Abstract
After 40 years of searching for the eukaryotic replicator sequence, it is time to abandon the concept of ‘the’ replicator as a single genetic entity. Here I propose a ‘relaxed replicon model’ in which a positive initiator–replicator interaction is facilitated by a combination of several complex features of chromatin. An important question for the future is whether the positions of replication origins are simply a passive result of local chromatin structure or are actively localized to coordinate replication with other chromosomal activities.
In the summer of 1962, François Jacob and Sydney Brenner sketched in the sand of France’s La Tranche-sur-Mer a model (FIG. 1a) to explain the regulation of DNA synthesis in bacteria1. Their ‘replicon model’ proposed that replication was regulated by a positive interaction between an initiator protein, which is encoded by a structural gene, and a specific genetic element of recognition, which is known as the replicator. Although originally intended to explain the replication of circular bacterial chromosomes, the replicon model was quickly adopted as the paradigm for eukaryotic chromosomes as well (FIG. 1b). Proteins with the properties of initiators have been identified in species from Escherichia coli to man. By contrast, genetically defined replicators have yet to be identified in most eukaryotes and are clearly dispensable for replication in many eukaryotic systems. Even in its shortcomings, though, the model has been useful. Indeed, the replicon model was the driving force behind the discovery of bacterial and budding yeast replicators as well as initiator proteins — from dnaA in prokaryotes2 to the origin-recognition complex (ORC) in eukaryotes3. Indeed, all cellular chromosomes can be thought of as replicons, or clusters of replicons — the replication of which is regulated, at least in part, by a positive initiator–replicator interaction.
Figure 1. The replicon model.
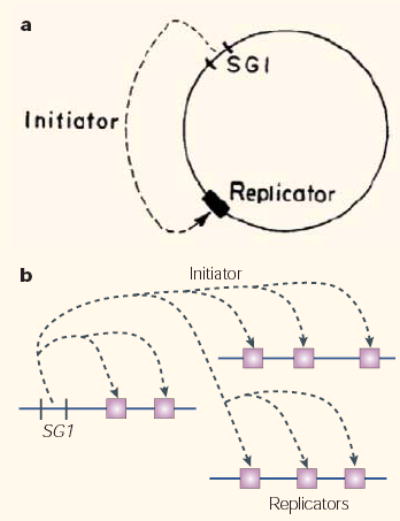
a | Original conception of the replicon model. “The replicon is assumed to be a circular structure carrying two specific genetic determinants. A structural gene (SG1) determines the synthesis of a diffusible active element, the initiator. The initiator acts on a replicator, allowing the beginning of the replication which proceeds along the circular structure”. Reproduced with permission from ref. 1 © (1964) Cold Spring Harbor Laboratory Press. b | The replicon model in the form that it was originally presumed to apply to eukaryotes.
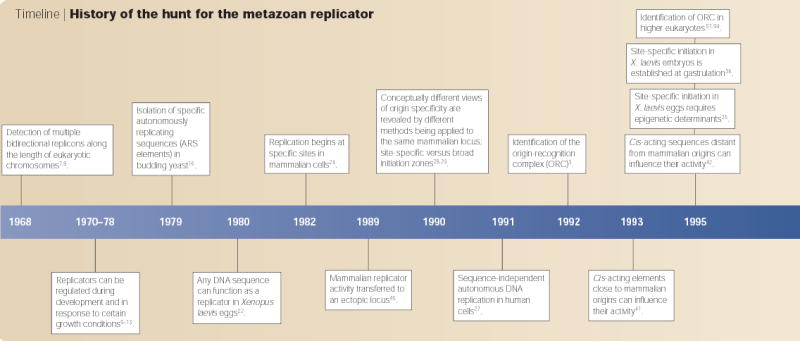
In this historical perspective, I will summarize the salient findings over the course of these four decades (TIMELINE), with a focus on strategies to identify metazoan replicators and the debate over their existence. I propose that we replace the concept of the replicator as a specific DNA sequence with that of a context-dependent element that is recognized by the initiator through several features, which can include DNA sequence. By embracing the complexity of replicators as being akin to that of promoter elements, future experiments can define these various features that influence the initiator–replicator interaction and address the biological significance of specifying replication-origin sites, given that site-specific initiation is dispensable for regulated genome duplication30.
Timeline. History of the hunt for the metazoan replicator.
The power of prediction
One of the beauties of the replicon model was the conceptually straightforward predictions that it provided. A replicator can confer the ability to replicate onto any DNA segments that are linked to it, and this became relatively simple to test after the development of recombinant-DNA technology in the early 1970s. By linking various segments of E. coli plasmids to a non-replicating DNA fragment that contained a selectable marker, Stanley Cohen and colleagues identified sequences that could confer, in cis, the ability of these molecules to replicate autonomously4. This straightforward assay (BOX 1) was later used to identify many different replicators, including oriC in E. coli5. Once a replicator sequence is defined, the initiator can be identified as a protein that binds to the replicator, and the dnaA protein in E. coli fulfilled this criterion2. Allowing for a few minor adjustments, it is clear that the replicon model is not only a useful paradigm, but that it accurately describes all bacterial replication systems under normal physiological selective pressures. Moreover, the replicator DNA sequence and the initiator protein are highly conserved across different bacterial species6.
Box 1 | The autonomously replicating sequence (ARS) assay
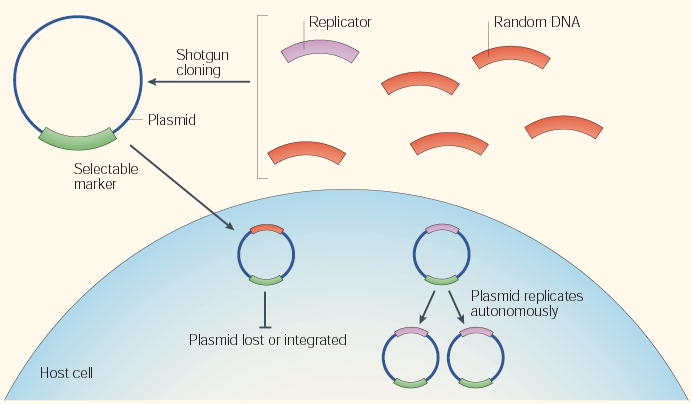
The ARS assay is used to identify replicators in genomes or DNA sequences that can range from those that are as simple as small bacterial plasmids and animal viruses to those that are as complex as budding and fission yeast genomes — the principle is very similar in all cases. The test genome is fragmented and the fragments are cloned into a vector that contains a selectable marker that can function in the appropriate host. The appropriate host is a cell type that is known to contain all the proteins that are necessary to support replication of the desired replicator, including the initiator. The collection of plasmids containing these genome fragments is then introduced into the host in such a way that each cell obtains one or fewer plasmids. Selection for the presence of the plasmid is applied, and the resulting recipient clonal colonies are expanded and evaluated for the ability of the (usually circular) plasmid to be maintained and to replicate as an independent unit, autonomously, within the host cell. In some variations of this assay, replicating plasmids can be detected or enriched without cell-growth selection. Myriad permutations of this assay have been applied to metazoan systems, but have generally failed to identify any specific DNA sequences that confer ARS activity.
Together, these studies show that initiation of replication in mammalian cells requires specific DNA sequences that are both close to and distant from the site of initiation, but no single consensus DNA-sequence motif is either necessary or sufficient for replicator activity. The simplest interpretation of these studies is that there is specificity to the initiator–replicator interaction, but the features that are recognized by the initiator are only indirectly determined by DNA sequences.
Eukaryotes: a different problem
It did not take long for investigators to appreciate that eukaryotic chromosomes had fundamentally different problems to solve, which could not be accommodated by the replicon model in its simplest form. Whereas bacteria usually have a single circular chromosome with only one replicator per chromosome, eukaryotes have several linear chromosomes that are each many times larger. Early autoradiography studies of DNA synthesis along the lengths of isolated DNA fibres showed that these chromosomes replicate using hundreds to thousands of replication origins7,8 (TIMELINE). Moreover, there was clearly flexibility in the usage of replication origins at different developmental stages9–11 and under different growth conditions12,13. Also, replication origins were shown to fire asynchronously throughout the DNA-synthesis phase of the cell cycle14,15. So, even though the existence of a common eukaryotic replicator sequence was proposed (FIG. 1b), it was already clear that additional layers of regulation would be needed to explain such flexibility.
Given these known complexities, perhaps it was not fair to hold the bacterial replicon model to the standard of eukaryotic replication. But the advent of budding yeast genetics in the 1970s provided the ideal model system in which to use the autonomous-replication assay, which had been so successful in bacterial systems, to identify eukaryotic replicators. The immediate success of this approach16, which resulted in the cloning of several ‘autonomously replicating sequences’ (ARS elements), all but confirmed the replicon model as a useful paradigm for eukaryotes. Indeed, ARS elements have since been shown to be sites of replication initiation in their native context17–19, and to contain a common DNA sequence that is required for replication initiation both on plasmids20 and within chromosomes21. Hence, this proof-of-principle experiment that was carried out 25 years ago set the stage for the hunt for replicators in higher eukaryotic organisms. Ironically, budding yeast has turned out to be an exceptional eukaryote with respect to the importance of a specific consensus replicator sequence.
The mystery of the metazoa
As they have genomes that are up to 100 times larger in size than those of yeast, mapping replication origins in higher eukaryotes was expected to be difficult. But no one could have predicted the degree of disparity that was uncovered between different systems, and even within individual systems that were studied using different methodologies. In 1980, Richard Harland and Ron Laskey demonstrated that any of several DNA fragments that were microinjected into Xenopus laevis eggs, even those containing mostly bacterial DNA, replicated with equal efficiency and in a regulated once-per-cell-cycle fashion22. The results were compelling, and they were reproducible23. In stark contrast, Nick Heintz and Joyce Hamlin demonstrated shortly thereafter that initiation of replication could be localized to specific sites in mammalian chromosomes24. So, the great debate had begun. The X. laevis experiments showed that once-per-cell-cycle replication could be accomplished without the need for specific replicator sequences. But mammalian cells clearly did not initiate replication at random sites, and X. laevis embryos are a specialized system, which is designed to duplicate the entire genome within 20 minutes. For most, this was sufficient reason to disregard the X. laevis embryo results, and the quest for the ‘holy grail’ of the human replicator shifted into high gear.
ARS assays: all or nothing
The obvious first strategy of choice was the autonomously replicating sequence (ARS) assay that had worked so well in bacteria and yeast (BOX 1). It is impossible to say how many clever permutations of this approach were tried and failed or found to be non-reproducible25. In most cases, very poor transient replication activity26 quickly resulted in selection for plasmids that had stably integrated into chromosomes25. Then, in the late 1980s, Michelle Calos and colleagues demonstrated that the efficiency of ARS assays could be greatly improved by preventing the loss of plasmids from the nucleus using DNA sequences that tether plasmids to segregating chromosomes. The results of these experiments were astonishingly clear. Plasmids could be maintained for many generations, but replication was independent of DNA sequence and was initiated at random sites within these plasmids27. The dirty ‘random’ word had reared its ugly head again, placing a nasty stigma on the ARS assay. For those bent on finding the holy replicator, it was clear that some nuance of mammalian cell culture precluded the ability of ARS assays to reveal physiological replicator activity.
Religion, politics and replication origins
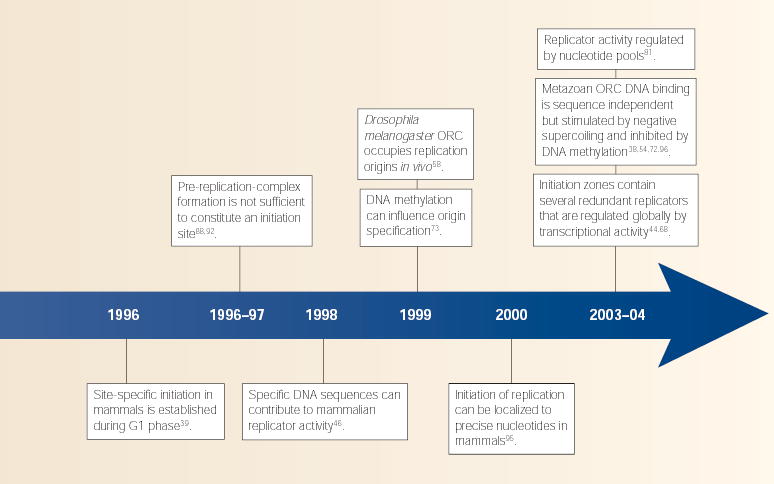
By the early 1990s, most investigators had conceded that classic genetic approaches, including those that employed promising viral models (BOX 2), were insufficient for identifying metazoan replicators, and many sought a more complete characterization of replicators in their native context. Finding replication intermediates in a genome of 3 billion basepairs required new technological developments, and several new methods for mapping replication origins emerged. Rather than clarifying things, however, the ‘random versus site specific’ debate escalated to a whole new level. In 1990, almost simultaneously, two reports in which different methods were used to map initiation sites within the Chinese hamster dihydrofolate reductase (DHFR) locus arrived at diametrically opposed conclusions. One study claimed that >80% of initiations within a 27-Kb region emanated from a single site28, whereas the other claimed that replication initiated at sites distributed virtually at random throughout the very same region29. These results shifted the debate from a philosophical comparison of different biological systems to a detailed technical evaluation of different methodological approaches, leaving investigators unable to reach a consensus as to the basic structure of metazoan replication origins nor the best means to evaluate this structure.
Box 2 | The contribution of viral replicators
The small double-stranded DNA viruses (for example, polyoma and papilloma viruses) were the preferred model for studying eukaryotic replication in the 1980s, and because they use the host’s machinery for replication, they were particularly promising models for mammalian chromosome replication. In fact, most of our knowledge of the events that occur downstream of replication-origin unwinding comes from studies with simian virus-40 (SV40), in which defined in vitro replication systems identified the proteins that are essential for DNA synthesis in human cells83,84. Initiation of replication clearly requires the interaction of a virally encoded initiator protein with a defined replicator sequence that could be genetically dissected with classic autonomously replicating sequence (ARS) assays85. But these viruses do not replicate once per cell cycle, they supply their own virally encoded initiator, and they are not subjected to the same selective pressures as cellular chromosomes, so they are limited in their use as models for mammalian replicator function.
By contrast, the large Epstein–Barr virus (EBV) does not supply its own initiator, but instead requires ORC86. Latent (non-lytic) replication of EBV is restricted to one round per cell cycle87, making this virus a good potential model for cellular replication. Interestingly though, a region known as oriP, which was shown to have ARS activity in the presence of a virally encoded protein, is dispensable for latent viral replication87, and origin-mapping studies have shown that initiation-site selection in the full-sized EBV genome is just as degenerate as it is within cellular chromosomes, whether or not oriP is present82.
Nearly 15 years have passed and the distribution of initiation sites at this and many other metazoan loci has still not been resolved. As more replication origins were mapped, some were localized to very defined sites, whereas others mapped indiscriminately throughout ‘initiation zones’ of 10–50 Kb (reviewed in ref. 30). Most of these sites have been mapped by only one method, so the extent to which these differences can be accounted for by methodological considerations is not clear. On the other hand, nearly every replication-origin mapping method has been applied to the DHFR locus, and there is still no consensus on the pattern of initiation at this locus. Some methods detect specific ‘replicator’ sites within the non-transcribed region downstream of the DHFR locus31, whereas other methods reveal initiation sites throughout every interrogated sequence within the same region32. At present, the best explanation for these discrepancies is that initiation can take place at many sites throughout this 50-Kb region, but that some methods are only capable of detecting a subset of these sites.
In the meantime, people who worked with the X. laevis system had all but proven that any DNA sequence can function as a replicator. After the development of cell-free replication systems that were derived from X. laevis eggs, many investigators were able to demonstrate the promiscuity of replication in this system, and the apparently random choice of initiation sites within each DNA molecule33–35. The same was found to be true within the native chromosomes of early X. laevis36 and Drosophila melanogaster37 embryos. Importantly, these latter studies revealed that replication initiation is eventually focused to specific sites after the blastula stage of development. These results were interpreted along existing party lines. Proponents of site-specific initiation assumed that random site selection was a special adaptation for rapid replication in early embryos, which was mediated either by a specialized embryonic initiator protein or by the high concentration of maternally accumulated initiator proteins that become diluted during development. These possibilities were ruled out by reconstituting ORC-depleted X. laevis egg extracts with human ORC, which restored sequence-independent initiation of replication in a concentration-independent manner38. The other party declared that developmentally regulated features of chromatin, rather than a specific DNA sequence, dictate origin specification. This interpretation was supported by the fact that X. laevis egg extracts are capable of initiating replication at specific sites when provided with chromatin from somatic cells as a template35, and that the defining epigenetic structure is re-assembled early in G1 phase of each cell cycle39. In fact, simply pre-loading transcription factors onto naked DNA templates is sufficient to direct site-specific initiation in pre-blastula X. laevis embryos40.
DNA sequence: a clear contributor
New hope in the hunt for the holy replicator arrived when Howard Cedar and colleagues demonstrated that a naturally occurring 8-Kb deletion that encompassed a region of replication-initiation activity within the human β-globin locus (HBB) eliminated bidirectional replication in this region41. Hailed as a triumph that would deliver us from the dark ages, this result inspired further deletion studies that soon revealed a more complex picture. At both the HBB and the DHFR locus, deletions 25–50 Kb away from the region of replication-initiation activity also completely eliminated its activity42,43. Deletions within the large region of activity at the DHFR locus actually augmented activity at the remaining sites44, implying a high degree of redundancy of potential replicators throughout this region.
A critical test of ‘replicator’ activity is to show that a DNA segment can direct the initiation of replication when placed in different sequence contexts. In the absence of convenient ARS assays, the next best option was to move putative replicators to other locations within cellular chromosomes. The first such study provided evidence that a 16-Kb segment from within the Chinese hamster ovary (CHO) DHFR replication-initiation zone could function as a replicator when integrated at random ectopic chromosomal sites45. However, this result was not universally accepted because negative-control fragments could not be directly compared at the same chromosomal sites. This limitation was overcome by using microbial recombinases to integrate test fragments into the same site, which provided persuasive evidence that specific DNA sequences are required for replicator activity46. More recent studies have revealed conceptually similar results, whether the sequences are derived from highly localized replication origins47–49 or from the broad DHFR initiation zone50. In all cases, ‘replicator’ activity can be transferred to many (but not all) ectopic sites. Importantly, however, these studies have not identified any specific DNA sequences or structural motifs that are shared by different replicators.
Replicator–initiator interactions
The discovery of a hetero-hexameric ORC in all eukaryotes from budding yeast to man3,51 provided a whole new strategy to hunt for replicators. Instead of searching for poorly defined sequences with laborious methods, the sequences to which the initiator binds, logically, should define the replicator. In fact, budding yeast ORC binds to replication-origin DNA sequences substantially better than to random templates52, and fission yeast ORC has evolved a specialized domain to recognize asymmetric stretches of adenines and thymidines that define its origins of replication53. So, it was quite revealing to learn that purified human and D. melanogaster ORC have little or no sequence-specific binding activity38,54. Interestingly though, D. melanogaster ORC has a strong preference for binding to negatively supercoiled DNA54. A preference for superhelical tension could explain why both local and distant sequences can affect initiation activity without the requirement for a specific consensus-sequence recognition element.
Chromatin immunoprecipitation (ChIP) experiments can potentially identify ORC-binding sites regardless of their DNA sequence, and recent studies suggest that ORC occupies replication origins in all eukaryotes, although its relative binding affinity compared to other sites is unknown. As expected, budding yeast55 and fission yeast56 ORCs show significant enrichment at origins, but some preference for origin occupation is also seen in flies57,58 and humans59. One drawback of most ChIP studies is that they interrogate only a few carefully chosen sites, which does not provide an unbiased view of the sequences to which ORC binds. Hybridization of DNA sequences that co-immunoprecipitate with ORC in complete-genome arrays (ChIP-chip) can provide this information and have already proved successful in budding yeast60. Interestingly, not all ARS consensus sequences are identified, which could be due to a lack of sensitivity of the method, to a requirement for further undefined sequences61, or to features of chromatin that prevent ORC from binding to some sites. Another confounding issue with these studies is that, counter-intuitively, stronger ORC binding sites have been shown to be less efficient replicators89. In any case, these studies require arrays that achieve complete unbiased coverage of the genome, which, at present, are available for very few species. An alternative approach is to clone and sequence DNA segments that co-immunoprecipitate with ORC, which was recently done for a human cell line62. In both the human and yeast studies, many (but not all) ORC-binding sites coincide with replication-initiation activity (BOX 3).
Box 3 | Regulation downstream of the initiator–replicator interaction
In addition to regulating the initiator–replicator interaction, the activity of a potential replication origin in metazoans is also strongly influenced by events downstream of the ORC–DNA interaction. In budding yeast, there are many origins that do not fire in every cell cycle and some that do not fire at all in a normal cell cycle, but these silent origins are occupied by ORC and other pre-RC proteins88. In fact, ORC is even more tightly bound to silent origins89. Many of these silent origins are activatable under the appropriate conditions, which implies that functional pre-RCs are assembled at these sites. The conditions that silence some budding yeast origins or activate others are beginning to be understood, and they include regulators of chromatin structure77, nucleosome-free transcriptional-control elements69, proximity to nucleosomes76, or simply proximity to other replication origins90,91. In X. laevis embryos, there are clearly many more pre-RCs assembled than are used92, and the relative preference for initiating replication at specific sites can be influenced downstream of ORC binding by the presence of transcription factors40. In D. melanogaster, binding of a protein complex that contains the oncoprotein Myb is required for efficient initiation of replication at the chorion gene cluster during selective gene amplification, but is not required for ORC localization to this origin93. For the time being, it is important to appreciate that in many cases it has not been determined whether the factors that influence origin activity are upstream or downstream of ORC binding, and that this will be an important distinction for our future understanding of replication origins.
If purified ORC does not preferentially bind to specific sequences, and replicators do not share a common sequence element, then what directs ORC to specific chromosomal sites in vivo? Several mechanisms can be imagined (FIG. 2). One possibility is that other proteins interact with ORC to direct it to specific sequences. This could occur through the direct interaction of ORC with chromatin proteins, or such proteins could alter the local topological state of DNA. For example, the EBNA1 protein of Epstein–Barr virus (EBV) interacts with ORC and might recruit ORC to the EBV origin63. In D. melanogaster follicle cells, an interaction of ORC with E2F transcription factors64,65 — possibly in conjunction with a developmentally regulated Myb-oncoprotein-containing complex66 and the histone de-acetylase Rpd3 (ref. 67) — seems to orchestrate a developmentally regulated redistribution of ORC to facilitate amplification of the chorion gene cluster. Chromosomal activities such as transcription can also destabilize nucleosomes and create superhelical tension, which might explain the transcription-dependent origin activity that has been described in several systems68–70. In short, any mechanism that removes a nucleosome and results in negative supercoiling could create a high-affinity binding site for ORC54.
Figure 2. Relaxed replicon model.
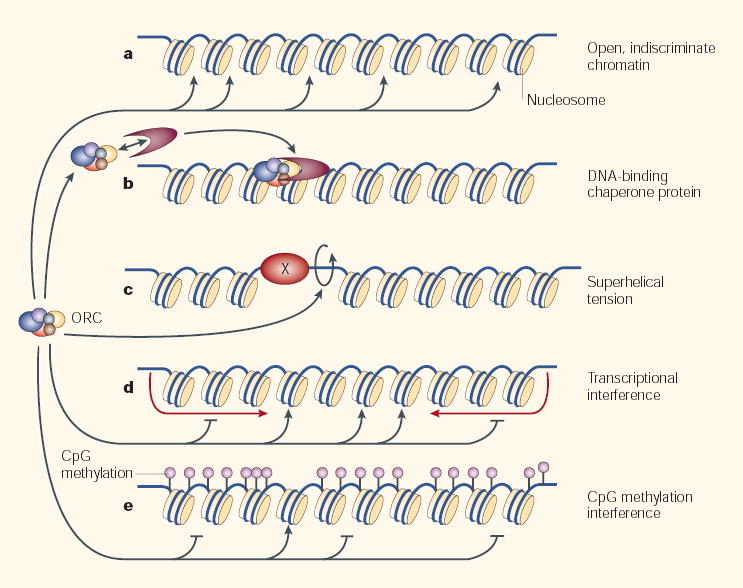
The hetero-hexameric origin-recognition complex (ORC) binds to naked DNA indiscriminately on its own, but the specificity of its binding to cellular chromatin in vivo is influenced by many factors. When presented with functionally inert (open or ‘indiscriminate’) chromatin (a), ORC binds nonspecifically38. However, ORC might be directed to specific sites by (b) interacting proteins that chaperone ORC to specific sites63 or repress its binding to others64–67. Or (c), by superhelical tension54, which can be created by nearby binding proteins (X) that might also position or remove nucleosomes, or otherwise create favourable ORC-binding sites. Superhelical tension can also be created by transcriptional activity, either upstream or downstream of gene promoters69,70. ORC can also be excluded from specific sites by (d) transcription68,71 or by (e) general CpG DNA methylation72, which can be quite prevalent73. Additional features of chromatin that lie downstream of the initiator–replicator interaction also influence origin specification (BOX 2). This model necessarily invokes complexity — origin specification results from a different combination of targeting mechanisms for each chromosomal site, making it no longer useful to think of the replicator as a single common genetic entity.
Another way to direct a nonspecific binding protein to specific sites is to prevent it from binding to other sites. Exclusion can occur at a global level, for example by transcriptional interference, which antagonizes the initiation of replication68,71, or by the interaction with global repressors, as is suggested during amplification of the chorion gene cluster in D. melanogaster64–67. There can also be more localized exclusion effects. A recent study shows that methylated DNA will neither bind ORC nor initiate replication in the otherwise omnipotent X. laevis egg extracts72. By cleverly constructing plasmid templates that contain a small segment of unmethylated DNA, ORC binding was focused to a specific segment of the plasmid and replication was initiated within 2 Kb of this segment. This intriguing result could explain an observed loss of localized initiation in methylation-deficient cell lines73, as well as the correlation between unmethylated CpG islands and replication origins74.
Several less well-defined features of chromosomes also influence the usage of replication origins, but in most cases it is not known whether they are upstream or downstream of the initiator–replicator interaction (BOX 3). The proximity of nucleosomes can have both inhibitory75 and stimulatory76 effects on the activity of budding yeast replication origins, although, at least in some cases, these effects are known to be downstream of ORC binding76. There is evidence that histone modifications can influence the usage of replication origins in budding yeast77, D. melanogaster 67 and X. laevis40. However, the effects of histone acetylation are not likely to be straightforward, as different early firing origins at the chicken β-globin locus have different patterns of epigenetic histone modifications78. Certainly, the fact that several origins are regulated during development70,79,80 and under different growth conditions81,82 implies that there are many complex factors that influence origin specification (FIG. 2).
A preview of the 50th anniversary
What can we expect in the next decade? First and foremost, the initiator–replicator interaction will become appreciated as an interaction with far greater complexity than was previously expected. The concept of the replicator as a specific genetic element will cease to be useful, and will be replaced by that of a context-dependent element akin to our present concept of promoters. New methodologies to map replicators more rapidly and confidently will continue to drive discovery. The structure of metazoan ORC and the mechanisms by which it recognizes DNA will be revealed. Proteins that interact with ORC will be identified and their effects on ORC binding to specific DNA sequences will be evaluated. Comparative analysis of the many different factors that influence ORC–replicator interactions will reveal unique and common features of recognition. Further factors downstream of ORC binding and/or pre-RC assembly will also be found to influence the usage of replication origins. Perhaps most importantly, we will begin to address the biological significance of origin localization. Ultimately, the regulation of replication-origin usage during development and in response to different growth conditions will become integrated into our knowledge of the cell cycle and cell differentiation.
Concluding remarks
The findings of the past decade (BOX 4) warrant a revision of the original concept of a replicator as it applies to eukaryotic cells. The idea that replication is initiated by the binding of an initiator to a specific DNA-sequence recognition element should be replaced by a ‘relaxed replicator’ model in which DNA sequence constitutes only one of several features that influence where replication will initiate. DNA topology, DNA methylation, transcriptional activity, ancillary protein chaperones, and long- and short-distance chromatin effects might each be absolutely critical or totally dispensable to origin specification under different conditions. This revision does not detract from the fundamental value and spirit of the replicon model. Jacob and Brenner intended the model to describe bacterial systems, and we should no more expect the replicon model to apply to metazoans than we do the operon model. Replication is clearly initiated by the positive interaction of an initiator with a replicator, but we must come to accept that the replicator can no longer be defined by a single specific DNA sequence. Our present, widely accepted, model for restricting replication to precisely once-per-cell-cycle does not invoke any requirement for specific DNA-sequence elements30. So, it is arguably the presence of specific initiation sites that is the greater mystery. In this regard, a highly significant question now is whether the ‘relaxed replicator’ arose by default or by design. Is the replicator simply any available ORC-binding site, or are origins actively positioned, possibly to coordinate the direction of replication forks with other chromosomal functions30? If so, selective pressures might have moulded a different blend of targeting mechanisms for each chromosomal site, and replicators might turn out to be no less complex than transcriptional promoters, where it is now a familiar concept that a common machinery can be targeted by many different mechanisms.
Box 4 | The status quo: a list of acceptable statements
Replicators
There are many more replicators (that is, potential replication origins that are occupied by the origin-recognition complex (ORC)) than are used during S phase.
More replicators are used than are needed to complete S phase.
Replicators fire asynchronously throughout S phase.
Replicator usage can be regulated during development and under different metabolic conditions.
Once-per-cell-cycle genome duplication does not require specific replicator sequences.
Different replicators have different DNA-sequence requirements; some have no DNA-sequence requirements.
Many different features of chromatin structure and function can influence the activity of replicators.
Initiators
ORC homologues are found in all eukaryotes.
ORC occupies replicators in vivo.
The factors that influence the DNA-binding activity of ORC are highly divergent.
Acknowledgments
I would like to apologize to those whose work was not cited due to space limitations. I would also like to thank R. Cross, C. Cvetic, J. Huberman, P. Kane, T. Melendy, C. Schildkraut, J. Walter and R. West for critical reading of the manuscript. D.M.G. is supported by the National Institutes of Health, the National Science Foundation and American Cancer Society.
Footnotes
Competing interests statement
The author declares no competing financial interests.
References
- 1.Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harb Symp Quant Biol. 1964;28:329–348. [Google Scholar]
- 2.Chakraborty T, Yoshinaga K, Lother H, Messer W. Purification of the E. coli dnaA gene product. EMBO J. 1982;1:1545–1549. doi: 10.1002/j.1460-2075.1982.tb01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 4.Timmis K, Cabello F, Cohen SN. Cloning, isolation, and characterization of replication regions of complex plasmid genomes. Proc Natl Acad Sci USA. 1975;72:2242–2246. doi: 10.1073/pnas.72.6.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yasuda S, Hirota Y. Cloning and mapping of the replication origin of Escherichia coli. Proc Natl Acad Sci USA. 1977;74:5458–5462. doi: 10.1073/pnas.74.12.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita MQ, Yoshikawa H, Ogasawara N. Structure of the dnaA region of Pseudomonas putida: conservation among three bacteria, Bacillus subtilis, Escherichia coli and P. putida. Mol Gen Genet. 1989;215:381–387. doi: 10.1007/BF00427033. [DOI] [PubMed] [Google Scholar]
- 7.Taylor JH. Rates of chain growth and units of replication in DNA of mammalian chromosomes. J Mol Biol. 1968;31:579–594. doi: 10.1016/0022-2836(68)90429-4. [DOI] [PubMed] [Google Scholar]
- 8.Huberman JA, Riggs AD. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- 9.Stambrook PJ, Flickinger RA. Changes in chromosomal DNA replication patterns in developing frog embryos. J Exp Zool. 1970;174:101–113. doi: 10.1002/jez.1401740110. [DOI] [PubMed] [Google Scholar]
- 10.Callan HG. Replication of DNA in the chromosomes of eukaryotes. Proc R Soc Lond B. 1972;181:19–41. doi: 10.1098/rspb.1972.0039. [DOI] [PubMed] [Google Scholar]
- 11.Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- 12.Taylor JH. Increase in DNA replication sites in cells held at the beginning of S phase. Chromosoma. 1977;62:291–300. doi: 10.1007/BF00327029. [DOI] [PubMed] [Google Scholar]
- 13.Hand R. Eucaryotic DNA: organization of the genome for replication. Cell. 1978;15:317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- 14.Mueller GC, Kajiwara K. Early- and late-replicating deoxyribonucleic acid complexes in HeLa nuclei. Biochim Biophys Acta. 1966;114:108–115. doi: 10.1016/0005-2787(66)90258-9. [DOI] [PubMed] [Google Scholar]
- 15.Balazs I, Brown EH, Schildkraut CL. The temporal order of replication of some DNA cistrons. Cold Spring Harb Symp Quant Biol. 1973;38:239–245. doi: 10.1101/sqb.1974.038.01.027. [DOI] [PubMed] [Google Scholar]
- 16.Struhl K, Stinchcomb DT, Scherer S, Davis RW. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huberman JA, Spotila LD, Nawotka KA, El-Assouli SM, Davis LR. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 18.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 19.Huberman JA, Zhu J, Davis LR, Newlon CS. Close association of a DNA replication origin and an ARS element on chromosome III of the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:6373–6384. doi: 10.1093/nar/16.14.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celniker SE, Sweder K, Srienc F, Bailey JE, Campbell JL. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 22.Harland RM, Laskey RA. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980;21:761–771. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- 23.Mechali M, Kearsey S. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell. 1984;38:55–64. doi: 10.1016/0092-8674(84)90526-9. [DOI] [PubMed] [Google Scholar]
- 24.Heintz NH, Hamlin JL. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci USA. 1982;79:4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert D, Cohen SN. Autonomous replication in mouse cells: a correction. Cell. 1989;56:143–144. doi: 10.1016/0092-8674(89)90885-4. [DOI] [PubMed] [Google Scholar]
- 26.Masukata H, Satoh H, Obuse C, Okazaki T. Autonomous replication of human chromosomal DNA fragments in human cells. Mol Biol Cell. 1993;4:1121–1132. doi: 10.1091/mbc.4.11.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krysan PJ, Calos MP. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol Cell Biol. 1991;11:1464–1472. doi: 10.1128/mcb.11.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burhans WC, Vassilev LT, Caddle MS, Heintz NH, DePamphilis ML. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990;62:955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- 29.Vaughn JP, Dijkwel PA, Hamlin JL. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell. 1990;61:1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert DM. Making sense of eukaryotic DNA replication origins. Science. 2001;294:96–100. doi: 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi T, Rein T, DePamphilis M. Identification of primary initiation sites for DNA replication in the hamster DHFR gene initiation zone. Mol Cell Biol. 1998;18:3266–3277. doi: 10.1128/mcb.18.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dijkwel PA, Wang S, Hamlin JL. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol Cell Biol. 2002;22:3053–3065. doi: 10.1128/MCB.22.9.3053-3065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyrien O, Mechali M. Plasmid replication in Xenopus eggs and egg extracts: a 2D gel electrophoretic analysis. Nucleic Acids Res. 1992;20:1463–1469. doi: 10.1093/nar/20.7.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahbubani HM, Paull T, Elder JK, Blow JJ. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 1992;20:1457–1462. doi: 10.1093/nar/20.7.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert DM, Miyazawa H, DePamphilis ML. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol Cell Biol. 1995;15:2942–2954. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 37.Sasaki T, Sawado T, Yamaguchi M, Shinomiya T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol Cell Biol. 1999;19:547–555. doi: 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vashee S, et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu JR, Gilbert DM. A distinct G1 step required to specify the Chinese hamster DHFR replication origin . Science. 1996;271:1270–1272. doi: 10.1126/science.271.5253.1270. [DOI] [PubMed] [Google Scholar]
- 40.Danis E, et al. Specification of a DNA replication origin by a transcription complex. Nature Cell Biol. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- 41.Kitsberg D, Selig S, Keshet J, Cedar H. Replication structure of the human β-globin gene domain. Nature. 1993;368:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- 42.Aladjem M, et al. Participation of the human β-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 43.Kalejta RF, et al. Distal sequences, but not ori-β/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol Cell. 1998;2:797–806. doi: 10.1016/s1097-2765(00)80294-4. [DOI] [PubMed] [Google Scholar]
- 44.Mesner LD, Li X, Dijkwel PA, Hamlin JL. The dihydrofolate reductase origin of replication does not contain any nonredundant genetic elements required for origin activity. Mol Cell Biol. 2003;23:804–814. doi: 10.1128/MCB.23.3.804-814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Handeli S, Klar A, Meuth M, Cedar H. Mapping replication units in animal cells. Cell. 1989;57:909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- 46.Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. Genetic dissection of a mammalian replicator in the human β-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 47.Paixao S, et al. Modular structure of the human lamin B2 replicator. Mol Cell Biol. 2004;24:2958–2967. doi: 10.1128/MCB.24.7.2958-2967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, et al. The human β-globin replication initiation region consists of two modular independent replicators. Mol Cell Biol. 2004;24:3373–3386. doi: 10.1128/MCB.24.8.3373-3386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu G, Malott M, Leffak M. Multiple functional elements comprise a mammalian chromosomal replicator. Mol Cell Biol. 2003;23:1832–1842. doi: 10.1128/MCB.23.5.1832-1842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altman AL, Fanning E. Defined sequence modules and an architectural element cooperate to promote initiation at an ectopic mammalian chromosomal replication origin. Mol Cell Biol. 2004;24:4138–4150. doi: 10.1128/MCB.24.10.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gavin K, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 52.Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC–DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 56.Ogawa Y, Takahashi T, Masukata H. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol Cell Biol. 1999;19:7228–7236. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bielinsky AK, et al. Origin recognition complex binding to a metazoan replication origin. Curr Biol. 2001;11:1427–1431. doi: 10.1016/s0960-9822(01)00444-4. [DOI] [PubMed] [Google Scholar]
- 58.Austin RJ, Orr-Weaver TL, Bell SP. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 1999;13:2639–2649. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdurashidova G, et al. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 2003;22:4294–4303. doi: 10.1093/emboj/cdg404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyrick JJ, et al. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 61.Breier AM, Chatterji S, Cozzarelli NR. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol. 2004;5:R22. doi: 10.1186/gb-2004-5-4-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keller C, Ladenburger EM, Kremer M, Knippers R. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J Biol Chem. 2002;277:31430–31440. doi: 10.1074/jbc.M202165200. [DOI] [PubMed] [Google Scholar]
- 63.Schepers A, et al. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J. 2001;20:4588–4602. doi: 10.1093/emboj/20.16.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Royzman I, Austin RJ, Bosco G, Bell SP, Orr-Weaver TL. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 1999;13:827–840. doi: 10.1101/gad.13.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bosco G, Du W, Orr-Weaver TL. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nature Cell Biol. 2001;3:289–295. doi: 10.1038/35060086. [DOI] [PubMed] [Google Scholar]
- 66.Beall EL, Bell M, Georlette D, Botchan MR. Dm-myb mutant lethality in Drosophila is dependent upon mip130: positive and negative regulation of DNA replication. Genes Dev. 2004;18:1667–1680. doi: 10.1101/gad.1206604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- 68.Saha S, Shan Y, Mesner LD, Hamlin JL. The promoter of the Chinese hamster ovary dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries. Genes Dev. 2004;18:397–410. doi: 10.1101/gad.1171404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muller M, Lucchini R, Sogo JM. Replication of yeast rDNA initiates downstream of transcriptionally active genes. Mol Cell. 2000;5:767–777. doi: 10.1016/s1097-2765(00)80317-2. [DOI] [PubMed] [Google Scholar]
- 70.Maric C, Benard M, Pierron G. Developmentally regulated usage of Physarum DNA replication origins. EMBO Rep. 2003;4:474–478. doi: 10.1038/sj.embor.embor822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haase SB, Heinzel SS, Calos MP. Transcription inhibits the replication of autonomously replicating plasmids in human cells. Mol Cell Biol. 1994;14:2516–2524. doi: 10.1128/mcb.14.4.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harvey KJ, Newport J. CpG methylation of DNA restricts prereplication complex assembly in Xenopus egg extracts. Mol Cell Biol. 2003;23:6769–6779. doi: 10.1128/MCB.23.19.6769-6779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rein T, Kobayashi T, Malott M, Leffak M, DePamphilis ML. DNA methylation at mammalian replication origins. J Biol Chem. 1999;274:25792–25800. doi: 10.1074/jbc.274.36.25792. [DOI] [PubMed] [Google Scholar]
- 74.Delgado S, Gomez M, Bird A, Antequera F. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17:2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simpson RT. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 76.Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 77.Pasero P, Bensimon A, Schwob E. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 2002;16:2479–2484. doi: 10.1101/gad.232902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prioleau MN, Gendron MC, Hyrien O. Replication of the chicken β-globin locus: early-firing origins at the 5′ HS4 insulator and the ρ- and βA-globin genes show opposite epigenetic modifications. Mol Cell Biol. 2003;23:3536–3549. doi: 10.1128/MCB.23.10.3536-3549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lunyak VV, Ezrokhi M, Smith HS, Gerbi SA. Developmental changes in the Sciara II/9A initiation zone for DNA replication. Mol Cell Biol. 2002;22:8426–8437. doi: 10.1128/MCB.22.24.8426-8437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou J, et al. The origin of a developmentally regulated Igh replicon is located near the border of regulatory domains for Igh replication and expression. Proc Natl Acad Sci USA. 2002;99:13693–13698. doi: 10.1073/pnas.212392399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 82.Norio P, Schildkraut CL. Plasticity of DNA replication initiation in Epstein–Barr virus episomes. PLoS Biol. 2004;2:E152. doi: 10.1371/journal.pbio.0020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990;346:534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- 84.Weinberg DH, et al. Reconstitution of simian virus 40 DNA replication with purified proteins. Proc Natl Acad Sci USA. 1990;87:8692–8696. doi: 10.1073/pnas.87.22.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bergsma DJ, Olive DM, Hartzell SW, Subramanian KN. Territorial limits and functional anatomy of the simian virus 40 replication origin. Proc Natl Acad Sci USA. 1982;79:381–385. doi: 10.1073/pnas.79.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dhar SK, et al. Replication from oriP of Epstein–Barr virus requires human ORC and is inhibited by geminin. Cell. 2001;106:287–296. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- 87.Norio P, Schildkraut CL, Yates JL. Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein–Barr virus chromosome after infection of established cell lines. J Virol. 2000;74:8563–8574. doi: 10.1128/jvi.74.18.8563-8574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Santocanale C, Diffley J. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- 89.Palacios DeBeer MA, Müller U, Fox CA. Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev. 2003;17:1817–1822. doi: 10.1101/gad.1096703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Santocanale C, Sharma K, Diffley JF. Activation of dormant origins of DNA replication in budding yeast. Genes Dev. 1999;13:2360–2364. doi: 10.1101/gad.13.18.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vujcic M, Miller CA, Kowalski D. Activation of silent replication origins at autonomously replicating sequence elements near the HML locus in budding yeast. Mol Cell Biol. 1999;19:6098–6109. doi: 10.1128/mcb.19.9.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Walter J, Newport JW. Regulation of replicon size in Xenopus egg extracts. Science. 1997;275:993–995. doi: 10.1126/science.275.5302.993. [DOI] [PubMed] [Google Scholar]
- 93.Beall EL, et al. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- 94.Ehrenhofer-Murray A, Gossen M, Pak D, Botchan M, Rine J. Separation of origin recognition complex functions by cross-species complementation. Science. 1995;270:1671–1674. doi: 10.1126/science.270.5242.1671. [DOI] [PubMed] [Google Scholar]
- 95.Abdurashidova G, et al. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023–2026. doi: 10.1126/science.287.5460.2023. [DOI] [PubMed] [Google Scholar]
- 96.Schaarschmidt D, Baltin J, Stehle IM, Lipps HJ, Knippers R. An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. EMBO J. 2004;23:191–201. doi: 10.1038/sj.emboj.7600029. [DOI] [PMC free article] [PubMed] [Google Scholar]


