Abstract
Plant growth can be studied at different organizational levels, varying from cell, leaf, and shoot to the whole plant. The early growth of seedlings is important for the plant's establishment and its eventual success. Wheat (Triticum aestivum, genome AABBDD) seedlings exhibit a low early growth rate or early vigor. The germplasm of wheat is limited. Wild relatives constitute a source of genetic variation. We explored the physiological and genetic relationships among a range of early vigor traits in Aegilops tauschii, the D-genome donor. A genetic map was constructed with amplified fragment-length polymorphism and simple sequence repeat markers, and quantitative trait loci (QTL) analysis was performed on the F4 population of recombinant inbred lines derived from a cross between contrasting accessions. The genetic map consisted of 10 linkage groups, which were assigned to the seven chromosomes and covered 68% of the D genome. QTL analysis revealed 87 mapped QTLs (log of the odds >2.65) in clusters, 3.1 QTLs per trait, explaining 32% of the phenotypic variance. Chromosomes 1D, 4D, and 7D harbored QTLs for relative growth rate, biomass allocation, specific leaf area, leaf area ratio, and unit leaf rate. Chromosome 2D covered QTLs for rate and duration of leaf elongation, cell production rate, and cell length. Chromosome 5D harbored QTLs for the total leaf mass and area and growth rate of the number of leaves and tillers. The results show that several physiological correlations between growth traits have a genetic basis. Genetic links between traits are not absolute, opening perspectives for identification of favorable alleles in A. tauschii to improve early vigor in wheat.
Plant growth is a complex process that can be studied at different organizational levels. The early growth of seedlings is crucial for their establishment and hence for their eventual success in terms of biomass production or seed output. The early growth of wheat (Triticum aestivum) seedlings is slow compared to that of other temperate cereals (López-Castaneda et al., 1996). This may severely limit yield in dry regions with a Mediterranean climate. In these areas, wheat is usually sown in winter and harvested at the beginning of the hot dry summer. Rainfall is frequent in winter, before canopy closure and flowering, but declines thereafter, and water then becomes limiting for crop growth and grain filling. Rapid early development of leaf area and above-ground biomass, denoted as early vigor, contributes to a high yield due to shading of the soil surface, thereby reducing evaporation of water from the soil and leaving more water available for the crop. Greater early vigor can increase the crop's seasonal water use efficiency by as much as 25% (Siddique et al., 1990), and it is recognized as a trait to select for to improve yield under water stress (Richards, 2000; Botwright et al., 2002; Richards et al., 2002). In more favorable environments, high early vigor may be beneficial by increasing seedling competitiveness against weeds, resulting in less need for herbicide use (Lemerle et al., 2001).
Cultivated wheats exhibit some genetic variation in early vigor (Rebetzke and Richards, 1999; Richards and Lukacs, 2002); however, due to decades of repeated breeding, the germplasm of bread wheat is rather narrow. Wild relatives constitute a potential rich source of genetic variation (Feldman and Sears, 1981). Despite the fact that their overall performance is agronomically less desirable, favorable alleles are present in wild relatives (see Xiao et al. [1998] and Ellis et al. [2000] for examples in rice [Oryza sativa] and barley [Hordeum vulgare], respectively). Using molecular markers, it is possible to identify beneficial alleles in the wild germplasm (Boyko et al., 1999). By means of marker-assisted selection, those alleles can eventually be transferred into the elite breeding lines (Tanksley and Nelson, 1996).
Bread wheat (T. aestivum) is a hexaploid (genome AABBDD) and originated by natural hybridization of the tetraploid Triticum turgidum (AABB) and the diploid Aegilops tauschii (DD; Dvorak et al., 1998, and refs. therein). A. tauschii is the fastest growing of the 20 species in the Aegilops genus (Villar et al., 1998). Moreover, A. tauschii exhibits a very rapid leaf expansion rate in the early growth stage of the seedling, which is initially even faster than in T. aestivum (Bultynck et al., 2004). These observations indicate that A. tauschii might indeed be a valuable species for improvement of early vigor in wheat.
Early vigor is a complex trait that is the result of a range of growth traits at different organizational levels in the plant, ranging from cell characteristics within the leaves via individual leaf growth performance to whole-shoot leaf area expansion and even whole-plant traits. It is associated with long and broad primary leaves on the main shoot and with a high individual leaf expansion rate. Essential leaf characteristics are leaf elongation rate (LER), leaf width, and leaf elongation duration (LED). There are several lines of evidence indicating that the LER is primarily dependent on the cell production rate, pointing to a key role for meristematic activity in determining individual leaf growth rate. The rate of leaf area expansion of the whole shoot, however, depends not only on characteristics of individual leaves, but also on the rate at which new leaves and tillers emerge. Finally, a high specific leaf area (SLA; leaf area to leaf mass ratio), leaf area ratio (LAR; leaf area to total plant mass ratio), and leaf mass fraction (LMF, leaf mass per unit plant mass) contribute to early vigor (López-Castaneda et al., 1996; Becraft, 1999; Bultynck et al., 1999, 2003, 2004; Rebetzke and Richards, 1999; Fiorani et al., 2000; Richards, 2000; Richards and Lukacs, 2002).
Early vigor traits such as a high LAR, SLA, and biomass allocation to the leaves and/or shoot are often associated with a high relative growth rate (RGR; rate of increase in biomass per unit of biomass already present per unit of time) of the whole plant (Lambers and Poorter, 1992). Thus, high early vigor might coincide with a high RGR in the early stages of seedling development. Differences in RGR, however, are not invariably associated with differences in LAR, SLA, or LMF, but may also be linked with differences in unit leaf rate (ULR; rate of increase in plant mass per unit leaf area per unit of time; Poorter and Remkes, 1990; Poorter and van der Werf, 1998; Garnier et al., 1999). ULR is a complex trait comprising carbon gain in photosynthesis and carbon loss in shoot and root respiration as well as root exudation.
Final seedling mass not only depends on RGR but also on initial seedling mass, which may be determined by seed mass. Although in cultivated and wild barley seed mass rather than RGR determined final seedling mass (López-Castaneda et al., 1996; Van Rijn et al., 2000; Richards and Lukacs, 2002), in wheat and related species of the Aegilops genus both RGR and seed mass determined the size of a seedling at any time after germination (Van den Boogaard et al., 1996; Villar et al., 1998).
The aim of this study was to elucidate the physiological and genetic relationships among the above-mentioned early vigor traits in A. tauschii. This was done by quantitative trait loci (QTL) analysis, a technique that requires the combined study of physiological characteristics and molecular genetics. The study was carried out on a population of recombinant inbred lines (RILs) derived from a cross between accessions that contrast both in early growth performance and at the molecular level. Important research questions were: (1) Which are the essential traits at the different organizational levels; (2) how are these levels connected; and (3) how and to what extent are the traits genetically linked? The ultimate goal was to provide markers closely linked to QTLs for growth traits in A. tauschii that might be helpful to improve early vigor in bread wheat.
RESULTS
Variation in Phenotypic Data
The examined traits are explained and the mean value for each parent is listed per trait in Table I. Supplemental Figure 1 shows the frequency distribution of variation in the growth traits among the RILs in the F4 population. Supplemental Figure 1A shows whole-plant traits, Figure 1B shows whole-shoot traits, Figure 1C shows whole-leaf traits, and Figure1D shows subleaf and other traits. Except for the relative tillering rate (RTR), all other traits showed significant line differences in ANOVA (P < 0.001). Furthermore, transgressive segregation occurred for almost all traits; the only exceptions were seed mass and RGRdry.
Table I.
Abbreviations and units of all measured growth traits, categorized per level of organization within the plant, and the mean values for the parents of the cross PI603228 and Ciae4
| Abbreviation | Trait | Unit | Mean PI603228 | Mean Ciae4 |
|---|---|---|---|---|
| Whole plant | ||||
| RGR1a | Relative growth rate (week 1) | g g−1 d−1 | 0.247 | 0.264 |
| RGR2a | Relative growth rate (week 2) | g g−1 d−1 | 0.215 | 0.233 |
| RGRtotala | Relative growth rate (week 1 + 2) | g g−1 d−1 | 0.231 | 0.249 |
| LAR | Leaf area ratio | m2 gplant−1 | 13.1 | 13.7 |
| ULR | Unit leaf rate | g m−2 d−1 | 16 | 17 |
| RGRdryb | Relative growth rate | mg g−1 d−1 | 94 | 123 |
| Plant mass | Final total plant mass (dry weight) | g | 0.340 | 0.280 |
| LMF | Leaf mass fraction | gleaf gplant−1 | 0.42 | 0.44 |
| SMF | Stem mass fraction | gstem gplant−1 | 0.22 | 0.19 |
| RMF | Root mass fraction | groot gplant−1 | 0.35 | 0.37 |
| Whole shoot | ||||
| No. leaves | No. of leaves | – | 15.3 | 8.8 |
| Areatotal | Total leaf area | cm2 | 46.5 | 38.0 |
| Leaf mass | Total leaf mass | g | 0.143 | 0.123 |
| Shoot mass | Total shoot mass (dry weight) | g | 0.221 | 0.176 |
| D%L | Dry-matter percentage leaf | % | 18.2 | 15.7 |
| D%S | Dry-matter percentage stem | % | 12.7 | 12.0 |
| SLA | Specific leaf area | m2 kgleaf−1 | 32.6 | 30.8 |
| RTR | Relative tillering rate | d−1 | 0.200 | 0.160 |
| RLaR | Relative leaf appearance rate | d−1 | 0.150 | 0.121 |
| Whole leaf | ||||
| LER | Leaf elongation rate | mm h−1 | 1.30 | 1.66 |
| LED | Leaf elongation duration | h | 132 | 133 |
| Length | Final leaf length | mm | 147 | 179 |
| Width | Maximal leaf width | mm | 4.5 | 4.6 |
| Arealeaf3 | Area of lamina of leaf 3 | mm2 | 546 | 647 |
| Sheath | Final sheath length | mm | 26.5 | 37.3 |
| Subleaf | ||||
| Cell | Maximal cell length | μm | 178 | 215 |
| No. cells | No. of cells | – | 830 | 838 |
| Cell production | Cell production rate | cells day−1 | 175 | 185 |
| Other | ||||
| Seed | Seed mass | mg | 24 | 9 |
| DMR | Dry-matter percentage root | % | 7.3 | 7.0 |
RGR calculated on fresh-weight basis.
RGR calculated on the basis of dry-weight measurement of the seed mass and the final plant mass.
Figure 1.
Path diagram for leaf length (A), RGR (B), and plant mass (C) and the underlying traits. Only significant (P < 0.05) path coefficients (single-headed arrows) and correlation coefficients (double-headed arrows) are shown as solid lines. Nonsignificant correlations are indicated by dashed lines.
Relationships among Growth Traits
Many of the early growth traits were significantly correlated (Table II). At the whole-plant level, RGR1, RGRtotal, and RGRdry were positively correlated with each other. These measures of RGR positively correlated with final plant mass, as well as with a range of shoot traits (e.g. number of leaves, areatotal, and leaf and shoot mass). The latter plant traits were not only dependent on RGR1 and RGRtotal, but also on seed mass. There was no correlation between RGR1 and RGR2. RGR2 clearly deviated from the other RGR measures in many respects. It showed no correlation with final leaf, shoot, and plant mass, but it correlated positively with ULR, SLA, and relative leaf appearance rate (RLaR) and with the allocation of biomass to the roots (root mass fraction [RMF]). RGR2 correlated negatively with LMF and maximal cell length. Length and width of leaf 3 were not correlated. Leaf length correlated positively with sheath length, which in turn correlated negatively with the RGR of number of leaves and tillers (RLaR and RTR). The dry-matter percentage in all plant organs (D%L, D%S, and dry-matter percentage root [DMR]) correlated negatively with different measures of RGR. In other words, a relatively high water content in the plant contributed to a high RGR on a fresh-weight basis. There was no relationship between dry-matter percentage and RGRdry.
Table II.
Correlations between early vigor traits in A. tauschii as calculated with the mean trait values of 134 F4 RILs derived from the cross between PI603228 and Ciae4
±, Positive response/negative correlation; α < 0.05.
| Trait
|
Whole Plant
|
Whole Shoot
|
Whole Leaf
|
Subleaf
|
Other
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RGR1 | RGR2 | RGRtotal | RGRdry | Plant Mass | LAR | ULR | LMF | SMF | RMF | No. Leaves | Areatotal | Leaf Mass | Shoot Mass | SLA | D%L | D%S | RLaR | RTR | LER | LED | Length | Width | Arealeaf3 | Sheath | Cell | No. Cells | Cell Production | Seed | DMR | |
| RGR1 | ||||||||||||||||||||||||||||||
| RGR2 | ||||||||||||||||||||||||||||||
| RGRtotal | + | + | ||||||||||||||||||||||||||||
| RGRdry | + | + | ||||||||||||||||||||||||||||
| Plant mass | + | + | + | |||||||||||||||||||||||||||
| LAR | − | |||||||||||||||||||||||||||||
| ULR | + | + | − | |||||||||||||||||||||||||||
| LMF | − | − | ||||||||||||||||||||||||||||
| SMF | ||||||||||||||||||||||||||||||
| RMF | + | + | − | + | − | − | ||||||||||||||||||||||||
| No. leaves | + | + | + | + | ||||||||||||||||||||||||||
| Areatotal | + | + | + | + | + | − | + | |||||||||||||||||||||||
| Leaf mass | + | + | + | + | + | − | + | + | ||||||||||||||||||||||
| Shoot mass | + | + | + | + | + | + | − | + | + | + | ||||||||||||||||||||
| SLA | + | − | + | − | − | + | + | − | − | |||||||||||||||||||||
| D%L | − | − | − | + | − | − | ||||||||||||||||||||||||
| D%S | − | + | − | − | ||||||||||||||||||||||||||
| RLaR | + | + | + | + | − | |||||||||||||||||||||||||
| RTR | + | + | + | + | + | − | ||||||||||||||||||||||||
| LER | + | + | − | + | + | + | − | |||||||||||||||||||||||
| LED | − | − | − | − | + | − | ||||||||||||||||||||||||
| Length | + | − | − | + | + | |||||||||||||||||||||||||
| Width | + | + | + | + | − | + | + | + | − | + | ||||||||||||||||||||
| Arealeaf3 | + | + | − | − | + | + | + | + | + | + | + | |||||||||||||||||||
| Sheath | − | − | − | − | + | − | − | + | + | + | + | |||||||||||||||||||
| Cell | + | − | + | + | + | + | + | |||||||||||||||||||||||
| No. cells | − | − | − | − | − | − | − | + | + | + | + | + | − | |||||||||||||||||
| Cell production | − | + | + | + | + | − | + | |||||||||||||||||||||||
| Seed | − | − | + | + | + | + | + | |||||||||||||||||||||||
| DMR | − | − | − | + | − | + | + | |||||||||||||||||||||||
The relations among growth traits were further analyzed by path analysis. The purpose of path analysis is the quantification of the relative contributions of correlated causal sources of variance once a certain network of interrelated variables has been accepted (Lynch and Walsh, 1998). The starting points for the analysis were thus the known relationships (from literature) between growth traits and the observed correlations in this dataset. Path analysis was performed for leaf length (Fig. 1A), RGR2 (Fig. 1B), and final plant mass (Fig. 1C). In Figure 1, only significant (P < 0.05) path and correlation coefficients are shown. A path coefficient of 0.265 between LER and leaf length (Fig. 1A) means than an increase of LER of 1 sd unit results in an increase of 0.265 sd units in leaf length.
Leaf Length
The leaf length of an individual leaf was determined by the LER and LED, which were not correlated, and the underlying traits of cell length and number of cells (Fig. 1A). Although there was no overall significant correlation between cell length and leaf length (Table II), the path coefficient from cell length toward leaf length was significant. The explanation for this seemingly contrasting observation is the strong negative correlation between cell length and number of cells (−0.710; P < 0.01), suggesting a trade-off between cell number and cell length. Apparently, a leaf either has a large number of small cells or has a small number of large cells. The number of cells, in turn, is determined by the cell production rate and the time period during which cell production occurs, which is reflected in the LED. Since there was no negative correlation between LER and LED, or between cell production rate and LED (Table II), a fast cell production rate contributed to a large number of cells and a fast LER and in that way contributed to a long leaf.
RGR
The path analysis of RGR and its underlying traits was based on the trait measurements in the second week and at the final harvest. The analysis covered three levels of organization in the plant: from leaf (Fig. 1B, bottom) via shoot (Fig. 1B, top left) to whole plant (Fig. 1B, top right). Variation in RGR2 is determined mainly by SLA and ULR and, to a lesser extent, by LMF at the whole-plant level. These traits were negatively correlated with each other, and this explains the absence of significant correlations between traits at the shoot level and RGR2 (Table II). SLA was the main trait connecting shoot traits with whole-plant RGR and was determined by total leaf mass and total leaf area. The positive correlation between leaf number and total leaf mass and/or area was stronger than the correlation between individual leaf traits and total leaf mass and/or area. Apparently, a large individual leaf area only moderately contributed to a large total leaf area. The explanation for this observation is the strong negative path from leaf length toward number of leaves, indicating that long leaves with a relatively large individual leaf area coincide with a small number of leaves. Taken together, the path diagrams of leaf length (Fig. 1A) and RGR (Fig. 1B), and the observed correlations between the traits (Table II), indicate a trade-off between long leaves achieved by a high LER, a rapid cell production rate, a large cell number, and a short cell length, on the one hand, and a large number of shorter leaves, on the other. We also observed a negative path from individual leaf width toward ULR. This further illustrates the compensating mechanisms at higher organizational levels within the plant, diminishing the impact of individual leaf traits on whole-plant RGR.
Plant Mass
Figure 1C shows how seed and plant mass are related with growth rates of biomass (RGR1 and RGR2) and of number of leaves and tillers (RLaR and RTR) during the whole experimental period. It illustrates that plant mass at the end of the 2-week growth period did not correlate with the RGR in the last week of growth (RGR2), but depended on seed mass, RGR1, and RTR, traits that were not correlated with each other (Table II). This path diagram reveals that the key trait determining plant mass is shoot mass, which is determined by seed mass, RGR1, and RTR. RLaR correlated positively with RGR1 and RGR2, but did not correlate with shoot mass and final plant mass.
Genetic Map and QTL Analysis
The genotyping of 180 F3 and 134 F4 lines resulted in 273 markers, mostly amplified fragment-length polymorphisms (AFLPs) combined with simple sequence repeats (SSRs) as anchor markers. The linkage map consisted of 10 linkage groups, in which 223 markers were mapped (15 SSRs and 208 AFLPs) and covered in total 865 cM. Six groups with a length ranging from 74 to 174 cM consisted of 30 to 40 markers. The remaining four groups were smaller, covered a length of 24 to 46 cM, and consisted of three to seven markers. All linkage groups contained at least one SSR, so that all groups could be assigned to the D genome of T. aestivum. Based on the position of the SSRs and the clear clustering of AFLP markers in the centromeric regions, the orientation of the six larger linkage groups was determined (Röder et al., 1998). This was not possible for the four smaller groups; these could only be assigned to either the long or the short arms of the chromosomes. Only the total length of chromosome 4D was smaller than 100 cM, and the two linkage groups did not enclose the centromeric region but represented two parts of the long arm of chromosome 4D.
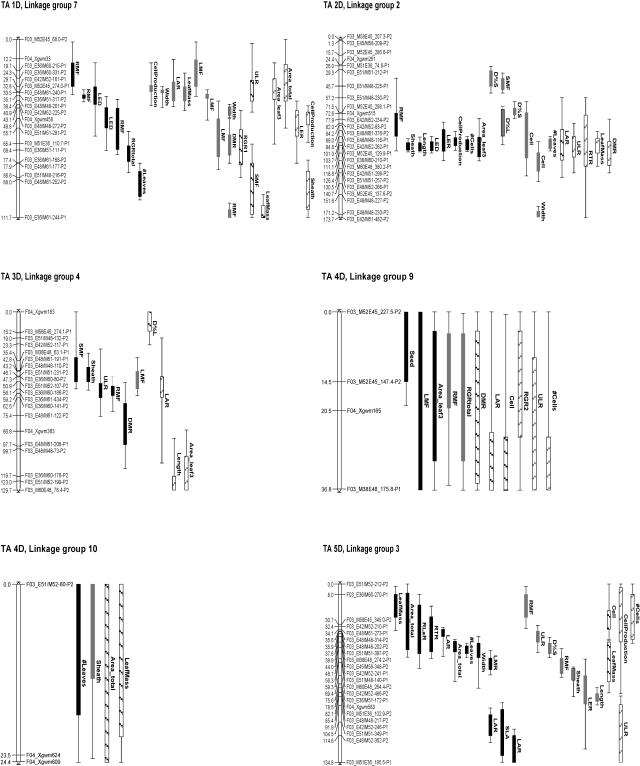
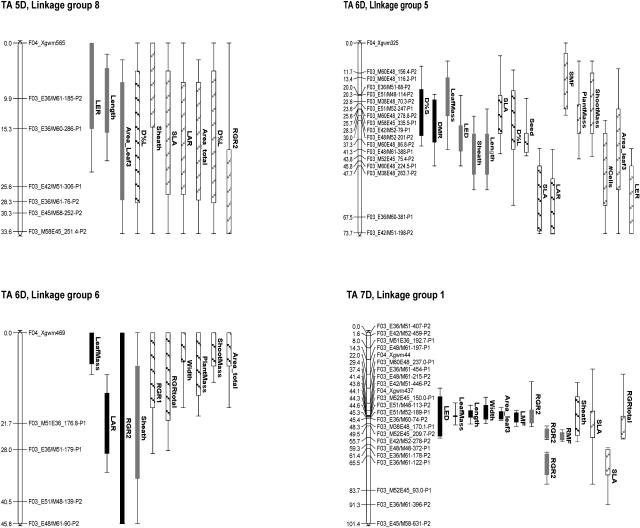
From this basic map, a core map was constructed for QTL analysis, as presented in Figure 2. QTL analysis resulted in 87 significant QTLs (log of the odds [LOD] > 2.65) and 63 putative QTLs (1.5 < LOD < 2.65; Fig. 2). Significant QTLs were found in every linkage group. On average, there were 3.1 significant QTLs per trait, together explaining 32% of the phenotypic variance per trait, i.e. almost 10% per QTL (Table III). There were large differences between traits: The explained percentage of phenotypic variance ranged from 8% for SLA (one QTL) to 82% for RMF (10 QTLs). There were some differences among the organizational levels: At the whole-plant level and at the leaf level, the average number of QTLs and the percent explained per trait were higher than at the other levels (5% and 40%, respectively, versus 2% and less than 30%, respectively). The lowest number of QTLs per trait was found at the subleaf level (2.0); however, these QTLs each explained a higher (15% versus 9%–10%) percentage of the phenotypic variance, resulting in a similar value of the percent explained phenotypic variance of the traits to that at the other organizational levels. Overall, the phenotypic variance explained by one QTL ranged from 5% for a QTL for RMF on chromosome 2D to 28% for a QTL for number of cells on chromosome 2D. For almost all traits, one or more significant QTLs were found; the only exceptions were plant mass and shoot mass, for which only two putative QTLs were detected on chromosome 6D (Table III). With the path analyses as a starting point, the relationship between the QTLs for the different growth traits was considered. For this purpose, Table IV presents an overview of the relevant QTLs per linkage group, arranged in order along the lines of the path diagrams presented in Figure 1, A to C.
Figure 2.
AFLP linkage map of A. tauschii chromosomes. Positions of QTLs above the LOD threshold of 2.65 are represented by solid bars, and QTLs with 1.5 < LOD < 2.65 by hatched bars. Black and gray bars represent QTLs with positive and negative additive effect, respectively. Bars and whiskers indicate QTL locations with a 1- and 2-LOD support interval, respectively. TA + number refers to the chromosome number in the D genome of T. aestivum. Linkage map drawing was made using MapChart (Voorrips, 2002).
Table III.
QTLs for early growth traits determined in the F4 RIL population of A. tauschii, derived from the cross between accession PI603228 and Ciae4
P1, Marker present in PI603228; P2, marker present in Ciae4; numbers within brackets, total number of significant QTLs for that trait; chromosome number, D genome of T. aestivum.
| Trait
|
Chromosome
|
Position
|
Peak LOD
|
Variance Explained
|
Additive Effecta
|
Cofactor Marker
|
||
|---|---|---|---|---|---|---|---|---|
| Per QTL | Average per Trait | Total per Trait | ||||||
| cM | % | % | ||||||
| Plant | ||||||||
| RGR2 (3) | 7D | 44.3 | 2.66 | 7 | 8 | 24 | −1.7 | E45M52-150-P1 |
| 7D | 55.7 | 3.07 | 8 | −1.9 | E42/M52-278-P2 | |||
| 7D | 65.5 | 3.59 | 9 | −2.1 | E36/M61-122-P1 | |||
| RGRtotal (2) | 1D | 42.2 | 3.42 | 10 | 10 | 20 | 1.6 | E36/M51-111-P1 |
| 4D | 19.5 | 3.57 | 10 | −1.6 | Xgwm165 | |||
| LAR (5) | 1D | 78.2 | 3.91 | 8 | 11 | 53 | −3.4 | E48/M61-240-P1 |
| 5D | 10 | 4.24 | 14 | 4.6 | E36/M51-196-P1 | |||
| 5D | 30.2 | 3.63 | 9 | 3.6 | E51/M51-349-P1 | |||
| 5D | 97.1 | 5.03 | 12 | 4.2 | E51/M51-397-P2 | |||
| 6D | 21.7 | 4.65 | 10 | 3.7 | E36/M51-177-P1 | |||
| ULR (2) | 3D | 56.1 | 3.25 | 8 | 9 | 18 | 4.1 | E36/M60-186-P2 |
| 5D | 97.1 | 4.08 | 11 | −4.6 | E51/M51-397-P2 | |||
| LMF (7) | 1D | 51.2 | 3.11 | 9 | 8 | 55 | −2.1 | E51/M51-291-P2 |
| 1D | 76.5 | 4.04 | 9 | −2.1 | E36/M51-317-P2 | |||
| 1D | 87.4 | 2.99 | 7 | −1.8 | E36/M60-331-P2 | |||
| 3D | 50.9 | 3.36 | 7 | −1.8 | E51/M52-107-P2 | |||
| 4D | 19.5 | 2.68 | 6 | 1.8 | Xgwm165 | |||
| 5D | 75.5 | 3.70 | 8 | 2.0 | E48M60-265-P2 | |||
| 7D | 45.4 | 4.43 | 9 | 2.1 | E36/M60-74-P2 | |||
| SMF (2) | 2D | 44.3 | 6.00 | 18 | 13 | 25 | 4.0 | E51/M48-225-P1 |
| 3D | 46.7 | 3.01 | 8 | 2.5 | E51/M51-231-P2 | |||
| RMF (10) | 1D | 0 | 3.49 | 7 | 8 | 82 | −2.3 | E36/M61-244-P1 |
| 1D | 61 | 3.85 | 8 | 2.7 | E48/M48-272-P2 | |||
| 1D | 76.5 | 6.12 | 12 | 3.1 | E36/M51-317-P2 | |||
| 1D | 87.4 | 3.72 | 8 | 2.5 | E36/M60-331-P2 | |||
| 2D | 77.9 | 2.94 | 5 | −1.9 | E42/M52-234-P2 | |||
| 3D | 59.2 | 2.92 | 5 | 2.0 | E36/M61-434-P2 | |||
| 4D | 14.5 | 4.91 | 8 | −2.9 | E45/M52-147-P2 | |||
| 5D | 75.5 | 4.93 | 9 | −2.7 | E48/M60-265-P2 | |||
| 5D | 119.1 | 6.15 | 14 | −3.5 | E36/M60-270-P1 | |||
| 7D | 55.7 | 4.52 | 7 | −2.4 | E42/M52-278-P2 | |||
| Mean (plant): | 9 | 40 | ||||||
| Shoot | ||||||||
| No. leaves (4) | 1D | 25 | 4.17 | 9 | 11 | 43 | 5.5 | E51/M48-216-P2 |
| 2D | 101 | 4.87 | 11 | −6.4 | E45/M52-130-P1 | |||
| 4D | 10 | 3.67 | 11 | 6.6 | E51/M52-80-P2 | |||
| 5D | 86.7 | 5.89 | 13 | 6.8 | E42/M52-241-P1 | |||
| Areatotal (2) | 5D | 86.7 | 3.93 | 10 | 13 | 26 | 6.2 | E42/M52-241-P1 |
| 5D | 114.1 | 4.56 | 16 | 8.5 | E45/M58-345-P2 | |||
| Leaf mass (5) | 1D | 76.5 | 2.77 | 6 | 7 | 36 | −5.1 | E36/M51-317-P2 |
| 5D | 119.1 | 3.72 | 11 | 6.8 | E36/M60-270-P1 | |||
| 6D | 0 | 2.97 | 6 | 5.9 | Xgwm469 | |||
| 6D | 25.7 | 2.69 | 5 | −4.5 | E45/M58-336-P1 | |||
| 7D | 44.1 | 3.81 | 8 | 5.1 | Xgwm437 | |||
| DML (1) | 2D | 72.6 | 3.93 | 11 | 11 | 11 | 2.2 | Xgwm515 |
| DMS (4) | 2D | 39.3 | 3.91 | 12 | 12 | 46 | 3.0 | E51/M48-225-P1 |
| 2D | 72.6 | 4.18 | 10 | 2.3 | Xgwm515 | |||
| 5D | 86.7 | 5.5 | 13 | −2.8 | E42/M52-241-P1 | |||
| 6D | 23.6 | 4.82 | 11 | 2.5 | E51/M52-247-P1 | |||
| SLA (1) | 5D | 20.1 | 2.92 | 8 | 8 | 8 | 3.4 | E48/M52-362-P2 |
| RTR (1) | 5D | 98.9 | 2.98 | 9 | 9 | 9 | 9.5 | E48/M48-202-P2 |
| RlaR (1) | 5D | 99.1 | 2.78 | 9 | 9 | 9 | 3.8 | E48/M48-314-P2 |
| Mean (shoot): | 10 | 24 | ||||||
| Leaf | ||||||||
| LER (3) | 2D | 101 | 3.07 | 8 | 10 | 31 | 2.9 | E45/M52-130-P1 |
| 5D | 5 | 5.59 | 15 | −3.8 | Xgwm565 | |||
| 5D | 52.7 | 3.32 | 9 | −2.9 | E36/M51-103-P2 | |||
| LED (5) | 1D | 61.9 | 2.76 | 6 | 9 | 42 | 2.4 | E48/M48-272-P2 |
| 1D | 78.2 | 3.3 | 7 | 2.5 | E48/M61-240-P1 | |||
| 2D | 108.7 | 6.01 | 15 | 3.7 | E36/M60-210-P1 | |||
| 6D | 37.3 | 3.53 | 8 | −2.5 | E48/M60-87-P2 | |||
| 7D | 43.8 | 2.9 | 6 | 2.2 | E42/M51-446-P2 | |||
| Length (5) | 2D | 103.7 | 8.45 | 15 | 10 | 50 | 4.8 | E36/M60-210-P1 |
| 5D | 9.9 | 5.89 | 10 | −3.8 | E36/M61-185-P2 | |||
| 5D | 49.3 | 4.45 | 7 | −3.3 | E48/M48-217-P2 | |||
| 6D | 45.8 | 4.44 | 8 | −3.3 | E48/M60-225-P1 | |||
| 7D | 44.3 | 6.13 | 10 | 3.7 | E45/M52-150-P1 | |||
| Width (6) | 1D | 66.9 | 4.25 | 11 | 10 | 61 | −3.4 | Xgwm458 |
| 1D | 78.9 | 6.13 | 14 | −3.8 | E45/M52-274-P1 | |||
| 2D | 173.7 | 3.25 | 7 | −2.6 | E42/M51-482-P2 | |||
| 4D | 25.5 | 2.91 | 8 | 3.1 | Xgwm165 | |||
| 5D | 83.5 | 5.69 | 13 | 3.9 | E42/M52-241-P1 | |||
| 7D | 45.3 | 4.48 | 10 | 3.2 | E51/M52-189-P1 | |||
| Arealeaf3 (3) | 2D | 108.7 | 3.51 | 8 | 7 | 20 | 4.9 | E36/M60-210-P1 |
| 4D | 19.5 | 2.66 | 5 | 4.0 | Xgwm165 | |||
| 5D | 20.3 | 3.59 | 7 | −4.6 | E42/M51-306-P1 | |||
| Sheath (5) | 2D | 103.7 | 7.63 | 12 | 8 | 38 | 6.6 | E36/M60-210-P1 |
| 3D | 47.3 | 3.64 | 5 | 4.3 | E36/M60-80-P2 | |||
| 5D | 65.4 | 6.57 | 10 | −5.8 | E42/M52-486-P2 | |||
| 6D | 41.3 | 3.93 | 6 | −4.4 | E48/M61-388-P1 | |||
| 4D | 0 | 3.58 | 5 | −4.3 | E51/M52-80-P2 | |||
| Mean (leaf): | 9 | 41 | ||||||
| Subleaf | ||||||||
| Cell (3) | 2D | 84.2 | 3.35 | 11 | 11 | 33 | −3.8 | E48/M61-376-P2 |
| 2D | 99.8 | 3.43 | 12 | −4.1 | E45/M52-130-P1 | |||
| 2D | 123.6 | 2.94 | 9 | −3.6 | E51/M51-257-P2 | |||
| No. cells (1) | 2D | 103.7 | 10.94 | 28 | 28 | 28 | 9.2 | E36/M60-210-P1 |
| Cell production (2) | 1D | 81.9 | 3.29 | 9 | 14 | 28 | −4.4 | E42/M52-161-P1 |
| 2D | 101 | 7.09 | 20 | 6.7 | E45/M52-130-P1 | |||
| Mean (subleaf): | 15 | 30 | ||||||
| Other | ||||||||
| Seed (1) | 4D | 5 | 2.94 | 9 | 9 | 9 | 6.2 | E45/M52-228-P2 |
| DMR (3) | 1D | 46.2 | 3.49 | 10 | 10 | 30 | −2.7 | E36/M51-111-P1 |
| 3D | 80.4 | 3.38 | 10 | 2.7 | E48/M61-122-P2 | |||
| 6D | 28.3 | 3.78 | 10 | 2.5 | E42/M52-79-P1 | |||
| Mean (other): | 10 | 20 | ||||||
| Mean (overall): | 10 | 32 | ||||||
Additive effect in percentage is calculated as follows: [(mean of the PI603228 genotype − mean of the Ciae4 genotype)/2]×100/mean of the heterozygous genotype.
Table IV.
Overview of peak LOD positions of QTLs (LOD >2.65) for various early vigor traits in the F4 RIL population of A. tauschii, derived from the cross between accession PI603228 and Ciae4
LK, Linkage group; TA + number, chromosome number in T. aestivum; positions in italic, positions of putative QTLs (1.5 < LOD < 2.65); ± sign in brackets, positive response/negative additive effect of the QTL; text in bold, the target traits of the path diagrams of Figure 1, A to C.
| Trait
|
LK7 | LK2 | LK4 | LK9 | LK10 | LK3 | LK8 | LK5 | LK6 | LK1 |
|---|---|---|---|---|---|---|---|---|---|---|
| TA 1D | TA 2D | TA 3D | TA 4D | TA 4D | TA 5D | TA 5D | TA 6D | TA 6D | TA 7D | |
| Length | 103.7 (+) | 129.7 (−) | 49.3 (−) | 9.9 (−) | 45.8 (−) | 44.3 (+) | ||||
| LER | 61.9 (−) | 101.0 (+) | 52.7 (−) | 5.0 (−) | 62.7 (−) | |||||
| LED | 61.9 (+) | 108.7 (+) | 37.3 (−) | 43.8 (+) | ||||||
| 78.2 (+) | ||||||||||
| No. cells | 103.7 (+) | 36.8 (−) | 104.1 (−) | 41.3 (−) | ||||||
| Cell | 84.2 (−) | 36.8 (+) | 104.1 (+) | |||||||
| 99.8 (−) | ||||||||||
| 123.6 (−) | ||||||||||
| Cell production | 61.9 (−) | 101.0 (+) | 114.1 (−) | |||||||
| 81.9 (−) | ||||||||||
| RGR2 | 14.5 (−) | 25.6 (−) | 45.8 (+) | 44.3 (−) | ||||||
| RGRtotal | 42.2 (+) | 19.5 (−) | 0 (+) | 55.7 (−) | ||||||
| 55.7 (−) | ||||||||||
| 65.2 (−) | ||||||||||
| RGR1 | 42.2 (+) | 5 (+) | ||||||||
| LMF | 51.2 (−) | 50.9 (−) | 19.5 (+) | 75.5 (+) | 45.4 (+) | |||||
| 56.0 (−) | ||||||||||
| 76.5 (−) | ||||||||||
| 87.4 (−) | ||||||||||
| ULR | 81.9 (+) | 103.7 (−) | 56.1 (+) | 36.8 (−) | 30.2 (−) | |||||
| 97.1 (−) | ||||||||||
| SLA | 20.1 (+) | 25.6 (+) | 44.1 (−) | |||||||
| 62.7 (+) | 49.5 (−) | |||||||||
| 65.5 (−) | ||||||||||
| LAR | 78.2 (−) | 103.7 (+) | 56.1 (−) | 36.8 (+) | 10.0 (+) | 15.3 (−) | 67.5 (+) | 21.7 (+) | ||
| 30.2 (+) | ||||||||||
| 97.1 (+) | ||||||||||
| Leaf mass | 0 (+) | 103.7 (−) | 0 (+) | 86.7 (+) | 25.7 (−) | 0 (+) | 44.1 (+) | |||
| 76.5 (−) | 119.1 (+) | |||||||||
| Areatotal | 78.2 (−) | 15 (+) | 86.7 (+) | 25.6 (−) | 0 (+) | |||||
| 114.1 (+) | ||||||||||
| No. leaves | 25.0 (+) | 101.0 (−) | 10.0 (+) | 86.7 (+) | ||||||
| Arealeaf3 | 78.9 (−) | 108.7 (+) | 129.7 (−) | 19.5 (+) | 20.3 (−) | 45.8 (−) | ||||
| Width | 66.9 (−) | 173.7 (−) | 25.5 (+) | 83.5 (+) | 0 (+) | 45.3 (+) | ||||
| 78.9 (−) | ||||||||||
| Plant mass | 25.7 (−) | 0 (+) | ||||||||
| Shoot mass | 25.7 (−) | 0 (+) | ||||||||
| Seed | 5.0 (+) | 41.3 (+) | ||||||||
| RLaR | 99.1 (+) | |||||||||
| RTR | 101.0 (−) | 98.9 (+) |
QTLs for Leaf Length
Leaf length is determined by the underlying traits LER, LED, cell length, number of cells, and cell production rate (Fig. 1A). Significant QTLs for leaf length always coincided with at least one QTL for one of the underlying traits (Table IV) and were detected on chromosomes 2D, 5D, 6D, and 7D. On chromosome 5D, QTLs for leaf length colocated with significant QTLs for LER. On chromosomes 6D and 7D, QTLs for leaf length colocated with significant QTLs for LED. On chromosome 2D, the QTL for leaf length colocated with QTLs for all underlying traits, and it explained a larger part of the phenotypic variance (15% versus <10%) of leaf length than the QTLs on the other chromosomes. Some of the QTLs of the different traits had the peak LOD score at the same position (e.g. LER and cell production rate on chromosome 2D). Others were close to each other (e.g. length and LED on chromosome 7D). Overlapping QTLs for LER and LED were observed on chromosome 2D and also on chromosomes 1D and 6D. The latter chromosomes both harbored a putative QTL for LER and a significant QTL for LED. In most cases, the additive effect of the QTLs for LER and LED had the same direction. The only exception was chromosome 1D, where the putative QTL for LER had a negative additive effect and the overlapping QTLs for LED had a positive effect. This opposite effect might explain why no QTL for leaf length was identified on chromosome 1D.
QTLs for RGR
Only a few significant QTLs for RGRtotal and RGR2 were detected and none for RGR1. The significant QTLs for RGRtotal were at the same position as the putative QTLs for RGR1 on chromosome 1D, and for RGR2 on chromosome 4D. Furthermore, a significant QTL for RGR2 on chromosome 7D colocated with a putative QTL for RGRtotal. These data show that RGRtotal was mainly determined by either RGR1 or RGR2. There were no overlapping putative QTLs for RGR1 and RGR2, confirming the absence of correlation between RGR1 and RGR2 observed earlier (Table II; Fig. 1C).
On chromosome 7D, significant QTLs for RGR2 were detected on positions that overlapped with significant and putative QTLs of the underlying traits LMF, SLA, leaf mass, leaf length, and LED. The additive effect of the QTLs for SLA was negative, just like the additive effect of the QTLs for RGR2, supporting the observed correlation between SLA and RGR2. Surprisingly, we did not identify QTLs for ULR in regions where QTLs for RGR were detected. Rather, QTLs for ULR were detected at positions where QTLs with opposite additive effects for LAR were located, and in those regions no QTLs for RGR were found. In some cases, there was overlap between QTLs for ULR and LMF (chromosomes 3D, 5D, and 7D), pointing at the importance of biomass allocation to leaves for the ULR. In general, this dataset showed a negative correlation between LMF and RGR, as well as between LMF and ULR, that is confirmed by the opposite additive effects of the overlapping QTLs for these traits. QTLs for LMF partly covered QTLs for leaf mass and/or areatotal on chromosomes 1D, 5D, and 7D. On each of these chromosomes, the colocating QTLs for LMF, leaf mass, and areatotal had additive effects with the same sign, illustrating the positive correlation between these leaf and shoot traits (Table II). The leaf and shoot QTLs on chromosome 7D have a positive additive effect and colocate with a negative QTL for RGR, demonstrating the negative correlation between RGR and biomass allocation to the leaves in this RIL population.
QTLs for Plant Mass
There were no significant QTLs detected for plant and shoot mass. Only in the two linkage groups that form a part of chromosome 6D were a few overlapping putative QTLs identified: in linkage group 5 with negative additive effect and in linkage group 6 with positive additive effect. Putative QTLs for plant and shoot mass, on the one hand, and RGR1, on the other, overlapped on chromosome 6D, which illustrates the importance of RGR1 for final plant mass. The other QTLs for RGR did not colocate with the QTLs for plant mass. For seed mass, one significant QTL was detected on chromosome 4D in the same region of QTLs for RGR, but with opposite additive effect, confirming the negative correlation between seed mass and RGR2 (Table II; Fig. 1C). A putative QTL for seed mass was detected on chromosome 6D, which did not overlap with QTLs for plant and shoot mass.
Significant QTLs for RLaR and RTR were detected on chromosome 5D in a region with QTLs for a range of shoot and whole-plant traits. This chromosome obviously harbored QTLs for many growth traits, covering all organizational levels, but no QTLs for plant mass or RGR were identified on chromosome 5D. Apparently, the effects of QTLs for the underlying growth traits counterbalanced each other so that no QTL for plant mass or RGR could be detected.
DISCUSSION
Clusters of QTLs: Genetic Links between Early Vigor Traits
This study provides insight into the genetic and physiological relationships among a range of growth traits at different organizational levels that contribute to early vigor of A. tauschii seedlings. Although many QTLs were identified for most of the early vigor traits, we did not detect significant QTLs for plant mass and shoot mass, and only a few for the different RGR measures. These characters are very complex composite physiological traits that presumably are under control of many loci on the genome. QTLs with small effects on the overall complex trait are difficult to detect so that, for such traits, usually only a few major QTLs are found (Kearsy and Farquhar, 1998). In this study, we found on average 3.1 QTLs per trait that explained together, on average, 32% (range 8%–82%) of the phenotypic variance of the characters. These results are similar to results obtained in wild barley (Van Rijn, 2001; Elberse, 2002). Considering the complexity of the growth traits studied, we regarded it as meaningful to present the putative QTLs with LOD scores between 1.5 and 2.65 in Figure 2. Further research is required to elucidate whether these putative QTLs represent chromosomal regions that are important for the traits under consideration or not. On all seven chromosomes, QTLs for early vigor traits were detected, and, in some cases, there was a tendency for clustering of the QTLs for related traits. For example, on chromosome 2D, there was a cluster of QTLs for leaf and subleaf traits that colocated with QTLs with opposite additive effect at the shoot level. Interpretation of these clusters is difficult in the sense that it is presently impossible to determine whether one gene affects a range of traits pleiotropically or whether there are several genes clustered in the same region that act upon different related traits. Clustering of QTLs for related characters has been observed in other studies (e.g. in rice, maize [Zea mays], and wheat; Khavin and Coe, 1997; Cai and Morishima, 2002; Sourdille et al., 2003), and it was suggested that these QTL clusters represent gene clusters that are separated by regions with noncoding sequences. Within such gene islands of chromosome 1DS of wheat and A. tauschii, the gene density was similar to that of Arabidopsis (Arabidopsis thaliana; Brooks et al., 2002). The gene clusters were considered as functional units of networks of genes that are expressed in concert and in that way contribute to the regulation of plant development and responses to the environmental conditions. Considering the nature of the studied traits in this work, we hypothesize that the QTL clusters might well represent such functional units.
An alternative explanation for clustering of QTLs for traits at different organizational levels in the plant is that of a mechanistic dependency rather than a genetic dependency between the traits, as visualized in the path diagrams (Fig. 1, A–C). For example, the colocation of QTLs for leaf length with QTLs for the underlying traits LER and/or LED and/or cell production rate, might be due to the fact that one or more genes in that region affect underlying traits and, consequently, this chromosomal region pleiotropically also affects the resulting leaf length. Likewise, the colocation on several chromosomes of putative and significant QTLs for LER, on the one hand, and number of cells and cell production rate, on the other, also suggests that QTLs for the latter traits substantially affect LER. It might indicate that the colocating QTLs in these cases do not represent different QTLs, but that a QTL affecting an important underlying trait in so doing also affects the higher level trait. Likewise, on chromosome 7D, a cluster was apparent of QTLs for leaf and shoot traits, which were recognized as important underlying traits for RGR2 (Fig. 1B) and which indeed colocated with a QTL for RGR2.
Leaf Area, Biomass Allocation, and RGR
Rapid development of total leaf area and leaf mass is important for the growth performance of the seedlings. The expansion of the total leaf area depends on the growth of the individual leaves as well as on the growth of the number of leaves. These characters are not independent of each other. A rapid leaf expansion of individual leaves is the result of a high LER and is accompanied by long leaves (Table II). Differences in cell production rate account for variation in LER (Fiorani et al., 2000; Bultynck et al., 2003). These differences were related to a difference in the number of dividing cells and not to a different rate of cell division. The faster cell production rate presumably increased the number of cells that were elongating at the same time and in that way stimulated LER. It was accompanied by a larger meristem that resulted in a larger number of cells per cell file. At the same time, it coincided with more cell files in parallel so that the leaf width was also larger in longer leaves, suggesting differences in the size of the leaf meristem in more dimensions. In this study, QTLs for LER and cell production rate colocated on chromosomes 1D and 2D, confirming the importance of meristematic activity for LER. However, a large number of cells in the leaves correlated negatively with the number of leaves and total leaf area and mass. This indicates a trade-off between meristematic activity within a leaf, determining the final leaf length of individual leaves, and meristematic activity associated with leaf initiation. Consequently, an increment of the total leaf area expansion rate cannot be achieved by focusing only on an increase of the LER of individual leaves despite its positive correlation with RGR and plant, leaf, and shoot mass. We observed no correlation between leaf length and width (Table II), and very little overlap in QTLs for leaf length and leaf width (Table IV). Accordingly, this dataset does not provide evidence for a strong genetic link between leaf length and width via the size of the meristem. This opens perspectives for increments of leaf area by further enlarging leaf width, thereby perhaps bypassing the counteractive response of the number of leaves on longer leaves.
Although LAR, and more specifically SLA, has been recognized as the main factor explaining variation in RGR in many herbaceous species (Poorter and van der Werf, 1998), in a comparison of 20 Aegilops species ULR turned out to be the main factor (Villar et al., 1998). In this study, ULR and SLA were both positively correlated with RGR, but the correlation between ULR and RGR was stronger (0.41 versus 0.19 for SLA). This was caused by the negative correlation between SLA and LMF that reduces the positive impact of a high SLA on RGR (Table II; Fig. 1B). Apparently, a lower biomass allocation to the leaves coincides with a relatively larger leaf area per unit of leaf mass and vice versa. The QTL analysis revealed some evidence for a genetic basis for the observed interdependency between SLA and LMF: On chromosome 7D, a QTL for LMF mapped in the same region as a putative QTL for SLA (position 44 cM) with opposite additive effect.
Although we detected only a few QTLs for RGR, in general, the QTL analysis showed that many relations between the different growth traits might have a genetic basis. Yet, the nature of this genetic basis cannot be resolved and needs further research.
Seed Mass, Plant Mass, and Early Vigor
Final plant and shoot mass was primarily influenced by RGR1, seed mass, and RTR. Because we found very few QTLs for these highly complex traits, the QTL analysis does not allow for conclusions on the genetic basis for observed correlations. The absence of overlapping QTLs for these traits is probably more due to the fact that only a few QTLs were detected than that it is a representation of absence of genetic relations between the traits. The QTL analysis does confirm, however, the observed negative correlation between seed mass and RGR2 (Table II): On chromosome 4D, overlapping QTLs with contrasting additive effects for these traits were found (Fig. 2; Table IV).
In accordance with studies with wheat and several species of the Aegilops genus (Van den Boogaard et al., 1996; Villar et al., 1998), in this study both RGR in the first period after germination (RGR1) and seed mass positively influenced the size of a seedling at the final harvest. Postgerminative growth is essential for the seedling's establishment. It ensures efficient colonization of the soil and results in a rapid development to autotrophy. Seed size often correlates negatively with RGR (Van Rijn et al., 2000, and refs. therein), but there are also examples showing the opposite (López-Castaneda et al., 1996; Treymayne and Richards, 2000; Van Rijn et al., 2000), or without correlation between seed mass and RGR (Clevering, 1999). We observed no correlation between seed mass and RGR1 and a negative correlation between seed mass and RGR2 (Table II). RGR2 did not correlate with plant mass after 14 d of growth (Table II; Fig. 1C), but it is feasible that RGR becomes important for seedling vigor in a later stage of plant development.
After the first period of growth, RTR became important. Tillers only started to emerge by the end of the first week of the experimental period (data not shown), and RTR and RGR1 were not correlated. A high RTR contributes to a large number of leaves, and thus to a high total leaf area and high shoot mass (Table II; Fig. 1C). A high RTR probably contributes to a slower decline of RGR of the plants after the initial establishment of the plant, as was also observed in Triticum seedlings (Bultynck et al., 2004). In that way, RTR can positively contribute to early vigor of the seedlings.
Thus, improvement of seedling vigor in wheat requires stimulation of postgerminative growth. For this purpose, smart combinations of QTLs with positive effects on leaf and shoot growth traits might stimulate the rapid development of the seedling. For example, E42/M52-241-P1 on 5D (width and areatotal) might be interesting in combination with E36/M60-186-P2 on 3D (ULR), but other combinations are also worthwhile to consider, depending on the objective of the breeder. Furthermore, QTLs on chromosome 6D might be interesting (e.g. those connected to marker Xgwm469 [leaf, shoot, and plant mass leaf area], E48/M60-225-P1 [leaf length], and/or E48/M60-87-P2 [LED]). In addition, in wheat it might be worthwhile to focus on seed mass to further strengthen the seedling's ability to rapidly develop shoot and leaf biomass and leaf area immediately after germination. Whatever the goal, the fact that, in most cases, genetic linkages between traits do not seem to be absolute opens perspectives for new combinations of beneficial leaf and shoot growth traits.
In this respect, it is important to note that almost all traits exhibited transgressive segregation (see Supplemental Fig. 1), indicating that both parents harbored positive and negative alleles. This is confirmed by the fact that the estimated QTL effects were both positive and negative. Accordingly, when introgression of alleles from A. tauschii into bread wheat is undertaken, one has to determine which parent carries the most favorable ones.
Relationship with the D Genome of Wheat and Other Grass Genomes
The parents of the cross, PI603228 and Ciae4, are representatives of two contrasting groups of accessions within the species A. tauschii. These groups were recognized in a screening of the variation in early vigor and AFLP fingerprints among 46 accessions that originated from the whole area of natural distribution of the species (data not shown). AFLP fingerprints of the two groups were strikingly different, and this was accompanied by differences in growth performance. To assign the 10 linkage groups of the D genome of A. tauschii to chromosomes of the D genome of T. aestivum, we used SSRs following the protocol described by Röder et al. (1998). Fifteen out of 16 SSRs showed polymorphism between the parents of the cross. These observations illustrate two important points. First, the fact that almost all SSRs exhibited polymorphisms between the parents strengthens the outcome of the AFLP fingerprints, showing substantial differences in genetic makeup of the two groups. Second, in accordance with other studies with microsatellites, it reveals the existence of a high level of correspondence between the D genome of T. aestivum and the genome of A. tauschii (Lelley et al., 2000; Guyomarc'h et al., 2002). This is essential if efforts are to be made to transfer favorable alleles from the wild relative to elite wheat lines.
The total length of the 10 linkage groups in our study was 865 cM, which represents 68% of a recently published D-genome map in wheat (Sourdille et al., 2003). Compared with the latter map, this map covered approximately 90% (86%–94%) of chromosomes 1D, 5D, and 6D, approximately 60% (57%–66%) of chromosomes 2D, 3D, and 4D, and 43% of chromosome 7D. Thus, substantial coverage has been achieved, although some parts of the genome require additional work.
Huang et al. (2003) reported QTLs for yield and seed mass close to the centromeric region of chromosome 4D in wheat. In accordance, we detected QTLs for seed mass in A. tauschii on linkage group 9, which is part of the long arm of chromosome 4D close to the centromere. Furthermore, we identified a QTL for RGR2 in that region. For seed mass, QTLs were also reported on chromosome 7D and on orthologous positions near the centromere on chromosomes 7A and 7B in wheat (Huang et al., 2003). We did not detect a QTL for seed mass in that chromosomal region, but we did identify QTLs for RGR2 in the centromeric region of 7D. Given the negative correlation between seed mass and RGR2 in our study, it is tempting to speculate that parts of chromosomes 4 and 7 affect seed mass and thereby also RGR2 (or vice versa). In rice, QTLs for seed mass and plant mass colocated on chromosomes 1, 3, 5, and 6 (Cui et al., 2002). Since rice chromosomes 3 and 6 represent parts of wheat chromosomes 4 and 7, respectively (Gale and Devos, 1998), these data support the observations of this study. Accordingly, the studies with rice, wheat, and A. tauschii elucidate the importance of chromosomes 4 and 7 for seed mass, yield, and growth.
We observed a trade-off between leaf and sheath length, on the one hand, and number of leaves, on the other. The growth rate of the number of leaves and tillers apparently depended on the length of individual leaves and sheaths. Huang et al. (2003) reported QTLs for tiller number (expressed as number/m2) on chromosomes 2D, 5D, and 6D in wheat. In A. tauschii, we identified a QTL for number of leaves and a putative QTL for RTR, as well as a QTL for leaf length at the same location on chromosome 2D (i.e. on the long arm about 15–20 cM from the centromere). Furthermore, on 6D we found a QTL for leaf length at the same position as the wheat tiller number QTL. Finally, on chromosome 5D, we detected QTLs for RTR, RLaR, and number of leaves. However, these were on different locations compared with the QTL for tiller number on 5D in wheat (centromeric region in A. tauschii and distal end of long arm in wheat), and it is thus uncertain whether these represent the same underlying genes. Together, both the QTL analysis in this study and the comparison with wheat QTL data point toward a genetic basis for the negative correlation between leaf length and growth of number of leaves and/or tillers, with underlying genes located on chromosomes 2, 5, and 6.
Studies of the genetic basis of RGR and underlying traits are scarce. We only know of a few studies with barley (Van Rijn, 2001; Elberse, 2002; Poorter et al., 2005) and one with Arabidopsis (El-Lithy et al., 2004). Comparative genetics revealed that the genomes of wheat and barley show almost complete colinearity (Gale and Devos, 1998), which makes comparison of our data of Aegilops with those of Hordeum meaningful. It reveals a high level of correspondence between the species, which can be summarized as follows. Chromosome 1 harbors QTLs for RGR, shoot traits (e.g. LMF, LAR), and leaf traits (leaf length and width); chromosome 2 has QTLs for number of leaves and leaf traits; chromosome 3 comprises few early growth-related QTLs in both species; chromosome 4 covers QTLs for seed mass, LMF, LAR, and number of leaves; chromosome 5 harbors QTLs for number of leaves, leaf length and width, LAR, and LMF; chromosome 6 has QTLs for RGR and several leaf traits; and, finally, chromosome 7 has the traits for seed mass. The lack of anchor markers between the AFLP maps of barley (Qi et al., 1998) and A. tauschii hampers a solid comparison of the positions of the QTLs in barley and in A. tauschii, but a quick scan shows that several, but not all, QTLs for the same traits might indeed be located in the same chromosomal region. More research is required to examine further details.
Based on this study, it is difficult to speculate on candidate genes for the growth traits under consideration. However, a few remarks can be made. As far as leaf (and sheath) length and underlying traits are concerned, it is striking that the genes, encoding enzymes of the early steps of the GA biosynthetic pathway in rice, are all positioned on chromosomes that harbor QTLs for leaf length and underlying traits in A. tauschii (Gale and Devos, 1998; Sakamoto et al., 2004). Since GA affects leaf expansion, SLA, and RGR in several species (Dijkstra et al., 1990; Nagel et al., 2001; Bultynck and Lambers, 2004), it is worthwhile to consider the involvement of GA-related genes in early vigor traits. Alternative candidate genes can perhaps be found in the carbon metabolism pathways. For instance, in rice, the gene for Suc phosphate synthase has been recognized as the gene controlling plant height (Ishimaru et al., 2004) through its effect on the amounts of Suc translocated to the leaves. It has also been suggested that invertase activity may play a role in rapid leaf growth (Causse et al., 1995). Comparative mapping has opened ways to search in model plant systems such as rice for candidate genes. However, whatever the candidate gene to search for may be, one has to take into account the numerous rearrangements within and among chromosomes that have been discovered by comparative sequence analysis of rice and wheat (La Rota and Sorrells, 2004).
MATERIALS AND METHODS
Plant Material
Accession PI603228 of Aegilops tauschii was crossed with accession Ciae4 (seeds were obtained from the National Small Grains Collection [NSGC], U.S. Department of Agriculture-Agricultural Research Service [USDA-ARS]), and the progeny was progressed until the F4 by single-seed descent at Zelder B.V. In order to obtain seeds for phenotyping, proliferation of the F4 seeds was done in the greenhouse at Cebeco B.V. Seeds were stored at 4°C in the dark at dry conditions.
Phenotyping F4 RILs: Growth Conditions and Experimental Design
For the phenotyping, 134 F4 lines were used. Because of the time required on the harvest day, germination and growth of the plants were staggered in time. In each batch of plants, nine to 14 RILs were grown, and two individuals per RIL were measured. Of each RIL, in total four plants were measured for all growth traits; thus each RIL was represented in two different batches. In each batch, the two parental lines were grown as controls.
Per batch, four to six randomly chosen seeds of each RIL were weighed individually and then surface sterilized with 2.5% (v/v) NaHClO3 and stratified for 7 d in petri dishes on moistened filter paper at 4°C in the dark. For germination, petri dishes were transferred to a germination cabinet with the following conditions: 16-h light, photosynthetically active radiation 25 μmol m−2 s−1, and day/night temperatures of 20°C/15°C. After 3 d, seedlings were transferred to a growth room (16-h d/8-h night, photosynthetically active radiation 400 ± 25 μmol m−2 s−1, temperature 20°C/20°C, relative humidity 70%) and to containers with washed river sand saturated with demineralized water to allow further growth of the roots for 5 d. After that period, the roots were rinsed with demineralized water and two randomly chosen seedlings per RIL were transferred to an aerated modified Hoagland solution (eight plants on 33-L containers; day 0). The composition of the nutrient solution was 0.6 mm Ca(NO3)2, 0.8 mm KNO3, 0.19 mm KH2PO4, 0.27 mm MgSO4, 2 μm MnSO4, 0.85 μm ZnSO4, 0.15 μm CuSO4, 20 μm H3BO3, 0.25 μm Na2MoO4, and 0.08 mm Fe-EDTA. The pH was adjusted regularly to 5.7, and the solution was renewed once a week. Plants were rotated at least three times per week within the growth room to minimize the variation in environmental conditions for individual plants.
Growth Trait Measurements
A range of growth traits was measured during 14 d after transfer to the nutrient solution (Table I). From day 0 onward, leaf and tiller emergence was recorded daily, and leaves and tillers were identified according to Klepper et al. (1982). RLaR (increase in number of leaves per number of leaves already present per day) and RTR (increase in number of tillers per number of tillers present per day) were calculated as the slope of the regression line through the log-transformed number of leaves and tillers, respectively, versus time. Individual leaf growth measurements were conducted nondestructively on leaf 3 of the main shoot by daily measurement of the leaf length from the leaf tip to the base of the whorl of the leaf sheaths with a ruler. LER (mm h−1) was calculated as the slope of the linear regression line fitted through the data points in the interval from 20% to 80% of the final leaf length. LED (h) was calculated as final leaf length divided by LER. Maximal leaf width and sheath length were measured when the leaf was fully elongated. Measurements were done on leaf 3 only, because earlier experiments showed very high correlations between individual leaf growth characteristics of different leaves within a plant (data not shown).
Whole-plant RGR was measured nondestructively by determination of individual plant fresh mass on days 0 and 7, and a destructive harvest on day 14. For the nondestructive measurement of plant mass, roots were blotted gently with tissue paper; plants were weighed and returned to the nutrient solution. Separate experiments showed no measurable effect of these handlings on RGR (data not shown). RGR was calculated as the slope of the regression line through the log-transformed fresh plant mass versus time for week 1 (RGR1), week 2 (RGR2), and for the whole growth period (RGRtotal). On day 14, length and width of the lamina of one fully expanded leaf blade were measured, and this leaf blade was harvested to make an imprint of the epidermal cells (imprint leaf) for maximal cell-length measurement. The rest of the plant was divided into leaf blades, stems (leaf sheaths), and roots, and the fresh weight of each portion was determined. Total leaf area of all leaves except the imprint leaf was measured with a Li-3100 area meter (LI-COR). Dry mass was measured after drying at 70°C for at least 48 h. RGRdry was calculated using seed mass and final plant dry mass. Leaf area of the imprint leaf was calculated as 0.9×length×width, and this value was added to the measured leaf area to obtain total leaf area. The correction factor 0.9 was based on experimental data (data not shown). From these data, the following parameters were calculated: SLA (leaf area per unit leaf mass), LAR (leaf area per unit plant mass), ULR (increment of plant mass per unit of leaf area per unit of time), LMF (leaf mass per unit plant mass), stem mass fraction (SMF; stem mass per unit plant mass), RMF (root mass per unit plant mass), and dry-matter percentages of leaves, stems, and roots.
The imprint of the epidermal cells for measurement of the maximal cell length was made according to Schnyder et al. (1990). On the abaxial side of the distal part of the leaf, a solution of polyvinylformaldehyde (Formvar) in chloroform (4%, w/w) was spread with a brush. After evaporation of the chloroform, a fine transparent film was left on the epidermis. With a transparent cellotape, this film was transferred to a clean microscopic slide. Dr. M. Terlou (Image Processing and Design, Faculty of Biology, Utrecht University, the Netherlands) developed a cell-length measurement program in the IBAS image analysis system (Zeiss). Microscopic images were recorded with a Panasonic black/white CCD camera (type WC-CD50 mounted on a Zeiss microscope equipped with a scanning table controlled by a joystick [objective was 6.3× with Optovar 1.25]). The images were digitized (frame size 768×512 pixels; 256 gray levels), the contrast was enhanced, and the cell boundaries of a series of cells in the image were indicated with the mouse pointer, followed by automatic calculation of the cell lengths. Cell lengths of two epidermal cell files (100 cells per file) adjacent to stomatal cell files were measured. The data for both files of a leaf were combined, and the average cell length per leaf was calculated. From these data, the cell production rate (cells day−1) during steady-state leaf elongation was estimated as LER divided by the cell length. The number of cells in the whole cell file was estimated as final leaf length divided by cell length.
Genetic Linkage Map
A genetic linkage map of AFLP markers was constructed using 180 F3 RILs. The AFLP protocol was according to Vos et al. (1995). DNA was isolated from frozen leaves according to the cetyl-trimethyl-ammonium bromide protocol (Saghai-Maroof et al., 1984). The DNA was digested with restriction enzymes EcoRI and MseI. The analysis was performed by Keygene B.V. for 12 primer combinations (M51E36, M60E36, M61E36, M58E45, M51E42, M52E42, M48E48, M52E48, M61E48, M48E51, M51E51, and M52E51 [Supplemental Table I]), which were selected on the basis of the occurrence of more than 10 polymorphisms between the parents of the cross. Keygene scored the AFLP fragments codominantly with AFLP-Quantar.
In addition, five more primer combinations (M52E36, M52E45, M38E48, M60E48, M44E51; see Supplemental Table I) were applied in the laboratory at Utrecht University using the 134 F4 lines, which were phenotyped. For this purpose, of each F4 RIL, a plant was grown for 14 d, as described above. Young leaves were collected for DNA isolation, quickly frozen in liquid N2, and stored at −80°C. DNA was isolated from 150 to 200 mg of frozen leaf samples with the GenomicPrep cells and tissue DNA isolation kit (Amersham-Pharmacia Biotech). The A260/280 of the DNA preparations was 1.7 to 1.8. The amplification reactions were done in an Eppendorf Mastercycler Gradient. The E + 3 primers were Cy5 labeled. Amplification products were separated and analyzed with an ALFexpress II, according to the manufacturer's protocol. UV-polymerized gels were used (ReproGel High Resolution) together with the UV box (ReproSet; all these materials from Amersham-Pharmacia Biotech). The presence or absence of AFLP fragments was scored by eye (dominant scoring) with the help of Cross Checker (Buntjer, 1999) using a digital image. Furthermore, using the 134 F4 lines, a set of 15 SSR markers was analyzed to assign the linkage groups to the D genome of wheat. SSRs analyzed were Xgwm458 and Xgwm33 for chromosome 1D, Xgwm 261 and Xgwm515 for chromosome 2D, Xgwm183 and Xgwm383 for chromosome 3D, Xgwm165, Xgwm624, and Xgwm609 for chromosome 4D, Xgwm583 and Xgwm565 for chromosome 5D, Xgwm325 and Xgwm469 for chromosome 6D, and Xgwm44 and Xgwm437 for chromosome 7D. Primers and PCR conditions were applied according to Röder et al. (1998). Amplification reactions and analysis were carried out on the same equipment as used for the AFLP described above. Primers were supplied by Isogen Life Science. The forward primers were Cy5 labeled. With the exception of Xgwm609 and Xgwm624, the SSRs were scored codominantly.
The genetic linkage map was constructed with Joinmap version 3.0 (Van Ooijen and Voorrips, 2001) with a supplementary module developed by P. Stam to deal with the mixed-generation (F3 /F4) setup of the dataset. A minimum LOD score of 5.0 was used and the Kosambi mapping function was applied for calculation of map distances.
Since clustering of markers was manifest, for QTL mapping a core map was made: Where possible, every 5 cM the most informative marker was chosen (i.e. preferably codominantly scored and with the least number of missing values).
Data Analysis
Phenotypic growth trait data were analyzed with SPSS for Windows statistical software (release 10.0; SPSS), using the mean values per RIL. Frequency histograms were calculated as well as simple product moment correlations (Pearson correlations) between traits. The relationship between growth traits was further studied by path analysis (Lynch and Walsh, 1998) on the standardized means per RIL. Multiple regression analysis was carried out to calculate the partial regression coefficients necessary for path analysis.
QTL analyses were done with MapQTL version 4.0 (Van Ooijen et al., 2002) on the means per RIL, using the core map described above. The mean trait values of the F4 RILs were regressed on the marker genotypes of the F3. QTL analysis was started by interval mapping (Jansen, 1993; Jansen and Stam, 1994). In the regions of QTLs and subsignificant QTLs (LOD > 1.5), markers with the highest LOD score were assigned as cofactors. The Automatic Cofactor Selection module was run, and the resulting selection of cofactors was used in the following restricted multiple-QTL model analysis. If LOD values for markers in other regions became significant, they were added as cofactors, and cofactors without an effect were left out again. This procedure was repeated until the LOD profile stabilized. In restricted multiple-QTL model mapping, all selected cofactors were used except the ones on the linkage group on which the QTL was fitted. The significant threshold for the LOD value was 2.65 (permutation test on the dataset, 1,000 permutations), giving the 95% confidence level of a significant QTL, which was close to the threshold value of 2.7 when calculated according to Van Ooijen (1999).
Supplementary Material
Acknowledgments
We thank Ineke Stulen, Hendrik Poorter, and Rens Voesenek for their valuable comments on the manuscript, and Maarten Terlou for developing a cell-length measurement software program. We thank Zelder B.V. and Cebeco B.V. for their efforts in making the cross and growing the RIL lines, and Keygene B.V. for their contribution to the AFLP analysis.
This work was supported by the Technology Foundation STW, Applied Science division of The Netherlands Organization for Scientific Research (NWO), and the technology program of the Ministry of Economic Affairs, The Netherlands.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.063263.
References
- Becraft PW (1999) Development of the leaf epidermis. Curr Top Dev Biol 45: 1–40 [DOI] [PubMed] [Google Scholar]
- Botwright TL, Condon AG, Rebetzke GJ, Richards RA (2002) Field evaluation of early vigor for genetic improvement of grain yield in wheat. Aust J Agric Res 53: 1137–1145 [Google Scholar]
- Boyko E, Gill KS, Mickelson-Young L, Nasuda S, Raupp WJ, Ziegle JN, Singh S, Hassawi DS, Fritz AK, Namuth D, et al (1999) A high-density genetic linkage map of Aegilops tauschii, the D-genome progenitor of bread wheat. Theor Appl Genet 99: 16–26 [Google Scholar]
- Brooks SA, Huang L, Gill BS, Fellers JP (2002) Analysis of 106 kb of contiguous DNA sequence from the D genome of wheat reveals high gene density and a complex arrangement of genes related to disease resistance. Genome 45: 963–972 [DOI] [PubMed] [Google Scholar]
- Bultynck L, Fiorani F, Lambers H (1999) Control of leaf growth in an ecological perspective. Plant Biol 1: 14–18 [Google Scholar]
- Bultynck L, Fiorani F, van Volkenburgh E, Lambers H (2003) Epidermal cell division and cell elongation in two Aegilops species with contrasting leaf elongation rates. Funct Plant Biol 30: 425–432 [DOI] [PubMed] [Google Scholar]
- Bultynck L, Lambers H (2004) Effects of applied gibberellic acid and paclobutrazol on leaf expansion and biomass allocation in two Aegilops species with contrasting leaf elongation rates. Physiol Plant 122: 143–151 [Google Scholar]
- Bultynck L, ter Steege MW, Schortemeyer M, Poot P, Lambers H (2004) From individual leaf elongation to whole shoot leaf area expansion; a comparison of three Aegilops and two Triticum species. Ann Bot (Lond) 94: 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buntjer JB (1999) Cross Checker Fingerprint Analysis Software v2.9. Wageningen University and Research Centre, Wageningen, The Netherlands
- Cai HW, Morishima H (2002) QTL clusters reflect character associations in wild and cultivated rice. Theor Appl Genet 104: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Causse M, Rocher JP, Henry AM, Charcosset A, Prioul J, Devienne D (1995) Genetic dissection of the relationship between carbon metabolism and early growth in maize, with emphasis on key-enzyme loci. Mol Breed 1: 259–272 [Google Scholar]
- Clevering OA (1999) Between- and within-population differences in Phragmites australis. The effects of nutrients on seedling growth. Oecologia 121: 447–457 [DOI] [PubMed] [Google Scholar]
- Cui KH, Peng SB, Xing YZ, Xu CG, Yu SB, Zhang Q (2002) Molecular dissection of seedling-vigor and associated physiological traits in rice. Theor Appl Genet 105: 745–753 [DOI] [PubMed] [Google Scholar]
- Dijkstra P, Reegen HT, Kuiper PJC (1990) Relation between relative growth rate, endogenous gibberellins, and the response to applied gibberellic acid for Plantago major. Physiol Plant 79: 629–634 [DOI] [PubMed] [Google Scholar]
- Dvorak J, Luo MC, Yang ZL (1998) Restriction fragment length polymorphism and divergence in the genomic regions of high and low recombination in self-fertilizing and cross-fertilizing Aegilops species. Genetics 148: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elberse IAM (2002) Genetic analysis of growth characteristics. Hordeum spontaneum under nutrient limitation. PhD thesis. Utrecht University, Utrecht, The Netherlands
- Ellis RP, Forster BP, Robinson D, Handley L, Gordon DC, Russel JR, Powell W (2000) Wild barley: a source of genes for crop improvement in the 21st century? J Exp Bot 51: 9–17 [PubMed] [Google Scholar]
- El-Lithy ME, Clerkx EJM, Ruys GJ, Koornneef M, Vreugdenhil D (2004) Quantitative trait locus analysis of growth-related traits in a new Arabidopsis recombinant inbred population. Plant Physiol 135: 444–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Sears ER (1981) The wild gene resources of wheat. Sci Am 244: 98–108 [Google Scholar]
- Fiorani F, Beemster GTS, Bultynck L, Lambers H (2000) Can meristematic activity determine variation in leaf size and elongation rate between four Poa species? A kinematic study. Plant Physiol 124: 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Devos KM (1998) Comparative genetics in grasses. Proc Natl Acad Sci USA 95: 1971–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier E, Salager JL, Laurent G, Sonié L (1999) Relationships between photosynthesis, nitrogen and leaf structure in 14 grass species and their dependence on the basis of expression. New Phytol 143: 119–129 [Google Scholar]
- Guyomarc'h H, Sourdille P, Charmet G, Edwards KJ, Bernard M (2002) Characterisation of polymorphic microsatellite markers from Aegilops tauschii and transferability to the D-genome of bread wheat. Theor Appl Genet 104: 1164–1172 [DOI] [PubMed] [Google Scholar]
- Huang XQ, Cöster H, Ganal MW, Röder MS (2003) Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat. Theor Appl Genet 106: 1379–1389 [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Ono K, Kashiwagi T (2004) Identification of a new gene controlling plant height in rice using the candidate-gene strategy. Planta 218: 388–395 [DOI] [PubMed] [Google Scholar]
- Jansen RC (1993) Interval mapping of multiple quantitative trait loci. Genetics 135: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, Stam P (1994) High resolution of quantitative traits into multiple loci via interval mapping. Genetics 136: 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsy MJ, Farquhar AGL (1998) QTL analysis in plants; where are we now? Heredity 80: 137–142 [DOI] [PubMed] [Google Scholar]
- Khavin E, Coe E (1997) Mapped genomic locations for developmental functions and QTLs reflect concerted groups in maize (Zea mays L.). Theor Appl Genet 95: 343–352 [Google Scholar]
- Klepper B, Rickman RW, Peterson CM (1982) Quantitative characterization of vegetative development in small cereal grains. Agron J 74: 789–792 [Google Scholar]
- La Rota M, Sorrells ME (2004) Comparative DNA sequence analysis of mapped wheat ESTs reveals the complexity of genome relationships between rice and wheat. Funct Integr Genomics 4: 34–46 [DOI] [PubMed] [Google Scholar]
- Lambers H, Poorter H (1992) Inherent variation in growth rate between higher plants: a search for physiological causes and ecological consequences. Adv Ecol Res 23: 187–261 [Google Scholar]
- Lelley T, Stachel M, Grausgruber H, Vollmann J (2000) Analysis of relationships between Aegilops tauschii and the D genome of wheat utilizing microsatellites. Genome 43: 661–668 [PubMed] [Google Scholar]
- Lemerle D, Gill GS, Murphy CE, Walker SR, Cousens RD, Mokhtari S, Peltzer SJ, Coleman R, Luckett DJ (2001) Genetic improvement and agronomy for enhanced wheat competitiveness with weeds. Aust J Agric Res 52: 527–548 [Google Scholar]
- López-Castaneda C, Richards RA, Farquhar GD, Williamson RE (1996) Seed and seedling characteristics contributing to variation in early vigor among temperate cereals. Crop Sci 36: 1257–1266 [Google Scholar]
- Lynch M, Walsh B (1998) Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA
- Nagel OW, Konings H, Lambers H (2001) Growth rate and biomass partitioning of wild type and low-gibberellin tomato (Solanum lycopersicum) plants growing at a high and low nitrogen supply. Physiol Plant 111: 33–39 [Google Scholar]
- Poorter H, Remkes C (1990) Leaf area ratio and net assimilation rate of 24 wild species differing in relative growth rate. Oecologia 83: 553–559 [DOI] [PubMed] [Google Scholar]
- Poorter H, van der Werf A (1998) Is inherent variation in RGR determined by LAR at low irradiance and by NAR at high irradiance? A review of herbaceous species. In H Lambers, H Poorter, M Vuuren, eds, Inherent Variation in Plant Growth, Physiological Mechanisms and Ecological Consequences. Backhuys Publishers, Leiden, The Netherlands, pp 309–336
- Poorter H, van Rijn CPE, Vanhala TK, Verhoeven KJF, de Jong YEM, Stam P, Lambers H (2005) A genetic analysis of relative growth rate and underlying components in Hordeum spontaneum. Oecologia 142: 360–377 [DOI] [PubMed] [Google Scholar]
- Qi X, Stam P, Lindhout P (1998) Use of locus-specific AFLP markers to construct a high-density molecular map in barley. Theor Appl Genet 96: 376–384 [DOI] [PubMed] [Google Scholar]
- Rebetzke GJ, Richards RA (1999) Genetic improvement of early vigor in wheat. Aust J Agric Res 50: 291–301 [Google Scholar]
- Richards RA (2000) Selectable traits to increase crop photosynthesis and yield of grain crops. J Exp Bot 51: 447–458 [DOI] [PubMed] [Google Scholar]
- Richards RA, Lukacs Z (2002) Seedling vigor in wheat—sources of variation for genetic and agronomic improvement. Aust J Agric Res 53: 41–50 [Google Scholar]
- Richards RA, Rebetzke GJ, Condon AG, van Herwaarden AF (2002) Breeding opportunities for increasing the efficiency of water use and crop yield in temperate cereals. Crop Sci 42: 111–121 [DOI] [PubMed] [Google Scholar]
- Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149: 2007–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc Natl Acad Sci USA 81: 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnyder H, Seo S, Rademacher IF, Kübauch W (1990) Spatial distribution of growth rates and of epidermal cell lengths in the elongation zone during leaf development in Lolium perenne L. Planta 181: 423–431 [DOI] [PubMed] [Google Scholar]
- Siddique KHM, Tennant D, Perry MW, Belford RK (1990) Water use and water use efficiency of old and modern wheat cultivars in a Mediterranean type environment. Aust J Agric Res 41: 431–447 [Google Scholar]
- Sourdille P, Cadalen T, Guyomarc'h H, Snape JW, Perretant MR, Charmet G, Boeuf C, Bernard S, Bernard M (2003) An update of the Courtot × Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor Appl Genet 106: 530–538 [DOI] [PubMed] [Google Scholar]
- Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92: 191–203 [DOI] [PubMed] [Google Scholar]
- Treymayne M, Richards AJ (2000) Seed weight and seed number affect subsequent fitness in outcrossing and selfing Primula species. New Phytol 148: 127–142 [DOI] [PubMed] [Google Scholar]
- Van den Boogaard R, Veneklaas EJ, Peacock J, Lambers H (1996) Relative growth rate, biomass allocation pattern and water use efficiency of three wheat cultivars during early ontogeny as dependent on water availability. Physiol Plant 98: 493–504 [Google Scholar]
- Van Ooijen JW (1999) LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83: 613–624 [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW, Boer MP, Jansen RC, Maliepaard C (2002) MapQTL Version 4.0, Software for the Calculation of QTL Positions on Genetic Maps. Plant Research International, Wageningen, The Netherlands
- Van Ooijen JW, Voorrips RE (2001) JoinMap 3.0, Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands
- Van Rijn CPE (2001) A physiological and genetic analysis of growth characteristics in Hordeum spontaneum. PhD thesis. Utrecht University, Utrecht, The Netherlands
- Van Rijn CPE, Heersche I, van Berkel YEM, Nevo E, Lambers H, Poorter H (2000) Growth characteristics in Hordeum spontaneum populations from different habitats. New Phytol 146: 471–481 [Google Scholar]
- Villar R, Veneklaas EJ, Jordano P, Lambers H (1998) Relative growth rate and biomass allocation in 20 Aegilops (Poaceae) species. New Phytol 140: 425–437 [DOI] [PubMed] [Google Scholar]
- Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93: 77–78 [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al (1995) AFLP; a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Li J, Grandillo S, Nag Ahn S, Yuan L, Tanksley SD, McCouch SR (1998) Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 150: 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.