Abstract
Cdc18/Cdc6 and Cdt1 are essential initiation factors for DNA replication. In this paper we show that expression of Cdc18 in fission yeast G2 cells is sufficient to override the controls that ensure one S phase per cell cycle. Cdc18 expression in G2 induces DNA synthesis by re-firing replication origins and recruiting the MCM Cdc21 to chromatin in the presence of low levels of Cdt1. However, when Cdt1 is expressed together with Cdc18 in G2, cells undergo very rapid, uncontrolled DNA synthesis, accumulating DNA contents of 64C or more. Our data suggest that Cdt1 may potentiate re-replication by inducing origins to fire more persistently, possibly by stabilizing Cdc18 on chromatin. In addition, low level expression of a mutant form of Cdc18 that cannot be phosphorylated by cyclin-dependent kinases is not sufficient to induce replication in G2, but does so only when co-expressed with Cdt1. Thus, regulation of both Cdc18 and Cdt1 in G2 plays a crucial role in preventing the re-initiation of DNA synthesis until the next cell cycle.
Keywords: Cdc18/Cdt1/cell cycle/DNA replication/licensing
Introduction
In all eukaryotes, cells have evolved control mechanisms to restrict DNA synthesis to once per cell division cycle. These mechanisms ensure that DNA synthesis is confined to the S phase of the cell cycle, and prevented during the G2 phase when the chromosomes have been fully replicated. Abrogation of this control leads to increases in ploidy and genomic instability, which are ultimately lethal to the cell. The central feature of replication control is the licensing of chromosomes for DNA replication. Licensing establishes whether the chromosomes are competent to undergo replication and is determined by the availability of factors that are required for the initiation step of DNA synthesis (Blow and Laskey, 1988). A hallmark of replication-competent chromosomes is the pre-replicative complexes (pre-RCs) that assemble onto origins of replication in G1 (Diffley et al., 1994). These protein complexes are targeted to origin DNA via association with the origin recognition complex (ORC) (Bell and Stillman, 1992; Diffley and Cocker, 1992; Gavin et al., 1995; Romanowski et al., 1996; Rowles et al., 1996; Tugal et al., 1998; Moon et al., 1999). In yeast, as cells exit mitosis and enter G1, the replication initiation factor Cdc18/Cdc6 (called Cdc18 in fission yeast and Cdc6 in budding yeast) accumulates and binds to ORC (Liang et al., 1995; Grallert and Nurse, 1996; Rowles et al., 1996; Saha et al., 1998). In conjunction with the Cdt1 protein and possibly other proteins, Cdc18/Cdc6 recruits the minichromosome maintenance (MCM) complex to origin DNA (Aparicio et al., 1997; Donovan et al., 1997; Tanaka et al., 1997; Ogawa et al., 1999; Maiorano et al., 2000; Nishitani et al., 2000). At this stage, the DNA is licensed for replication. At the onset of S phase, Cdc45 binds to pre-RCs, which are then activated by the Cdc7/Dbf4 and cyclin-dependent kinases (CDKs), and origins fire (reviewed by Leatherwood, 1998; Donaldson and Blow, 1999).
The assembly of the pre-RC is critical to the G1 control regulating the onset of S phase. ORC is bound to origins throughout the cell cycle in yeast and the recruitment of pre-RC components in G1 appears to be dependent on the prior association of Cdc18/Cdc6 with ORC (Diffley et al., 1994; Aparicio et al., 1997; Liang and Stillman, 1997; Tanaka et al., 1997; Lygerou and Nurse, 1999; Ogawa et al., 1999). In all eukaryotes, Cdc18/Cdc6 is strictly regulated during the cell cycle, although how it is regulated varies between organisms. In the yeasts, Cdc18/Cdc6 accumulates in mitosis and G1, and is targeted for proteolysis at the onset of S phase (Kelly et al., 1993; Zwerschke et al., 1994; Nishitani and Nurse, 1995; Piatti et al., 1995; Detweiler and Li, 1997; Jallepalli et al., 1997; Kominami and Toda, 1997; Baum et al., 1998). In higher eukaryotes, Cdc18/Cdc6 is present in both interphase and mitosis, but is modified during the cell cycle by phosphorylation (Coleman et al., 1996; Saha et al., 1998; Jiang et al., 1999; Petersen et al., 1999). In human cells, a fraction of chromatin-bound Cdc18/Cdc6 persists during S phase and G2 (Coverley et al., 2000; Mendez and Stillman, 2000), but phosphorylation of nucleoplasmic Cdc18/Cdc6 relocalizes the protein from the nucleus to the cytoplasm during S phase (Jiang et al., 1999; Petersen et al., 1999). Therefore, Cdc18/Cdc6 may be one of the limiting factors for pre-RC assembly in all organisms.
Once cells have replicated their DNA and enter the G2 phase, the re-firing of replication origins is inhibited until the next cell cycle. In the fission yeast Schizosaccharomyces pombe, the targets of this cell cycle control have not been identified and it is not known which initiation factors are limiting for the re-licensing of origins in G2. Two candidates are Cdc18 and Cdt1 since they are both extensively downregulated in G2 (Nishitani and Nurse, 1995; Nishitani et al., 2000). Cdc18/Cdc6 has an essential function in loading the MCM complex onto chromatin (Nishitani et al., 2000); this implies that it may be necessary to dissociate Cdc18/Cdc6 from chromatin once initiation occurs to prevent further re-loading of MCMs. In addition, re-replication in the absence of intervening mitoses can be induced by overexpression of Cdc18 in exponentially growing cells and potentiated by co-expression of Cdt1 (Nishitani and Nurse, 1995; Nishitani et al., 2000). While it is not known from which stage of the cell cycle re-replication is induced, it is possible that high levels of Cdc18 trap cells in S phase and, together with other proteins such as Cdt1, allow persistent loading of the MCMs and the replication machinery onto chromatin. However, if Cdc18 and Cdt1 are bona fide licensing factors, they should also be sufficient to override the cell cycle controls that prevent the re-firing of origins in G2. In this paper, we have expressed Cdc18 and Cdt1 in G2-arrested cells to test the capability of these proteins to serve as licensing factors. We have shown that they are sufficient to re-initiate DNA synthesis from G2 by specifically re-firing origins and loading the MCM Cdc21 onto chromatin. Thus, their proper regulation is essential to prevent DNA replication in G2.
Results
Cdc18 induces replication in G2
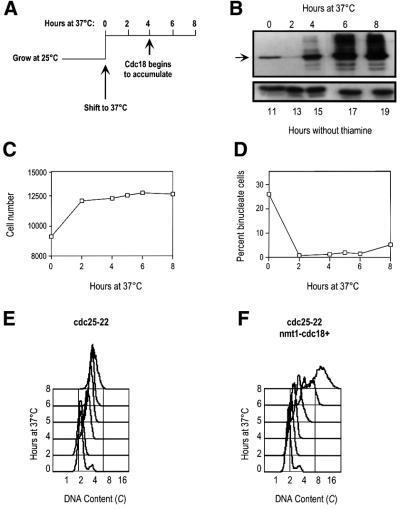
To determine whether downregulation of Cdc18 is important to block replication after the completion of S phase, we expressed Cdc18 ectopically in cells arrested in G2 and assessed whether these cells could re-replicate. Cdc18 was expressed under the control of the regulatable nmt1 promoter in cdc25-22 cells (cdc25-22 nmt1-cdc18+; kindly provided by H.Nishitani). These cells block the cell cycle at the G2/M boundary when shifted to the restrictive temperature of 37°C. To ensure that the inducible Cdc18 was expressed only when all cells had arrested in G2, the nmt1 promoter was derepressed 11 h before shifting the culture to 37°C. Under these conditions, the endogenous Cdc18 was degraded after 2 h at 37°C while the ectopically expressed Cdc18 accumulated after 4 h at 37°C (Figure 1A and B). These cells maintained an effective block over mitosis throughout the incubation at 37°C; after 2 h at 37°C, there was no further increase in cell number and the percentage of binucleate cells remained <5% (Figure 1C and D). The control strain cdc25-22 exhibited a similar block over mitosis (data not shown).
Fig. 1. Overexpression of Cdc18 in cdc25-22 cells induces DNA synthesis. (A) Schematic of the experimental system. cdc25-22 nmt1-cdc18+ cells were grown at 25°C for 11 h in the absence of thiamine to induce the expression of Cdc18, then shifted to 37°C for 8 h. (B) The expression of Cdc18 was monitored by western blotting. The arrow indicates the Cdc18 protein. Tubulin serves as a loading control. (C) Measurements of cell number in the cdc25-22 nmt1-cdc18+ culture following incubation at 37°C. (D) Percentage of binucleates in the experiment described in (C). (E) FACS analysis of cdc25-22 cells shifted to 37°C. (F) FACS analysis of cdc25-22 nmt1-cdc18+ cells shifted to 37°C in the absence of thiamine.
We next assessed whether high levels of Cdc18 in G2 cells were sufficient to induce replication. Using fluorescence-activated cell sorter (FACS) analysis, we measured the DNA content of the cdc25-22 and cdc25-22 nmt1-cdc18+ strains when shifted to 37°C in the absence of thiamine. In the control strain cells arrested with a 2C DNA content, as expected for G2 cells, and there was no further increase in DNA content during the 8 h incubation at 37°C (Figure 1E). It should be noted that the FACS peak in these cells drifted towards the right as they persisted in the block due to an autofluorescence artefact as a result of cell elongation (Sazer and Sherwood, 1990). In contrast, when Cdc18 was overexpressed in the cdc25-22 block, the DNA content began to increase by 6 h at 37°C and significant re-replication was evident after 8 h at 37°C (Figure 1F, note the log scale). Taking into account the effect of cell elongation on apparent DNA content, we estimate that these cells had undergone at least two doublings of DNA content by the end of the time course. This increase in DNA content coincides with the accumulation of Cdc18 protein (Figure 1B), suggesting that overexpression of Cdc18 is driving these cells to re-replicate from G2.
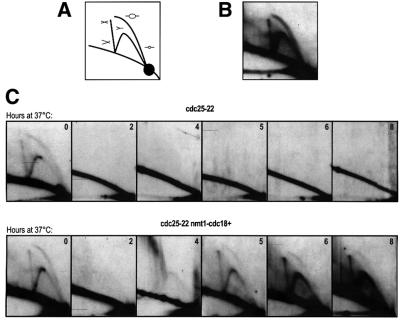
Replication origins re-fire illegitimately in G2 cells overexpressing Cdc18
While FACS analysis revealed a bulk increase in DNA content, we next determined whether this was arising due to de novo replication initiation or origin firing in these G2 cells. Using neutral–neutral two-dimensional (2D) gel electrophoresis to detect replication intermediates in a specific region of the genome (Friedman and Brewer, 1995), we tested whether origin firing occurred at the ars3001 replication origin when Cdc18 was overexpressed in G2. In wild-type cells, 2D gels probed for ars3001 detected both a bubble arc and a fork arc, representing initiation and continuing replication, respectively (Figure 2A and B). This is consistent with previous 2D gel results for this origin (Sanchez et al., 1998). To test whether origin firing occurs in G2 cells overexpressing Cdc18, genomic DNA was extracted from the cdc25-22 control and cdc25-22 nmt1-cdc18+ cells before the shift to 37°C and every 2 h thereafter. Digested DNA was run on 2D gels and probed for ars3001 (Figure 2C). In both strains, the typical pattern of replication intermediates (Figure 2B) was detected in cells before the shift to 37°C. In the control strain, replication intermediates were not detected by 2 h after the shift to 37°C and for the remainder of the block, consistent with all cells arresting in G2 (Figure 2C, upper panel). In the strain overexpressing Cdc18, replication intermediates were not detected at 2 and 4 h at 37°C. However, as Cdc18 began to accumulate by 5 h, both replication forks and initiation bubbles appeared and persisted as cells re-replicated (Figure 2C, lower panel). This shows unequivocally that origins are induced to fire illegitimately in G2 and that the mechanism that blocks origin firing is sensitive to the levels of Cdc18 in the cell.
Fig. 2. Cdc18 promotes origin firing in G2 cells. (A) Schematic of replication intermediates detectable by 2D gel electrophoresis. The bubble arc corresponds to origin firing, the fork arc corresponds to passive replication, and the X-spike corresponds to recombination intermediates or termination events. (B) 2D gel of genomic DNA extracted from exponentially growing wild-type cells and probed with a 3 kb HindIII–KpnI fragment that recognizes the origin ars3001. (C) 2D gel of genomic DNA extracted from cdc25-22 cells and cdc25-22 nmt1-cdc18+ cells, probed for ars3001. Samples were collected at the indicated times (hours) following the shift to 37°C in the absence of thiamine.
We consistently observed that the ratio between the bubble and fork arcs changed as cells re-replicated. At 5 h, the ratio was less than that observed in exponentially growing cells (Figure 2C, 0 and 5 h time points in the lower panel) and, as cells increased their DNA content, the bubble arc became progressively weaker and was dominated by a stronger fork arc. This suggests that the origin firing in G2 is less efficient than during a normal S phase.
Association of initiation factors with chromatin during re-replication
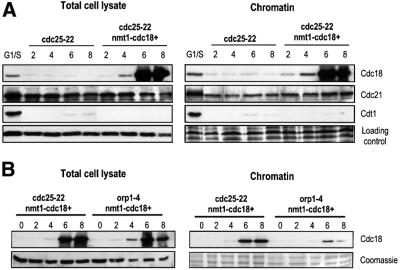
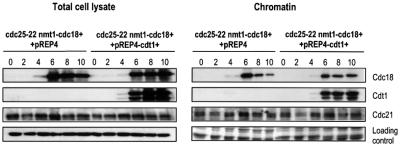
During G1, components of the pre-replicative complex become sequentially associated with chromatin in preparation for origin firing at the onset of S phase (Ogawa et al., 1999; Kearsey et al., 2000; Nishitani et al., 2000). We were interested in determining whether components of the pre-RC become re-associated with chromatin in cdc25-22 nmt1-cdc18+ arrested cells that are induced to re-replicate. Total cell lysates and chromatin-associated fractions were prepared from cdc25-22 and cdc25-22 nmt1-cdc18+ cells every 2 h throughout the incubation at 37°C. Equal amounts of each, as judged by the respective loading controls, were run on SDS–PAGE (Figure 3A). As a control for the increased chromatin association of pre-RC components that is observed in preparation for a normal S phase, cdc25-22 cells were arrested for 4 h at 37°C, then shifted back to the permissive temperature of 25°C to allow cells to re-enter the cell cycle synchronously. Cells were collected 60 min after the shift to 25°C, which corresponds to the G1/S phase. Fractionation of these cells and western blotting confirmed that increased Cdc18, the MCM Cdc21 and Cdt1 became associated with chromatin in G1/S cells relative to the cdc25-22 cells arrested in G2.
Fig. 3. Chromatin association of replication initiation factors during re-replication. (A) Total cell lysates and chromatin fractions were extracted from cdc25-22 or cdc25-22 nmt1-cdc18+ cells collected every 2 h following the shift to 37°C as described in Figure 1A and western blotted with antibodies to Cdc18, Cdc21 and Cdt1. The lane G1/S refers to cdc25-22 cells arrested at 37°C for 4 h, shifted down to 25°C and collected 60 min later. Tubulin and Coomassie Blue staining serve as loading controls for the total cell lysate and chromatin fractions, respectively. Note that equal amounts of protein were loaded from each fraction but not the same ratio of the chromatin fraction relative to the total cell lysate. (B) Cdc18 was overexpressed in cdc25-22 and orp1-4 temperature-sensitive mutants as described in Figure 1A. Cells were shifted to 37°C in the absence of thiamine and samples collected every 2 h for 8 h. Total cell lysates and chromatin fractions were analysed by western blotting using anti-Cdc18 antibodies. Tubulin and Coomassie Blue staining serve as loading controls for the total cell lysate and chromatin fractions, respectively.
We first examined whether Cdc18 itself becomes associated with chromatin when overexpressed (Figure 3A). Concurrent with expression of high levels of Cdc18 (6 and 8 h at 37°C), we detected a significant increase in the amount of Cdc18 that was associated with chromatin. This corresponds to the time when origin firing had re-initiated (Figure 2C), and cells began to increase their DNA content as measured by FACS (Figure 1F). To address the possibility that this apparent increase in Cdc18 binding to chromatin was an artefact due to excess Cdc18 protein in the cell, we followed a similar time course with a strain overexpressing Cdc18 in an orp1-4 temperature-sensitive mutant (kindly provided by H.Nishitani). Orp1+ encodes one of the subunits of the ORC and is required for the initiation of DNA synthesis (Grallert and Nurse, 1996). When we compared the amount of Cdc18 in the total cell lysate of the orp1-4 mutant with the cdc25-22 mutant, we found that it was overexpressed to approximately similar levels in both strains when compared with the tubulin loading control (Figure 3B). However, in the orp1-4 mutant only a small fraction of the Cdc18 in the total cell lysate was associated with chromatin. This indicates that the binding of high levels of Cdc18 to chromatin in the cdc25-22 strain that was detected in our assay is specific, and confirms that binding of Cdc18 to chromatin requires Orp1.
We next examined whether the chromatin association of one of the MCMs, Cdc21, was increased when Cdc18 was overexpressed during the cdc25-22 arrest (Figure 3A). We found that the levels of Cdc21 in the total cell lysate remained constant in both the control and Cdc18-overexpressing cells. In the chromatin fraction, we detected a low level of Cdc21 binding during the G2 block, which increased upon overexpression of Cdc18 to nearly the same level detected in G1/S cells. When the total cell lysates and chromatin fractions were blotted for Cdt1, none was detected in the cdc25-22 arrested cells (Figure 3A), consistent with the protein being downregulated in G2. Surprisingly, no Cdt1 was detected in either fraction during re-replication. This was in stark contrast to the accumulation of Cdt1 in cells and on chromatin that normally occurs in G1/S cells. From these results, we conclude that there is a large increase in the amounts of Cdc18 and a more modest increase in the amounts of Cdc21 that become re-associated with chromatin during re-replication, and that high levels of Cdc18 may recruit Cdc21 to chromatin with apparently undetectable amounts of Cdt1.
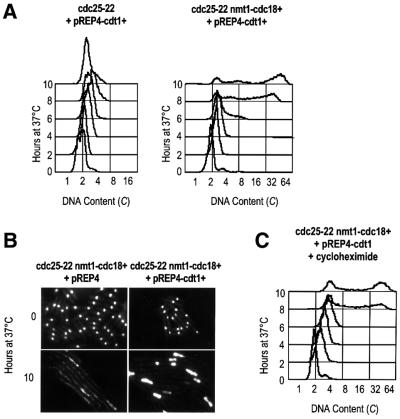
Cdt1 potentiates the re-replication induced by Cdc18 in G2
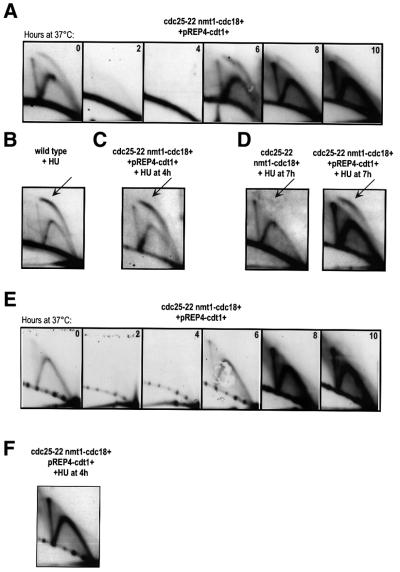
To assess whether the inefficient re-replication observed in response to high levels of Cdc18 results from limited amounts of Cdt1 present in G2 cells, we tested the effect of co-expressing Cdt1 with Cdc18 in the cdc25-22 arrest. In these experiments, Cdt1 was expressed on a pREP4 plasmid under the control of the strongest nmt1 promoter in the same way as Cdc18. By FACS analysis, we found that overexpression of Cdt1 alone in G2 cells failed to induce DNA synthesis (Figure 4A). This result shows that while Cdc18 is sufficient to redirect a G2 cell back into S phase and induce all the necessary events leading to the initiation of DNA synthesis, Cdt1 cannot overcome the inhibitory controls that prevent replication in G2. However, when Cdt1 was co-expressed with Cdc18, cells underwent massive re-replication from G2 (Figure 4A). These cells contained large amounts of DNA (perhaps as much as 64C) within 4 h of the accumulation of Cdc18 and Cdt1 (see the western blot in Figure 5). Consistent with this, 4′,6-diamidino-2-phenylindole (DAPI) staining of these nuclei detected significantly more DNA in cells expressing both Cdc18 and Cdt1 compared with those expressing Cdc18 alone (Figure 4B). From the FACS profiles, we estimate that the presence of Cdt1 reduced the replication doubling time from 2 h to ∼30 min. While it has been shown that Cdt1 can potentiate re-replication in cells expressing lower levels of Cdc18, such rapid DNA synthesis was not observed and cells only proceeded to 8–16C within a similar time period (Nishitani et al., 2000).
Fig. 4. Cdt1 potentiates re-replication induced by Cdc18. (A) FACS analysis of cdc25-22 cells and cdc25-22 nmt1-cdc18+ cells expressing Cdt1 on a pREP4 plasmid, shifted to 37°C in the absence of thiamine. (B) DAPI staining of cells expressing either Cdc18 alone or both Cdc18 and Cdt1 at 0 and 10 h after the shift to 37°C in the absence of thiamine. (C) FACS analysis of cells expressing both Cdc18 and Cdt1 shifted to 37°C in the absence of thiamine and treated with 100 µg/ml cycloheximide at 6 h.
Fig. 5. Association of pre-RC components upon co-expression of Cdc18 and Cdt1. Total cell lysates and chromatin fractions were extracted from cdc25-22 nmt1-cdc18+ cells transformed with either pREP4 or pREP4-cdt1+ every 2 h following the shift to 37°C and western blotted for Cdc18, Cdt1 and Cdc21. Tubulin and Coomassie Blue staining serve as loading controls for the total cell lysates and chromatin fractions, respectively.
Our results demonstrate that Cdc18 can overcome the block to replication in G2, and establish that Cdt1 is important for determining the extent to which re-replication can occur. We found that the re-replication induced by Cdc18 and Cdt1 was insensitive to cycloheximide, suggesting that replication from G2 and the acceleration of re-replication in response to Cdt1 can occur in the absence of continuing protein synthesis (Figure 4C). This implies that other proteins required for re-initiation must be stable in G2. In fission yeast, the MCMs are very abundant proteins, which are expressed constitutively during the cell cycle (Maiorano et al., 1996; Sherman and Forsburg, 1998). Since Cdc18 and Cdt1 are normally both required to recruit the MCMs to chromatin in preparation for initiation (Ogawa et al., 1999; Kearsey et al., 2000; Nishitani et al., 2000), we assessed whether Cdt1 accelerates re-replication by increasing the levels of chromatin-associated Cdc21. Total cell lysates and fractions enriched for chromatin-associated proteins were prepared from G2-arrested cells expressing Cdc18 either alone or with Cdt1 (Figure 5). In the strain expressing Cdc18 and Cdt1, both proteins accumulated with the same timing and became associated with chromatin. Interestingly, in the absence of any detectable Cdt1, Cdc18 initially bound to chromatin efficiently, but then began to dissociate at the later time point of 10 h. However, in the presence of Cdt1, the levels of Cdc18 associated with chromatin remained constant. While a slight decrease in Cdc18 protein levels was observed in the total cell lysate in the absence of Cdt1, the effect was more striking in the chromatin fraction. This suggests that Cdt1 may stabilize Cdc18 on chromatin, which may contribute to more efficient and rapid re-replication. When Cdc21 levels were examined, we did not detect a significant difference in the association of Cdc21 with chromatin in cells co-expressing Cdt1 compared with the increase observed in cells expressing Cdc18 alone. However, Cdc18 and Cdt1 failed to induce re-replication when overexpressed in a strain carrying a temperature-sensitive mutation in the Cdc21 gene (data not shown). This suggests that while Cdc21 is required for Cdc18-induced re-replication from G2, Cdt1 may potentiate re-replication by a mechanism independent of increased chromatin association of Cdc21. Alternatively, the association of Cdc21 with chromatin during re-replication may be more transient during the rapid DNA synthesis that occurs in the presence of Cdt1. Thus, our chromatin assay may not be sensitive enough to detect large changes in the rates of association and dissociation of Cdc21 during re-replication.
Origin firing in response to Cdt1
We next tested whether Cdt1 accelerated re-replication in G2 by increasing the efficiency of origin firing in cells co-expressing Cdc18 and Cdt1 (Figure 6A). Replication intermediates disappeared when cells arrested in G2 at 2 and 4 h at 37°C, and then re-appeared when Cdc18 and Cdt1 accumulated at 6 h. We found that the ratio of bubbles to forks at ars3001 was initially higher than that observed in cells expressing Cdc18 alone (Figure 6A; compare with Figure 2C), although not as high as in a normal S phase (Figure 6A, zero time point). Once again, as cells continued to re-replicate we observed a decrease in the ratio between the bubble arc and the fork arc. This suggests that Cdt1 is not increasing the number of origins that fire, but instead may be affecting the rate at which origins can re-fire to initiate another round of DNA synthesis or allowing more persistent origin firing. This latter hypothesis is supported by the 2D gels of cells treated with hydroxyurea (HU), an inhibitor of DNA synthesis. In wild-type cells, replication bubbles can expand for several kilobases in the presence of HU and then arrest (Santocanale and Diffley, 1998). Using 2D gels, we detected a more intense bubble arc in wild-type cells treated with HU, which facilitated the detection of origin firing (Figure 6B). In G2 cells expressing Cdc18 and Cdt1, the bubble arc was clearly detectable after 2 h in the presence of HU (Figure 6C). We also observed a strong bubble arc in cells expressing both Cdc18 and Cdt1 treated with HU at 7 h at 37°C and collected 1 h later, after which point no bubbles were detected in cells expressing only Cdc18 (Figure 6D). This is consistent with origin firing persisting for an extended period in the presence of Cdt1. Another possible explanation for the accelerated re-replication in response to Cdt1 is that Cdt1 may allow initiation to occur at non-origin sequences. To address this, we hybridized the 2D gels from cells expressing both Cdc18 and Cdt1 with a probe recognizing a non-origin region flanking ars3001 (Figure 6E) (Sanchez et al., 1998). We detected only replication forks in this region, even in the presence of HU (Figure 6F), suggesting that the specificity of origin firing is maintained throughout re-replication within the ars3001 region. However, we cannot exclude the possibility that the 2D gel assay may not be sensitive enough to detect occasional initiation events elsewhere in the rDNA repeat.
Fig. 6. Origin firing in response to Cdt1. (A) 2D gels of genomic DNA extracted from cdc25-22 nmt1-cdc18+ pREP4-cdt1+ cells probed for ars3001. Samples were collected every 2 h following the shift to 37°C in the absence of thiamine. (B) 2D gel of wild-type cells treated with HU (11 mM) for 3.5 h at 32°C, probed for ars3001. The arrow indicates the stronger bubble arc. (C) 2D gel of cdc25-22 nmt1-cdc18+ pREP4-cdt1+ cells shifted to 37°C in the absence of thiamine and treated with HU (24 mM) at 4 h. Samples were collected at 6 h at 37°C, after 2 h of treatment with HU. Gels were probed for ars3001. (D) 2D gels of cdc25-22 nmt1-cdc18+ pREP4 and cdc25-22 nmt1-cdc18+ pREP4-cdt1+ cells shifted to 37°C in the absence of thiamine, treated with HU (24 mM) at 7 h and collected 1 h later. Gels were probed for ars3001. (E) 2D gel of genomic DNA extracted from cdc25-22 nmt1-cdc18+ pREP4-cdt1+ cells and probed for a non-origin region downstream of ars3001. Samples were collected every 2 h following the shift to 37°C in the absence of thiamine. (F) 2D gel of cdc25-22 nmt1-cdc18+ pREP4-cdt1+ cells shifted to 37°C in the absence of thiamine and treated with HU (24 mM) at 4 h. Cells were collected at 10 h after 6 h of treatment with HU. The 2D gel was probed with the same fragment as described in (E).
Cdt1 and stable Cdc18 induce replication in G2
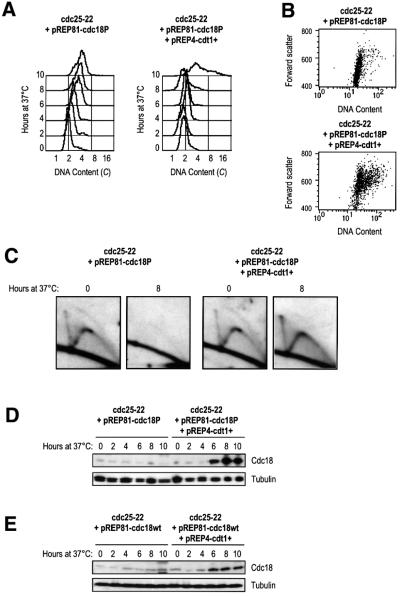
The results described above have identified Cdc18 as a factor that must be downregulated in G2 cells, and Cdt1 as an auxiliary factor that helps Cdc18 promote re-replication. Normally, Cdc18 is strictly regulated during the cell cycle by periodic transcription and proteolysis (Kelly et al., 1993; Kominami and Toda, 1997; Baum et al., 1998). Transcription of Cdc18 is not active in G2 and the available protein is degraded by Cdc2-dependent phosphorylation, which targets the protein for ubiquitin-mediated proteolysis (Jallepalli et al., 1997; Kominami and Toda, 1997; Baum et al., 1998). If Cdc18 is a licensing factor, then stabilization of Cdc18 should promote re-replication. However, mutation of the six Cdc2 phosphorylation sites, which renders the protein stable, fails to induce re-replication when expressed at low levels in exponentially growing cells (Jallepalli et al., 1997; Baum et al., 1998). Our results suggest that Cdt1 may faciliate re-replication induced by Cdc18, particularly if Cdc18 levels are low, and thus may be important in resolving this paradox. We tested whether co-expression of Cdt1 with the non-phosphorylatable form of Cdc18 (Cdc18P) expressed under the control of a weak promoter on a plasmid could replicate in G2. We found that low level expression of the non-phosphorylatable form of Cdc18 could not induce replication in G2, but co-expression of Cdt1 induced DNA synthesis, with some cells accumulating DNA contents of 8–16C (Figure 7A). To circumvent the effects of plasmid loss, elongated cells were selected and plotted against their DNA content (Figure 7B). These plots show that in the presence of Cdt1, but not in cells expressing only Cdc18P, the majority of elongated cells had a DNA content >2C. This was confirmed by 2D gel analysis, which showed that after 8 h at 37°C replication intermediates were detected only in those cells co-expressing Cdt1 (Figure 7C).
Fig. 7. Expression of Cdt1 in G2 induces replication in the presence of stable Cdc18. (A) FACS analysis of cdc25-22 cells transformed with pREP81-cdc18P (left) and pREP4-cdt1+ (right), shifted to 37°C in the absence of thiamine. (B) Dot plots of cells shown in (A), with forward scatter plotted against DNA content. (C) 2D gels of cells shown in (A) collected at 0 and 8 h after the shift to 37°C and probed for ars3001. (D) Boiled extracts from cells in (A) were western blotted for Cdc18. Tubulin serves as a loading control. (E) Boiled extracts were prepared from cdc25-22 cells transformed with pREP81-cdc18wt (left) and pREP4-cdt1+ (right), shifted to 37°C in the absence of thiamine. Samples were run on SDS–PAGE and western blotted for Cdc18. Tubulin serves as a loading control.
When we examined the protein levels of Cdc18 and Cdt1 in these cells, we found that Cdc18 accumulated significantly in the presence of Cdt1 (Figure 7D). Expression of the wild-type Cdc18 (Cdc18wt) in cdc25-22 arrested cells resulted in a small increase in the protein level when Cdt1 was co-expressed (Figure 7E). This suggests that Cdt1 may protect both the wild-type and non-phosphorylatable forms of Cdc18, although the mutant form can accumulate to higher levels. These data lead us to two conclusions. First, that Cdt1 allows low levels of the non-phosphorylatable Cdc18 to induce replication in G2. Secondly, that mechanisms that prevent the accumulation of Cdc18 in G2 include one dependent on Cdc2 phosphorylation, and another that only occurs when Cdt1 is absent.
Discussion
In this paper we have identified an essential role for the downregulation of Cdc18 and Cdt1 in preventing licensing of chromatin in the G2 phase of the cell cycle. We have found that the presence of Cdc18 and Cdt1 in G2 cells is sufficient to induce DNA synthesis and reverse the inhibition of origin firing. The implications of this finding are that a G2 cell, which should be in a state incompetent to undergo DNA synthesis, can be transformed into a G1-like cell and re-initiate DNA synthesis. This suggests that the global cell cycle controls that define a cell as being in G2 or G1 are dependent on the availability of factors such as Cdc18 and Cdt1. This is consistent with the requirement for Cdc10-dependent transcription of Cdc18 and Cdt1 for re-replication in response to depletion of mitotic CDK activity (Kelly et al., 1993; Hayles et al., 1994; Hofmann and Beach, 1994). These cells re-set the cell cycle from G2 back to G1 and undergo multiple complete rounds of G1–S–G2. An important distinction in the re-replication described here is that Cdc18 and Cdt1 are unlikely to be promoting repeated rounds of a normal S phase. By FACS analysis we detected broad peaks of DNA content, not like the discrete peaks corresponding to complete genome duplications (Hayles et al., 1994), and cell viability was severely compromised (data not shown). Further characterization of this re-replication by 2D gel analysis revealed that origin firing was initially less efficient than in a normal S phase and the ratio of bubbles to forks decreased further as cells increased their DNA content, suggesting that origins re-fired progressively less efficiently. When Cdt1 was co-expressed with Cdc18, the origins initially fired with greater efficiency than with Cdc18 alone, but the efficiency also declined as DNA content increased. However, re-replication was greatly potentiated in the presence of Cdt1, thus identifying a previously undescribed function for Cdt1 in accelerating the re-replication induced by Cdc18. While Cdt1 did not appear to increase the number of origins that fired or allow initiation to occur in non-origin regions, Cdt1 may play a role in facilitating the continuous re-firing of origins, possibly by stabilizing Cdc18 on chromatin.
These results indicate that Cdc18 and Cdt1 are sufficient to initiate DNA synthesis in G2 and form the core of the control which licenses chromosomes for replication, although other replication factors are also likely to be important. We have shown that only a modest amount of the MCM Cdc21 becomes re-associated with chromatin during re-replication, in comparison with the more significant increase in the re-association of Cdc18. Other pre-RC components may be limiting for the association of Cdc21 with chromatin, which could contribute to inefficient and incomplete genome duplication. In budding yeast, overexpression of Cdc6, the homologue of Cdc18, does not initiate DNA replication in G2 (Piatti et al., 1996; Tanaka et al., 1997). This may be because the localization of the MCMs is cell cycle regulated and activation of the mitotic kinases promotes the export of MCMs from the nucleus during S phase (Labib et al., 1999; Nguyen et al., 2000). Therefore, additional mechanisms may exist to repress re-replication in budding yeast cells, possibly reflecting their short or non-existent G2 period, after which the mitotic kinases accumulate to high levels for much of the cell cycle, preventing re-replication (Dahmann et al., 1995; Piatti et al., 1996). This is in contrast to fission yeast cells, which spend a larger portion of the cell cycle in G2 when the kinase activity is not high enough either to initiate mitotic events or to block re-replication if Cdc18 and Cdt1 are present.
We propose that at least three mechanisms are important to repress replication in G2 in fission yeast. Transcriptional repression of cdc18 and cdt1 in G2 ensures that these genes are not expressed (Kelly et al., 1993; Hofmann and Beach, 1994; Baum et al., 1998). Another mechanism acts by destabilizing remaining Cdc18 protein through Cdc2-dependent phosphorylation (Jallepalli et al., 1997; Baum et al., 1998). A third mechanism functions to downregulate Cdt1, which may be particularly important when Cdc18 is not fully phosphorylated. These pathways are analogous to the mechanisms that prevent licensing in Xenopus metaphase extracts, which require both CDK activity and geminin to block re-replication (Hua et al., 1997; Mahbubani et al., 1997; McGarry and Kirschner, 1998; Wohlschlegel et al., 2000; Tada et al., 2001). High CDK activity inhibits the association of ORC and Cdc6 with chromatin, while geminin is a specific inhibitor of Cdt1 that prevents the chromatin association of MCMs (Tada et al., 2001). Inhibition of CDKs and depletion of geminin in metaphase extracts stimulate licensing of sperm nuclei to the same level as in interphase extracts, allowing replication to initiate from a G2-like state (Tada et al., 2001). Since there is no obvious sequence homologue of geminin in fission yeast to inhibit Cdt1, it is likely that fission yeast and metazoans have evolved different mechanisms by which to regulate the expression of Cdt1, as was shown for Cdc18 (Coleman et al., 1996; Saha et al., 1998; Jiang et al., 1999; Petersen et al., 1999; Coverley et al., 2000; Mendez and Stillman, 2000). However, inhibition of both initiation factors is essential to prevent licensing in G2, emphasizing the strong parallels between the controls that restrict S phase to once per cell cycle in fission yeast and metazoa.
Materials and methods
Schizosaccharomyces pombe strains and methods
All strains were derived from the wild types 972h– and 975h+. Media and growth conditions were as previously described (Moreno et al., 1991). The cdc25-22 nmt1-cdc18+ strain was generated by crossing the cdc25-22 strain (Fantes, 1979) with one containing multiple integrations of nmt1-cdc18+ (Nishitani and Nurse, 1995). The orp1-4 nmt1-cdc18+ strain was generated by crossing the orp1-4 strain (Fantes, 1979) with one containing multiple integrations of nmt1-cdc18+ (Nishitani and Nurse, 1995). Temperature-sensitive mutants were cultured at the permissive temperature of 25°C or at the restrictive temperature of 37°C. All cdc25-22 nmt1-cdc18+ integrants were maintained in EMM plus thiamine (final concentration 5 µg/ml), and expression from the nmt1 promoter was induced after washing cells three times with EMM and resuspending in EMM. Yeast transformations were performed as described previously (Bähler et al., 1998). Cells were fixed and processed for FACS as previously described (Sazer and Sherwood, 1990), counted in a Coulter counter, or rehydrated and stained with DAPI.
2D gel electrophoresis
Approximately 8 × 108 cells were harvested by filtration and washed once with 50 ml of ice-cold buffer (50 mM MOPS pH 7.2, 150 mM potassium acetate, 2 mM magnesium chloride) with 0.1% sodium azide, then washed again with 50 ml of buffer alone. Genomic DNA was purified from cells as described by Wu and Gilbert (1995) and digested with 80 U of restriction enzymes. Precipitated DNA was run on a 0.4% agarose gel in the first dimension and a 1.1% agarose gel in the second dimension as described previously (Friedman and Brewer, 1995). Gels were transferred to GeneScreen Plus membranes (Amersham) using the Posiblot system (Stratagene). Hybridizations were carried out with ∼2 × 106 c.p.m. of random primed probe per millilitre of QuickHyb hybridization solution (Stratagene). After washing, the membranes were exposed to BioMax film (Kodak), scanned and processed in Photoshop. To detect the replication origin ars3001 within the tandem rDNA repeats, genomic DNA was digested with HindIII and KpnI, and probed with the same 3 kb HindIII–KpnI fragment from the rDNA repeat. This fragment recognizes the 100 or more copies of ars3001 within the tandem array of repeats. To detect non-origin DNA within the rDNA repeats, genomic DNA was digested with EcoRI and KpnI, and probed with the same 3.4kb EcoRI–KpnI fragment from the rDNA repeat.
Chromatin association assay
Spheroplast preparation, lysis and chromatin isolation were performed as described previously (Lygerou and Nurse, 1999). Chromatin-associated proteins were released from chromatin by incubation with 277 U of DNase I (Sigma) for 10 min at 25°C in a buffer containing 20 mM HEPES pH 7.9, 1.5 mM magnesium acetate, 50 mM potassium acetate, 1% glycerol, 0.5 mM dithiothreitol (DTT), 150 mM sodium chloride, protease and phosphatase inhibitors. The chromatin fraction was then analysed by SDS–PAGE. Coomassie Blue staining of total proteins in this fraction served as a loading control.
Cell extract preparation
Boiled cell extracts were prepared from 2 × 108 cells as described (Nishitani and Nurse, 1995). Cells were washed once in STOP buffer (150 mM sodium chloride, 50 mM sodium fluoride, 10 mM EDTA, 1 mM sodium azide pH 8.0), resuspended in 150 µl of HB buffer (25 mM MOPS pH 7.2, 60 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, 15 mM EGTA, 1 mM DTT, 1% Triton X-100) and boiled for 5 min. Cells were broken using glass beads and extracts recovered by centrifugation at 13 000 r.p.m. for 15 min at 4°C in an Eppendorf microfuge, and boiled in 5× sample buffer (400 mM Tris–HCl pH 6.8, 50% glycerol, 10% SDS, 500 mM DTT, 0.02% bromophenol blue).
Western blotting
Western blotting was carried out as described previously (Hayles et al., 1994). The antibodies used were anti-Cdc18 (1:1000 dilution) (Nishitani et al., 2000), anti-Cdt1 (1:1000 dilution) (Nishitani et al., 2000), anti-Cdc21 (1:1000 dilution) (Nishitani et al., 2000) and anti-α-tubulin monoclonal antibody (1:10 000; Sigma). Proteins were detected using horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibody and an enhanced chemiluminescence (ECL) detection kit (Amersham).
Acknowledgments
Acknowledgements
We wish to thank Damien Hermand, Hiroshi Murakami, Jacky Hayles and Takashi Toda for helpful comments and suggestions on the manuscript. We thank Hideo Nishitani for providing strains and antibodies. S.K.Y. is supported by the Natural Sciences and Engineering Research Council of Canada and the British Council with an Athlone–Vanier doctoral fellowship, and a grant from the Association for International Cancer Research to P.N.
References
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu,J.-Q., Longtine,M.S., Shah,N.G., McKenzie,A.,III, Steever,A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Baum B., Nishitani,H., Yanow,S. and Nurse,P. (1998) Cdc18 transcription and proteolysis couple S phase to passage through mitosis. EMBO J., 17, 5689–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P. and Stillman,B. (1992) Nucleotide dependent recognition of chromosomal origins of DNA replication by a multi-protein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- Blow J.J. and Laskey,R.A. (1988) A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature, 332, 546–548. [DOI] [PubMed] [Google Scholar]
- Coleman T.R., Carpenter,P.B. and Dunphy,W.G. (1996) The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell, 87, 53–63. [DOI] [PubMed] [Google Scholar]
- Coverley D., Pelizon,C., Trewick,S. and Laskey,R.A. (2000) Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A–cdk2 dependent process. J. Cell Sci., 113, 1929–1938. [DOI] [PubMed] [Google Scholar]
- Dahmann C., Diffley,J.F. and Nasmyth,K.A. (1995) S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol., 5, 1257–1269. [DOI] [PubMed] [Google Scholar]
- Detweiler C. and Li,J. (1997) Cdc6p establishes and maintains a state of replication competence during G1 phase. J. Cell Sci., 110, 753–763. [DOI] [PubMed] [Google Scholar]
- Diffley J.F. and Cocker,J.H. (1992) Protein–DNA interactions at a yeast replication origin. Nature, 357, 169–72. [DOI] [PubMed] [Google Scholar]
- Diffley J.F., Cocker,J.H., Dowell,S.J. and Rowley,A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- Donaldson A.D. and Blow,J.J. (1999) The regulation of replication origin activation. Curr. Opin. Genet. Dev., 9, 62–68. [DOI] [PubMed] [Google Scholar]
- Donovan S., Harwood,J., Drury,L.S. and Diffley,J.F. (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA, 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes P. (1979) Epistatic gene interactions in the control of division in fission yeast. Nature, 279, 428–430. [DOI] [PubMed] [Google Scholar]
- Friedman K.L. and Brewer,B.J. (1995) Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol., 262, 613–627. [DOI] [PubMed] [Google Scholar]
- Gavin K., Hidaka,M. and Stillman,B. (1995) Conserved initiator proteins in eukaryotes. Science, 270, 1667–1677. [DOI] [PubMed] [Google Scholar]
- Grallert B. and Nurse,P. (1996) The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev., 10, 2644–2654. [DOI] [PubMed] [Google Scholar]
- Hayles J., Fisher,D., Woollard,A. and Nurse,P. (1994) Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2/mitotic B cyclin complex. Cell, 78, 813–822. [DOI] [PubMed] [Google Scholar]
- Hofmann J.F. and Beach,D. (1994) cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J., 13, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X.H., Yan,H. and Newport,J. (1997) A role for Cdk2 kinase in negatively regulating DNA replication during S phase of the cell cycle. J. Cell Biol., 137, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli P.V., Brown,G.W., Muzi-Falconi,M., Tien,D. and Kelly,T.J. (1997) Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev., 11, 2767–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Wells,N.J. and Hunter,T. (1999) Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl Acad. Sci. USA, 96, 6193–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey S.E., Montgomery,S., Labib,K. and Lindner,K. (2000) Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J., 19, 1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T.J., Martin,G.S., Forsburg,S.L., Stephen,R.J., Russo,A. and Nurse,P. (1993) The fission yeast cdc18 gene product couples S phase to start and mitosis. Cell, 74, 371–382. [DOI] [PubMed] [Google Scholar]
- Kominami K. and Toda,T. (1997) Fission yeast WD-repeat protein Pop1 regulates genome ploidy through ubuiquitin-proteosome-mediated degradation of the CDK inhibitor Rum1 and the S phase initiator Cdc18. Genes Dev., 11, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Labib K., Diffley,J.F.X. and Kearsey,S.E. (1999) G1 phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nature Cell Biol., 1, 415–422. [DOI] [PubMed] [Google Scholar]
- Leatherwood J. (1998) Emerging mechanisms of eukaryotic DNA replication initiation. Curr. Opin. Cell Biol., 10, 742–748. [DOI] [PubMed] [Google Scholar]
- Liang C. and Stillman,B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev., 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Weinreich,M. and Stillman,B. (1995) ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell, 81, 667–676. [DOI] [PubMed] [Google Scholar]
- Lygerou Z. and Nurse,P. (1999) The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J. Cell Sci., 112, 3703–3712. [DOI] [PubMed] [Google Scholar]
- Mahbubani H.M., Chong,J.P., Chevalier,S., Thommes,P. and Blow,J.J. (1997) Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J. Cell Biol., 136, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D., van Assendelft,G.B. and Kearsey,S.E. (1996) Fission yeast cdc21, a member of the MCM protein family, is required for onset of S phase and is located in the nucleus throughout the cell cycle. EMBO J., 15, 861–872. [PMC free article] [PubMed] [Google Scholar]
- Maiorano D., Moreau,J. and Mechali,M. (2000) XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature, 404, 622–625. [DOI] [PubMed] [Google Scholar]
- McGarry T.J. and Kirschner,M.W. (1998) Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell, 93, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Mendez J. and Stillman,B. (2000) Chromatin association of human origin recognition complex, cdc6 and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol., 20, 8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K., Kong,D., Lee,J., Raychaudhuri,S. and Hurwitz,J. (1999) Identification and reconstitution of the origin recognition complex from Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA, 96, 12367–12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nguyen V.Q., Co,C., Irie,K. and Li,J.J. (2000) Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2–7. Curr. Biol., 10, 195–205. [DOI] [PubMed] [Google Scholar]
- Nishitani H. and Nurse,P. (1995) p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell, 83, 397–405. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Lygerou,Z., Nishimoto,T. and Nurse,P. (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature, 404, 625–628. [DOI] [PubMed] [Google Scholar]
- Ogawa Y., Takahashi,T. and Masukata,H. (1999) Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol., 19, 7228–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B.O., Lukas,J., Sorensen,C.S., Bartek,J. and Helin,K. (1999) Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J., 18, 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Lengauer,C. and Nasmyth,K. (1995) Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J., 14, 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Bohm,T., Cocker,J.H., Diffley,J.F. and Nasmyth,K. (1996) Activation of S phase-promoting CDKs in late G1 defines a ‘point of no return’ after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev., 10, 1516–1531. [DOI] [PubMed] [Google Scholar]
- Romanowski P., Madine,M.A., Rowles,A., Blow,J.J. and Laskey,R.A. (1996) The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol., 6, 1416–1425. [DOI] [PubMed] [Google Scholar]
- Rowles A., Chong,J.P., Brown,L., Howell,M., Evan,G.I. and Blow,J.J. (1996) Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell, 87, 287–296. [DOI] [PubMed] [Google Scholar]
- Saha P., Chen,J., Thome,K.C., Lawlis,S.J., Hou,Z., Hendricks,M., Parvin,J.D. and Dutta,A. (1998) Human CDC6/Cdc18 associates with Orc1 and Cyclin/cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol., 18, 2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J.A., Kim,S. and Huberman,J.A. (1998). Ribosomal DNA replication in the fission yeast Schizosaccharomyces pombe. Exp. Cell Res., 238, 220–230. [DOI] [PubMed] [Google Scholar]
- Santocanale C. and Diffley,J.F.X. (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature, 395, 615–617. [DOI] [PubMed] [Google Scholar]
- Sazer S. and Sherwood,S.W. (1990) Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci., 97, 509–516. [DOI] [PubMed] [Google Scholar]
- Sherman D.A. and Forsburg,S.L. (1998) Schizosaccharomyces pombe Mcm3p, an essential nuclear protein, associates tightly with Nda4p (Mcm5p). Nucleic Acids Res., 26, 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S., Li,A., Maiorano,D., Mechali,M. and Blow,J.J. (2001) Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nature Cell Biol., 3, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Knapp,D. and Nasmyth,K. (1997) Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Tugal T., Zou-Yang,X.H., Gavin,K., Pappin,D., Canas,B., Kobayashi,R., Hunt,T. and Stillman,B. (1998) The orc4p and orc5p subunits of the Xenopus and human origin recognition complex are related to orc1p and cdc6p. J. Biol. Chem., 273, 32421–32429. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Dwyer,B.T., Dhar,S.K., Cvetic,C., Walter,J.C. and Dutta,A. (2000) Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science, 290, 2309–2312. [DOI] [PubMed] [Google Scholar]
- Wu J. and Gilbert,D.M. (1995) Rapid DNA preparation for two-dimensional gel analysis of replication intermediates. Nucleic Acids Res., 23, 3997–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerschke W., Rottjakob,H.W. and Kuntzel,H. (1994) The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J. Biol. Chem., 269, 23351–23356. [PubMed] [Google Scholar]