various duplex formations between Oxa-ODNs and several complementary ODNs (the total concentrations of ODNs; 4 µM) were analyzed. In the cases of -COG-/-GNC- type DNA duplexes, Tm of O:C, O:T, O:G and O:A were 64.49, 61.15, 62.57 and 62.68°C, respectively. Compared with a perfectly matched DNA duplex (Tm of -CGG-/-GCC-; 71.32°C), the differences of Tm [ΔTm = Tm (G:C) − Tm (O:N)] were 6.83°C (O:C), 10.17°C (O:T), 8.75°C (O:G) and 8.64°C (O:A). In the cases of -TOA-/-ANT- types, Tm values were 56.80°C (O:C), 55.37°C (O:T), 53.31°C (O:G) and 54.51°C (O:A). Compared with Tm of G:C (66.72°C), Tm values of O:C, O:T, O:G and O:A were lower by 9.92, 11.35, 13.41 and 12.21°C, respectively. These differences are similar to the common difference reported for one mismatch (i.e. the cases of G:T, G:G and G:A). Among them, O:C duplexes made relatively stable formations compared with other combinations of Oxa base-pairings.
After repeatedly measuring the Tm values in various total concentrations (Ct), the thermodynamic parameters were obtained for each duplex, as listed in Table 4. ΔH0 and ΔS0 were calculated on the basis of the linear van't Hoff equation using versus ln(Ct/4) plot (Supplementary Figure S-6). Then, [=ΔH0− (310.15 × ΔS0)] were compared for analyzing the stability patterns of DNA duplex in 37°C according to Oxa base-parings. In cases of -COG-/-GNC- type, values were 15.293 (O:C), 13.494 (O:T), 11.889 (O:G) and 13.073 kcal mol−1 (O:A). Their differences [ (G:C) − (O:N)] were 2.643 (O:C), 4.442 (O:T), 6.047 (O:G) and 4.863 (O:A) kcal mol−1. The order of Oxa-paring stabilities in -COG-/-GNC-DNA duplex are O:C > O:T >O:A > O:G. The cases of -TOA-/-ANT- types showed the similar results to those of -COG-/-GNC. values of O:C, O:T, O:G and O:A are 12.332, 11.118, 10.191 and 10.563 kcal mol−1, respectively, and were 3.648, 4.862, 5.789 and 5.417 kcal mol−1, respectively. Oxa-pairing stabilities in -TOA-/-ANT- type follows the same orders of -COG-/-GNC- type as O:C > O:T > O:A > O:G.
Although all the base-matches of Oxa are less stable than a G:C perfect match in DNA duplex, Oxa forms relatively stable base-paring with Cyt compared with other matches with Thy, Gua and Ade. As proposed in previous reports (13,14), Oxa with an acceptor–acceptor–donor configuration can form two hydrogen bondings with Cyt. This result suggested that Oxa would maintain relatively stable interaction with Cyt without severe influence on the local structure of DNA duplex even after Oxa is formed from Gua. It is probable that such a base-paring stability of O:C would account for the recent results of repair enzyme responses, i.e. the general BER system is not effective for repairing Oxa in DNA strands (15,21). This is because BER enzyme might not easily recognize (and hydrolyze) Oxa due to the relative stable base-pairing with Cyt in DNA duplex (15,21).
In a previous paper, O:T pairs had been expected to show similar stability to that of O:C because Oxa might also have two possible hydrogen bondings with Thy. However, the expected hydrogen bonding between ring oxygen of Oxa and the imino proton of Thy would be weak or could not be formed because the ring oxygen atom of Oxa base has an sp3 hybrid orbital and the lone pairs of electrons of oxygen exist out of plane. As represented in Table 4, O:T pairs in DNA duplex showed low thermodynamic stability compared to O:C pairs. In the cases of O:A and O:G base-parings, low stabilities were also observed compared with O:C pairs indicating that Oxa has a very low chance to make hydrogen-bonding with Ade and Gua. However, O:A was observed to show better thermodynamic stability than O:G and similar stabilityto O:T.
In the previous reports involving base-incorporation and relevant mutagenesis, Oxa in DNA strand has been found to induce the misincorporation of Thy and Ade opposite Oxa, which may mediate the mutagenic conversion of G:C to A:T during the DNA polymerization (14,19,20). Thermodynamic stabilities of Oxa base-pairing, presented here, show a similar trend to the incorporation efficiency of four normal bases opposite Oxa in a DNA strand; O:C > O:T > O:A >O:G (14,19).
CONCLUSION
Since Oxa is one of the major damaged bases generated from guanine by NO- or HNO2-induced nitrosative deamination, Oxa has been analyzed in terms of its biochemical and biophysical properties (1,8,9,13–21,26). Although several papers have been reported previously, Oxa-related research still needs to be further investigated, especially, Oxa-related genotoxicity and repair mechanisms in cellular systems. To achieve these, there has been a need to establish a conventional method by which Oxa-ODN with desired base sequence can be prepared in large quantities. In this study, we have developed chemical synthesis procedures for Oxa-ODNs. For obtaining DMT-protected phosphoramidite of Oxa (DMT-dOxo-amidite), we have established the following three-steps procedures: (i) nitrosation of dGuo by acidic nitrite condition (dOxo; 15% yield), (ii) 5′-O-selective tritylation of dOxo on the basis of the method reported previously (22) (DMT-dOxo; 70% yield), (iii) phosphoramidation of DMT-dOxo (DMT-dOxo-amidite; 72.5% yield). Prepared DMT-dO-amidite was successfully used for synthesizing Oxa-ODN with a coupling efficiency of over 93%. Enzymatic digestion and spectroscopic data including NMR and mass spectroscopy supported that the incorporated Oxa is in an intact form in the ODN. Also, we have analyzed the thermodynamic properties of Oxa in DNA strands. Melting temperature (Tm) and thermodynamic parameters (, and ) were obtained to observe the thermodynamic stability of DNA duplexes containing Oxa-base pairings. All the duplexes containing O:N base pairs showed Tm values lower than that of a G:C perfect match by 6.83∼10.17°C in 5′-COG-3′ type duplexes and 9.92∼13.41°C in 5′-TOA-3′ types. Using , it was observed that all the DNA duplexes with O:N base pairs showed less thermodynamic stability than that of G:C by 2.643∼6.047 kcal mol−1. By comparing Tm and values, it has been found that O:C pairs have a relatively higher stability in DNA duplex than O:T, O:A and O:G. The orders are O:C > O:T > O:A > O:G.
Xan, another main damaged base of nitrosated Gua, has been investigated in terms of its biochemical role in DNA strands using the chemically synthesized Xan-containing ODN (27,28). However, in the case of Oxa, there has been no efficient method for the preparation of Oxa-ODN. To reveal the whole mechanism of NO-induced genotoxicity or cytotoxicity, it is also essential to study more detailed roles of both Oxa and Xan in cellular systems. Especially, a large amount of the designed Oxa-ODNs is required for analyzing Oxa-induced DPC formation and Oxa-response cellular events. Therefore, this paper introduces the total chemical synthesis procedure of Oxa-ODNs and thermodynamic characteristics of Oxa-ODNs for the first time. These results shown here will be helpful for elucidating the biological significance of Oxa related to genotoxic and repair mechanisms. Moreover, since Oxa possesses a kind of carboxylate function, Oxa-ODN can be used as one of the functional DNA oligomers, which can make covalent cross-linkage with amine or amine-containing biomolecules. Therefore, the results obtained here will be applied to extending the biotechnological application of Oxa-ODN, for example, novel biomolecule conjugations or nano-biostructure fabrication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work is partially supported by the Grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan to K.M. (No. 15350099), and by the Center of Excellence (COE) program for the ‘Establishment of COE on Sustainable-Energy System’, Grand-in-aid for Science and Technology Promotion from the Ministry of Education, Science, Sports and Culture, Japan. This work was also supported by CREST of the Japan Science and Technology Corporation. Funding to pay the Open Access publication charges for this article was provided by MEXT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Suzuki T., Yamaoka R., Nishi M., Ide H., Makino K. Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligodeoxyribonucleotide, and calf thymus DNA treated by nitrous acid and nitric oxide. J. Am. Chem. Soc. 1996;118:2515–2516. [Google Scholar]
- 2.Karran P., Lindahl T. Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry. 1980;19:6005–6011. doi: 10.1021/bi00567a010. [DOI] [PubMed] [Google Scholar]
- 3.Peng W., Shaw B.R. Accelerated deamination of cytosine residues in UV-induced cyclobutane pyrimidine dimers leads to CC→TT transitions. Biochemistry. 1996;35:10172–10181. doi: 10.1021/bi960001x. [DOI] [PubMed] [Google Scholar]
- 4.Tamir S., Burney S., Tannenbaum S.R. DNA damage by nitric oxide. Chem. Res. Toxicol. 1996;9:821–827. doi: 10.1021/tx9600311. [DOI] [PubMed] [Google Scholar]
- 5.Spencer J.P., Whiteman M., Jenner A., Halliwell B. Nitrite-induced deamination and hypochlorite-induced oxidation of DNA in intact human respiratory tract epithelial cells. Free Radic. Biol. Med. 2000;28:1039–1050. doi: 10.1016/s0891-5849(00)00190-8. [DOI] [PubMed] [Google Scholar]
- 6.Lucas L.T., Gatehouse D., Shuker D.G. Efficient nitroso group transfer from N-nitrosoindoles to nucleotides and 2′-deoxyguanosine at physiological pH. A new pathway for N-nitrosocompounds to exert genotoxicity. J. Biol. Chem. 1999;274:18319–18326. doi: 10.1074/jbc.274.26.18319. [DOI] [PubMed] [Google Scholar]
- 7.Glaser R., Son M.S. Pyrimidine ring opening in the unimolecular dediazoniation of guanine diazonium ion. An ab initio theoretical study of the mechanism of nitrosative guanosine deamination. J. Am. Chem. Soc. 1996;118:10942–10943. [Google Scholar]
- 8.Hernandez B., Soliva R., Luque F.J., Orozco M. Misincorporation of 2′-deoxyoxanosine into DNA: a molecular basis for NO-induced mutagenesis derived from theoretical calculations. Nucleic Acids Res. 2000;28:4873–4883. doi: 10.1093/nar/28.24.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki T., Ide H., Yamada M., Endo N., Kanaori K., Tajima K., Morii T., Makino K. Formation of 2′-deoxyoxanosine from 2′-deoxyguanosine and nitrous acid: mechanism and intermediates. Nucleic Acids Res. 2000;28:544–551. doi: 10.1093/nar/28.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayat S., Glaser R. 5-Cyanoimino-4-oxomethylene-dihydroimidazole and nitrosative guanine deamination. A theoretical study of geometries, electronic structures and N-Protonation. J. Org. Chem. 2003;68:9882–9892. doi: 10.1021/jo0351522. [DOI] [PubMed] [Google Scholar]
- 11.Qian M., Glaser R. 5-Cyanoamino-4-imidazolecarboxamide and nitrosative guanine deamination: Experimental evidence for pyrimidine ring-opening during deamination. J. Am. Chem. Soc. 2004;126:2274–2275. doi: 10.1021/ja0389523. [DOI] [PubMed] [Google Scholar]
- 12.Lucas L.T., Gatehouse D., Jones G.D., Shuker D.G. Characterization of DNA damage at purine residues in oligonucleotides and calf thymus DNA induced by the mutagen 1-nitrosoindole-3-acetonitrile. Chem. Res. Toxicol. 2001;14:158–164. doi: 10.1021/tx000179r. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki T., Matsumura Y., Ide H., Kanaori K., Tajima K., Makino K. Deglycosylation susceptibility and base-pairing stability of 2′-deoxyoxanosine in oligodeoxynucleotide. Biochemistry. 1997;36:8013–8019. doi: 10.1021/bi970166l. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki T., Yoshida M., Yamada M., Ide H., Kobayashi M., Kanaori K., Tajima K., Makino K. Misincorporation of 2′-deoxyoxanosine 5′-triphosphate by DNA polymerases and its implication for mutagenesis. Biochemistry. 1998;37:11592–11598. doi: 10.1021/bi980971f. [DOI] [PubMed] [Google Scholar]
- 15.Nakano T., Terato H., Asagoshi K., Masaoka A., Mukuta M., Ohyama Y., Suzuki T., Makino K., Ide H. DNA–protein cross-link formation mediated by oxanine. A novel genotoxic mechanism of nitric oxide-induced DNA damage. J. Biol. Chem. 2003;278:25264–25272. doi: 10.1074/jbc.M212847200. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T., Yamada M., Ishida T., Morii T., Makino K. Reactivity of 2′-deoxyoxanosine, a novel DNA lesion. Nucleic Acids Symp. Ser. 1999;42:7–8. doi: 10.1093/nass/42.1.7. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T., Yamada M., Ide H., Kanaori K., Tajima K., Morii T., Makino K. Identification and characterization of a reaction product of 2′-deoxyoxanosine with glycine. Chem. Res. Toxicol. 2000;13:227–230. doi: 10.1021/tx990164x. [DOI] [PubMed] [Google Scholar]
- 18.Terato H., Masaoka A., Asagoshi K., Honsho A., Ohyama Y., Suzuki T., Yamada M., Makino K., Yamamoto K., Ide H. Novel repair activities of AlkA (3-methyladenine DNA glycosylase II) and endonuclease VIII for xanthine and oxanine, guanine lesions induced by nitric oxide and nitrous acid. Nucleic Acids Res. 2002;30:4975–4984. doi: 10.1093/nar/gkf630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hitchcock T.M., Gao H., Cao W. Cleavage of deoxyoxanosine-containing oligodeoxyribonucleotides by bacterial endonuclease V. Nucleic Acids Res. 2004;32:4071–4080. doi: 10.1093/nar/gkh747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hitchcock T.M., Dong L., Connor E.E., Meira L.B., Samson L.D., Wyatt M.D., Cao W. Oxanine DNA glycosylase activity from mammalian alkyladenine glycosylase. J. Biol. Chem. 2004;279:38177–38183. doi: 10.1074/jbc.M405882200. [DOI] [PubMed] [Google Scholar]
- 21.Nakano T., Katafuchi A., Shimizu R., Terato H., Suzuki T., Tauchi H., Makino K., Skorvaga M., Van Houten B., Ide H. Repair activity of base and nucleotide excision repair enzymes for guanine lesions induced by nitrosative stress. Nucleic Acids Res. 2005;33:2181–2191. doi: 10.1093/nar/gki513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kataoka M., Hayakawa Y. A conventional method for the synthesis of N-free 5′-O-(p,p′-dimethoxytrityl)-2′-deoxyribonucleodises via the 5′-O-selective tritylation of the parent substances. J. Org. Chem. 1999;64:6087–6089. [Google Scholar]
- 23.Santa Lucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Napoli L., Di Fabio G., Messere A., Montesarchio D., Piccialli G., Varra M. A new synthesis of oxanosine and 2′-deoxyoxanosine. Tetrahedron Lett. 1998;39:7397–7400. [Google Scholar]
- 25.Hayakawa Y., Kataoka M. Facile synthesis of oligodeoxynucleotides via the phosphoramidite method without nucleoside base protection. J. Am. Chem. Soc. 1998;120:12395–12401. [Google Scholar]
- 26.Suzuki T., Yamada M., Ide H., Kanaori K., Tajima K., Morii T., Makino K. Influence of ring opening-closure equilibrium of oxanine, a novel damaged nucleobase, on migration behavior in capillary electrophoresis. J. Chromatogr. A. 2000;877:225–232. doi: 10.1016/s0021-9673(00)00178-3. [DOI] [PubMed] [Google Scholar]
- 27.Wuenschell G.E., O'Connor T.R., Termini J. Stability, miscoding potential, and repair of 2′-deoxyxanthosine in DNA: Implications for nitric oxide-induced mutagenesis. Biochemistry. 2003;42:3608–3616. doi: 10.1021/bi0205597. [DOI] [PubMed] [Google Scholar]
- 28.Vongchampa V., Dong M., Gingipalli L., Dedon P. Stability of 2′-deoxyxanthosine in DNA. Nucleic Acids Res. 2003;31:1045–1051. doi: 10.1093/nar/gkg177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.