Abstract
The in vivo intracellular location of components of the Caulobacter replication apparatus was visualized during the cell cycle. Replisome assembly occurs at the chromosomal origin located at the stalked cell pole, coincident with the initiation of DNA replication. The replisome gradually moves to midcell as DNA replication proceeds and disassembles upon completion of DNA replication. Although the newly replicated origin regions of the chromosome are rapidly moved to opposite cell poles by an active process, the replisome appears to be an untethered replication factory that is passively displaced towards the center of the cell by the newly replicated DNA. These results are consistent with a model in which unreplicated DNA is pulled into the replication factory and newly replicated DNA is bidirectionally extruded from the complex, perhaps contributing to chromosome segregation.
Keywords: chromosome segregation/DNA replication/localization/replication factory/replisome
Introduction
Efficient replication and segregation of chromosomes to daughter cells is an essential function for all living cells. Most bacterial chromosomes are circular, and bidirectional DNA replication is initiated from a single origin. The elaborate mitotic apparatus of eukaryotic cells has not been observed in bacterial cells, but their chromosomes are structured and regions of the chromosome move during the cell cycle in specific and dynamic patterns (Jensen and Shapiro, 1999b; Gordon and Wright, 2000). Chromosome segregation occurs gradually during DNA replication, since newly replicated regions of the chromosome are separated before other regions are replicated. Duplicated origins of replication move immediately to the opposite ends of the cell, whereas the terminus region of the chromosome eventually resides at midcell (Gordon et al., 1997; Webb et al., 1997, 1998; Niki and Hiraga, 1998; Jensen and Shapiro, 1999a). Regions of the chromosome that lie between the origin and the terminus on the genetic map are located between them in the cell (Teleman et al., 1998; Niki et al., 2000).
The organization and positioning of the DNA replication apparatus have major implications for how bacterial chromosomes can be separated from each other. Currently, two different models for the organization of DNA replication are considered (Cook, 1999; Falaschi, 2000). The tracking model for DNA replication proposes that DNA polymerase moves along the DNA template from the origin to the terminus. The factory model for DNA replication proposes that the DNA polymerase is in a fixed position and the DNA template is moved through the replication factory (Dingman, 1974; Pardoll et al., 1980; Lemon and Grossman, 1998). According to the factory model, unreplicated DNA is pulled into the stationary replisome and the daughter strands are then extruded from the complex. If DNA replication is based purely on freely moving molecules, as proposed in the tracking model, DNA replication would result in chaotic entanglement of the newly replicated DNA strands, which would have to be disentangled by topoisomerases before they could be separated. The factory model for DNA replication, on the other hand, allows the newly replicated DNA strands to be extruded in opposite directions from the stationary replisome, thereby minimizing entanglement. Addition ally, both DNA polymerase and DNA helicase appear to be powerful molecular motors, so if the two newly replicated DNA strands are extruded bidirectionally towards opposite poles of the cell, the process of DNA replication itself may provide the force responsible for movement of the bulk of the chromosome (Lemon and Grossman, 1998; Cook, 1999).
The idea that the DNA polymerase complex is stationary and the DNA is moved through the replisome complex was originally proposed more than 25 years ago (Dingman, 1974), however, only recently has experimental evidence supporting this model been obtained. The organization of DNA replication in eukaryotic cells has been examined by labeling newly replicated DNA or by examining the intracellular localization of components of the DNA replication apparatus. Initiation of DNA replication appears to take place at a limited number of positions (between five and 20), and later during the S phase a couple of hundred replication sites are distributed throughout the nuclei (Cook, 1999; Kennedy et al., 2000). Time-lapse microscopy of living cells expressing green fluorescent protein (GFP) fused to one of the components of the DNA replication apparatus (GFP–PCNA) shows that the replication foci assemble and disassemble during DNA replication, but they do not appear to exhibit directed movement (Leonhardt et al., 2000). The presence of discrete sites where DNA replication takes place supports the factory model of DNA replication, and the replication factories in eukaryotic cells may be immobilized by interacting with a component of the nucleoskeleton (Cook, 1999; Falaschi, 2000). The location of the DNA replication apparatus in the bacterium Bacillus subtilis was examined using GFP fusions to different DNA polymerase subunits (Lemon and Grossman, 1998, 2000; Losick and Shapiro, 1998). In these cells, the replisome resides predominantly near midcell or sites that become midcell after cell division. Additionally, simultaneous visualization of the position of the replisome and specific regions of the chromosome revealed that the DNA template is pulled to the replication factory for replication and moved away from it after duplication (Lemon and Grossman, 2000). Thus, the replisome in B.subtilis appears to be stationary and the DNA moves during replication, supporting the factory model for DNA replication. The replisome may be anchored to a cellular structure (e.g. the cytoplasmic membrane or the cell wall) to prevent movement during DNA replication (Lemon and Grossman, 2000), but no direct evidence for anchoring has been obtained.
Caulobacter crescentus is a useful system for studying bacterial chromosome replication and segregation because cells are easily synchronized and chromosome replication is initiated only once per cell cycle at a well defined time. An asymmetric division yields two distinct daughter cells: a replication-competent stalked cell and a replication-repressed swarmer cell (Hung et al., 2000). In swarmer cells, the origin of replication is located at the flagellated pole of the cell and the terminus is located near the opposite end of the cell (Jensen and Shapiro, 1999a). The motile swarmer cell differentiates into a stalked cell by shedding its flagellum, synthesizing a stalk at the same pole, and initiating chromosome replication. Upon initiation of DNA replication at the swarmer-to-stalked cell transition, one of the origin copies is rapidly moved to the opposite end of the cell, whereas the terminus only gradually moves to the middle of the cell during the S phase (Jensen and Shapiro, 1999a).
In B.subtilis, the origin of replication appears to be located at midcell when DNA replication is initiated, the same position as the replication apparatus (Lewis and Errington, 1997; Lemon and Grossman, 1998, 2000; Teleman et al., 1998). In Caulobacter, the origin is located at a cell pole when DNA replication is initiated (Jensen and Shapiro, 1999a), so the position of the replisome in Caulobacter is expected to be different from that observed in B.subtilis. To examine the position of the replisome during the Caulobacter cell cycle, we constructed fusions between GFP or yellow fluorescent protein (YFP) and replisome components. The intracellular position of these fusions were then examined in live cells. Surprisingly, a single replisome focus was observed that gradually moved from near the stalked pole to the cell division plane during progression of DNA replication. Both the formation and movement of the replisome depend on DNA replication, but neither protein synthesis nor cell elongation. We propose that the replisome is an untethered replication factory.
Results
Fusions between GFP or YFP and Caulobacter replication proteins
To visualize the intracellular location of the replisome in live Caulobacter cells, we constructed in-frame fusions between GFP or YFP and different subunits of the DNA polymerase III complex, which is responsible for the bulk of DNA replication (Kornberg and Baker, 1992). The gene fusions replaced the wild-type alleles in single copy in the Caulobacter chromosome under the control of their endogenous promoters. The holB and holC genes encode the δ′ and the χ subunits of DNA polymerase III, respectively, and both proteins are parts of the β clamp loader complex (Davey and O’Donnell, 2000; Hingorani and O’Donnell, 2000). The dnaB gene encodes DNA helicase, which is responsible for separating the two DNA strands during DNA replication (Kornberg and Baker, 1992). Strains expressing HolB–GFP, HolC–YFP or DnaB–YFP fusion proteins as the only source of the respective replisome components are viable and no growth or cell cycle defects were observed. Thus, these observations suggest that the fusion proteins are fully functional.
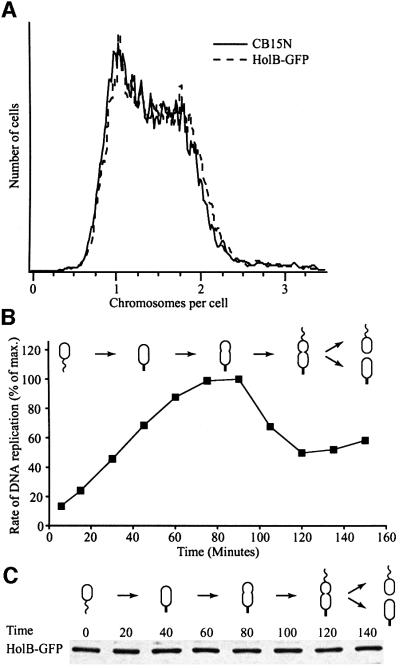
The DNA content profile of a non-synchronized population of HolB–GFP-expressing cells, examined by flow cytometry, is identical to that of cells from the parent strain CB15N (Figure 1A). Similarly, when DNA replication in a synchronized cell population was measured directly by incorporation of [α-32P]dCTP into DNA (Figure 1B), the cell cycle timing of DNA replication was found to be similar to that previously observed in the wild-type strain (Marczynski and Shapiro, 1992). Thus, the HolB–GFP strain does not appear to have any DNA replication defects.
Fig. 1. Characterization of the HolB–GFP fusion protein strain. (A) The DNA contents profiles of mixed cultures of the HolB–GFP strain (dashed line) and the parent strain CB15N (solid line) were compared using flow cytometry of chromamycin A3-stained cells. Chromosome equivalents are indicated on the x-axis and relative number of cells on the y-axis. (B) DNA replication during the cell cycle in synchronized populations of HolB–GFP-expressing cells, measured as incorporation of [α-32P]dCTP into chromosomal DNA during a 1 min period. The degree of DNA replication is expressed as a percentage of maximal incorporation of radioactivity into DNA. (C) The abundance of the HolB–GFP fusion protein during the cell cycle analyzed by western blotting using anti-GFP antibodies. Samples were withdrawn at the indicated time points and equal amounts of total protein were loaded in all lanes.
Western blots of cell samples taken from cultures progressing synchronously through the cell cycle show that HolB–GFP, HolC–YFP and DnaB–YFP are present in approximately equal amounts at all time points (Figure 1C and data not shown). Thus, the replisome components are present in swarmer and late pre-divisional cells where no DNA replication takes place.
Localization of the HolB, HolC and DnaB replication proteins during the cell cycle
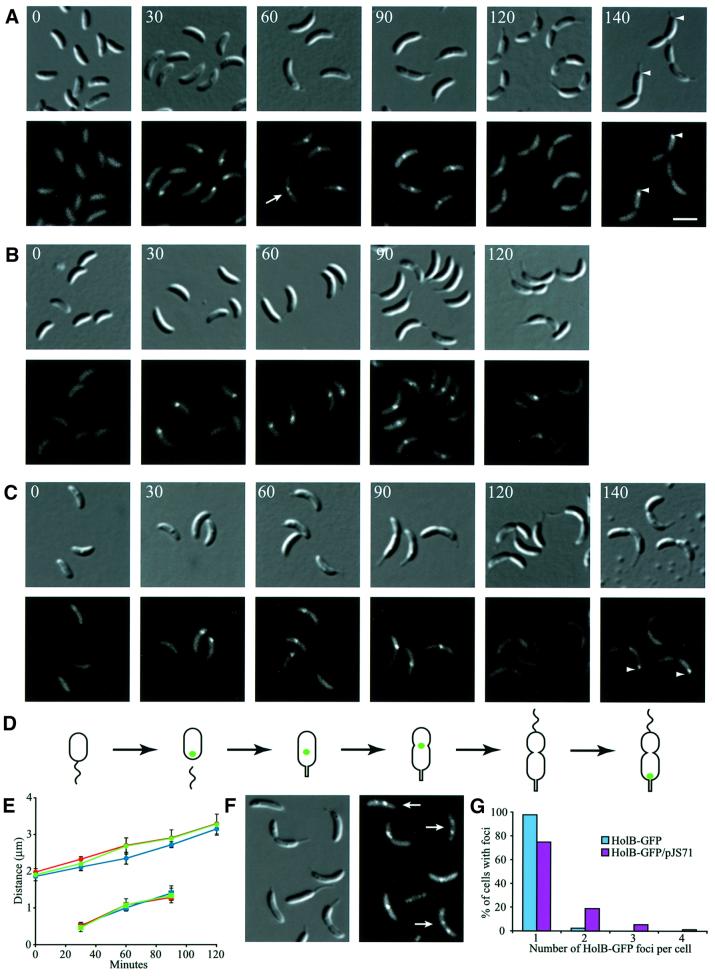
To examine the intracellular location of the replication complex during the Caulobacter cell cycle, we isolated pure populations of swarmer cells from strains bearing the holB-gfp (Figure 2A), holC-yfp (Figure 2B) or dnaB-yfp (Figure 2C) alleles. When incubated in growth medium, the cells progress synchronously through the cell cycle. At different time points during the cell cycle, the intracellular location of the replisome components was examined by fluorescence microscopy (Figure 2). Swarmer cells, which do not replicate their DNA, had no fluorescent foci, but showed faint diffuse fluorescence throughout the cells (Figure 2A–C, 0 min). At the swarmer-to-stalked cell transition, when DNA replication is initiated, a fluorescent focus formed near one pole of the cell (Figure 2A–C, 30 min). In stalked and early pre-divisional cells, the fluorescent focus was located closer to the cell division plane (Figure 2A–C, 60 and 90 min). Late pre-divisional cells, which have completed replicating their chromosome, had no fluorescent foci, indicating that the replisome disassembles upon completion of DNA replication (Figure 2A–C, 120 min). Finally, in dividing cells, foci formed specifically at the stalked pole of the asymmetric cell (Figure 2A and C, 140 min). DNA replication initiates only in the progeny stalked cell during or immediately after cell division. Measurements of the intracellular positions of HolB–GFP, HolC–YFP and DnaB–YFP during the cell cycle show that these proteins have identical localization patterns during the cell cycle (Figure 2E). Thus, replisome assembly occurs only in cells initiating replication and those in the process of replicating their DNA.
Fig. 2. Intracellular localization of replisome proteins. HolB–GFP (A), HolC–YFP (B) or DnaB–YFP (C) expressing swarmer cells were isolated and allowed to progress synchronously through the cell cycle. Samples for microscopy were taken at the indicated time points (in minutes). Top panels show Nomarski DIC microscopy images of the cells and the lower panels show GFP or YFP fluorescence. A white arrow indicates a cell with two closely spaced HolB–GFP foci. White arrowheads indicate the stalked pole of the dividing cells. The white scale bar represents 2 µm. (D) Schematic of cell cycle progression of the strains and the intracellular positions of HolB–GFP, HolC–YFP or DnaB–YFP (green dots). Time-lapse microscopy of HolB–GFP-expressing cells (Figure 4) shows that HolB–GFP specifically assembles at the stalked pole during the swarmer-to-stalked cell transition, as indicated in the schematics. (E) Measurements of the average cell length (circles) and distance from the stalked pole to the middle of the replisome protein foci (squares) at different time points of the cell cycle of the HolB–GFP (blue), HolC–YFP (green) and DnaB–YFP (red) expressing cells. (F) Localization of replisome foci in HolB–GFP-expressing cells containing a freely replicating plasmid (pJS71). Approximately 30–50 copies of the plasmid are present per cell and plasmid replication is initiated by a plasmid-encoded replication-control system. White arrows indicate cells with multiple replisome foci or foci located at positions where they are not observed in plasmid-free cells. (G) Analysis of the number of replisome foci per cell in non-synchronized HolB–GFP-expressing cells in the absence (blue bars) or presence (purple bars) of the pJS71 plasmid. At least 250 cells with clearly visible foci were counted for each strain.
In synchronized cell populations, 80–90% of the cells have foci at the time points where DNA replication takes place. The vast majority of these cells (∼98% of the cells with foci; Figure 2G) have only a single fluorescent focus and the two foci observed in the remaining ∼2% of the cells are always located very close to each other (see the cell labeled with a white arrow in Figure 2A). No cells with more than two foci were observed. However, a strain containing a freely replicating plasmid (pJS71), in the presence of a chromosomal copy of HolB–GFP, had cells with multiple foci (Figure 2F and G). The additional foci probably originate from replisomes formed on replicating plasmids. Up to four foci per cell were observed, and some of the foci were located at positions where none was observed in plasmid-free cells (e.g. foci close to the flagellated pole or two foci far from each other). Only a few additional foci are observed in the plasmid-carrying strain even though the plasmid is present in 30–50 copies per cell. This is most likely because initiation of DNA replication on plasmid origins takes place throughout the cell cycle and the plasmids are so small that they are replicated very fast. Thus, only a few plasmids are being replicated at any given time during the cell cycle. Alternatively, the plasmids could be clustered in a few places (Pogliano et al., 2001).
Formation of replisome foci requires DNA replication
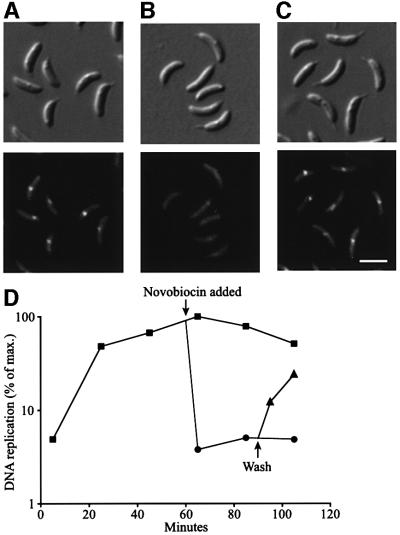
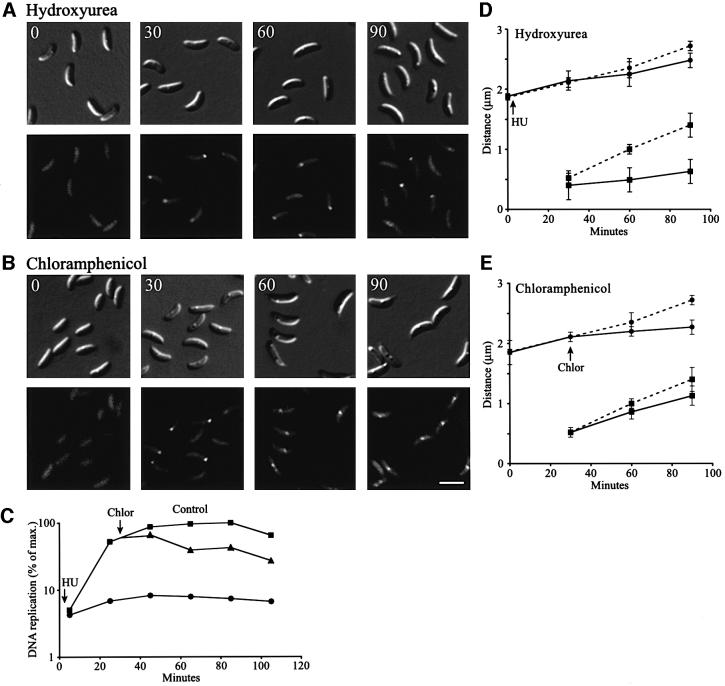
Fluorescent foci were observed only during the periods of the cell cycle when DNA replication takes place; no foci were observed in swarmer cells before DNA replication is initiated or in late pre-divisional cells when chromosome replication has been completed. Thus, active DNA replication appears to be required for the formation of replisome foci. To determine whether this is the case, we treated a population of replication-competent stalked cells (Figure 3A) with the DNA gyrase inhibitor novobiocin and examined cells for replisome foci (Figure 3B). DNA gyrase is a topoisomerase that introduces negative supercoils, a requirement for procession of DNA replication (Kornberg and Baker, 1992). Measurements of DNA replication show that addition of novobiocin resulted in a complete block of DNA replication within 5 min of the addition of the drug (Figure 3D). Upon inhibition of DNA replication, HolB–GFP foci were observed in <0.2% of the cells (Figure 3B), supporting the argument that the presence of replisome foci requires ongoing DNA replication. Replisome foci already present disappear rapidly when DNA replication is inhibited. The inhibition of replication by novobiocin is reversible, since replication restarts and HolB–GFP foci reappear at the expected intracellular positions shortly after the cells are resuspended in growth medium without novobiocin (Figure 3C and D).
Fig. 3. Effect of DNA replication inhibition on the presence of replisome foci. HolB–GFP-expressing cells were synchronized and novobiocin (final concentration 100 µg/ml) was added at the stalked cell stage (60 min into the cell cycle). Some cells were washed and resuspended in fresh growth medium 30 min later. Shown are DIC (top panel) and fluorescence (lower panel) images of (A) untreated stalked cells 60 min into the cell cycle, (B) cells 10 min after novobiocin addition and (C) cells treated with novobiocin for 30 min, washed and incubated in fresh growth medium for 10 min. The scale bar represents 2 µm. (D) Direct measurement of DNA replication using [α-32P]dCTP pulse-labeling in control and novobiocin-treated cells. Squares indicate untreated control cells, circles indicate novobiocin-treated cells and triangles represent novobiocin-treated cells that were washed and resuspended in fresh medium.
Gradual movement of the replisome from near the stalked pole to midcell during DNA replication
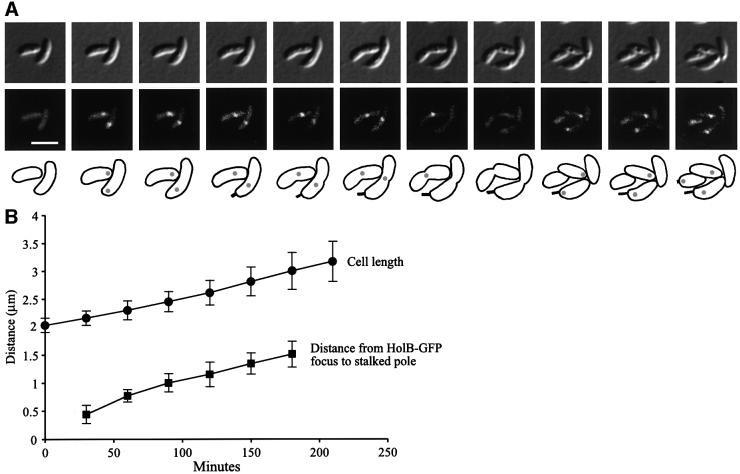
The position of the replisome proteins in cells examined at different stages of the cell cycle (Figure 2A–C) is consistent with the replisome moving from the stalked pole to near the middle of the cell. To distinguish between replisome movement and replisome disassembly and reassembly, we performed time-lapse microscopy of living cells progressing through the cell cycle (Figure 4). Swarmer cells from the HolB–GFP-expressing strain were isolated and placed on a thin layer of nutrient-containing agarose, and images of these cells were recorded every 30 min during their entire cell cycle. Figure 4A shows a representative series of time-lapse images of cells progressing through the cell cycle, and Figure 4B shows measurements of the positions of the HolB–GFP foci during the cell cycle in a large number of cells. The time-lapse microscopy confirmed that the non-replicating swarmer cells do not have HolB–GFP foci. At the swarmer-to-stalked cell transition, a HolB–GFP focus appears specifically at the stalked pole of the new stalked cells, and as they progress through the cell cycle the HolB–GFP focus moves gradually from near the stalked pole to the middle of the cell. The HolB–GFP focus disappears in late pre-divisional cells, but a focus is reformed at the stalked pole of the replication-competent stalked cell progeny immediately after cell division, but not in the progeny swarmer cell. Gradual movement of the HolB–GFP focus was observed in all cells examined by time-lapse microscopy. Measurements of the intracellular positions of HolB–GFP during the cell cycle (Figure 4B) confirm that the movement is gradual. The speed of HolB–GFP movement is 0.45 ± 0.11 µm/h and the cell elongates at 0.34 ± 0.08 µm/h under these growth conditions, so replisome movement is a continual and slow process.
Fig. 4. Time-lapse microscopy analysis showing gradual movement of HolB–GFP foci from the pole to midcell. HolB–GFP-expressing swarmer cells were placed on a thin layer of agarose containing nutrients, and images of the same cells were acquired every 30 min as the cells progressed synchronously through the cell cycle. Cell division occurs at ∼235 min under these growth conditions. (A) DIC images (top panel), fluorescence images (middle panel) and schematics (lower panel) of the same cells progressing through the cell cycle. Grey circles in the schematics indicate the intracellular positions of HolB–GFP. The white scale bar represents 2 µm. A QuickTime movie of this time-lapse sequence is available as Supplementary data at The EMBO Journal Online. (B) The distance from the stalked pole to the middle of the HolB–GFP focus (squares) and the cell length (circles) were measured at different time points in cells from time-lapse sequences. The graph shows averages and the error bars show the standard deviation. Gradual movement of the HolB–GFP focus from near the stalked pole to midcell was observed for all cells analyzed.
Movement of replisome foci depends on DNA replication
Movement of the replisome focus towards the middle of the cell occurs concomitantly with replication of the bulk of the chromosome. To examine whether replication of the majority of the chromosome is required for replisome movement, we slowed DNA replication by treating synchronized HolB–GFP-expressing cells with hydroxyurea (HU) (Figure 5A). This compound inhibits ribonucleotide reductase, an enzyme required for the production of dNTPs. The concentration of HU used here partially inhibits the enzyme, resulting in low dNTP pools and therefore a slower rate of DNA replication. General RNA and protein synthesis are only weakly affected (Degnen and Newton, 1972; Stephens and Shapiro, 1993). Measurements of DNA replication show that HU reduced the rate of DNA replication 15- to 20-fold, but did not completely block DNA replication (Figure 5C). Flow cytometry analysis confirmed that <5% of the chromosome is replicated 90 min after the cells are treated with HU (data not shown). When HU was added to synchronized HolB–GFP-expressing swarmer cells at the beginning of the cell cycle (Figure 5A, 0 min), normal assembly of HolB–GFP fluorescent foci near the stalked pole was observed at the swarmer-to-stalked cell transition (Figure 5A, 30 min). However, the fluorescent foci remained close to the pole at later time points (Figure 5A, 60 and 90 min). Measurements of the intracellular positions of HolB–GFP foci confirm that replisome movement from the pole to midcell is delayed and that cell elongation is only slightly affected (Figure 5D). Thus, replisome movement appears to require ongoing replication.
Fig. 5. Effect of slowing DNA replication or inhibiting protein synthesis on replisome movement. HolB–GFP-expressing swarmer cells were isolated and allowed to progress synchronously through the cell cycle. (A) To slow down DNA replication, HU (final concentration 3 mg/ml) was added at the beginning of the cell cycle. DIC and fluorescence images show cells at the indicated time points (in minutes) during the cell cycle. (B) Chloramphenicol (final concentration 25 µg/ml) was added to the synchronized cells to inhibit protein synthesis. Since chloramphenicol inhibits initiation of new rounds of DNA replication, but not completion of ongoing DNA replication (Winzeler and Shapiro, 1995), the drug was added at 30 min when DNA replication has been initiated in the majority of the cells. DIC and fluorescence images of cells at the indicated time points are shown. The white scale bar represents 2 µm. (C) The degree of DNA replication in control (squares), HU-treated (circles) and chloramphenicol-treated cells (triangles). (D and E) Measurements of the average distances from the stalked pole to the middle of the HolB–GFP focus (squares) or the cell length (circles) in control (broken lines), HU-treated (D, line) or chloramphenicol-treated (E, line) cells at the indicated time points.
In the absence of protein synthesis, initiation of new rounds of DNA replication in Caulobacter is inhibited, but once initiated, replication continues (Winzeler and Shapiro, 1995). To examine the effect of inhibiting protein synthesis on replisome movement after replication initiation, we added chloramphenicol just after initiation of DNA replication at the swarmer-to-stalked cell transition (30 min) of synchronized HolB–GFP-expressing cells (Figure 5B). The HolB–GFP focus appeared to move normally towards the middle of the cell when protein synthesis was inhibited, even though cell elongation was significantly reduced (Figure 5B, 60 and 90 min; Figure 5E). Measurements of [α-32P]dCTP incorporation into DNA show high levels of DNA replication in chloramphenicol-treated cells (Figure 5C). DNA replication is reduced compared with the control cells because some cells did not initiate replication before the chloramphenicol addition, resulting in blocking of DNA replication in these cells. Thus, new protein synthesis and cell elongation are not required for the movement of the replisome from the pole to the division plane.
Newly replicated DNA is moved away from the replication apparatus
To examine the positions of the replicating DNA during the cell cycle, we pulse-labeled replicating DNA with the nucleoside analog bromodeoxyuridine (BrdU) (Figure 6). Synchronized cells were pulse-labeled with BrdU for 15 min at different time points during the cell cycle, followed by fixation and indirect immunofluorescence using anti-BrdU antibodies. The appearance of a specific immunofluorescence signal required both BrdU labeling of DNA and the presence of anti-BrdU antibodies. Thus, the BrdU immunofluorescence signals in Figure 6A show the site in the cell where newly replicated DNA is positioned during the 15 min labeling period. Unreplicated DNA and DNA replicated before the addition of BrdU are not labeled. No immunofluorescence signals were observed when non-replicating swarmer cells were pulse-labeled with BrdU (Figure 6A, 15 min), confirming that BrdU incorporation into DNA is dependent on replication. In cells labeled during the swarmer-to-stalked cell transition, commensurate with replication of the origin-proximal part of the chromosome, two BrdU signals located at both poles of the cell were observed (Figure 6A, 30 min). This shows that the origin-proximal parts of the chromosome are moved towards the extreme poles of the cells shortly after they are replicated. As DNA replication continues in the stalked cells, the BrdU foci, representing the DNA replicated during the pulse, were located away from the extreme poles of the cells (Figure 6A, 60 min). In early pre-divisional cells either two BrdU foci close to the middle of the cell or a single midcell BrdU focus were observed (Figure 6A, 90 min). The single BrdU focus may represent two BrdU foci that are so close to each other that they cannot be resolved. A possible explanation for the progressively centralized localization of the BrdU foci is that after the origin regions are replicated and moved to opposite poles, the replicated origin-proximal DNA occupies the polar regions. This forces the DNA that is replicated next to be positioned closer to the center of the cell (see the schematic in Figure 6A). Finally, in late pre-divisional cells when DNA replication is completed (Figure 6A, 120 min), either no BrdU signal or a single BrdU focus at the division site was observed. This shows that the terminus region of the chromosome is positioned near the middle of the cell when the last part of the chromosome is replicated.
Fig. 6. Intracellular localization of DNA replicated at particular time points in the cell cycle determined using BrdU pulse-labeling. Synchronized Caulobacter cells were exposed to BrdU for 15 min prior to the indicated times (in minutes). Cells were then fixed and the sites of BrdU incorporation were visualized by indirect immunofluorescence microscopy using anti-BrdU antibodies. (A) DIC images (top panel), immunofluorescence images (middle panel) of the BrdU-labeled cells, and an overlay of these images (lower panel), where the BrdU immunofluorescence signals were pseudocolored red. The scale bar represents 2 µm. Black arrows indicate typical BrdU foci positions in cells at that stage of the cell cycle. White arrows indicate cells where a weak third BrdU focus is observed, probably at the same intracellular position as the replisome. The schematics show cell cycle progression, the intracellular position of the replisome (green dot) and the organization of the chromosome. Red lines indicate the position of DNA labeled with BrdU during the 15 min pulse-labeling, dark blue lines indicate unreplicated DNA and light blue lines indicate DNA replicated earlier in the cell cycle, prior to addition of BrdU. (B) Measurements of the average intracellular positions of the center of the BrdU foci relative to the cell pole (red circles) at the indicated time points. The schematic shows how the distances were measured. Red dots indicate the BrdU foci and a green dot indicates the replisome focus.
Two BrdU foci are observed during most of the cell cycle, indicating that daughter strands are moved away from the replisome in opposite directions shortly after replication. In some cells (labeled with white arrows in Figure 6A), an additional weak BrdU focus is present, probably at the position of the replisome, perhaps representing newly replicated DNA strands that have not yet been moved away from the replisome.
Discussion
To examine the organization of the replication apparatus in Caulobacter, we constructed GFP fusions to three different components of the replisome and examined their intracellular localization during the cell cycle in live cells. The holB-gfp, holC-yfp and dnaB-yfp strains express the fusion proteins as the only source of these replisome components. They are expressed from their normal promoter in the same chromosomal context as the normal genes, so they are expressed with the same cell cycle timing as the normal proteins. Since these proteins are essential for the functions of the DNA polymerase III complex, and no growth, cell cycle or DNA replication defects are observed, the proteins appear to be fully functional and therefore are expected to localize normally. Identical localization patterns were observed in cells expressing HolB–GFP, HolC–YFP and DnaB–YFP (Figure 2).
No replisome-component foci were observed after the completion of chromosome replication or when DNA replication is inhibited (Figures 2, 3 and 4). Since the replisome components are present at all times in the cell cycle (Figure 1C), the replisome focus most likely contains multiple replication proteins that become distributed throughout the cell in the absence of DNA replication. The intracellular concentrations of the replisome components in Caulobacter are not known, but the abundance is expected to be similar to that in Escherichia coli, where it is estimated that each cell has 10–20 molecules of the DNA polymerase holoenzyme and ∼140 molecules of the clamp loader complex (Wu et al., 1984; Leu et al., 2000). Both holB and holC encode components of the clamp loader complex, which loads the ring-shaped β-clamps onto DNA. Clamp loader complexes are associated with the DNA polymerase holoenzyme. The free clamp loader complexes unload β-clamps from the DNA, a process that takes place continuously on the lagging DNA strand (Leu et al., 2000). The DnaB helicase unwinds the DNA strands, thereby allowing the DNA polymerase holoenzyme to replicate DNA. Since most of the clamp loader complexes are expected to be located near the replication fork and DNA unwinding is coupled with DNA replication, the position of the HolB–GFP, HolC–YFP and DnaB–YFP fusion proteins most likely reflects the position of the replisome. Identical localization of clamp loader subunits and DNA helicase, proteins that are part of the DNA replication machinery but have different functions, supports the conclusion that the position of the fusion proteins reflects the location of the replisome in Caulobacter.
The majority of the cells with foci (98%) have only a single replisome focus, and when two foci are observed, they are always closely spaced (Figure 2). Thus, the two replication forks present during bidirectional replication of the chromosome must be located very close to each other during the entire DNA replication cycle. The close proximity of the two replication forks is easiest to explain if the replication machines of the two forks are associated with each other, forming a double replisome. Footprinting of the E.coli DnaB helicase binding to oriC shows that two helicases are positioned head to head during initiation of DNA replication (Fang et al., 1999). If the helicases remain together during replication of the bulk of the chromosome, they would keep the two replication forks close together. The archeal MCM helicase and the SV40 T-antigen helicase form double helicases that remain attached to each other (Wessel et al., 1992; Chong et al., 2000). Electron microscopy shows that two SV40 T-antigen helicase units remain joined in a single complex that reels in duplex DNA from opposite directions and extrudes two single-stranded DNA loops. Similar formation of a double helicase in Caulobacter or other interactions between components of the replication machinery could ensure the observed close proximity of the two replication forks. Linking the two replication forks together would result in the formation of a replication factory where unreplicated DNA is pulled into the complex and replicated daughter strands are extruded (Cook, 1999). Anchoring the replication apparatus to a cellular structure is not required to form a replication factory; the different polarity of the linked DNA polymerase complexes at the two replication forks would ensure that unreplicated DNA is pulled into the complex. The benefits of forming a DNA replication factory are that the energy released by the DNA replication process can be harnessed and used to power the movement of the bulk of the DNA to ensure proper chromosome segregation. Additionally, spooling of newly replicated DNA strands in opposite directions away from the replication factory reduces entanglement of the replicated sister chromosomes in the crowded nucleoid mass, making it easier to segregate the chromosomes properly (Lemon and Grossman, 1998; Losick and Shapiro, 1998; Cook, 1999).
If independent DNA polymerases track along the DNA instead of forming a replication factory, we would expect to see two replisome foci in the majority of the cells and not one as observed. Because of the limited resolution of light microscopy, two adjacent replisome foci might not be resolved and may therefore appear to be a single focus. However, the bacterial chromosome appears to breathe, similar to the constrained diffusional motion of chromosomes in eukaryotic interphase nuclei (Marshall et al., 1997; Roos et al., 2001). Therefore, if the DNA polymerase tracks along the DNA, two replication foci should separate sufficiently to form separable replisome foci with a much higher frequency than observed here. Thus, it would be unlikely to observe the replisome localization pattern described here unless the two replication forks are linked to each other in a replication factory.
Unlike the replication factories in eukaryotic cells and in B.subtilis, the Caulobacter replisome is not stationary. Time-lapse microscopy of HolB–GFP-expressing cells shows a gradual movement of the replisome from near the stalked pole to the middle of the cell (Figure 4). The movement is delayed when DNA replication is slowed down and is unaffected by inhibition of protein synthesis (Figure 5). Thus, the process of DNA replication itself, or the accumulation of replicated DNA, probably has a direct role in moving the replication factory. This is to be contrasted with the rapid movement of the newly replicated origin region to the opposite pole of the cell (Jensen and Shapiro, 1999a), which is probably moved by a separate and active process. Movement of the replisome in the absence of cell elongation, when cells are treated with chloramphenicol (Figure 5B), shows that zonal growth cannot be responsible for the movement of the replisome.
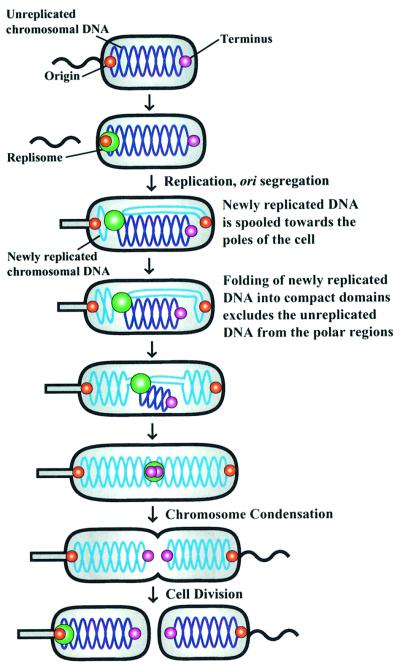
We propose that newly replicated DNA accumulating near the stalked pole displaces the replisome, thereby passively moving it from near the stalked pole to midcell (Figure 7). Newly replicated DNA strands are extruded bidirectionally from the replication factory towards both ends of the cell. Shortly after initiation of chromosome replication, the newly replicated origin-proximal parts of the chromosome are positioned at both cell poles. Refolding of newly replicated DNA into compact chromosomal domains may exclude other parts of the chromosome from the regions of the cell already occupied by the newly replicated DNA. As DNA replication progresses, the next portion of the DNA to be replicated is forced to positions closer to midcell because the origin-proximal regions of the chromosome have occupied the polar regions. The continual accumulation of replicated DNA near the cell poles could push the unreplicated DNA away from those regions, resulting in net movement of the unreplicated DNA towards the middle of the cell (Figure 7). The terminus region of the chromosome appears to move gradually towards midcell, as expected with this model (Jensen and Shapiro, 1999a). Since the replication factory is associated with the unreplicated part of the chromosome, exclusion from the cell poles consequentially displaces the replisome towards midcell, resulting in the passive movement of the replication factory.
Fig. 7. Model describing chromosome segregation and replication factory localization in Caulobacter. Swarmer cells do not replicate their DNA (dark blue lines) and replisome components are distributed throughout the cell. The origin of replication (red dot) is located at the flagellated pole of the cell and the terminus (purple dot) is at the opposite end of the cell (Jensen and Shapiro, 1999a). At the swarmer-to-stalked cell transition, the replication factory (green dot) assembles at the origin and DNA replication is initiated. As DNA replication proceeds, the newly replicated DNA strands (light blue lines) are moved towards both poles of the cell and the replication apparatus gradually migrates to midcell. This model proposes that the unreplicated part of the chromosome and the replication factory is passively displaced by the accumulating bulk of replicated DNA near the poles. When DNA replication is completed, the replication apparatus disassembles.
This model for replisome movement and chromosome segregation is consistent with the observed gradual replication-dependent movement of HolB–GFP. Further support was obtained by pulse-labeling synchronized cells with the thymine analog BrdU at different times during the cell cycle. Examining the location of BrdU-containing DNA provided a snapshot of the cellular location of the DNA replicated during the 15 min BrdU pulse-labeling time periods. Two BrdU foci are observed during replication of the bulk of the chromosome, showing that daughter DNA strands are bidirectionally extruded from the replication apparatus and moved in opposite directions, as assumed in the model. Figure 7 shows a schematic of the spatial deployment of unreplicated (dark blue) and newly replicated DNA (light blue) as derived from these BrdU incorporation studies. The earliest replicated DNA regions are moved towards the extreme poles of the cell. This is consistent with previous fluorescence in situ hybridization (FISH) results, showing bipolar localization of the origins in stalked cells (Jensen and Shapiro, 1999a). As replication progresses in the stalked and early pre-divisional cells, the DNA regions replicated during that part of the cell cycle are located at intermediate positions between the poles and midcell (Figures 6 and 7). The last part of the chromosome to be replicated, the terminus region, is positioned close to midcell when DNA replication is completed, in agreement with previous FISH results (Jensen and Shapiro, 1999a). Thus, newly replicated regions close to the origin on the genetic map are located close to the cell poles, newly replicated regions close to the terminus are located close to midcell, and regions in between the origin and the terminus on the genetic map are located at intermediate positions in the cell. Thus, BrdU pulse-labeling shows that positioning of replicating DNA in the cell is highly organized and the observed chromosome organization was included in our model describing chromosome segregation and replisome movement (Figure 7).
Both DNA polymerase and helicase appear to be powerful molecular motors, so the energy released during DNA replication may be harnessed for chromosome movement (Lemon and Grossman, 1998, 2000; Cook, 1999). The released energy could be used to pull the unreplicated DNA into the replication factory. Alternatively, the harnessed energy could be used for moving newly replicated DNA in opposite directions away from the replication factory, thereby providing the energy required for movement of the bulk of the DNA towards the poles. The structural maintenance of chromosomes (SMC) protein most likely has a role in organizing and folding newly replicated DNA, creating new compact chromosomal domains near the cell poles (Jensen and Shapiro, 1999a; Holmes and Cozzarelli, 2000). SMC-mediated condensation could pull newly replicated DNA towards the poles (Holmes and Cozzarelli, 2000; Sawitzke and Austin, 2001). Thus, spooling of the newly replicated DNA away from the replication factory and SMC-mediated pulling of DNA together could be responsible for segregation of the bulk of the chromosome.
It is not known how the origin regions of the chromosome are moved to the extreme poles after initiation of DNA replication. An as yet uncharacterized mitotic-like apparatus could be responsible for directed movement of the origin regions and/or the anchoring of these regions to the extreme cell poles. Thus, anchoring of the origins would provide the initial polarity to the chromosome segregation process. Subsequent spooling of replicated DNA away from the replication factory and organized refolding, perhaps mediated by SMC, could be responsible for segregating the rest of the chromosome.
The observed movement of the replication factory suggests that it is not anchored to any cellular structure in Caulobacter, as suggested for the replication factories in eukaryotic cells and B.subtilis (Cook, 1999; Falaschi, 2000; Lemon and Grossman, 2000; Sawitzke and Austin, 2001). This is supported by the observation of additional replisome foci when freely replicating plasmids are present in the cell. The additional foci probably originate from replisomes assembled on the plasmids when they are replicated. Some of the replisome foci in plasmid-containing cells are located at positions where no foci are observed in plasmid-free cells (Figure 2). Thus, replisome proteins can diffuse freely and assemble on DNA to replicate it anywhere in the cell. This implies that origin localization and movement determine the spatial organization of the replication factory.
Materials and methods
Strains and growth conditions
Caulobacter crescentus strain CB15N and derivatives were grown at 28°C in M2-glucose minimal medium (Ely, 1991). Synchronization was performed as described (Evinger and Agabian, 1977). The HolB–GFP, HolC–YFP and DnaB–YFP fusion strains were constructed as follows. The 3′-ends of the holB, holC or dnaB genes were PCR amplified using the primers HolB-1 (TATAAAGCTTAAGTTGCTGCCGACCATC) and HolB-2 (TATAGGATCCATCGCCGTCTTGACCGC), HolC-L (TATAGGATCCTGTGCCTGCTTCTCCCAACCC) and HolC-U (TATAAAGCTTCCCTGCGAGGTCTGGTTCTA), or DnaB-L (ATAGGATCC CTCGTCCGACGGAATATTCC) and DnaB-U (TATAAAGCTTCAGGTGGGCCTTGATCTCAT). The BamHI- and HindIII-digested PCR products and the BamHI–XbaI gfp fragment from pEGFP (Clontech) or yfp fragment from pEYFP (Clontech) were cloned into the integration vector pNPT228 (HindIII–XbaI). The resulting plasmids were integrated into the chromosome of CB15N by a single cross-over event. The integrity of the HolB–GFP-, HolC–YFP- or DnaB–YFP-expressing strains was confirmed using PCR, and western blotting show that a GFP fusion protein of the expected size was expressed. The pJS71 plasmid is a small high-copy-number cloning vector (Skerker and Shapiro, 2000). When relevant, cultures were treated with 100 µg/ml novobiocin, 3 mg/ml HU or 25 µg/ml chloramphenicol.
Measurement of DNA replication
DNA synthesis rates were measured by [α-32P]dCTP pulse-labeling as described (Marczynski and Shapiro, 1992). Samples for flow cytometry were prepared and data were analyzed as described (Winzeler and Shapiro, 1995).
Western blotting
Western blot analysis was performed as described (Domian et al., 1997). Samples were normalized so that equal amounts of total protein were loaded in all lanes. Mouse monoclonal anti-GFP antibodies (Covance) were used at a 1:500 dilution and horseradish peroxidase-conjugated sheep anti-mouse antibodies (Roche Molecular Biochemicals) were used at a 1:10 000 dilution. The Renaissance chemiluminescence kit (New England Nucleotides) was used for detection.
Live cell microscopy
Cells were immobilized using a thin layer of agarose as described (Webb et al., 1998). For time-lapse microscopy, isolated swarmer cells were placed on a pad of agarose containing M2-glucose medium. Nomarski differential interference contrast (DIC) and fluorescence microscopy images were taken on a Nikon E800 microscope with a 100× DIC objective and a 5 MHz Micromax 5600 cooled CCD camera controlled through Metamorph (Universal Imaging Corp.). Images were processed using Metamorph and Photoshop (Adobe). Distances were measured using Metamorph and at least 50 cells were measured for each time point; error bars in the graphs with positional data show the standard deviation.
BrdU labeling of newly replicated DNA
Synchronized cells at different stages of the cell cycle were pulse-labeled by adding 65 µM BrdU (Sigma). After 15 min of BrdU labeling, the cells were fixed using 3% formaldehyde. Immunofluorescence microscopy was then performed as described (Domian et al., 1997) with the following modifications: filtration was used to remove the fixation solution and cells were treated with 4 M HCl for 1 h and washed with phosphate-buffered saline before the blocking step. Mouse monoclonal anti-BrdU antibodies (Roche Molecular Biochemicals) were used at a 1:10 dilution and Fluorescein isothiocyanate-conjugated goat anti-mouse antibodies (Jackson ImmunoResearch) were used at a 1:200 dilution.
Supplementary data
Supplementary data for this paper (QuickTime movie) are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank K.Ryan, L.S.Kahng and A.Reisenauer for critical reading of the manuscript. R.B.J. was supported by post-doctoral fellowships from EMBO and the Carlsberg foundation. S.C.W. was supported by a pre-doctoral fellowship from National Science Foundation. This work was supported by National Institutes of Health grants GM32506/5120MZ and GM51426, and Office of Naval Research grant N00014-96-1-0564.
References
- Chong J.P., Hayashi,M.K., Simon,M.N., Xu,R.M. and Stillman,B. (2000) A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl Acad. Sci. USA, 97, 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P.R. (1999) The organization of replication and transcription. Science, 284, 1790–1795. [DOI] [PubMed] [Google Scholar]
- Davey M.J. and O’Donnell,M. (2000) Mechanisms of DNA replication. Curr. Opin. Chem. Biol., 4, 581–586. [DOI] [PubMed] [Google Scholar]
- Degnen S.T. and Newton,A. (1972) Dependence of cell division on the completion of chromosome replication in Caulobacter. J. Bacteriol., 110, 852–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingman C.W. (1974) Bidirectional chromosome replication: some topological considerations. J. Theor. Biol., 43, 187–195. [DOI] [PubMed] [Google Scholar]
- Domian I.J., Quon,K.C. and Shapiro,L. (1997) Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell, 90, 415–424. [DOI] [PubMed] [Google Scholar]
- Ely B. (1991) Genetics of Caulobacter crescentus. Methods Enzymol., 204, 372–384. [DOI] [PubMed] [Google Scholar]
- Evinger M. and Agabian,N. (1977) Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol., 132, 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaschi A. (2000) Eukaryotic DNA replication: a model for a fixed double replisome. Trends Genet., 16, 88–92. [DOI] [PubMed] [Google Scholar]
- Fang L., Davey,M.J. and O’Donnell,M. (1999) Replisome assembly at oriC, the replication origin of E.coli, reveals an explanation for initiation sites outside an origin. Mol. Cell, 4, 541–553. [DOI] [PubMed] [Google Scholar]
- Gordon G.S. and Wright,A. (2000) DNA segregation in bacteria. Annu. Rev. Microbiol., 54, 681–708. [DOI] [PubMed] [Google Scholar]
- Gordon G.S., Sitnikov,D., Webb,C.D., Teleman,A., Straight,A., Losick,R., Murray,A.W. and Wright,A. (1997) Chromosome and low copy plasmid segregation in E.coli: visual evidence for distinct mechanisms. Cell, 90, 1113–1121. [DOI] [PubMed] [Google Scholar]
- Hingorani M.M. and O’Donnell,M. (2000) Sliding clamps: a (tail)ored fit. Curr. Biol., 10, R25–29. [DOI] [PubMed] [Google Scholar]
- Holmes V.F. and Cozzarelli,N.R. (2000) Closing the ring: links between SMC proteins and chromosome partitioning, condensation and supercoiling. Proc. Natl Acad. Sci. USA, 97, 1322–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung D., McAdams,H. and Shapiro,L. (2000) Regulation of the Caulobacter cell cycle. In Brun,Y.V. and Shimkets,L.J. (eds), Prokaryotic Development. American Society for Microbiology, Washington, DC, pp. 361–377.
- Jensen R.B. and Shapiro,L. (1999a) The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl Acad. Sci. USA, 96, 10661–10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.B. and Shapiro,L. (1999b) Chromosome segregation during the prokaryotic cell division cycle. Curr. Opin. Cell Biol., 11, 726–731. [DOI] [PubMed] [Google Scholar]
- Kennedy B.K., Barbie,D.A., Classon,M., Dyson,N. and Harlow,E. (2000) Nuclear organization of DNA replication in primary mammalian cells. Genes Dev., 14, 2855–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A. and Baker,T.A. (1992) DNA Replication. Freeman, New York, NY.
- Lemon K.P. and Grossman,A.D. (1998) Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science, 282, 1516–1519. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman,A.D. (2000) Movement of replicating DNA through a stationary replisome. Mol. Cell, 6, 1321–1330. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Rahn,H.P., Weinzierl,P., Sporbert,A., Cremer,T., Zink,D. and Cardoso,M.C. (2000) Dynamics of DNA replication factories in living cells. J. Cell Biol., 149, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu F.P., Hingorani,M.M., Turner,J. and O’Donnell,M. (2000) The δ subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J. Biol. Chem., 275, 34609–34618. [DOI] [PubMed] [Google Scholar]
- Lewis P.J. and Errington,J. (1997) Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein. Mol. Microbiol., 25, 945–954. [DOI] [PubMed] [Google Scholar]
- Losick R. and Shapiro,L. (1998) Bringing the mountain to Mohammed. Science, 282, 1430–1431. [DOI] [PubMed] [Google Scholar]
- Marczynski G.T. and Shapiro,L. (1992) Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J. Mol. Biol., 226, 959–977. [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Straight,A., Marko,J.F., Swedlow,J., Dernburg,A., Belmont,A., Murray,A.W., Agard,D.A. and Sedat,J.W. (1997) Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol., 7, 930–939. [DOI] [PubMed] [Google Scholar]
- Niki H. and Hiraga,S. (1998) Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev., 12, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Yamaichi,Y. and Hiraga,S. (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev., 14, 212–223. [PMC free article] [PubMed] [Google Scholar]
- Pardoll D.M., Vogelstein,B. and Coffey,D.S. (1980) A fixed site of DNA replication in eucaryotic cells. Cell, 19, 527–536. [DOI] [PubMed] [Google Scholar]
- Pogliano J., Ho,T.Q., Zhong,Z. and Helinski,D.R. (2001) Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl Acad. Sci. USA, 98, 4486–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos M., Lingeman,R., Woldringh,C.L. and Nanninga,N. (2001) Experiments on movement of DNA regions in Escherichia coli evaluated by computer simulation. Biochimie, 83, 67–74. [DOI] [PubMed] [Google Scholar]
- Sawitzke J. and Austin,S. (2001) An analysis of the factory model for chromosome replication and segregation in bacteria. Mol. Microbiol., 40, 786–794. [DOI] [PubMed] [Google Scholar]
- Skerker J.M. and Shapiro,L. (2000) Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J., 19, 3223–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C.M. and Shapiro,L. (1993) An unusual promoter controls cell-cycle regulation and dependence on DNA replication of the Caulobacter fliLM early flagellar operon. Mol. Microbiol., 9, 1169–1179. [DOI] [PubMed] [Google Scholar]
- Teleman A.A., Graumann,P.L., Lin,D.C.H., Grossman,A.D. and Losick,R. (1998) Chromosome arrangement within a bacterium. Curr. Biol., 8, 1102–1109. [DOI] [PubMed] [Google Scholar]
- Webb C.D., Teleman,A., Gordon,S., Straight,A., Belmont,A., Lin,D.C., Grossman,A.D., Wright,A. and Losick,R. (1997) Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B.subtilis. Cell, 88, 667–674. [DOI] [PubMed] [Google Scholar]
- Webb C.D., Graumann,P.L., Kahana,J.A., Teleman,A.A., Silver,P.A. and Losick,R. (1998) Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol., 28, 883–892. [DOI] [PubMed] [Google Scholar]
- Wessel R., Schweizer,J. and Stahl,H. (1992) Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol., 66, 804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. and Shapiro,L. (1995) Use of flow cytometry to identify a Caulobacter 4.5 S RNA temperature-sensitive mutant defective in the cell cycle. J. Mol. Biol., 251, 346–365. [DOI] [PubMed] [Google Scholar]
- Wu Y.H., Franden,M.A., Hawker,J.R. and McHenry,C.S. (1984) Monoclonal antibodies specific for the α subunit of the Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem., 259, 12117–12122. [PubMed] [Google Scholar]