Abstract
The 2.6 Å resolution crystal structure of an inactive complex between yeast tRNAAsp and Escherichia coli aspartyl-tRNA synthetase reveals the molecular details of a tRNA-induced mechanism that controls the specificity of the reaction. The dimer is asymmetric, with only one of the two bound tRNAs entering the active site cleft of its subunit. However, the flipping loop, which controls the proper positioning of the amino acid substrate, acts as a lid and prevents the correct positioning of the terminal adenosine. The structure suggests that the acceptor stem regulates the loop movement through sugar phosphate backbone– protein interactions. Solution and cellular studies on mutant tRNAs confirm the crucial role of the tRNA three-dimensional structure versus a specific recognition of bases in the control mechanism.
Keywords: aminoacylation reaction/crystal structure/protein–RNA recognition/species specificity
Introduction
The quality of the translation of genetic information relies upon the fidelity of the aminoacylation reaction, i.e. the attachment of the correct amino acid to its tRNA by the cognate aminoacyl-tRNA synthetase (aaRS). Two main sources of error are possible, associated with inaccurate substrate recognition. The binding of the wrong amino acid is a rare event, except in the case of those that are stereochemically close, such as isoleucine and valine or threonine and serine and for which proofreading mechanisms exist. As for tRNAs, the situation is more complex since a given synthetase can bind different isoacceptor tRNA with similar affinities. Two main factors have been found to be responsible for the accuracy of the tRNA recognition process: the identity determinants that anchor the tRNA to the protein and the anti-determinants that prevent and disturb its positioning. The crystal structures of tRNA–synthetase complexes revealed that each identity determinant makes specific contact with the protein and that both the substrates and the protein undergo conformational changes for mutual adaptation.
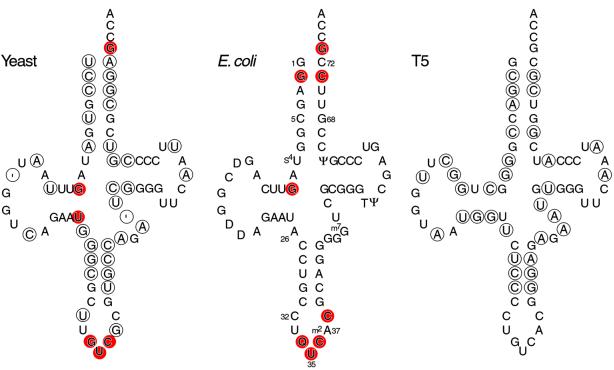
Yeast tRNAAsp harbors six identity elements (Figure 1), which are located in the anticodon loop (G34, U35 and C36), in the D stem (G10 and U25) and in the acceptor stem (the discriminator base G73). These nucleotides are necessary and sufficient elements for recognition by the cognate aaRS (Pütz et al., 1991; Nameki et al., 1992).
Fig. 1. Cloverleaf representation of wild-type tRNAAsp from yeast, E.coli and T5 phage. For the cellular experiments reported here, the anticodon loop of yeast tRNAAsp was modified to contain C32 and A37 rather than the wild-type U32 and G37 residues (see text). Standard position numbers are shown in the E.coli molecule. Circles show base positions in yeast and T5 tRNAs that differ from the respective base in E.coli tRNAAsp; the ′ symbol represents an alignment gap. Substitutions of red-shaded nucleotides in yeast or E.coli tRNAAsp diminish the kinetics of aminoacylation by homologous AspRS (Pütz et al., 1991; Nameki et al., 1992). Nucleotide modifications are shown for E.coli tRNAAsp (Q is modified G), although the modification states of tRNAs reported here were not investigated.
tRNA mischarging was used very early as a tool to understand the aminoacylation reaction. Biochemical studies revealed the kinetic nature of the control of the reaction and the importance of conformational changes as shown by the influence of the solvent on the mischarging capacities of a given system (Giegé et al., 1982). The involvement of the tRNA as an active partner in the process was first demonstrated by an experiment showing that a modified yeast tRNAPhe lacking the 5′ quarter of the molecule could no longer be aminoacylated by PheRS, but upon removal of the 7-methylguanine base at position 46 in this tRNA fragment, a significant aminoacylation could be restored (Renaud et al., 1979). To study the role of tRNA, the aspartic acid system presents an interesting peculiarity: while the yeast enzyme can charge both Escherichia coli and yeast tRNAs, the bacterial enzyme can only charge its cognate tRNA, although it binds both molecules with the same affinity. Since both tRNAs share many of the same identity elements (Figure 1), the E.coli enzyme is a good candidate for investigating the molecular mechanism that controls the activation process. In the aspartic acid system, we already know the molecular structure of both cognate complexes (Ruff et al., 1991; Cavarelli et al., 1994; Eiler et al., 1999). Here we present the crystal structure of the inactive E.coli AspRS–yeast tRNAAsp complex. The functional implications of our structural observations are corroborated by solution studies on mutant tRNAs. Biochemical and genetic approaches stress the role of the structure of the partner in the control of the specificity of the reaction.
Results
Solution studies
A comparison of E.coli and Saccharomyces cerevisiae (yeast) tRNAAsp sequences revealed the presence of different base pairs at the end of the acceptor helix: E.coli tRNAAsp contains G1–C72 and G2–C71, whereas yeast tRNAAsp contains U1–A72 and C2–G71 (Figure 1). The proximity of these residues to the 3′ acceptor end of yeast tRNAAsp may impair its aminoacylation by E.coli AspRS. We tested this possibility by in vitro aminoacylation after redesigning yeast tRNAAsp to contain the corresponding E.coli base pairs. Genes for three mutants were constructed so the transcribed molecules contained the following sequences: (i) yeast tRNAAsp with G1–C72; (ii) yeast tRNAAsp with G2–C71; and (iii) yeast tRNAAsp with G1–C72 and G2–C71. The mutant genes were inserted separately in a plasmid under the control of a strong constitutive lpp promoter and expressed in E.coli. Crude bacterial tRNA extracts were fractionated by high-performance liquid chromatography (HPLC) on a DEAE column. Pooled fractions of the column eluate were analyzed for overexpression by polyacrylamide gel electrophoresis and also tested for aspartic acid-accepting activity catalyzed by E.coli AspRS.
The G1–C72, G2–C71 mutant tRNAAsp was overexpressed, and its aspartic acid-accepting activity represented as much as 30% of the crude tRNA preparation. The G1–C72 mutant was overexpressed similarly, but it showed no aspartic acid acceptance in excess of the background activity from E.coli tRNAAsp. The other mutant, G2–C71, was not overexpressed. We also observed that wild-type yeast tRNAAsp was not overexpressed. Perhaps yeast tRNAs containing U1–A72 are not optimal sequence contexts for precursor RNA processing and/or tRNA stability in E.coli. We note that >80% of the E.coli-type tRNA genes start with a guanine residue at position 1.
We determined the kinetic parameters of E.coli AspRS attachment of aspartic acid to tRNAAsp for the E.coli and yeast tRNAAsp and the G1–C72, G2–C71 mutant tRNAAsp. This revealed that the kcat/Km of yeast wild-type tRNAAsp was reduced ∼4 × 103-fold relative to that of E.coli tRNAAsp. However, the corresponding reduction for the G1–C72, G2–C71 mutant was just 26-fold (Table I). Thus, redesigning the end of the acceptor helix of yeast tRNAAsp improved its kinetics of aspartic acid acceptance ∼150-fold.
Table I. Kinetic parameters of tRNA aminoacylation by E.coli AspRS.
| kcat/Km (µM–1) | Fold reduction | ||
|---|---|---|---|
| E.coli tRNAAsp | kcat = 9000 U | 1.5 × 1010 | 1 |
| Km = 0.6 × 10–6 M | |||
| Yeast tRNAAsp | kcat = 10–15 U | ∼4.2 × 106 | ∼4 × 103 |
| Km = 3 × 10–6 M | |||
| Yeast mutant tRNAAsp | kcat = 2000 U | 5.7 × 108 | 26 |
| G1–C72, G2–C71 | Km = 3.5 × 10–6 M |
Escherichia coli AspRS was purified to homogeneity as described (Eriani et al., 1990). The kcat values were calculated for saturating concentrations of aspartic acid. The kcat values are given in nmol/min/mg. To obtain yeast mutant G1–C72, G2–C71 tRNAAsp, its gene was expressed in E.coli from plasmid pBSTNAV1.
Cellular studies
We determined the functional capacity of mutant tRNAAsp in an E.coli tRNAAsp UVT knockout strain in which all three chromosomal aspartic acid-accepting tRNA genes are interrupted and rendered non-functional so that growth depends on a plasmid-encoded tRNAAsp gene. Two test systems were used to evaluate mutant activity in knockout cells. In the two-plasmid system, E.coli wild-type tRNAAsp was expressed from a regulated promoter in pGAD2 and a mutant tRNAAsp was expressed constitutively from a second plasmid, pSU81. When expression of the wild-type tRNAAsp gene is repressed by glucose, cell viability depends on the mutant tRNAAsp. Alternately, the one-plasmid system involved a plasmid switch in which pSU81 producing a mutant tRNAAsp was switched for a resident plasmid (pGAD2) producing wild-type tRNAAsp. Both test systems maintained the physiological balance (Swanson et al., 1988) of tRNAAsp, since the relative amount of mutant tRNAAsp (1.26 ± 0.19 SD) produced from pSU81 approximated that present in wild-type cells.
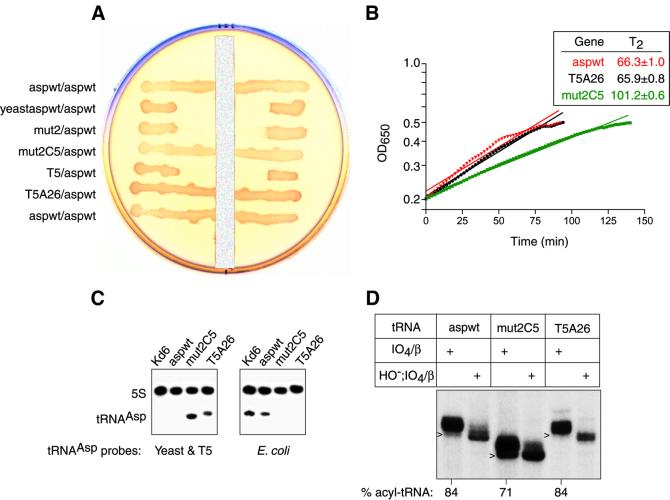
Synthetic yeast wild-type and mutant tRNAAsp genes were tested for their functional capacity in UVT knockout cells. All yeast tRNAAsp genes were designed so that the tRNA contains the E.coli C32 and A37 nucleotides in the anticodon loop in order to optimize the tRNA’s translation performance in E.coli (Yarus, 1982). The yeast wild-type tRNAAsp was inactive in UVT knockout cells (Figure 2A) as expected from the solution studies. Likewise, a double mutant of yeast tRNAAsp containing G1–C72 and G2–C71 (mutant 2) did not support knockout cell growth (Figure 2A and data not shown). The inactivity of mutant 2 is consistent with the solution studies that showed a 26-fold decrease in kcat/Km relative to E.coli wild-type tRNAAsp (Table I). In light of this modest kinetic decrease relative to the 4 × 103-fold decrease for the yeast wild-type molecule, we reasoned that activation of mutant 2 might be achieved by the substitution of a few additional E.coli bases. We focused on the U–G wobble pair at level 5 in the interior of the acceptor helix of yeast tRNAAsp as a potential target. This choice was made not only because the corresponding pair is C–G in E.coli tRNAAsp, but also because the wobble pair introduces structural differences relative to Watson–Crick pairs (Varani and McClain, 2000) that could affect the overall acceptor stem conformation and thereby influence the ability of residue A76 to enter the enzyme active site. We modified the yeast mutant 2 gene so that the tRNAAsp contains a C–G rather than a U–G wobble pair at level 5 (mutant 2C5). Remarkably, mutant 2C5 tRNA supported knockout cell growth (Figure 2A and B), although the growth rate was reduced. The steady-state levels of aspartyl-tRNA for mutant 2C5 and E.coli wild-type tRNAAsp were 71 and 84%, respectively (Figure 2D).
Fig. 2. Functional analyses in knockout cells. (A) Spread plate of UVT knockout cells carrying two plasmids with a vertical strip of 20% glucose. The plasmid–gene constructs are labeled as pSU81 tRNA gene/pGAD2 wild-type tRNAAsp gene. The tRNA genes in pSU81 are: E.coli wild-type tRNAAsp (aspwt); yeast wild-type tRNAAsp (yeastaspwt); yeast mutant 2 tRNAAsp (mut2); yeast mutant 2C5 tRNAAsp (mut2C5); T5 phage wild-type tRNAAsp (T5); and T5 phage mutant A26 tRNAAsp (T5A26). Cell growth in the zone of inhibition near the glucose strip depended on pSU81 producing a functional tRNAAsp since glucose repressed expression of E.coli wild-type tRNAAsp from pGAD2. (B) Plot of exponential growth phase of UVT knockout cells carrying only plasmid pSU81 expressing either aspwt, T5A26 or mut2C5. Determinations were made in LB medium plus chloramphenicol at 37°C using a microtiter plate reader with OD650 nm readings at 2 min intervals. The inset gives the doubling time T2 at the 95% confidence interval for the fitted line. Two representations are shown for each tRNA: raw data that are the average of five replicates between the OD650 nm range of 0.2 and 0.5, and a straight line fitted by linear regression to the raw data. The goodness of fit r2 value of each fitted line was >0.95. A Scheffé analysis indicated that the slopes of aspwt and T5A26 are not statistically different at the 95% confidence interval, whereas that of mut2C5 differs from the other two. On solid medium, both T5A26 and mut2C5 are heterogeneous and form smaller colonies than aspwt. (C) Northern blot analysis of wild-type Kd6 and knockout strains hybridized to the indicated tRNA and 5S RNA probes. The hybridization results confirm that mut2C5 and T5A26 UVT clones contain no wild-type E.coli tRNAAsp (right) and that the latter mutant tRNAs are made (left). (D) Steady-state level of aspartyl-tRNAAsp in UVT cells. Portions of acidified tRNA preparations were treated by periodate oxidation and β elimination to identify deacyl tRNAAsp (the N –1 length chains indicated by arrowheads) and aminoacyl-tRNA (the N length chains) on a sequencing gel. As multiple bands were observed reproducibly, we confirmed the accuracy of the determinations by an acid gel analysis of the tRNA preparations.
To probe further the structural basis of specificity, we looked for naturally occurring variability in tRNAAsp sequences. While the three isoacceptors of E.coli tRNAAsp contain identical sequences, that of T5 phage tRNAAsp differs from E.coli at 33 base positions and from yeast tRNAAsp at 35 positions (Figure 1). Our knowledge that phage-coded tRNAs function in protein synthesis in E.coli (Foss et al., 1979) prompted us to test T5 tRNAAsp in knockout cells. When we attempted to construct a T5 tRNAAsp gene by selection in UVT knockout cells, we obtained a mutant with a U to A substitution at position 26 (T5A26) in the transcribed molecule. The T5 wild-type tRNAAsp gene was isolated subsequently in wild-type E.coli cells. To our surprise, the cell growth rate in liquid media and steady-state level of aspartyl-tRNAAsp of T5A26 tRNAAsp were indistinguishable from those of E.coli wild-type tRNAAsp, whereas T5 wild-type tRNAAsp was inactive (Figure 2).
Structure determination and overview
Escherichia coli AspRS is an α2 dimer, each monomer containing 590 amino acids. AspRS was cloned, expressed and purified as described previously (Eriani et al., 1990; Boeglin et al., 1996). Yeast tRNAAsp was purified according to Dock et al. (1984). Crystals were grown from a solution containing AspRS, tRNAAsp, adenylyl[β,γ-methylen]-diphosphonat (AMP-PCP, an ATP analog) and aspartic acid with ammonium sulfate as precipitant. The structure of the complex was solved by molecular replacement using the Thermus thermophilus enzyme (Poterszman et al., 1994) as a model. The good quality of the electron density map at 2.6 Å resolution allowed us to build the complete model of the complex except the CCA end of one tRNA molecule and the last five residues of the protein. The final model comprises one AspRS dimer, two tRNA molecules, two aspartyl-adenylate substrates and 402 water molecules, and corresponds to an R-factor of 20.4% and an Rfree of 25.7% (Table II).
Table II. Data collection and refinement statistics for E.coli AspRS–yeast tRNAAsp–aspartyl-adenylate complex.
| Data set | |
| Resolution (Å) | 11.5–2.6 |
| No. of unique reflections | 72 094 |
| Redundancya | 2.3 (2.1) |
| Completeness (%)a | 92 (80) |
|
Rsym (I)a,b |
3.7 (21.6) |
| Refinement statistics | |
| R-factor (%) | 20.4 |
| No. of reflections in working set | 60 220 |
| Rfree (%) | 25.7 |
| No. of reflections in test set | 3084 |
| No. of water molecules | 402 |
| Average B-factor (Å2) | |
| monomer 1 | 44.4 |
| monomer 2 | 46.4 |
| R.m.s.d. | |
| bond lengths (Å) | 0.007 |
| bond angles (°) | 1.447 |
| dihedral (°) | 26.33 |
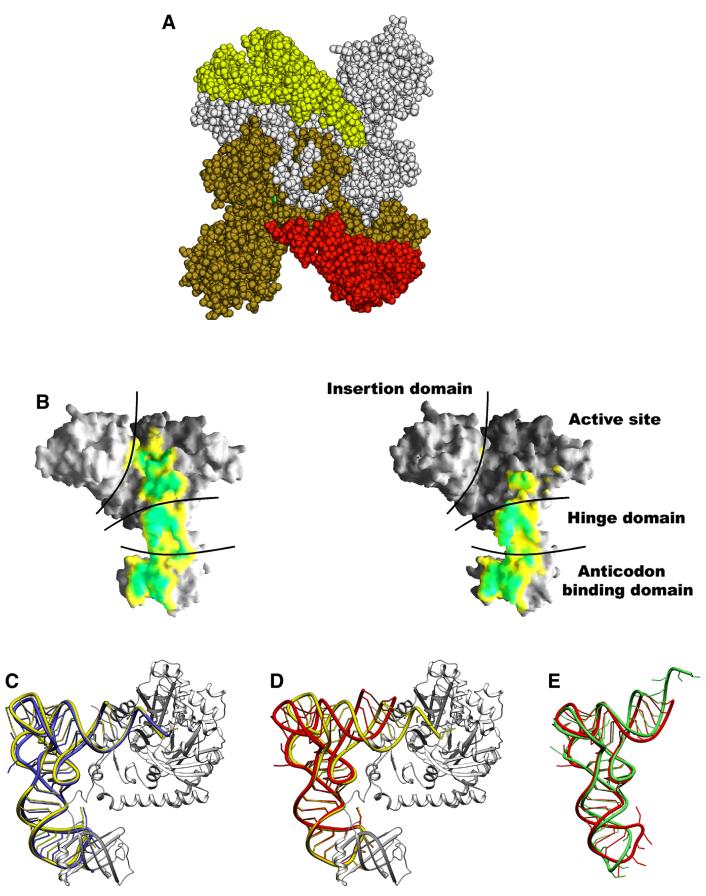
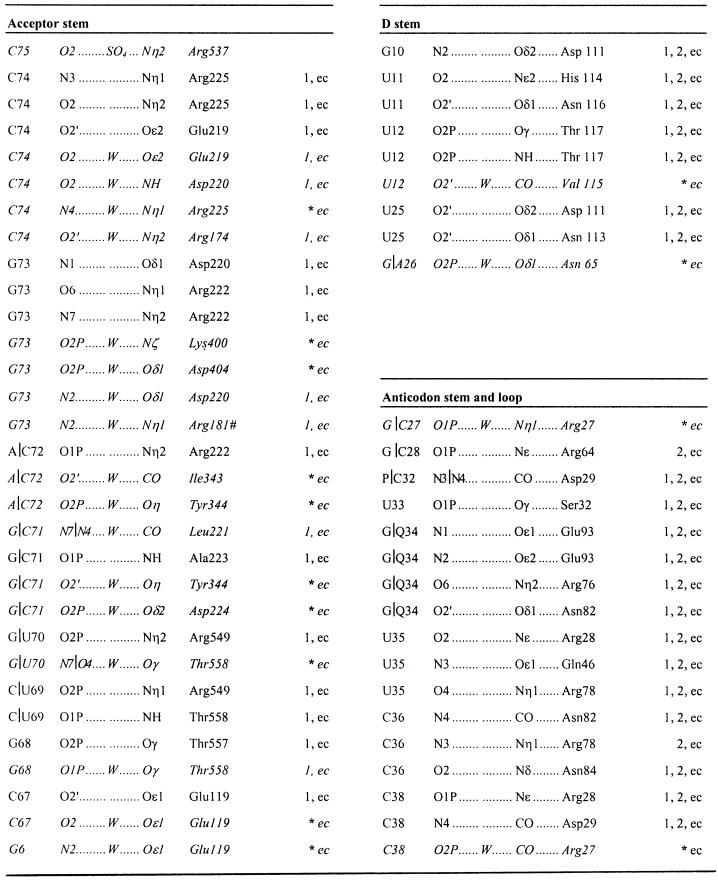
The dimeric heterologous E.coli AspRS–yeast tRNAAsp complex is shown in Figure 3A. While the two tRNA molecules are asymmetric, the structure of the enzyme is close to that observed in the homologous E.coli AspRS–tRNAAsp complex (Eiler et al., 1999). Each monomer is made of four modules. The catalytic domain and the C-terminal end of the protein form the core of the molecule that contains the three class II signature motifs. The N-terminal domain, responsible for the tRNA anticodon recognition, is connected to the catalytic core through a small hinge module. The last module, characteristic of eubacterial AspRSs, is inserted between motifs 2 and 3 of the catalytic domain; its function is not yet well understood.
Fig. 3. (A) General view of the dimeric aspartyl-tRNA synthetase from E.coli complexed with yeast tRNAAsp and aspartyl-adenylate. The tRNAAsp molecules, colored in red and yellow, are bound to one protein subunit shown in brown and white, respectively. (B) AspRS surface buried by the tRNA in monomer 1 (left) and monomer 2 (right) calculated and displayed using GRASP (Nicholls and Honig, 1991). The surface is colored according to a distance array between the two molecular surfaces: distances <2.5 Å between the tRNA and the enzyme are drawn in green, and distances between 2.5 and 3.5 Å are in yellow. The interaction surfaces are highly similar for the protein N-terminal domain in both monomers but vary through the rest of the complex. (C) Ribbon representation of one AspRS subunit in gray (monomer 1) of the heterologous complex with the bound yeast tRNAAsp in yellow. The E.coli tRNAAsp as seen in the cognate complex is drawn in blue after superposition of the enzymes on their active sites. (D) Relative position of the two tRNAs of the heterologous complex. Superposition was optimized on the two subunit active sites. The tRNAAsp from monomer 1 is drawn in yellow, the tRNA from monomer 2 in red. (E) Comparison of the tRNA molecule from monomer 2 of the heterologous complex (red) and the free (uncomplexed) yeast tRNAAsp (green) (Moras et al., 1980). Figures 3–5 were generated using the Program SETOR (Evans, 1998).
Comparison of subunits 1 and 2
In order to compare the two monomers of the asymmetric unit, the AspRS catalytic core domains have been superposed. The main difference between the two monomers consists of a rigid body displacement of the eubacterial insertion domain: a rotation of ∼5° is required to superpose them. Part (Leu 110–Ala 120) of the hinge domain and a few loops, located in the catalytic core, exhibit some conformational differences with displacements of up to 2 Å. These residues are involved mainly in tRNA recognition. In contrast, the N-terminal anticodon-binding domains are perfectly superposed, with an r.m.s. displacement of 0.3 Å for the Cα atoms, which is less than half that of the complete protein.
Each protein monomer binds one tRNA molecule with no cross-subunit interactions. Whereas both tRNAs have their anticodon stem–loop bound to the protein N-terminal domain, only one tRNA molecule interacts through its amino acceptor stem with the synthetase catalytic core. This asymmetry is highlighted in Figure 3B, which shows a surface representation from the footprint of the tRNA on AspRS. In the first monomer, 17% (2250 Å2) of the tRNAAsp solvent-accessible surface is buried, while in the second subunit only 10% (1300 Å2) of the surface is buried. The most striking feature is the large conformational difference between the two tRNA molecules. While the structure of subunit 1 tRNA resembles that of the E.coli tRNA in the homologous complex (Eiler et al., 1999) with an angle of 95° between the two arms of the L-shaped molecule (Figure 3C), the tRNA from subunit 2 resembles the free yeast tRNAAsp (Moras et al., 1980) with an angle of 110° (Figure 3E). The two helical arms are held together at the elbow through tertiary interactions between the D and T stem that are maintained in both molecules. The conformation of the anticodon loop and stem of tRNAAsp is almost identical in both monomers (r.m.s.d. of 17 superposed phosphates is 0.4 Å).
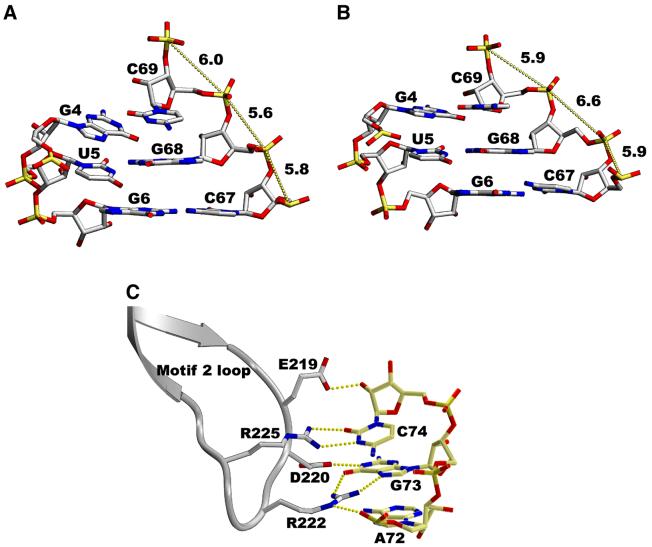
Protein-induced conformational change of the tRNA acceptor stem can be evidenced in particular at the wooble U5–G68 base pair. Binding of the tRNA introduces a series of rotations in the backbone that result in the displacement of phosphate of C67 (Figure 4A). The backbone dihedral angles α and γ of G68 in the second monomer (the ‘unbound’ tRNA) are trans/trans, while they are gauche–/gauche+, a regular RNA conformation, in the first one (Figure 4A). This conformation is stabilized by water molecules in a geometry similar to that observed in the homologous E.coli complex.
Fig. 4. Yeast tRNAAsp acceptor stem in monomer 1 (A) and monomer 2 (B) showing the U–G mismatches. The tRNA acceptor stem in monomer 1 is bound to the enzymes and shows a regular RNA conformation for the backbone dihedral angles α and γ (gauche–/gauche+); the distance between the phosphorus atoms of G68 and C69 is 5.6 Å. The tRNA molecule in monomer 2 shows no contact between the acceptor stem and the protein. As a consequence, the dihedral angles α and γ are trans/trans for G68, and the distance between the phosphorus atoms of G68 and C69 is 6.6 Å. ‘Accommodation’ of U–G mismatches has already been observed for the yeast AspRS–tRNAAsp complex (equivalent to tRNA 1) and for the free tRNA (equivalent to tRNA 2). (C) Recognition of the discriminator base G73 of yeast tRNAAsp by the E.coli AspRS. The hydrogen bonds between the protein and the nucleic acid are shown as yellow dotted lines. They are similar to those observed in the homologous E.coli AspRS–tRNAAsp complex.
Comparison with the E.coli cognate complex
We will focus our analysis on the first monomer, i.e. that with the tRNA acceptor stem bound to the catalytic core of the enzyme, and compare it with the homologous E.coli complex. In order to do so, the catalytic domains from both complexes have been superposed. Rigid body rotations of 4 and 6° are required to superpose the N-terminal and the eubacterial insertion domains, respectively. The overall r.m.s. distance between both enzymes is 0.95 Å for 585 Cα (r.m.s.d. of 0.59 Å when rigid body rotations are applied to the N-terminal and the insertion domains) and 1.23 Å for 65 phosphate atoms common to yeast and E.coli tRNAAsp. The D and variable loops, being of different length in the two tRNAs, have been excluded for the calculation of the r.m.s.d.
The binding of the anticodon stem–loop of the tRNA to the N-terminal β-barrel domain of AspRS is very similar to that observed in the E.coli complex (Table III). In both structures, the five bases 33–37 are splayed out towards the exterior of the anticodon loop. The loop conformation is stabilized by intra-backbone hydrogen bonds (2′OH U35–O4′ C38 and 2′OH C38–O2P C36). Base-specific interactions are restricted almost exclusively to the three anticodon bases which are essential recognition elements of AspRS. The hydrogen bonding pattern is identical in both complexes. One difference between the two organisms concerns base 37. This nucleotide, involved in the tight bulge of the loop, is an m2A37 in E.coli and an m1G37 in yeast tRNA. When the tRNA is bound to the E.coli AspRS, no interaction is observed between this base, which is completely exposed to the solvent, and the enzyme or tRNA. In contrast, when the tRNA is bound to the yeast enzyme, base 37 is tilted slightly and the N2 atom of m1G37 interacts with the phosphate group of U25.
Table III. Direct and water-mediated hydrogen bond interactions in the E.coli AspRS–yeast tRNAAsp–aspartyl-adenylate complex.
Interactions that occur between the tRNA and the protein in the first monomer from the heterologous complex are labeled ‘1’, those present in the second monomer ‘2’ and the hydrogen bonds observed in the homologous E.coli AspRS–E.coli tRNAAsp–aspartyl-adenylate complex are indicated by ‘ec’. Arg181 belongs to the second monomer. The protein anchors the anticodon loop of the tRNA through direct interactions while the acceptor stem binding involves both direct and water-mediated hydrogen bonds. An asterisk indicates that the missing bonds in monomer 1 correspond to water-mediated interactions in E.coli. The corresponding water molecules are not visible in the electron density map of the heterologous complex. However, the tRNA and the synthetase parts are located at the same position. Thus, the absence of the water molecules is most likely to be the consequence of the lower resolution of the diffraction data.
At the elbow of the L-shaped tRNA molecule, the small hinge domain (residues 108–131) contacts the D-stem (Table III). Base-specific hydrogen bonds occur with G10 (a minor identity element) and U11, while ribose and phosphate backbone interactions involve U12 and U25. One additional hydrogen bond between Glu119 and nucleotide C67 of the acceptor stem helps to anchors the tRNA acceptor arm and is only observed in monomer 1. When comparing the first monomer with the second one, a rigid body displacement of ∼1.5 Å is observed for the hinge module. This rigid body movement is propagated to the tRNA and all interactions but one are conserved in the second monomer. The missing hydrogen bond is the one that involves C67 of the acceptor stem, a direct consequence of the open conformation of the two arms of the L-shaped tRNA molecule of the second monomer.
The tRNA acceptor stem of monomer 1 comes into contact with the catalytic core domain and forms extensive interactions with the protein (Figure 3) in a recognition pattern almost identical to that observed in the homologous E.coli complex. The contacts occur all along the double-stranded acceptor stem with the tRNA sugar– phosphate backbone. In contrast, the discriminator base G73 and nucleotide C74 are involved in base-specific interactions with residues from the motif 2 loop (Figure 4C). The structures of the homologous yeast and E.coli complexes have shown two different recognition patterns of the tRNAs acceptor stem. In yeast, the protein–RNA interactions are concentrated at the acceptor end of the stem, while in E.coli they occur all along the double-stranded stem. These features are characteristic of eukaryotes and eubacteria, respectively, as deduced from sequence alignments. Yeast tRNAAsp bound to the E.coli AspRS adopts a eubacterial recognition scheme which induces conformational changes to the acceptor stem. As a result, the structure of yeast tRNAAsp in the present complex deviates from that found in the complex with its own native enzyme and resembles more the E.coli tRNA in its native complex. The r.m.s.d. between the heterologous and the yeast complex for the acceptor stem is 0.85 Å and the r.m.s.d. between the heterologous and the E.coli complex is 0.55 Å.
About 25% of water molecules are conserved between the present complex and the homologous E.coli complex. These are located mainly at the interface of the dimeric enzyme and between the N-terminal anticodon-binding domain of one subunit and the catalytic domain of the other. Almost all water molecules of the active site that involve the tRNA acceptor stem, the motif 2 loop, one extremity of the flipping loop and residues of the second subunit are conserved between the two complexes. The flipping loop adopts an open conformation in the E.coli complex while it is closed in the heterologous complex. The footprint left at the ‘open position’ of this loop is fully occupied by water molecules in the present complex. Finally, we should mention that almost no water is present between the eubacterial insertion domain and the tRNA acceptor stem, in contrast to the layer of water molecules observed in the cognate E.coli complex.
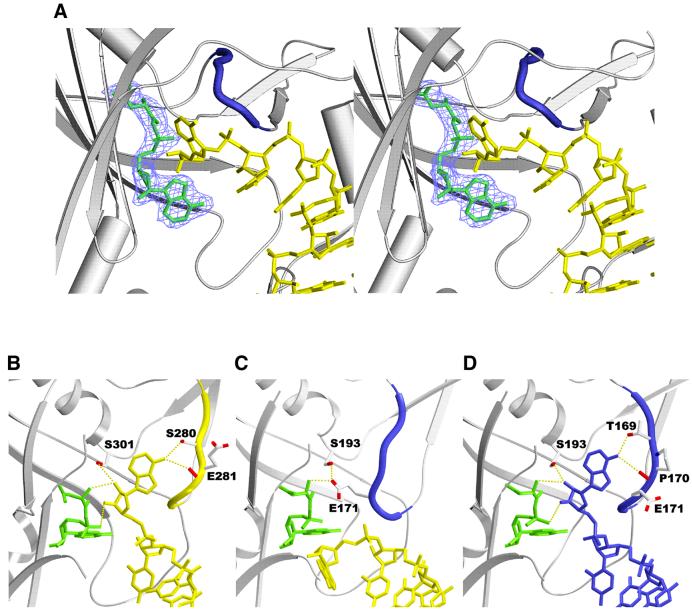
The active site pocket
The electron density map shows unambiguously the density for an aspartyl-adenylate molecule formed in the crystal (Materials and methods; Figure 5A). The overall recognition pattern of interactions is similar to that observed in the homologous complex. The adenylate substrates superpose with an r.m.s.d. of 0.16 Å (0.3 Å when the superposition is optimized for the β-sheet of the AspRS catalytic domain). At the tRNA acceptor end, the terminal adenosine is positioned at the entry of the catalytic site, siting on the adenylate molecule and surrounded by loops (the flipping loop and the histidine loop). Compared with the yeast (Figure 5B) or E.coli (Figure 5D) homologous active complexes, A76 is rotated by 110° around the backbone dihedral angle ζ (defined by C3′–O3′–P–O5′) and points outwards from the active site (Figure 5C). The terminal adenosine is stabilized by hydrogen bonds with Met447 and Glu482, while in the cognate E.coli complex A76 hydrogen binds to Thr169, Pro170 (residues located in the flipping loop) and the amino group of the aspartyl-adenylate. As for the protein conformation, the main difference concerns the flipping loop (residues 167–174), located immediately after motif 1. In the active complexes (Figure 5B and D), A76 is inserted between the aspartyl-adenylate and the flipping loop, which adopts the so-called ‘open’ conformation. Also, the 3′ oxygen atom of A76 sits above the carbonyl carbon of the aspartyl-adenylate at the proper distance for interaction. In the present complex (Figure 5C), the flipping loop is in a ‘closed’ conformation as in the binary AspRS–aspartyl-adenylate complex, maintaining the hydrogen bond between the aspartic moiety of the adenylate and Glu171 (Schmitt et al., 1998). A76 positioned at the opposite side of the substrate cannot react. The flipping loop acts as a door that only opens when the right tRNA is bound to the synthetase.
Fig. 5. (A) Stereoview of the active site area of E.coli AspRS with bound yeast tRNAAsp and aspartyl-adenylate. The (2Fo – Fc) electron density map contoured at 1σ shows the aspartyl-adenylate substrate sitting on the antiparallel β-sheet of the catalytic domain. The flipping loop (residues 167–174 colored in blue) adopts a closed conformation. The contribution of this loop is essential to the binding of the aspartyl-adenylate and tRNA terminal adenosine substrates. (B–D) Conformation of the flipping loop and the tRNA terminal adenosine in (B) the yeast AspRS–yeast tRNAAsp complex, (C) the heterologous E.coli AspRS–yeast tRNAAsp complex and (D) the E.coli AspRS–E.coli tRNAAsp complex. The aspartyl-adenylate substrate is present in all three complexes.
Discussion
The present structure between E.coli AspRS and yeast tRNAAsp is that of an inactive heterologous complex. In the E.coli and yeast homologous complexes, both tRNA molecules are bound to their respective dimeric enzyme in a symmetrical manner, with A76 oriented such that the transfer of the aspartic acid on the 3′OH of the ribose is possible. In the heterologous complex, the tRNA molecule in monomer 1 is bound properly to the protein, except for A76, which points away from its catalytic site. The situation in monomer 2, where the tRNA molecule interacts with the enzyme only through its anticodon stem–loop and D stem, is reminiscent of that observed in the T.thermophilus complex (Briand et al., 2000). In the case of T.thermophilus, the temperature of crystallization, far below the optimal one, led to the stabilization of an inactive complex where the tRNA molecule is frozen at the entrance of the active site. The correlation of these two independent observations suggests a sequential order of binding, monomer 2 being in an intermediate position awaiting further clearance to proceed.
Two main points differentiate the active sites of monomer 1 of the inactive heterologous complex from that of a monomer in an active homologous complex of E.coli. These are the positions of the terminal adenosine and the conformation of the flipping loop located immediately after motif 1 (residues 167–173). The position of this flipping loop depends on the nature of the substrates bound to the enzyme; its role has already been discussed (Schmitt et al., 1998; Eiler et al., 1999). In apo AspRS, the loop adopts an open conformation allowing the aspartic acid to reach its binding site. When properly positioned, the amino acid is maintained by the loop flipping over and covering it in the pocket. The reopening of the active site through a lift of the flipping loop takes place when the cognate tRNA is bound to the enzyme. The loop is then responsible for the recognition and correct positioning of the terminal adenine. A76 intercalates between the aspartyl-adenylate substrate and the flipping loop in a position that allows the transfer of aspartic acid to its ribose. In the present heterologous complex, the flipping loop is closed and A76 points in a direction opposite to the active site. However, no particular contact hinders the movements of the flipping loop or A76 in this structure, suggesting that the triggering signal could be transmitted either through intersubunit protein contacts and/or through the tRNAs.
Kinetic analyses are in agreement with the proposal that AspRS monomer subunits are not functionally independent. For example, there are two affinity constants for ATP binding (Kern et al., 1985). In addition, the interdependence of the monomer subunits has clearly been established by the mutation of an invariant proline in signature motif 1 of class II aaRS (Eriani et al., 1993). The present crystal structure and those of the active homologous yeast and E.coli complexes show extensive interactions between the active site domain of one monomer and the anticodon-binding region of the other monomer. These interactions could channel a recognition message from one monomer to the other.
An analysis of tRNAAsp primary sequences reveals many differences in the acceptor stem. The first two base pairs are purine–pyrimidine in E.coli while they are pyrimidine–purine in yeast. Moreover, the second base pair of the acceptor stem is a minor identity element in E.coli tRNAAsp (Nameki et al., 1992). When the two first base pairs of yeast tRNAAsp are changed for those from E.coli (U1–A72→G1–C72 and C2–G71→G2–C71), then the system gains substantial activity in vitro (the kcat of aminoacylation relative to E.coli improves from 900 with yeast tRNA to 4.5 with the mutant tRNA). However, the double-stranded part of the acceptor stem shows no base-specific interactions with the enzyme, even for the first two base pairs. When complexed with E.coli AspRS, the tRNAs from both yeast and E.coli exhibit a similar overall conformation, except for the wobble base pairs in the acceptor stem, which display local shifts. An explanation for the gain of activity with the mutant tRNA could be that the G–C stacking effect of the two E.coli base pairs favors a more pronounced helical bend in the mutant tRNA that results in a better positioning of the acceptor end in the active site.
The role of the tRNA via its structure in the control of the fidelity of the reaction is confirmed further by our cellular studies. A cellular analysis of tRNAAsp structure and function is notable because it is conducted under conditions where the aspartylation reaction reflects the net outcome of 20 aaRS competing for the mutant tRNA. The tRNA not only must be a good substrate for its cognate AspRS synthetase, but also a poor substrate for non-cognate synthetase enzymes to avoid matching the wrong amino acid to the tRNA. This approach allows us to refine our analysis and assess the biological relevance of the structural differences. As expected, the yeast wild-type tRNAAsp is inactive in UVT knockout cells, but so is the double mutant 2 with E.coli base pairs in the first two positions of the acceptor stem. A plausible explanation for this inactivity could be that the kinetic decrease, although very modest compared with that of the wild-type E.coli tRNAAsp, is still too large (26-fold in kcat/Km) for the cellular competition. This confirms the subtlety of the molecular mechanism of recognition.
Two tRNA mutants strengthen the structural hypothesis. The structural analysis clearly shows the importance of the U–G wobble pair in the adaptability of the acceptor stem (Figure 4A and B). The functional improvement observed with mutant 2C5 most probably correlates with the stabilization of the A-form RNA g–/g+ conformation provided by the canonical C–G base pair. The T5 phage tRNAAsp mutant with a change at position 26 may also be enlightening. In the E.coli homologous AspRS–tRNAAsp complex, nucleotide 26 interacts with the anticodon-binding domain of AspRS whereas the adjacent tRNA nucleotide 25 interacts with the hinge domain whose additional interactions fix the angle between the anticodon helix and acceptor helix of the tRNA. This series of interactions may be part of a long-range functional communication or coupling pathway between the anticodon and residue A76 that is critical for aminoacylation.
The predominance of tRNA backbone-mediated interactions observed with the acceptor stem, the D stem and anticodon stem in the E.coli homologous complex suggest that much of the corresponding wild-type tRNA base sequence should be replaceable by alternative sequences so long as they preserve the initial structure. A corollary of structure-based recognition is that the smallest unit of function is a group of nucleotides rather than a single nucleotide or base pair. An aminoacylation specificity that depends on molecular structure rather than base sequence can explain why molecules as dissimilar as E.coli tRNAAsp and particular mutants of yeast and T5 tRNAAsp can be functionally active.
What can be said about the mechanism that controls aminoacylation? The influence of intra- and/or intermonomer subunit communication on aminoacylation is discussed above. In addition, for monomer 1 and the active complexes, the structural data identify in the tRNA a continuous patch of interactions with the anticodon-binding domain, the hinge domain and the acceptor stem-binding domain. In monomer 2 and other inactive complexes, the acceptor stem interaction is missing. One way to think about this system is to consider a zipper, in a hierarchical sense. In order to have an active complex, interactions first form with the anticodon domain, then the hinge domain, then the acceptor stem beginning around residues 67 and 68, then the rest of the acceptor end, with finally A76 in the active site. This dynamic process emphasizes the importance of the fine tuning of the tRNA conformation for the proper positioning of A76. A mutual adaptation of tRNA and enzyme through an induced fit mechanism would allow A76 to enter the active site. Whether this scenario of interactions occurs in a temporally sequential sense is not proven, but it may be a useful way to think about the system.
Materials and methods
Construction of the synthetic genes coding for yeast tRNAAsp and variants
The gene encoding yeast tRNAAsp was assembled from four complementary pairs of synthetic oligonucleotides. The resulting EcoRI–PstI fragment was cloned between the same restriction sites of plasmid pBSTNAV-1, behind the strong constitutive lpp promoter (Meinnel et al., 1988). Strain TB1 {F– ara _(lac-proAB) hsdR (rk– mk+) rpsL(Strr) [_80, dlac_(lacZ)M15]} was used as recipient for DNA transformation and tRNA extraction.
Overproduction and purification of the overproduced tRNA
Cells overproducing tRNA were grown in LB medium containing 100 µg/ml ampicillin. Bacterial cells were harvested by centrifugation, washed with sterile water, resuspended in 150 ml of 20 mM Tris–HCl pH 7.5, 20 mM magnesium acetate, and then extracted by an equal volume of phenol saturated with 200 mM Tris–HCl pH 8; after 20 min of vigorous shaking at room temperature, the aqueous phase was collected by low-speed centrifugation. This phase, containing small RNAs, was extracted again under the same conditions, ethanol precipitated, resuspended in 20 ml of 500 mM Tris–HCl pH 8.8, and incubated at 37°C for 45 min in order to deacylate the extracted tRNAs. Finally, this solution was neutralized by addition of 4 ml of 1 M sodium acetate pH 5.1, and the tRNA recovered by ethanol precipitation.
The tRNA pellet obtained after high speed centrifugation (15 min at 12 000 g) was dried, redissolved in 4 ml of 50 mM Tris–HCl pH 7.5, 258 mM NaCl, 10% methanol, and was fractionated on a Spherogel DEAE-TSK 2 SW HPLC column using an isocratic elution mode at pH 7.5 (Martin et al., 1993). The elution profile was analyzed by aminoacylation reaction and denaturing polyacrylamide gel electrophoresis.
Enzyme assays
The aminoacylation assays were performed in the following reaction mixture: 50 mM Tris–HCl pH 7.5, 30 mM KCl, 0.1 mg/ml bovine serum albumin, 5 mM glutathione, 10 mM ATP, 0.25 mM l-[14C]aspartic acid (220 Ci/mol), 15 mM MgCl2 and 20 µM pure tRNAAsp. The reaction was initiated by the addition of enzyme and conducted at 37°C. At varying time intervals (usually 1 min), aliquots of 40 µl were spotted onto Whatman 3MM discs, which finally were measured for radioactivity. For tRNAAsp Km measurements, the substrate concentrations were varied from 0.5 to 4 Km values. We did not detect any variation of affinity of aspartic acid and ATP with the different AspRS–tRNA complexes. Their values are: for aspartic acid 60 µM, for Km ATP = 60 µM.
Knockout cell analysis
For the spread plate analysis, pSU81 gene constructs were transformed separately into UVT cells (McClain and Gabriel, 2001) harboring pGAD2 containing wild-type tRNAAsp. The transformed cells were plated on Min A arabinose agar plates containing ampicillin to select for pGAD2 and chloramphenicol to select for pSU81. After incubating for 20 h at 37°C, a single colony was resuspended in 75 µl of Min A and a 6 µl aliquot was spread across a Min A arabinose agar plate. A 3MM filter paper strip saturated with 20% glucose was laid in the center of the plate and the plate was incubated overnight at 37°C (Gabriel and McClain, 2001).
Single-plasmid UVT strains maintained by mutant tRNAs were generated as follows. UVT/pGAD2::Aspwt cells were grown overnight at 37°C in Min A arabinose medium. The cells were pelleted, washed in cold sterile water and electroporated to lose much of the pGAD2::Aspwt resident plasmid. After recovering for 20 min in SOC broth at 37°C, they were pelleted and washed as before. Plasmid pSU81 carrying a gene for either wild-type or mutant tRNAAsp was electroporated into the partially cured UVT competent cells, which again were allowed to recover in SOC media. Cells were subcultured overnight in LB broth containing chloramphenicol and 0.1% glucose to select for pSU81 and to repress expression of wild-type tRNAAsp from any residual pGAD2 plasmid. After streaking on LB agar, individual UVT colonies were tested for the loss of ampicillin resistance (indicating loss of plasmid pGAD2::Aspwt), for the inserted gene sequence and for the absence of wild-type E.coli tRNAAsp.
For both northern blot and aminoacylation analyses, cells were grown to 0.5 OD550 nm in rich media, pelleted, resuspended in 0.3 M NaOAc pH 5, extracted with acid phenol and precipitated with ethanol. Northern blot samples were electrophoresed on a 10% polyacrylamide, 7 M urea minigel and electroblotted onto Nytran SuperCharge membrane in 1× TAE. Membranes were hybridized overnight at 37°C in Sigma PerfectHyb™ Plus buffer to 5′-32P-labeled probes. The probes were complementary to the following residue positions: 5S RNA, 34–53; aspwt tRNA, 30–46; yeast wild-type and mutant tRNA, 29–45; and T5 A26 tRNA, 29–46. The steady-state level of aspartyl-tRNA was determined by both acid gel analysis and periodate oxidation and β elimination as described (Chang et al., 1999). The prepared acidified samples were electroblotted and hybridized as above. Quantitations of the steady-state level of aspartyl-tRNAAsp in UVT cells used a Molecular Dynamics Storm 860 instrument and ImageQuaNT 4.2a software. The percentage aspartyl-tRNAAsp for each sample was calculated as {100 × [N/(N + N – 1)]}.
Crystallization and data collection
Crystals were grown using the hanging drop method from a solution buffered at pH 6.7 containing AspRS, tRNAAsp, AMPPCP and aspartic acid, using ammonium sulfate as precipitant (Boeglin et al., 1996). Single crystals suitable for X-ray analysis were obtained by macroseeding and grown for a few weeks. Although AMPPCP was used to slow down the reaction, the time required for crystal growth is long enough for the reaction to take place. This is confirmed by the observation of aspartyl-adenylate in the crystals. The crystals belong to space group P21 with unit cell dimensions a = 75.8 Å, b = 222.8 Å, c = 80.8 Å and β = 111.8°. For data collection, the crystals were quickly transferred to a solution containing 20% glycerol (as well as 2.2 M ammonium sulfate and BisTrisPropane buffer pH 6.7) for 30 s before flash cooling in a nitrogen gas stream. A native data set has been collected with a single crystal using synchrotron radiation from DESY (Hambourg, Germany). Diffraction images (183 frames of 0.4° oscillation and 66 frames of 0.6° with a crystal–image plate distance of 400 mm) were analyzed with the MARXDS program (Kabsch, 1988) and the data processed further using programs from the Collaborative Computational Project No. 4 (CCP4, 1994). The merging R-value for all measurements was 3.7% (Table II).
Structure determination and refinement
The structure was solved by the molecular replacement method using the T.thermophilus AspRS as search model (Poterszman et al., 1994). The E.coli and T.thermophilus enzymes share 50% sequence identity. A self-rotation function was computed using the program POLARRFN (CCP4, 1994; W.Kabsch), the cross-rotation using the program ALMN (Dodson, 1985) and the translation function with the TSFGEN program (CCP4, 1994). Two solutions related by a 2-fold axis and consistent with the self-rotation function were obtained. This axis corresponds to the dimer axis and thereby indicates that the asymmetric unit comprises one dimer. Two yeast tRNAAsp molecules were added to the enzyme using the yeast AspRS–tRNAAsp complex as reference. This new model, correctly oriented and translated, was then submitted to rigid body refinement using FROG (Urzhumtsev et al., 1989). At this point, the crystallographic R-factor was 40.3% with data between 10 and 3.5 Å. The model was then improved further by several rounds of model building using the program O (Jones et al., 1991) and simulated annealing with the program CNS (Brünger et al., 1998). Towards the end, cartesian coordinates refinement followed by individual B-factor refinement was performed. The refinement statistics are given in Table II. The resulting crystallographic R-factor is 20.4% using all reflections between 11.5 and 2.6 Å (Rfree = 25.7%). The final model contains one AspRS dimer, two tRNA molecules, two aspartyl-adenylate substrates, two SO42– ions and 502 water molecules. The enzyme in both subunits is missing five residues at the C-terminal end (each monomer contains 590 amino acid residues). The tRNA molecule of one subunit is missing the GCCA extremity of the acceptor stem, while the tRNA from the other subunit is complete. The model shows good stereochemistry and geometry, as analyzed using the program PROCHECK (Laskowski et al., 1993). All residues have φ and ψ angles within the allowed regions of the Ramachandran plot, with 13.9% in the most favored region.
Structure deposition
Coordinates for the X-ray structure of the E.coli AspRS–yeast tRNAAsp–aspartyl-adenylate have been deposited in the Brookhaven Protein Data Bank under accession code 1IL2.
Acknowledgments
Acknowledgements
We thank André Mitschler for his contribution to the cryocooling experiments and the staff of LURE (Orsay) and DESY (Hambourg) for their assistance during data collection. We thank Jay Schneider for experimental help, Sharee Otten and Gabriele Varani for comments on the manuscript, and Hyunsic Choi for discussions. This work was supported by institutional grants from the Université Louis Pasteur de Strasbourg, the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale and a grant from the Fond de Recherche Hoechst Marion Roussel. The work at the University of Wisconsin was supported by US Public Health Service grant GM42123 from NIGMS.
References
- Boeglin M., Dock-Bregeon,A.-C., Eriani,G., Gangloff,J., Ruff,M., Poterszman,A., Mitschler,A., Thierry,J.-C. and Moras,D. (1996) Crystallization of Escherichia coli aspartyl-tRNA synthetase in its free state and in complex with yeast tRNAAsp. Acta Crystallogr. D, 52, 211–214. [DOI] [PubMed] [Google Scholar]
- Briand C., Poterszman,A., Eiler,S., Webster,G., Thierry,J.-C. and Moras,D. (2000) An intermediate step in the recognition of tRNAAsp by aspartyl-tRNA synthetase. J. Mol. Biol., 299, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cavarelli J. et al. (1994) The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J., 13, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) Collaborative computational project No. 4. The CCP4 suite: programs from protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chang K.-Y., Varani,G., Bhattacharya,S., Choi,H. and McClain,W.H. (1999) Correlation of deformability at a tRNA recognition site and aminoacylation specificity. Proc. Natl Acad. Sci. USA, 96, 11764–11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dock A.-C., Lorber,B., Moras,D., Pixa,G., Thierry,J.-C. and Giégé,R. (1984) Crystallization of transfer ribonucleic acids. Biochimie, 66, 179–201. [DOI] [PubMed] [Google Scholar]
- Dodson E.G. (1985) Molecular replacement: the method and its problems. In Machin,P.A. (ed.), Molecular Replacement: Proceedings of the Daresbury Study Weekend. SERC Daresbury Laboratory, Warrington, UK, pp. 33–45.
- Eiler S., Dock-Bregeon,A.-C., Moulinier,L., Thierry,J.-C. and Moras,D. (1999) Synthesis of aspartyl-tRNAAsp in E.coli—a snapshot of the second step. EMBO J., 18, 6532–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Dirheimer,G. and Gangloff,J. (1990) Aspartyl-tRNA synthetase from E.coli: cloning and characterisation of the gene, homologies of its translated amino acid sequence with asparaginyl- and lysyl-tRNA synthetases. Nucleic Acids Res., 18, 7109–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Cavarelli,J., Martin,F., Dirheimer,G., Moras,D. and Gangloff,J. (1993) Role of dimerization in yeast aspartyl-tRNA synthetase and importance of the class II invariant proline. Proc. Natl Acad. Sci. USA, 90, 10816–10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S. (1998) SETOR: hardware lighted three-dimensional model representations of macromolecules. J. Mol. Graphics, 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Foss K., Kao,S.-H. and McClain,W.H. (1979) Three suppressor forms of bacteriophage T4 leucine transfer RNA. J. Mol. Biol., 135, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Gabriel K. and McClain,W.H. (2001) Plasmid systems to study RNA function in Escherichia coli. J. Mol. Biol., 310, 543–548. [DOI] [PubMed] [Google Scholar]
- Giegé R., Lorber,B., Ebel,J.P., Moras,D. and Thierry,J.C. (1982) Formation of a catalytically active complex between tRNAAsp and aspartyl-tRNA synthetase from yeast in high concentrations of ammonium sulphate. Biochimie, 64, 357–362. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1988) Evaluation of a single crystal X-ray diffraction from a position-sensitive detector. J. Appl. Crystallogr., 21, 916–924. [Google Scholar]
- Kern D., Lorber,B., Boulanger,Y. and Giegé,R. (1985) A peculiar property of aspartyl-tRNA synthetase from baker’s yeast: chemical modification of the protein by the enzymatically synthesized aminoacyl adenylate. Biochemistry, 24, 1321–1332. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Martin F., Eriani,G., Eiler,S., Moras,D., Dirheimer,G. and Gangloff,J. (1993) Overproduction and purification of native and queuine-lacking Escherichia coli tRNAAsp, role of the wobble base in tRNAAsp acylation. J. Mol. Biol., 234, 965–974. [DOI] [PubMed] [Google Scholar]
- McClain W.H. and Gabriel,K. (2001) Construction of an Escherichia coli knockout strain for functional analysis of tRNAAsp. J. Mol. Biol., 310, 537–542. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Mechulam,Y. and Fayat,G. (1988) Fast purification of a functional elongator tRNAMet expressed from a synthetic gene in vivo. Nucleic Acids Res., 16, 8095–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D., Comarmond,M.B., Fischer,J., Weiss,R., Thierry,J.C., Ebel,J.P. and Giegé,R. (1980) Crystal structure of yeast tRNAAsp. Nature, 288, 669–674. [DOI] [PubMed] [Google Scholar]
- Nameki N., Tamura,K., Himeno,H., Asahara,H., Hasegawa,T. and Shimizu,M. (1992) Escherichia coli tRNAAsp recognition mechanism differing from that of the yeast system. Biochem. Biophys. Res. Commun., 189, 856–862. [DOI] [PubMed] [Google Scholar]
- Nicholls A. and Honig,B. (1991) A rapid finite difference algorithm utilizing successive over-relaxation to solve the Poisson–Boltzmann equation. J. Comp. Chem., 12, 435–445. [Google Scholar]
- Poterszman A., Delarue,M., Thierry,J.C. and Moras,D. (1994) Synthesis and recognition of aspartyl-adenylate by Thermus thermophilus aspartyl-tRNA synthetase. J. Mol. Biol., 244, 158–167. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi,J.D., Florentz,C. and Giegé,R. (1991) Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science, 252, 1696–1699. [DOI] [PubMed] [Google Scholar]
- Renaud M., Ehrlich,R., Bonnet,J. and Remy,P. (1979) Lack of correlation between affinity of the tRNA for the aminoacyl-tRNA synthetase and aminoacylation capacity as studied with modified tRNAPhe. Eur. J. Biochem., 100, 157–164. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy,S., Boeglin,M., Poterszman,A., Mitschler,A., Podjarny,A., Rees,B., Thierry,J.C. and Moras,D. (1991) Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science, 252, 1682–1689. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Moulinier,L., Fujiwara,S., Imanaka,T., Thierry,J.C. and Moras,D. (1998) Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation. EMBO J., 17, 5227–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R., Hoben,P., Sumner-Smith,M., Uemura,H., Watson,L. and Söll,D. (1988) Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science, 242, 1548–1551. [DOI] [PubMed] [Google Scholar]
- Urzhumtsev A.G., Lunin,V.Yu. and Vernoslova,E.A. (1989) Frog— high-speed restraint–constraint refinement program for macro molecular structure. J. Appl. Crystallogr., 22, 500–506. [Google Scholar]
- Varani G. and McClain,W.H. (2000) The G⋅U wobble base pair: a fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep., 1, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M. (1982) Translational efficiency of transfer RNAs: uses of an extended anticodon. Science, 218, 646–652. [DOI] [PubMed] [Google Scholar]