Abstract
Insertional mutagenesis of Arabidopsis (Arabidopsis thaliana) was used to identify a novel recessive mutant, designated resurrection1 (rst1), which possesses a dramatic alteration in its cuticular waxes and produces shrunken nonviable seeds due to arrested embryo development. The RST1 gene sequence associated with these phenotypes was verified by three independent, allelic, insertion mutants, designated rst1-1, rst1-2, and rst1-3, with inserts in the first exon, 12th intron, and fourth exon, respectively. These three rst1 allelic mutants have nearly identical alterations in their wax profiles and embryo development. Compared to wild type, the wax on rst1 inflorescence stems is reduced nearly 60% in total amount, has a proportional reduction in aldehydes and aldehyde metabolites, and has a proportional increase in acids, primary alcohols, and esters. Compared to wild type, the C29 alkanes on rst1 are nearly 6-fold lower, and the C30 primary alcohols are 4-fold higher. These results indicate that rst1 causes shunting of most wax precursors away from alkane synthesis and into the primary-alcohol-producing branch of the pathway. In contrast to stems, the wax on rst1 mutant leaves increased roughly 43% in amount relative to the wild type, with the major increase occurring in the C31 and C33 alkanes. Unique among known wax mutants, approximately 70% of rst1 seeds are shrunken and nonviable, with these being randomly distributed within both inflorescence and silique. Viable seeds of rst1 are slightly larger than those of wild type, and although the viable rst1 seeds contain more total triacylglycerol-derived fatty acids, the proportions of these fatty acids are not significantly different from wild type. Shrunken seeds contain 34% of the fatty acids of wild-type seeds, with proportionally more palmitic, stearic, and oleic acids, and less of the longer and more desaturated homologs. Histological analysis of aborted rst1 seeds revealed that embryo development terminates at the approximate heart-shaped stage, whereas viable rst1 and wild-type embryos develop similarly. The RST1 gene encodes a predicted 1,841-amino acid novel protein with a molecular mass of 203.6 kD and a theoretical pI of 6.21. The RST1 transcript was found in all tissues examined including leaves, flowers, roots, stems, and siliques, but accumulation levels were not correlated with the degree to which different organs appeared affected by the mutation. The new RST1 gene reveals a novel genetic connection between lipid synthesis and embryo development; however, RST1's exact role is still quite unknown. The degree to which RST1 is associated with lipid signaling in development is an important focus of ongoing studies.
Plant lipids, including phospholipids (Meijer and Munnik, 2003), sterols (He et al., 2003), sphingolipids (Sperling and Heinz, 2003), fatty acids, and their derivatives like jasmonate (Turner et al., 2002; Weber, 2002), can serve as important signal molecules in the regulation of plant development. Recent studies have brought to light the possibility that the very long chain aliphatic cuticular waxes found coating the aerial surface of plants may likewise regulate plant development (Bird and Gray, 2003). For example, the C30 primary alcohol (triacontanol) induced shoot growth, early flowering, and synthesis of isopentenyl adenosine in Arabidopsis (Arabidopsis thaliana L.) Heynh. (He and Loh, 2002), and also increased photosynthetic rates and the expression of photosynthetic genes while down-regulating abscisic acid- and other stress-associated genes in rice (Oryza sativa; Chen et al., 2002). Ectopic 35S-driven expression of FAE1, which causes high accumulation of very long chain fatty acids in Arabidopsis vegetative tissues, produced numerous dramatic changes in overall plant morphology (Millar et al., 1998). Moreover, several plant wax mutants display altered morphology, including changes in stomatal shape and index (Zeiger and Stebbins, 1972; Jenks et al., 1996, 2002; Todd et al., 1999; Gray et al., 2000; Chen et al., 2003). Mutations in Arabidopsis FDH (Yephremov et al., 1999; Pruitt et al., 2000) and HIC (Gray et al., 2000), which encode very long acyl chain β-ketoacyl synthase enzymes, likewise cause major changes in overall plant morphology, including new postgenital fusion phenotypes and altered stomatal index.
Although several wax mutants in Arabidopsis show reduced reproductive capacity due to suppressed pollen recognition by the stigma (Fiebig et al., 2000; Chen et al., 2003), a connection between aliphatic waxes and other aspects of reproduction, like embryo development, has not been reported. And despite numerous reports of embryo-lethal mutants in Arabidopsis (Helliwell et al., 2001; Tzafrir et al., 2003), only acc1 has been associated with changes in the synthesis of aliphatic lipids (Baud et al., 2003, 2004). Mutation in the acetyl-CoA carboxylase encoding gene ACC1 caused a deficiency in the very long chain fatty acids but a specific enrichment of the C18:1 acid homologs in the seeds, a change that caused embryos to be shortened and lack cotyledons (Baud et al., 2003). The ACC1 protein is predicted to function in the synthesis of malonyl-CoA, an important two-carbon donor in lipid metabolism, and may even play a role in wax metabolism. Whether the ACC1 gene product impacts the synthesis of the very long chain waxes is unknown.
We recently used insertion mutagenesis to identify a recessive mutant in Arabidopsis, designated resurrection1 (rst1), which exhibits altered cuticular wax synthesis and arrested embryo development. We describe here the novel RST1 gene, and how mutation in its sequence specifically alters both cuticle and embryo formation. The new ideas arising from this work regarding the role of aliphatic lipids in signaling plant development are also discussed.
RESULTS
Isolation and Genetic Analysis of the rst1 Mutants
Approximately 35,000 families from a T-DNA mutagenized T2 population of Arabidopsis ecotype C24 (created as in Weigel et al., 2000) were screened to identify mutants having reduced visible glaucousness (increased glossiness) of the inflorescence stem due to reduction in epicuticular wax crystals. One glossy-stemmed mutant arising from this screen also displayed a seed abortion phenotype, wherein most seeds within the inflorescence terminated prematurely, producing many wrinkled (shrunken), dark-reddish, nonviable seeds. This mutant, designated rst1-1, contained a single insert in the first exon of a putative Arabidopsis novel gene (At3g27670, MGF10.8). Using this sequence information, the SALK Institute's insertion-tag populations were screened to find two additional independent mutants, this time in the Columbia ecotype, which had inserts in the same open reading frame (ORF). Both SALK mutants, designated rst1-2 and rst1-3, produced glossy stems and a high frequency of dark-red, wrinkled seeds in every mutant, essentially identical to the original rst1-1. A total of 26 F1 plants from hybrids of rst1-1 by rst1-2, 27 plants from hybrids of rst1-1 by rst1-3, and 32 F1 plants from hybrids of rst1-2 by rst1-3 had the same cuticle wax and seed/embryo defects as both parents (i.e. no complementation), demonstrating that these three mutations were allelic. As such, we report here three independent mutant alleles of the RST1 locus, the plants of which exhibit cuticular wax deficiencies and seed developmental defects. Two of the rst1 alleles are in Columbia and one in the C24 ecotype, which demonstrates that the function of RST1 is conserved, even when occurring in two quite divergent genetic backgrounds.
To establish basic inheritance, all three rst1 alleles were backcrossed to their respective isogenic wild types (with rst1-2 and rst1-3 also being reciprocally crossed). The seed and wax phenotypes of all resulting F1s were clearly wild type (at least 25 F1s scored for each), indicating recessive inheritance. Seeds heterozygous for rst1 do not abort at elevated rates, whether growing on wild-type or homozygous rst1 mutant parents. Segregating F2 seeds on F1 plants heterozygous for rst1 have an elevated proportion of aborted seeds. This provides additional evidence that abortion is not affected by the maternal genotype since the heterozygous parent still displays seed abortion, presumably due to abortion of a portion of the seeds that are homozygous for rst1 within the segregating F2 seed population. Thus, a heterozygous parent does not complement the seed abortion defect of the rst1 homozygous seeds that it carries. Reduced inflorescence stem glaucousness, and the presence or absence of many dark-red, wrinkled seeds in the siliques, were used as a visual score for both the mutant wax and seed traits, respectively. The glossy stems and wrinkled seed phenotypes showed 100% cosegregation in three large F2 populations derived from rst1 backcrosses to wild type. There was no evidence in any one of these three alleles of environmental effects on these traits, even though we closely examined the effect of photoperiod, salt, and drought on rst1-1 wax and seed formation (data not shown). As such, monogenic recessive inheritance of the wax and seed defects for all rst1 alleles is thus clearly established. The wax and seed phenotypes of all three rst1 allelic mutants were found to be nearly identical, so only data from the rst1-2 mutant (in the third Columbia backcross generation) and its isogenic parent Columbia are presented below (unless noted).
Cuticular Wax Alteration
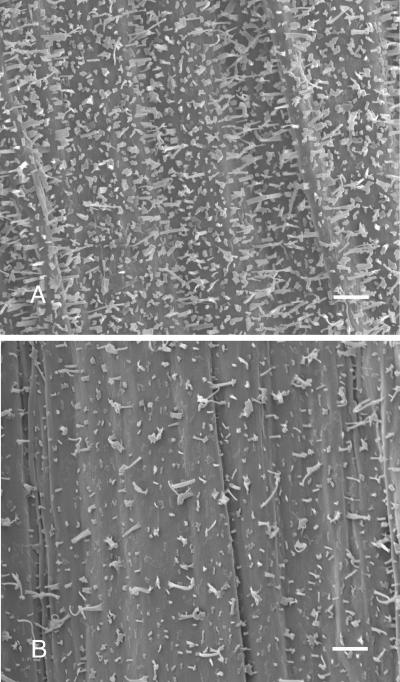
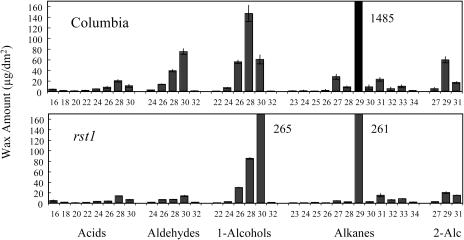
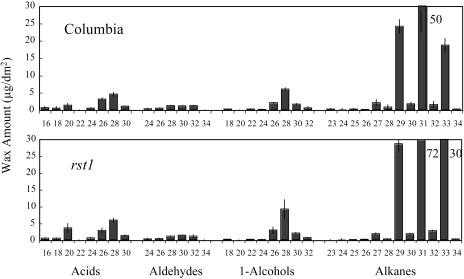
Stem surfaces of the rst1 mutants are much more glossy green than the respective isogenic wild-type stems, which display the normal glaucous stem wax coating. Leaf surfaces of both the wild type and respective rst1 mutants lack visible waxes. Scanning electron microscopy (SEM) was used to show that the density of wax crystals on stems of the rst1-2 mutant was reduced relative to the wild type; however, the shape and proportion of each of the wax crystal types was not altered dramatically (Fig. 1). Relative to wild type, total wax amount per area on rst1-2 stems was reduced 59.1% (Table I). Waxes of rst1-2 had a large proportional reduction in the aldehyde, alkane, secondary alcohol, and ketone classes, and a proportional increase in the acid, primary alcohol, and ester classes (Table I). For individual wax homologs, the largest changes on rst1-2 stems occurred in the C29 alkanes that decreased 5.7-fold and the C30 primary alcohols that increased 4.4-fold (Fig. 2). Interestingly, the total wax amount on rst1-2 leaves was elevated 43% above wild-type levels (Table I). Except for aldehydes, the amount of all rst1-2 leaf wax classes was elevated, with the largest increase occurring in the alkane class (Table I; Fig. 3). The C31 and C33 homologs of rst1-2 leaf alkanes were increased more than any other (Fig. 3).
Figure 1.
Epicuticular wax crystallization patterns on stem surfaces of Arabidopsis ecotype Columbia (A) and the rst1-2 mutant (B) visualized using SEM. Bar = 10 μm.
Table I.
Cuticular waxes on the inflorescence stems and leaves of wild-type Arabidopsis ecotype Columbia and the isogenic rst1-2 mutant
The mean total wax amount (Amt, μg dm−2 ± sd), total amount of each wax class (μg dm−2 ± sd), and the percentage (%) of each class within each sample extract are shown. nd, Not detectable.
| Stem
|
Leaf
|
|||
|---|---|---|---|---|
| Columbia | rst1-2 | Columbia | rst1-2 | |
| Acids | ||||
| Amt | 50.9 ± 5.0 | 32.9 ± 1.9 | 11.8 ± 1.0 | 15.3 ± 2.2 |
| % | 1.6 ± 0.1 | 2.5 ± 0.1 | 9.1 ± 0.3 | 8.2 ± 0.9 |
| Aldehydes | ||||
| Amt | 130.3 ± 5.8 | 28.7 ± 2.0 | 4.4 ± 0.1 | 4.4 ± 0.6 |
| % | 4.0 ± 0.2 | 2.1 ± 0.1 | 3.4 ± 0.3 | 2.3 ± 0.3 |
| 1-Alcohols | ||||
| Amt | 269.0 ± 3.7 | 382.7 ± 8.1 | 10.7 ± 0.9 | 15.4 ± 4.0 |
| % | 8.2 ± 0.4 | 28.6 ± 0.4 | 8.2 ± 0.2 | 8.2 ± 1.8 |
| Alkanes | ||||
| Amt | 1,571.1 ± 123.0 | 302.5 ± 8.3 | 99.7 ± 9.7 | 138.5 ± 4.6 |
| % | 48.0 ± 0.8 | 22.6 ± 0.2 | 76.8 ± 1.1 | 74.4 ± 1.1 |
| 2-Alcohols | ||||
| Amt | 81.1 ± 4.2 | 36.8 ± 1.9 | nd | nd |
| 2.5 ± 0.3 | 2.7 ± 0.2 | |||
| Ketones | ||||
| Amt | 761.3 ± 37.8 | 212.7 ± 8.2 | 0.7 ± 0.2 | 0.6 ± 0.1 |
| % | 23.3 ± 0.7 | 15.9 ± 0.1 | 0.5 ± 0.1 | 0.3 ± 0.1 |
| Esters | ||||
| Amt | 162.6 ± 17.2 | 146.8 ± 15.2 | 2.5 ± 1.2 | 12.1 ± 3.3 |
| % | 5.0 ± 0.8 | 11.0 ± 0.8 | 2.0 ± 1.0 | 6.5 ± 1.9 |
| Total | ||||
| Amt | 3,273.2 ± 203.6 | 1,339.0 ± 42.2 | 129.9 ± 10.8 | 186.2 ± 7.1 |
Figure 2.
Inflorescence stem cuticular wax constituents on Arabidopsis ecotype Columbia and the rst1-2 mutant. Values represent cuticular wax load in μg/dm2 of surface area ± sd. Chain lengths are labeled on the horizontal axis. Where cuticular wax constituent amount was off the scale, a number designating actual value is next to the bar. Secondary alcohols are abbreviated 2-Alc (see Table I for total quantities in each class).
Figure 3.
Rosette leaf cuticular wax constituents on Arabidopsis ecotype Columbia and the rst1-2 mutant. Values represent cuticular wax load in μg/dm2 of surface area ± sd. Chain lengths are labeled on the horizontal axis. Where cuticular wax constituent amount was off the scale, a number designating actual value is next to the bar. Secondary alcohols are abbreviated 2-Alc (see Table I for total quantities in each class).
Transmission electron microscopy with osmium stain and light microscopy with Sudan IV lipid stain were used to examine the cuticle membrane covering both the stem and embryo cotyledonary surface of rst1-2 and wild type. However, no visible differences were evident between rst1-2 and wild-type cuticles on these organs (data not shown). Additionally, transpiration rates of whole flowering plants growing in pots (as examined in Chen et al., 2003) of rst1-2 did not differ from those of wild type (data not shown), providing evidence that the leaf and inflorescence cuticle membrane permeability is normal in rst1 (cuticle membrane mutants reported to date show high transpiration; thus, rst1-2's cuticle membrane appears normal for this character).
Termination of Seed Development
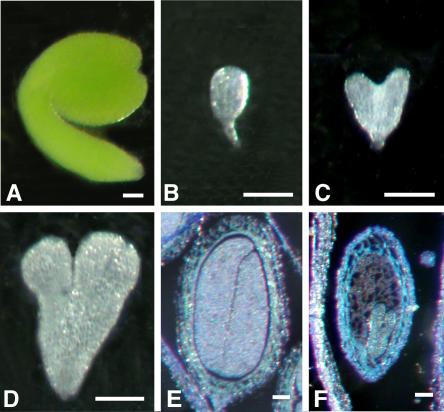
In a pool of 520 seeds from 30 rst1-2 plants, 70% of the seeds were visibly smaller, darker reddish-brown, and highly shrunken and wrinkled (Fig. 4A). When 100 each of wild-type, rst1-2 shrunken, and rst1-2 normal seeds were tested for seed germination, nearly all of the wild-type and normal rst1-2 seeds germinated, whereas none of the rst1-2 shrunken seeds germinated. Visual examination of siliques with mature seeds revealed both green and white seed types (Fig. 4B). Although embryos could be seen in mature green seeds of the rst1 mutants using light microscopy, an embryo could not be likewise delineated in the white seeds when viewed through the seed coat of cleared seeds (Fig. 4C). Additional studies using the seeds from five randomly selected siliques from six plants each of wild type and rst1-2 revealed that the occurrence of embryo abortion was completely random throughout the inflorescence and within the siliques (i.e. no differences between apical and basal sections of inflorescence or silique).
Figure 4.
Seed morphology of wild-type Arabidopsis Columbia and rst1-2 viewed using light microscopy. A, Desiccated wild-type seeds, normal rst1-2 seeds, and shrunken nonviable rst1-2 seeds. Normal rst1-2 seeds are slightly larger than wild-type seeds. B, Mature silique section showing viable dark-green (white arrows) and nonviable light-green seeds in an rst1-2 mutant. C, Mature wild-type seed, normal rst1-2 seed, and nonviable rst1-2 seed. Normal seeds show chlorophyll accumulation in the cotyledons visible through the seed coat. Bar = 0.25 mm.
The wrinkled coat trait of mature seeds was clearly linked with and resulted from embryo abortion, and more than 100 mature wrinkled seeds selected randomly from the F2 populations described above all possessed tiny, aborted embryos (visualized using light microscopy). Moreover, the embryo defective mutant seeds were easily identified when very young, as the young mutant seeds did not display the green coloration typical of normal immature seeds (due to chlorophyll accumulation in normal embryos). Well over 100 immature white seeds were carefully observed in siliques to develop into mature seeds having dark-red, wrinkled, and shrunken phenotypes and aborted embryos, whereas immature green seeds developed normal, nonwrinkled seed coats and fully formed embryos. Moreover, a calculation of percentage of immature white seeds from 275 total seeds (from five randomly selected siliques from six replicate plants) was the same as the percentage of dark-red, wrinkled seeds in a pool of 520 dried and matured seeds from a bulked rst1-2 seed population, showing that 70% of the seeds were defective.
Wild-type Columbia air-dried seeds had an average weight per seed of 16.9 ± 1.0 μg. The average weights per seed of the rst1-2 normal and wrinkled (shriveled) seeds grown under the same conditions were 25.7 ± 0.6 μg and 5.4 ± 0.3 μg, respectively. No seeds were produced in 30 emasculated wild-type and rst1-2 mutant flowers, indicating that, unlike previously described fertilization independent seed mutants (Kohler et al., 2003), pollination is required for both normal and shrunken seed development.
In a random sampling of 45 aborted embryos from siliques 12 d after anthesis, arrest of embryo development was observed as early as the late globular stage and as late as the early torpedo stage, with most embryos classified as aborting at the mid-heart stage (Fig. 5, A–D). Paraffin embedding and sectioning showed that 8-d-old wild-type (Fig. 5E) and normal rst1-2 (data not shown) seeds at the same developmental stage had visually similar embryos that completely filled the seed. Aborted heart-shaped embryos appeared to possess normal cell patterning up to the heart-shaped stage (Fig. 5, E and F). Duplication of the endosperm nuclei and cellularization of the endosperm tissue appeared normal, beginning 2 d after anthesis, in all rst1-2 seeds examined (data not shown).
Figure 5.
Embryo morphology of wild-type Arabidopsis Columbia and rst1-2 visualized with light microscopy. A to D, Wild-type embryo (12 d after anthesis) excised from the seed (A), excised rst1-2 embryo aborted at the globular stage (B), excised rst1-2 embryo aborted at the heart-shaped stage (C), and excised rst1-2 embryo aborted at the late heart stage (D). Eight-day-postanthesis wild-type and rst1-2 embryos were sectioned and viewed using light microscopy. E, Wild-type seed filled with embryo cotyledons; F, rst1-2 embryo that terminated growth at the heart-shaped stage. Bar = 100 μm.
Seed Oil Composition
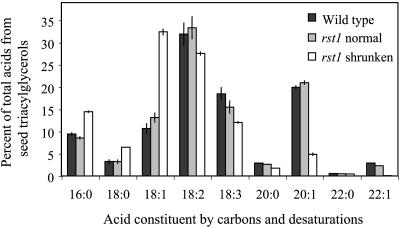
To determine whether the rst1 mutation affected lipids of the seed (triacylglycerols) in addition to lipids of the stem (waxes), detailed analysis of seed triacylglycerol-derived fatty acids was performed. Results indicated that the normal-type mature seeds on rst1-2 mutants had essentially identical acid proportions as seeds of wild type (Fig. 6); however, the total acid amount per seed was elevated from 3.35 μg (19.8% of seed weight) in wild type to 6.07 μg (23.6% of seed weight) in normal-type seeds of rst1-2. The mature but shrunken seeds of rst1-2 were severely deficient in these lipids, with total acid amount per seed decreasing to an average 0.4 μg (6.8% of seed weight) and with an especially large reduction in the longer and more desaturated acid constituents (Fig. 6). Rather than the C18:2 acid (linoleic acid) being the major constituent as in wild-type and normal-type rst1-2 seeds, the C18:1 acid (oleic acid) accumulated most in the aborted rst1 seeds (Fig. 6).
Figure 6.
Percent composition (±sd) of seed storage lipid acids derived from triacylglycerols for wild-type Arabidopsis seeds, seeds of rst1-2 that resemble wild type (rst1 normal), and the shrunken seeds of rst1-2 (rst1 shrunken).
Leaf Epidermal Morphology
Because many cuticle wax mutants are known to alter stomatal shape and stomatal index, the epidermal cell morphology of both the adaxial and abaxial surfaces of the rosette leaves of wild type and rst1-2 mutants was determined. No significant difference could be observed in leaf epidermal pavement or guard cell shape, leaf stomatal index, or trichome number between wild-type and rst1-2 plants (data not shown).
Molecular Cloning of the RST1-2 Gene
Southern-blot analysis using the BAR gene fragment as probe showed that the original rst1-1 mutant had at least two inserts (data not shown). However, thermal asymmetric interlaced PCR (Liu et al., 1995) and plasmid rescue later verified three inserts all on chromosome 5. PCR using primers designed from the T-DNA border sequences and insert-flanking plant DNA sequences were used to check the linkage of all three inserts to the rst1-1 phenotypes in 50 mutants of a backcross F2 population. One of the inserts was homozygous in all mutants, indicating a close linkage of the rst1-1 mutant phenotypes to this insert. The rst1-1 mutant having this insert only (checked by both PCR and Southern blot) was used for further characterization.
The rst1-1 insert occurred in the first exon of a putative Arabidopsis novel gene (At3g27670, MGF10.8). The expressed sequence tag (EST) H76516 was found to be identical to the putative 3′ cDNA of this gene, and the associated clone (197C20) was ordered from the Arabidopsis Biological Resource Center (ABRC) and sequenced. The full-length EST was 1,157 bp, and included sequence of the last four putative exons (17th to 20th exons) of At3g27670, a 139-bp 3′ untranslated region (UTR), and a 12-bp polyA. A 1,778-bp 5′ cDNA fragment was obtained by 5′ RACE using primers designed from the putative At3g27670 cDNA. The sequence included a 204-bp 5′ UTR and the first four putative exons of At3g27670. A 3,555-bp cDNA fragment was amplified from the cDNA using primers designed from the 5′ RACE and the EST clone fragment sequences. This reverse transcription (RT)-PCR fragment had a 294-bp overlap to the 5′ RACE fragment at the 5′ end and a 465-bp overlap to the sequenced EST clone at the 3′ end. Sequencing revealed an additional exon of 78 bp between the originally predicted 14th and 15th exons of the putative At3g27670 ORF. As such, the actual full-length RST1 ORF was thus found to span 8,164 bp of genomic DNA and have 21 exons and 20 introns. The full-length predicted transcript is 5,881 bp, and has a coding region of 5,526 bp, a 204-bp 5′ UTR, and a 139-bp 3′ UTR. The 5′ UTR contains a stop codon immediately before the start codon, and there were no other alternative start codons within it. The full RST1 nucleotide sequence and associated annotation is now available under GenBank accession number AY307371.
Two knockout rst1 mutants were obtained from the SALK Institute through the ABRC stock center. The first Columbia rst1 mutant (SALK_ 070359, designated rst1-2) had a single T-DNA insert in the 12th intron, and another Columbia rst1 mutant (SALK_129280, designated rst1-3) had a single T-DNA insert in the 4th exon. SALK assistance was obtained from the SIGnAL Web site at http://signal.salk.edu. Both rst1-2 and rst1-3 showed the same increase in stem glossiness, change in wax composition and structure, and wrinkled seed phenotype as did rst1-1. As for rst1-1, backcross F2 populations were used to select mutant lines that were homozygous for single rst1-2 and rst1-3 inserts, and cosegregation (at least 50 mutant plants each) of the inserts with the altered cuticle and seed phenotypes was verified. The RST1 locus was not closer than an estimated 20 cM to any of the existing cer loci on chromosome 5 (Rashotte et al., 2004), so tests of allelism were not performed.
Characterization of the Predicted RST1 Protein
The RST1 transcript encodes a predicted polypeptide of 1,841 amino acids with a molecular mass of 203.6 kD and a theoretical pI of 6.21. No alternative ORFs were identified. The RST1 protein does not show high identity to any protein of known function; however, it was 34% (636/1,841) identical and had 51% (964/1,841) positives to the 1,842-amino acid annotated rice protein OJ1276_B06.27 (GenBank BAB92518). Also, RST1 was 28% (32/113) identical and had 51% (51/113) positives to a 113-amino acid fragment of the 1,801-amino acid human hypothetical protein FLJ20357 (GenBank AAN17740). No integral membrane domain was found in the RST1 protein by the TMHMM program. Using TargetP (http://www.cbs.dtu.dk/services/TargetP/), RST1 was predicted to target the mitochondria with TargetP score of 0.550 and probable signal sequence length of 78 amino acids. The rice protein OJ1276_B06.27 (having highest identity to RST1) is predicted to target the chloroplast with a TargetP score of 0.554 and probable signal sequence length of 74 amino acids, whereas the rice RST1's targeting to mitochondria had only a 0.110 TargetP score. A PROSITE database (http://us.expasy.org/prosite/) search found three possible domains in RST1, the aldo/keto reductase family putative active site signature in residue 874-889 (accession no. PS00063), the cytochrome C family heme-binding site signature in residue 1512-1517 (accession no. PS00190), and the G-protein-coupled receptor family 1 signature (Rhodopsin like) in residue 1733-1749. Interestingly, the Oryza protein OJ1276_B06.27 has none of these three conserved domains.
Analysis of RST1 Transcript Expression
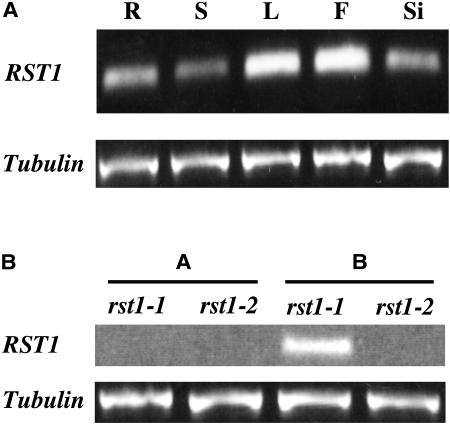
The 5′ RACE RST1 cDNA fragment was used as a probe in RNA gel-blotting experiments. Using RNA blotting, transcripts were not detected in stems, rosette leaves, siliques, and whole flowers of wild-type C24 and wild-type Columbia after 7-d exposure, indicating probable low expression in the whole plant (data not shown). However, one-step RT-PCR did reveal the presence of RST1 transcripts in these same tissues, and suggested that the amount of RST1 transcript was higher in leaves and flowers and lower in roots, stems, and siliques of wild type (Fig. 7A). RT-PCR with leaves using primers flanking the rst1-1 T-DNA insert site did not produce amplified product in any of the three allelic rst1 mutants. RT-PCR using primers to sequences downstream from the rst1-1 T-DNA insert site amplified products in the rst1-1 mutant, but not in rst1-2 (Fig. 7B) or rst1-3 (data not shown). Thus, both rst1-2 and rst1-3 were null alleles, appearing to have completely blocked transcript expression, whereas rst1-1 produced a truncated RST1 transcript driven by the 35S promoter on the activation insert. Interestingly, the rst1-1 mutant often displayed short, more rounded leaves that senesced early, and inflorescences that emerged later via axillary buds (appearing to “come back to life after senescence” and leading to our naming the mutant resurrection1 [rst1]). Linkage between this novel, slightly truncated rst1-1 allele and incomplete penetrance of these novel leaf developmental phenotypes will be described in subsequent reports. Verification that rst1-1's truncated transcript is responsible for the additional phenotypes and their inheritance will require substantial experimentation to verify.
Figure 7.
Expression of RST1 in wild-type Arabidopsis ecotypes C24 and Columbia using quantitative RT-PCR. A, RST1 transcript expression in various organs of wild-type C24, with tubulin gene expression as control. R, Root; S, stem; L, rosette leaf; F, flower; Si, silique. B, RST1 transcript expression in the two rst1 alleles, rst1-1 and rst1-2. Expression under A was done using primers designed from the first exon, whereas B represents expression using primer from the fourth exon. The rst1-1 allelic mutant showed expression of a truncated transcript, whereas the knockout mutant rst1-2 (and rst1-3, not shown) expressed no RST1 transcript.
DISCUSSION
Mutations in cuticle genes such as ATT1, CER1, CER6, WAX2, and others are known to influence various aspects of plant development, including fertility, postgenital fusion, epidermal architecture, and leaf size and shape (Gray et al., 2000; Jenks et al., 2002; Chen et al., 2003; Xiao et al., 2004). Gray et al. (2000) and others speculate that the connection between cuticle lipids and other aspects of development lies in the yet to be elucidated signaling functions of the cuticle lipids. We report here that mutation of the RST1 gene causes alterations in leaf and stem cuticular waxes and terminates embryo development in a majority of seeds. This analysis of the rst1 mutant raises new and intriguing questions about the possible role of lipids as signals in plant development.
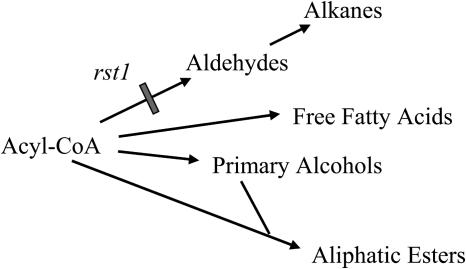
As a proportion of total waxes, rst1 inflorescence stems have a major deficiency in aldehydes and aldehyde metabolites, whereas all other wax classes are proportionally elevated. The very high elevation in C30 alcohols and reduction in C29 alkanes on rst1 stems leads to an interpretation that the C30 acyl-CoA precursors are being shunted toward the C30 primary alcohol branch (pool) of the wax pathway and away from the C29 aldehyde to C29 alkane generating pathway. The RST1 gene product may thus have a direct function in acyl-CoA reduction to aldehydes, but likely not acyl-CoA conversion to primary alcohols, free fatty acids, or esters (Fig. 8). Whether RST1's short, predicted 15-amino acid sequence homology to an aldo/keto reductase active site signature indicates a possible role in the acyl-CoA reduction required for aldehyde synthesis is yet to be established. The reason for the increase in wax amount on rst1 null mutant leaves is unknown. This condition is not without precedent, however, as cuticle mutants in Sorghum, such as bm4, that have reduced leaf sheath waxes have increased leaf blade waxes (Rich, 1994; Jenks et al., 2000). Further studies are needed to investigate whether RST1 may act as a negative regulator of the wax pathway in Arabidopsis leaves.
Figure 8.
Model pathway for acyl-CoA conversion in Arabidopsis stem cuticle wax metabolism. Long chain acyl-CoAs serve as substrates in the synthesis of cuticle lipids (Kolattukudy, 1996). The rst1 mutation primarily blocks synthesis of free aldehydes and its metabolites (represented by a gray bar), suggesting that the RST1 protein may function in acyl-CoA reduction. The secondary alcohols and ketones are metabolites of alkanes (not shown).
The aborted seeds of rst1 have greatly reduced amounts of triacylglycerol-derived fatty acids, with constituent profiles shifted toward elevations in shorter and more saturated chains. Viable seeds in rst1 plants were slightly larger but had normal proportions of these storage lipids. The reduced acids in aborted rst1 seeds may have been caused by the early termination of embryo formation. Accumulation of triacylglycerols (with their covalently linked fatty acids) begins rapidly with the onset of the torpedo stage when lipid bodies first appear in the expanding cotyledons (Bowman, 1994). Our lipid analysis corroborates our observations using light microscopy that embryo development is terminated in rst1 just before the torpedo stage, potentially then suppressing normal lipid accumulation. Whether embryos require normal lipid synthesis to complete development has not been thoroughly investigated. Potentially, a deficiency in certain lipid constituents at the onset of the torpedo stage might create a limited resource condition for embryo development, potentially even signaling an embryo abortion program. Interestingly, mutation in ACC1 involved in the synthesis of malonyl-CoA (the two-carbon donor in lipid synthesis) reduced seed lipid content overall and led to a proportional increase in the accumulation of C18:1 fatty acids in seeds of Arabidopsis (Baud et al., 2003, 2004), just as did the rst1 mutation (in aborted seeds). Like rst1, the acc1 mutation terminates embryo development at the heart stage when cotyledons begin to differentiate. In the case of acc1, a defect in synthesis of seed lipid precursors is the suspected cause of embryo abortion. Whether rst1 disrupts the function of acetyl-CoA carboxylase in the embryo or embryo lipid pathway utilization of malonyl-CoA is still uncertain.
Numerous embryo-lethal mutants in Arabidopsis show termination of embryo development during the preglobular to cotyledonary stages (Tzafrir et al., 2003). These embryo mutants commonly possess disruptions in cell patterning due to disruptions in signaling molecules, metabolic enzymes, transcription factors, or unknown proteins (Berleth and Chatfield, 2002; Lohe and Chaudhury, 2002). Mutation in the E2748 gene of pea (Pisum sativum) causes embryo abortion, with lethality preceded by disrupted differentiation of the embryo's epidermal cells into transfer cells. E2748 epidermal cells erroneously develop larger vacuoles, thicker cell walls, larger size, and also lack normal Suc-transporting capability. E2748 thus shows that differentiation of the epidermis is a critical phase of embryo development (Borisjuk et al., 2002). By comparison, the rst1 mutant has a defect in leaf and stem epidermal development expressed as an inability to synthesize normal surface waxes. We used light (with Sudan IV stain) and transmission electron microscopy to examine cuticles of defective rst1 embryos at seed maturity; however, we could not detect a change from wild type (data not shown). Plant cuticle mutants, like att1 (Xiao et al., 2004), wax2 (Chen et al., 2003) and bm2 (Jenks et al., 1994), possess altered leaf epidermal permeability to both large and small molecules (including water, chlorophyll, and herbicides). It was proposed by Borisjuk et al. (2002) that embryo abortion in the E2748 mutant in pea could be due to an inability to import necessary nutrients through the altered epidermal layer of the mutant embryos. Whether rst1 embryo epidermises (or cuticle membranes) show similar nutrient transport deficiency is a subject for future research.
The rst1 mutant in Arabidopsis shows incomplete penetrance of seed abortion, with 70% of seeds becoming shrunken and nonviable. The only other plant mutant showing similar incomplete penetrance of seed abortion is amp1 in Arabidopsis (Helliwell et al., 2001). The amp1 mutation causes an elevation in cytokinin levels, which are associated with altered cotyledon number in 30% of the seeds. Most other seeds of amp1 develop normally. The AMP1 gene is predicted to encode a Glu carboxypeptidase that Helliwell et al. (2001) speculate may modulate the level of a putative signaling molecule that interacts with cytokinin pathways. It is interesting to note that many plants (including crops like soybeans [Glycine max] and rice) commonly exhibit fruit, seed, and/or embryo abortion. In some cases, this thinning process may serve as a means of reallocating limited nutrient resources in suboptimal environments to promote survival and healthy growth in the remaining seeds. Although Arabidopsis rarely aborts seeds when grown in optimum conditions, it often aborts seeds during stress. The ability to terminate seed development is likely a precise regulatory process since excess thinning would limit (or eliminate) offspring and the subsequent representation of parental genes in the next generation. Future studies might show that RST1 is involved in the synthesis of lipids (or other growth regulatory molecules) that serve as signals in embryo development associated with the abortion program.
Considerable evidence has emerged in recent years that implicate epigenetic control in embryo development, including a role for gene imprinting (Chaudhury et al., 2001). Embryo development in Arabidopsis may be substantially influenced by genes such as LEC (Lotan et al., 1998), MEA (Grossniklaus et al., 1998; Kiyosue et al., 1999), FIE, and FIS2 (Gehring et al., 2004), which directly or indirectly control the maternal environment of the developing embryo and thereby embryo gene expression. Maternal gene effects on embryo development have, in a few cases, been examined in some detail, such as the SIN1 gene in Arabidopsis (Ray et al., 1996). The domain structure of SIN1 suggests it may be involved in RNA processing/metabolism (Schauer et al., 2002), including gene-silencing mechanisms like that of ARGONAUTE (Zilberman et al., 2003). Motifs found on RST1 are also indicative of a possible function in RNA metabolism, and such similarities may connect RST1 to embryo viability by affecting gene expression during embryo development. Analysis of seeds from reciprocal crosses using rst1 presented in this report, however, implies that RST1 has neither maternal nor paternal influence on embryo development.
The rst1 mutant reveals an unexpected connection between synthesis of cuticular waxes and formation of the embryo. The RST1 gene encodes a very large but novel protein with homologies to proteins in organisms as diverse as humans and rice. Future studies to examine RST1 gene expression and function in Arabidopsis could shed much light on very long chain lipid involvement in embryo development.
MATERIALS AND METHODS
Genetic Analysis of the rst1 Mutant
The activation T-DNA vector pSKI015 was used to generate an insertion mutant population (T1) in the genetic background of Arabidopsis (Arabidopsis thaliana L.) Heynhold ecotype C24 as in Weigel et al. (2000). Bialophos resistance was used as the selectable marker. Plants were grouped into pools of 10 families, and T2 progenies of roughly 200 plants from these pools were screened for mutants having reduced visible glaucousness of the inflorescence stem surface. Mutagenized populations were grown in a greenhouse at Purdue University using overhead irrigation, natural light, and a temperature varying around 22°C.
Analysis of Cuticular Waxes
SEM was used to analyze epicuticular wax crystallization patterns. Leaf and stem (first internode above the rosette) samples were collected from the Arabidopsis wild type and rst1 mutants after 6 weeks of growth. Four replicates of each sample were mounted on aluminum stubs and sputter coated with gold palladium using six 30-s bursts from the sputter coater. Previous research showed that air-dried samples coated in this way were similar to specimens prepared using low-temperature SEM with little evidence of artifacts in wax crystallization patterns observed (Jenks et al., 1992). Coated surfaces were viewed using a JEOL JSM-840 scanning electron microscope (JEOL) at 5 kV.
For compositional analysis, cuticular wax samples were extracted from the leaves and stems of flowering Arabidopsis wild type and rst1 mutants after 6 weeks of growth. Leaf and stem (representing the first through fifth internodes) samples were inserted into a 20-mL standard glass scintillation vial, and approximately 15 mL of GC-grade hexane added. The tissues were agitated for 30 s and the solvent decanted off into new scintillation vials. Tissues and vials were given a 1-s rinse with approximately 2 mL of hexane, and then the solution decanted into the sample vial. The leaf extracts thus contain waxes from both abaxial and adaxial leaf surfaces and, like stems, showed no coloration due to chlorophyll or other internal lipids. Wax compositional analysis was according to Jenks et al. (1995). The hexane-soluble cuticular wax extracts were evaporated to dryness under a nitrogen stream and the dried residue prepared for gas chromatography by derivatization using N,O-bis(trimethylsilyl)trifluoroacetamide. Derivatization was for 15 min at 100°C. After surplus N,O-bis(trimethylsilyl)trifluoroacetamide was evaporated under nitrogen, the sample was redissolved in hexane for analysis with a Hewlett-Packard 5890 series II gas chromatograph (GC) equipped with a flame ionization detector. The GC was equipped with a 12-m, 0.2-mm HP-1 capillary column with helium as the carrier gas. The GC was programmed with an initial temperature of 80°C and increased at 15°C min−1 to 260°C, where the temperature remained unchanged for 10 min. The temperature was then increased at 5°C min−1 to 320°C, where the temperature was held for 15 min. Quantification was based on flame ionization detector peak areas and the internal standard hexadecane. Specific correction factors were developed from external standards and applied in a standard way as in Jenks et al. (1995, 1996). The total amount of cuticular wax was expressed per unit stem or leaf surface area. Stem surface areas were calculated as the surface area of a right circular cylinder, and leaf areas were determined using computer digitization (NIH Image, version 1.62). All values represent the average of four replicate plant samples ± sd. Selected subsamples were used for injection in a GC-mass spectrometer (FinniganMAT/Thermospray) to produce electron ionization mass spectra to verify identity of all components.
Seed and Embryo Morphology
Seeds were scored visually and using a dissecting light microscope for darker red coloration, smaller size, and wrinkled seed coat. Live embryos were examined under light microscopy by immersing seeds in a few drops of 1 m KOH placed on a glass side, then placing a coverslip on top and applying gentle pressure to force the embryos out of the seed coat. Seeds were cleared using lactic acid. Paraffin embedding was done by immersing siliques in formaldehyde-acetic acid for 48 h, dehydrating siliques in a graded ethanol/tert-butanol series, and then immersing in graded tert-butanol/paraplast series. After sectioning of paraffin blocks, paraplast was removed with xylene, followed by hydrating the specimens with a graded ETOH/H2O series. Specimens were then stained with 1% toluidine blue. The quantitative and observational data presented for seed or embryo represented the average of at least four siliques each on at least six plants.
Seed Lipid Analysis
Seed lipid triacylglycerides were extracted by crushing 25 seeds per sample (100 seeds for shrunken seeds) in a 4-mL glass vial with 1 mL hexane and 50 ppm butylated hydroxytoluene (Sigma). After incubation at 50°C for 15 min, vials were centrifuged at 13,000g for 15 min to pellet seed debris. Hexane containing triacylglycerides were decanted to a new vial. Heptadecanoic acid was added as an internal standard, followed by blowing down the sample to dryness with nitrogen stream. Transesterification was done by adding 700 μL of 3 n HCl in methanol (Supelco) and incubating at 80°C for 2 h. After cooling, 1 mL hexane and 1.5 mL of 0.9% (w/v) NaCl were added, followed by gentle agitation for 1 min. After phase separation, 250 mL of the top organic phase was transferred to a new vial for injection into a Hewlett-Packard 5890 series II GC equipped with a flame ionization detector. The GC was equipped with a 12-m, 0.2-mm HP-1 capillary column with helium as the carrier gas. The GC was programmed with an initial temperature of 80°C and increased at 15°C min−1 to 200°C, then the temperature was increased at 2°C min−1 to 220°C, and then the temperature was increased at 30°C min−1 to 280°C. Quantification was based on flame ionization detector peak areas and the internal standard. Multilevel external standards for every lipid constituent were used to develop specific correction factors as in Jenks et al. (1995, 1996). Selected subsamples were used for injection in a GC-mass spectrometer (FinniganMAT/Thermospray) to produce electron ionization mass spectra to verify identity of all components.
Analysis of Stomatal Index
Stomatal density, epidermal pavement-cell density, and stomatal index for both the adaxial and abaxial surfaces were determined using light microscopy modified from Gray et al. (2000). Instead of dental rubber impressions, we created imprints using Duro Super Glue (cyanoacrylate) taken from the middle of the blade between the midrib and leaf margin. Leaves were selected having uniform size, being numbered 20 to 23 from the first true leaves, and the plants were 39 d old. Values for stomatal index for each surface represented the average of nine to 12 replicate imprints of 0.251 mm2 each.
Plasmid Rescue and Linkage Analysis
Thermal asymmetric interlaced PCR was performed according to Liu et al. (1995). Plasmid rescue was performed according to Weigel et al. (2000). Restriction enzymes EcoRI and HindIII were used to rescue the right T-DNA border flanking sequences, and BamHI was used for left border rescue. The rescued fragments were directly sequenced from both ends using primers designed from inside the restriction site sequences and the T-DNA border sequences. The flanking gene-specific DNA sequence was searched against the Arabidopsis genome database (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/ara.html).
5′ RACE and cDNA Amplification
5′ RACE using the SMART RACE cDNA amplification kit (BD Biosciences Clontech) was performed as described by the manufacturer. The gene-specific primers used for RACE were designed from the At3g27670 putative mRNA sequence. The gene-specific primers for RST1 and rst1-1 5′ RACE were EX3R (5′-GACAAGGGACGTTAGTTCGAG-3′) and 5′ RACE (5′-ACAGCTAGCAACAGCGGGACACTCA-3′). The cDNA sequence between the 5′ RACE and 3′ EST was obtained by PCR amplification from the RACE first-strand cDNA using primers designed from the 5′ RACE and the EST clone fragment sequences. The first round PCR used primer EX3F (5′-CGCACTGATGTCTTCTCCTTC-3′) and EX17R (5′-ACTCTTTCACCGAACCCATCT-3′). The second round PCR used primer 3′ RACE (5′-CACACCTTCCACGTCTTCCTTCCTCA-3′) and EX17R. The PCR fragments were cloned into the pGEM-T easy vector (Promega).
RT-PCR Analysis
RT-PCR was performed using the SuperScript One-Step RT-PCR with Platinum Taq kit as described by the manufacturer (Invitrogen). Two pairs of primers were used for RT-PCR. The first pair was designed from the first and second exons to flank the rst1-1 T-DNA left border insertion site (RST1F, 5′-TCTCTCCAGCCAAAGCGA-3′; RST1R, 5′-CAACGATGAAGACGAATCTG-3′). The second pair was designed to flank the rst1-2 T-DNA left border insertion site (RST2F, 5′-AAATGCGGAAATTCTGAATGCT-3′; RST2R, 5′-AATGCCTCCTCGTATTGAAAATG-3′). RT-PCR of a 517-bp Arabidopsis β-8 tubulin cDNA fragment was used as control. The primers for tubulin amplification are Tubulin5′, 5′-CGTGGATCACAGCAATACAGAGCC-3′, and Tubulin3′, 5′-CCTCCTGCACTTCCACTTCGTCTTC-3′.
DNA and RNA Gel-Blot Analysis
Genomic DNA was isolated from rosette leaves based on Dellaporta et al. (1983), with minor modifications. DNA was digested using three restriction enzymes, HindIII, BamHI, and EcoRI, and fractionated using electrophoresis. A BAR gene fragment was used as DNA probe. The probe was labeled with [α-32P]dCTP (NEN-DuPont) using the random prime labeling kit (Decaprime II kit, Ambion). Membranes were washed at 42°C in 2× SSC, 0.1% SDS for 2×5 min, followed by washing in 0.1×SSC, 0.1% SDS for 2×15 min.
Total RNA was isolated from various tissues using the RNeasy Plant Mini kit (Qiagen) as described by the manufacturer. The RNA gel blot was performed according to standard methods. The 1,778-bp 5′ RACE product was used as probe, with probe labeling and hybridization the same as in the DNA blot.
Bioinformatics
Multiple sequence alignment was performed with ClustalW using default parameters through BCM Search Launcher (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html). The box shading was created by BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html). The rooted phylogenetic tree was constructed using ClustalW (Thompson et al., 1994). The transmembrane analysis was done using the TMHMM program (http://www.cbs.dtu.dk/services/TMHMM/).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AY307371.
Acknowledgments
We thank the SALK Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants (SALK 070359 and 129280). We would also like to thank Debra Sherman of the Purdue University Electron Microscopy Center and Dr. Karl Wood of the Purdue University Mass Spectrometry Center for their assistance.
This work was supported by the U.S. Department of Agriculture National Research Initiative (grant no. 97–35301–5291). This is publication number 17199 of the Purdue University Office of Agricultural Research.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.066753.
References
- Baud S, Bellec Y, Miquel M, Bellini C, Caboche M, Lepiniec L, Faure JD, Rochat C (2004) gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl-CoA carboxylase. EMBO Rep 5: 515–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Guyon V, Kronenberger J, Wuilleme S, Miquel M, Caboche M, Lepiniec L, Rochat C (2003) Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Berleth T, Chatfield S (2002) Embryogenesis: pattern formation from a single cell. In C Somerville, E Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0051, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Bird SM, Gray JE (2003) Signals from the cuticle affect epidermal cell differentiation. New Phytol 157: 9–23 [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Wang TL, Rolletschek H, Wobus U, Weber H (2002) A pea seed mutant affected in the differentiation of the embryonic epidermis is impaired in embryo growth and seed maturation. Development 129: 1595–1607 [DOI] [PubMed] [Google Scholar]
- Bowman JL (1994) Arabidopsis: An Atlas of Morphology and Development. Springer-Verlag, New York
- Chaudhury AM, Koltunow A, Payne T, Luo M, Tucker MR, Dennis ES, Peacock WJ (2001) Control of early seed development. Annu Rev Cell Dev Biol 17: 677–699 [DOI] [PubMed] [Google Scholar]
- Chen X, Goodwin M, Boroff VL, Liu X, Jenks MA (2003) Cloning and characterization of Arabidopsis WAX2 involved in cuticle membrane synthesis. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yuan H, Chen R, Zhu L, Du B, Weng Q, He G (2002) Isolation and characterization of triacontanol-regulated genes in rice (Oryza sativa L.): possible role of triacontanol as a plant growth stimulator. Plant Cell Physiol 43: 869–876 [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood VP, Hicks JB (1983) A plant DNA mini-preparation: version II. Plant Mol Biol Rep 1: 19–21 [Google Scholar]
- Fiebig A, Mayfield JA, Miley NL, Chau S, Fischer RL, Preuss D (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12: 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Choi Y, Fisher RL (2004) Imprinting and seed development. Plant Cell (Suppl) 16: S203–S213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JE, Holroyd GH, van der Lee FM, Bahrami AR, Sijmons PC, Woodward FI, Schuch W, Hetherington AM (2000) The HIC signaling pathway links CO2 perception to stomatal development. Nature 408: 713–716 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada J, Hoeppner M, Gagliano WB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- He X, Zhang B, Tan H (2003) Overexpression of a sterol C-24 (28) reductase increases ergosterol production in Saccharomyces cerevisiae. Biotechnol Lett 25: 773–778 [DOI] [PubMed] [Google Scholar]
- He YW, Loh CS (2002) Induction of early bolting in Arabidopsis thaliana by triacontanol, cerium and lanthanum is correlated with increased endogenous concentration of isopentenyl adenosine (iPAdos). J Exp Bot 53: 505–512 [DOI] [PubMed] [Google Scholar]
- Helliwell C, Chin-Atkins AN, Wilson I, Chapple R, Dennis ES, Chaudhury A (2001) The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks MA, Eigenbrode S, Lemeiux B (2002) Cuticular waxes of Arabidopsis. In C Somerville, E Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0016, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Jenks MA, Joly RJ, Peters PJ, Rich PJ, Axtell JD, Ashworth EA (1994) Chemically-induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) Moench. Plant Physiol 105: 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks MA, Rashotte AM, Tuttle HA, Feldmann KA (1996) Mutants in Arabidopsis thaliana altered in epicuticular wax and leaf morphology. Plant Physiol 110: 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks MA, Rich PJ, Peters PJ, Axtell JD, Ashworth EN (1992) Epicuticular wax morphology of bloomless (bm) mutants in Sorghum bicolor. Int J Plant Sci 153: 311–319 [Google Scholar]
- Jenks MA, Rich PJ, Rhodes D, Ashworth EA, Axtell JD, Ding CK (2000) Chemical composition of leaf sheath cuticular waxes on bloomless and sparse-bloom mutants of Sorghum bicolor (L.) Moench. Phytochemistry 54: 577–584 [DOI] [PubMed] [Google Scholar]
- Jenks MA, Tuttle HA, Eigenbrode SD, Feldmann KA (1995) Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol 108: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada JJ, Goldberg RB, et al (1999) Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci USA 96: 4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U (2003) The polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolattukudy PE (1996) Biosynthetic pathways of cutin and waxes, their sensitivity to environmental stresses. In G Kersteins, ed, Plant Cuticles, An Integrated Functional Approach. BIOS Scientific Publishers, Oxford, pp 83–108
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Lohe AR, Chaudhury A (2002) Genetic and epigenetic processes in seed development. Curr Opin Plant Biol 5: 19–25 [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MAL, Lom R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Meijer HJG, Munnik T (2003) Phospholipid-based signaling in plants. Annu Rev Plant Biol 54: 265–306 [DOI] [PubMed] [Google Scholar]
- Millar AA, Wrische M, Kunst J (1998) Accumulation of very-long-chain fatty acids in membrane glycerolipids is associated with dramatic alteration in plant morphology. Plant Cell 11: 1889–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RE, Lemieux B, Yen G, Davis RW (2000) FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA 97: 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Jenks MA, Ross AS, Feldmann KA (2004) Novel eceriferum mutants in Arabidopsis thaliana. Planta 219: 5–13 [DOI] [PubMed] [Google Scholar]
- Ray A, Lang JD, Golden T, Ray S (1996) SHORT INTEGUMENT (SIN1), a gene required for ovule development in Arabidopsis, also controls flowering time. Development 122: 2631–2638 [DOI] [PubMed] [Google Scholar]
- Rich PJ (1994) Quantitative and qualitative characterization of epicuticular wax from chemically induced bloomless and sparse bloom mutants of Sorghum bicolor. PhD thesis. Purdue University, West Lafayette, IN
- Schauer SE, Jacobsen SE, Meinke DW, Ray A (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Sperling L, Heinz E (2003) Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. Biochim Biophys Acta 1632: 1–15 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski JG (1999) KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17: 119–130 [DOI] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell (Suppl) 14: S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D (2003) The Arabidopsis SeedGenes Project. Nucleic Acids Res 31: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H (2002) Fatty acid-derived signals in plants. Trends Plant Sci 7: 217–223 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Goodwin SM, Xiao Y, Sun Z, Baker D, Tang X, Jenks MA, Zhou JM (2004) Arabidopsis CYP86A2 negatively regulates Pseudomonas syringae type III genes and is required for cuticle development. EMBO J 23: 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H (1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger E, Stebbins GL (1972) Developmental genetics in barley: a mutant for stomatal development. Am J Bot 59: 143–148 [Google Scholar]
- Zilberman D, Cao XF, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]