Abstract
U7 snRNPs were isolated from HeLa cells by biochemical fractionation, followed by affinity purification with a biotinylated oligonucleotide complementary to U7 snRNA. Purified U7 snRNPs lack the Sm proteins D1 and D2, but contain additional polypeptides of 14, 50 and 70 kDa. Microsequencing identified the 14 kDa polypeptide as a new Sm-like protein related to Sm D1 and D3. Like U7 snRNA, this protein, named Lsm10, is enriched in Cajal bodies of the cell nucleus. Its incorporation into U7 snRNPs is largely dictated by the special Sm binding site of U7 snRNA. This novel type of Sm complex, composed of both conventional Sm proteins and the Sm-like Lsm10, is most likely to be important for U7 snRNP function and subcellular localization.
Keywords: Cajal/coiled bodies/histone pre-mRNA 3′ processing/Sm core structure/Sm-like protein/small nuclear ribonucleoprotein
Introduction
Small nuclear ribonucleoproteins (snRNPs) play important roles in mRNA metabolism. The abundant U1, U2, U4/U6 and U5 snRNPs have distinctive functions in pre-mRNA splicing (reviewed in Moore et al., 1993). The U1, U2, U4 and U5 small nuclear RNAs (snRNAs) present in these particles each contain a conserved single-stranded sequence element, the Sm binding site, with the consensus RAUUU/GUUGR (Branlant et al., 1982; Liautard et al., 1982). This element interacts with the seven Sm proteins, B, D1–D3, E, F and G, to form an Sm core structure (see below). The U1, U2 and U5 snRNPs, as well as more complex snRNP assemblies such as the U4/U6⋅U5 tri-snRNP, contain various additional proteins that interact either with other regions of the snRNAs or with proteins already associated with the particles (reviewed in Lührmann et al., 1990).
A less frequent group of introns are spliced using a different set of snRNPs, with the U11, U12 and U4atac/U6atac snRNPs replacing the U1, U2 and U4/U6 snRNPs, respectively (Burge et al., 1998). Only the U5 snRNP seems to be involved in the splicing of both intron classes. The U11 and U12 snRNPs contain the common Sm proteins, interacting with canonical Sm binding sites, certain U2 snRNP-specific proteins also present in the U12 snRNP, and new kinds of U11 and U12 snRNP-specific proteins that may be functional equivalents of U1- and U2-specific proteins (Will et al., 1999).
Another minor snRNP, the U7 snRNP, is an essential cofactor for 3′-end processing of the replication-dependent histone pre-mRNAs in metazoans (reviewed in Müller and Schümperli, 1997; Dominski and Marzluff, 1999). These histone transcripts lack introns and a poly(A) tail in the mature message. The endonucleolytic cleavage generating the mRNA 3′-end is distinct from the cleavage-polyadenylation reaction that processes all other mRNAs. The U7 snRNA is 58–63 nucleotides (nt) long, depending on the species. Its 5′-end is complementary to a purine-rich downstream element (DSE) in histone pre-mRNA. Base-pairing between U7 snRNA and this DSE is essential for the assembly of a functional processing complex (Strub et al., 1984; Schaufele et al., 1986; Bond et al., 1991). Following this base-pairing region, U7 snRNA contains a non-canonical Sm binding site, AAUUUGUCUAG, followed by a long hairpin structure and a very short single-stranded 3′ extension. Purified murine U7 snRNPs have been described to contain most or all of the common Sm proteins as well as two apparently U7-specific proteins of 14 and 50 kDa (Smith et al., 1991), which were not characterized further.
All Sm proteins share certain structural features, including two conserved amino acid motifs, Sm motifs 1 and 2 (Cooper et al., 1995; Hermann et al., 1995; Seraphin, 1995). Based on X-ray crystallographic structures of B–D3 and D1–D2 heterodimers, the seven Sm proteins were proposed to form a heptameric ring around the RNA Sm binding site (Kambach et al., 1999). Further evidence for this ring structure was recently obtained from UV-cross-linking studies (Urlaub et al., 2001), stoichiometry analyses in yeast (Walke et al., 2001) and cryo-electron microscopy of purified U1 snRNPs (Stark et al., 2001).
All eukaryotes contain additional Sm-like or Lsm proteins. Seven of these, Lsm2–Lsm8, are found in association with U6 snRNA in the U6 snRNP, the U4/U6 di-snRNP and the U4/U6⋅U5 tri-snRNP (Cooper et al., 1995; Seraphin, 1995; Achsel et al., 1999; Gottschalk et al., 1999; Mayes et al., 1999; Salgado-Garrido et al., 1999). They are also thought to form a heptameric ring around the U6 snRNA 3′-end (Achsel et al., 1999). Despite their being called Sm-like as opposed to Sm proteins, each of these Lsm proteins more closely resembles one of the conventional Sm proteins than the other Lsm proteins. Another member of this group, Lsm1, forms a different complex with Lsm2–Lsm7, which plays a role in mRNA degradation in the cytoplasm (Boeck et al., 1998; Bouveret et al., 2000; Tharun et al., 2000). Moreover, Sm-like proteins forming heptameric rings exist in archaea, indicating a very ancient origin for this protein family (Salgado-Garrido et al., 1999; Achsel et al., 2001; Collins et al., 2001; Mura et al., 2001; Toro et al., 2001).
Here, we characterize the polypeptide composition of U7 snRNPs. Surprisingly, the Sm proteins D1 and D2 are absent, but highly purified U7 snRNPs contain additional polypeptides of 14, 50 and 70 kDa. We identify the 14 kDa polypeptide as a new member of the Sm/Lsm protein family. Like U7 snRNA, this protein is enriched in Cajal or coiled bodies (CBs) in the nucleus. Its association with the U7 snRNP is at least in part dictated by the special Sm binding site of U7 snRNA. Thus, the U7 snRNP is so far unique in that it contains both conventional Sm proteins and a new type of Sm-like protein in the same particle.
Results
Purification of U7 snRNPs from HeLa cell nuclear extracts
To identify the proteins present in U7 snRNPs, we took advantage of a strategy allowing for the large-scale purification of spliceosomal snRNPs from HeLa cell nuclear extract (Bach et al., 1990). In the first steps we found U7 snRNA, along with total snRNPs, to bind to anti-trimethyl guanosine cap (anti-m3G) antibodies, and, upon glycerol gradient centrifugation, to co-sediment with the peak of 12S U1 and U2 snRNPs (data not shown). After further separation by ion-exchange chromatography on a Resource Q column, specific fractions eluting at 300–350 mM KCl, just ahead of the U1 peak, contained primarily U7 snRNA, along with small amounts of U1 snRNA, as revealed by RNA 3′ end-labelling with α-[32P]pCp; however, U7 snRNA also trailed into the U1 peak of the column (Figure 1A).
Fig. 1. Purification and polypeptide composition of U7 snRNPs. (A) RNA 3′ end-labelling of fractions enriched for U7 and U1 snRNPs. RNA samples were isolated from U7 and U1 peak fractions of a Resource Q column (see Materials and methods), 3′ end-labelled with α-[32P]pCp and analysed by denaturing PAGE. The positions and sizes (in bp) of relevant marker bands (HpaII-digested pBR322) are indicated on the left. (B) Polypeptide composition of the Resource Q U7 peak fraction from which the 14 kDa polypeptide (arrow) was identified by tandem mass spectrometry. A sample of the fraction was separated by 15% SDS–PAGE. M, protein marker, sizes in kDa are indicated on the left. (C) Polypeptide composition of affinity-purified U7 snRNPs. U7 (lane 2) or U1 snRNPs (lane 3) were affinity selected from a Resource Q fraction containing both U7 and U1 snRNPs (ResQ; lane 1) using biotinylated RNA-specific antisense oligonucleotides and magnetic streptavidin beads and resolved by 12% SDS–PAGE. The positions of the common Sm proteins present in the U1 snRNP and of streptavidin (SA) released from the magnetic beads are indicated on the right. Unidentified proteins present in the U7 snRNP purification are labelled with asterisks. (D) Identification of Sm proteins present in affinity-purified U7 snRNPs by western probing. ResQ (lane 1) and U7 (lane 3) are labelled as in (C); beads (lane 2) show the background precipitation by beads alone. The antibodies used were (from top to bottom): Y12 anti-Sm antibody, anti-Sm D2, anti-Sm D3, anti-Sm F and anti-Sm G. The Y12 antibody detects a double band corresponding to Sm B/B′, and a weak signal for Sm D1 (Hoch et al., 1999; Brahms et al., 2000).
An SDS–polyacrylamide gel analysis revealed that such a U7 peak fraction was composed of many polypeptides (Figure 1B). Based on their migration, some of the bands could be attributed to the common Sm proteins. Others seemed to correspond to the U1-specific 70K and A proteins, suggesting that the fraction was still impure. Nevertheless, candidates for the reported U7-specific proteins of 14 and 50 kDa (Smith et al., 1991) were also visible. Importantly, this fraction was able to complement a U7 snRNP-depleted K21 nuclear extract in histone RNA 3′ processing (Stauber et al., 1990 and data not shown).
Higher purification of a similar Resource Q fraction (but which contained both U7 and U1 snRNA) was achieved by affinity selection with a biotinylated oligonucleotide complementary to 10 nt at the 5′-end of U7 snRNA. Interestingly, these affinity-purified U7 snRNPs (Figure 1C, lane 2) contained bands co-migrating with the Sm proteins present in similarly affinity-purified U1 snRNPs (lane 3) and in the input Resource Q fraction (lane 1), with the notable exception of the Sm proteins D1 and D2. Moreover, additional bands of ∼70 and ∼50 kDa, respectively, that did not correspond to any of the common Sm proteins were seen in the affinity-purified U7 snRNPs (lane 2, asterisks). Note that the polypeptide composition of the input Resource Q fraction (lane 1) differed somewhat from that shown in Figure 1B.
Identification of Sm proteins present in highly purified U7 snRNP
To characterize the common Sm proteins present in U7 snRNPs, we subjected the affinity-purified fraction, its input Resource Q fraction, as well as a control precipitation with streptavidin beads alone, to SDS–PAGE and identified individual Sm proteins by western blotting (Figure 1D). The Y12 anti-Sm antibody is known to react primarily with Sm B/B′ (the two splicing variants of Sm B), more weakly with Sm D1 and very faintly with D3 (Hoch et al., 1999; Brahms et al., 2000). Probing of the western blot with Y12 antibody indeed revealed an Sm B/B′ doublet and a weaker band corresponding to D1 in the Resource Q fraction (Figure 1D top, lane 1; detection of D3 would have required higher concentrations of protein and antibody, data not shown). In contrast, only the B/B′ bands, but not D1, could be detected in the affinity-purified U7 preparation (lane 3). Similarly, a western blot with an Sm D2-specific antibody confirmed the absence of D2 in the purified U7 snRNPs. In contrast, Sm D3, F and G were each detected by specific antibodies in the affinity-purified U7 preparation. No specific antibodies were available for Sm protein E, but material isolated from the corresponding band of the affinity-purified U7 snRNPs could be identified as Sm E by MALDI-TOF analysis.
Although the relative recoveries of the B/B′, D3, F and G proteins were variable as judged from the western blots (Figure 1D), inspection of the Coomassie Blue-stained gel did not indicate any deviation from equimolarity, at least for D3, E, F and G (Figure 1C, lane 2).
Identification of a 14 kDa U7-specific protein
A U7-specific protein of 14 kDa, previously detected in affinity-purified U7 snRNPs by Smith et al. (1991), could not be seen in our SDS–PAGE of the purest fraction, because this part of the gel contained a strong band corresponding to streptavidin, which was present in both the U7 and U1 snRNPs (Figure 1C, lanes 2 and 3). However, a 14 kDa band present in the Resource Q fraction shown in Figure 1B (arrow) was isolated from a preparative gel, digested with trypsin, and the peptides were separated by reverse-phase HPLC and identified by nano-electrospray tandem mass spectrometry coupled to the Sequest database search algorithm (Chittum et al., 1998). All 14 peptides identified were found to be encoded by a human expressed sequence tag (EST; DDBJ/EMBL/GenBank accession No. AI147739). This EST belongs to a ubiquitously expressed transcription unit (Unigene cluster no. Hs.3496) that encodes a novel polypeptide of 123 amino acids (Figure 2A). The complete coding region is present in a genomic clone (AL358433) from human chromosome 1.
Fig. 2. Lsm10 is a novel Sm-like protein. (A) Amino acid sequence of Lsm10. Peptides identified by mass spectrometry are underlined. Arrows indicate the region that was deleted in the Lsm10ΔPstI construct. The cDNA sequence was deposited in the DDBJ/EMBL/GenBank (accession No. AF394685). (B) Alignment of Lsm10 with human Sm D1 and D3 and the related Lsm2 and Lsm4 proteins across the Sm motifs 1 and 2. Highlighted Lsm10 residues are present in some (black) or all (grey) other proteins. The consensus sequence was adapted from previous sources (Hermann et al., 1995; Achsel et al., 1999). h, hydrophobic amino acids.
A BLAST search (Altschul et al., 1990) of the protein databases with the predicted amino acid sequence revealed that the 14 kDa protein has low, but significant, homology to the common Sm proteins D1 and D3 found in spliceosomal snRNPs, as well as to their Lsm counterparts Lsm2 and Lsm4. An amino acid sequence alignment with these related proteins shows that it shares many conserved residues that define the Sm motifs 1 and 2 (Hermann et al., 1995; Seraphin, 1995; Figure 2B). This feature identifies the 14 kDa protein as a new member of the Sm/Lsm protein family, so we propose naming it Lsm10.
Seven putative homologues of human Lsm10 were identified in the EST/Unigene databases: mouse, Mus musculus (Unigene Mm.41865); rat, Rattus norwegicus (Unigene Rn.3612); cattle, Bos taurus (EST AW653562); and swine, Sus scrofa (EST AW347575 and others), whose encoded polypeptides are 91–93% identical to the human counterpart; the Xenopus laevis homologue (EST AW634754 and others) shows 74% identity; finally, there are incomplete cDNAs encoding homologues from the zebrafish, Danio rerio (EST AI105956), and the channel catfish, Ictalurus punctatus (EST BE212828), which are 62 and 67% identical within the known regions, respectively. In contrast, we could not detect any homologues outside the vertebrates.
Lsm10 is a bona fide component of the U7 snRNP
As Lsm10 had been isolated from a still impure Resource Q fraction, we tested whether it was indeed part of the U7 snRNP. To this end, polyclonal antibodies were raised in rabbits by immunization with recombinant glutathione S-transferase (GST)-tagged Lsm10. In a western blot of a Resource Q U7 peak fraction, this immune serum recognized a polypeptide of 14 kDa (Figure 3A, lane 2) that was not detected by the pre-immune serum (lane 1). Because both the pre-immune and immune sera stained other bands, some of which had electrophoretic mobilities similar to the Sm D proteins, we affinity purified the antibodies using immobilized Lsm10 or Lsm10ΔPstI, lacking a central piece of 75 amino acids encompassing the Sm motifs (Figure 2A), resulting in preparations named Lsm10full and Lsm10ΔPstI, respectively. The Lsm10ΔPstI antibody recognized only one 14 kDa band in the U7-enriched Resource Q fraction (Figure 3A, lane 3). The same band was also detected in an affinity-purified U7 snRNP preparation similar to the one shown in Figure 1C (Figure 3A, lane 5), but not in a sample from a mock purification with beads alone (lane 4).
Fig. 3. Lsm10 is in a complex with U7 snRNA and common Sm proteins. (A) Polyclonal antibodies to Lsm10 recognize a specific 14 kDa polypeptide. Samples of a Resource Q U7 peak fraction similar to that shown in Figure 1B were separated by 15% SDS–PAGE, western blotted and analysed with pre-immune serum (pre, lane 1), immune serum (im, lane 2) and affinity-purified Lsm10ΔPstI antibody (lane 3). A specific band of 14 kDa was also detected by the Lsm10ΔPstI antibody in affinity-purified U7 snRNPs (U7, lane 5), but not in a control precipitation with beads alone (bd, lane 4). (B) Native gel analysis of U7 snRNPs. Samples of a Resource Q U7 peak fraction were incubated with a radiolabelled oligonucleotide complementary to the 5′ end of U7 snRNA, followed by incubation with different antibodies as indicated, before separation on a non-denaturing gel (Melin et al., 1992). Only the region between the gel slots and the U7 snRNP complexes is shown. The positions of native U7 snRNPs, as well as that of the antibody-induced supershift, are indicated on the right. (C) Co-immunoprecipitation of U7 snRNA with Lsm10 from HeLa nuclear extracts. Equal amounts of HeLa nuclear extract were incubated with different antibodies as indicated. RNAs were extracted from the immunopellets and analysed by primer extension for U7 and U1 RNA. The migration of the unextended primers and of the U7- and U1-specific extension products are indicated on the right.
We next tested whether the purified antibody could react with U7 snRNPs in a U7-enriched Resource Q column fraction, as detected by an electrophoretic mobility shift in a native gel. As demonstrated previously (Melin et al., 1992), the U7 snRNP was detected by a radiolabelled oligonucleotide complementary to the 5′-end of U7 snRNA (Figure 3B, lane 1), and was shifted into the gel pockets by the anti-Sm antibody Y12 (lane 2). The pre-immune serum caused a slight and diffuse shift (lane 3), possibly due to its cross-reactivity with Sm D proteins (Figure 3A, lane 2). Most importantly, however, both the Lsm10 immune serum and the purified Lsm10ΔPstI antibody shifted the U7 complex to a specific position between the original position of the U7 snRNPs and the wells (Figure 3B, lanes 4 and 5). The specificity of this shift was underlined by the fact that affinity-purified antibodies directed against the histone hairpin binding protein (lane 6) or an unrelated RNA binding protein, p756 (lane 7), did not alter the electrophoretic mobility.
To provide additional evidence that the 14 kDa protein is a component of the U7 snRNP, immunoprecipitations were performed with HeLa nuclear extracts, and the presence of U7 and U1 snRNA was determined by primer extension. Antibodies against Lsm10 precipitated significant amounts of U7 snRNA, irrespective of whether crude immune serum (Figure 3C, lane 5), Lsm10full (lane 6) or Lsm10ΔPstI (lane 7) antibodies were used. In contrast, U7 snRNA co-precipitation was not observed with pre-immune serum (lane 4) or with the unrelated anti-p756 antibody (lane 8). As expected, the anti-Sm antibody Y12 co-precipitated both the U7 and U1 snRNAs tested in the primer extension assay (lane 9), but no co-precipitation of U1 snRNA was observed with any of the Lsm10-specific antibodies (lanes 5–7). Taken together, these results prove that Lsm10 is an integral component of the U7 snRNP.
Lsm10 is localized in CBs
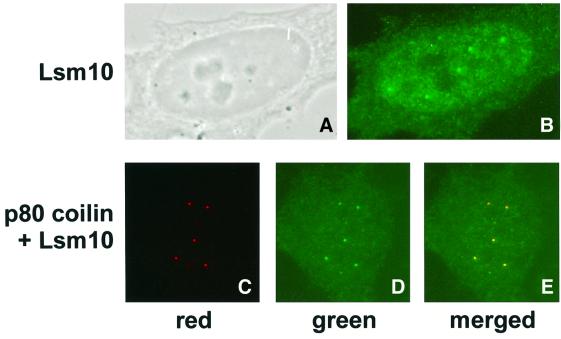
U7 snRNA was previously shown by fluorescence in situ hybridization to be concentrated in CBs in the nuclei of human and murine cells, with only low amounts being distributed throughout the nucleoplasm, but not in the nucleoli (Frey and Matera, 1995). Having shown that Lsm10 is a component of the U7 snRNP, we wanted to determine whether it has a similar localization in vivo. Analysis of HeLa cells by indirect immunofluorescence with the Lsm10ΔPstI antibody indeed revealed a faint nucleoplasmic staining with a few bright foci reminiscent of CBs (Figure 4B). Moreover, Lsm10 appeared to be excluded from the nucleoli. As this resembled the subcellular localization observed for U7 snRNA, we investigated whether the bright foci were indeed CBs. For this purpose, the cells were co-stained with the Lsm10ΔPstI antibody and a monoclonal antibody against human p80 coilin, a CB marker protein, and the signals from the two secondary antibodies were recorded separately (green for Lsm10 and red for p80 coilin). Consistent with previous observations (Sleeman et al., 1998), the p80 coilin antibody stained a similar number of bright nuclear foci (Figure 4C). Most importantly, these co-localized with the foci revealed by the Lsm10ΔPstI antibody (Figure 4D and E). Under the conditions used, the green signal indeed came from the Lsm10 antibody and not from bleedthrough of the p80 coilin signal (data not shown). From these experiments we conclude that Lsm10 is concentrated in CBs and shows a subcellular distribution similar to that previously described for U7 snRNA (Frey and Matera, 1995).
Fig. 4. Lsm10 co-localizes with p80 coilin. The top row shows the same HeLa cell viewed by phase contrast (A) and by indirect immunofluorescence microscopy (B) with Lsm10ΔPstI antibody followed by Oregon Green-labelled goat anti-rabbit antibody. Bottom row: co-staining for (C) p80 coilin, using monoclonal coilin-specific antibody SP10 followed by Texas Red-labelled anti-mouse antibodies and (D) Lsm10 [as in (B)]. In (E), the red and green images from (C) and (D) were merged to demonstrate the co-localization of the brightly stained CBs.
Association of Lsm10 with U7 snRNA depends on the U7-specific Sm binding site
The Sm binding site of U7 snRNA differs from that found in spliceosomal snRNAs and this feature is partly responsible for the low abundance of U7 snRNPs. In particular, when the U7 Sm binding site AAUUUGUCUAG (U7 Sm WT) was converted to the consensus sequence of spliceosomal snRNPs AAUUUUUGGAG (U7 Sm OPT; changes underlined), the resulting snRNPs accumulated in transfected cells or microinjected Xenopus oocytes 2–3 times more efficiently, but they were non-functional in histone RNA 3′ processing (Grimm et al., 1993; Stefanovic et al., 1995). A likely explanation for these results was that the U7 Sm WT sequence mediated the assembly of a U7-specific snRNP structure, whereas a standard Sm core structure was formed with U7 Sm OPT RNA.
To test this hypothesis, we wanted to co-express a tagged version of Lsm10 together with either tagged U7 Sm WT or U7 Sm OPT RNA. The tags should allow us to detect a stable association of the two components. First, plasmids encoding haemagglutinin (HA)-tagged Sm proteins or Lsm10 were expressed by transfection into human 293T cells, and their association with endogenous U7 or U1 snRNA was studied by precipitation of nuclear extracts with biotinylated antisense oligonucleotides specific for U7 and U1 snRNA (Figure 5A). HA-tagged versions of Sm B, D1, D2 and D3 were tested by transient transfection. For HA-tagged Lsm10, we used both transient transfections and a stably transfected cell line, but saw no differences between these two approaches. Importantly, the HA-tagged Sm B and D3 proteins associated with both U1 and U7 snRNAs, D1 and D2 interacted only with U1 RNA, and tagged Lsm10 bound exclusively to U7 RNA (Figure 5A). Thus, these experiments were consistent with the specific associations of the untagged endogenous proteins described in Figure 1, proving the validity of this experimental approach.
Fig. 5. Importance of the Sm binding site for the association of Lsm10 with U7 snRNA in vivo. (A) Association of HA-tagged proteins with U1 and U7 snRNAs. Human 293T cells were transiently transfected with plasmids encoding HA-tagged Sm proteins or stably transformed with a plasmid encoding HA-tagged Lsm10. Nuclear extracts were incubated with biotinylated oligonucleotides complementary to the 5′-ends of U7 or U1 snRNA and precipitated with magnetic streptavidin beads. The samples were subjected to SDS–PAGE and immunoblotted with anti-HA antibody. –, precipitation by beads without oligonucleotide; input, sample of original nuclear extract. (B) RNase mapping of 28-WT and 28-OPT RNAs in nuclear extract from 293T cell clone HA-Lsm10 #2 stably expressing HA-tagged Lsm10. These cells were transfected with plasmids encoding U7 Sm WT and U7 Sm OPT RNA (Grimm et al., 1993; Stefanovic et al., 1995), modified with a 28 nt sequence tag at the 5′ end. The riboprobe, derived from U7 Sm OPT RNA, was complementary to the resulting 28-OPT and 28-WT transcripts over ∼42 and ∼33 nt, respectively. Lanes: tRNA, control hybridization to tRNA; M, HpaII-digested pBR322 DNA (sizes of fragments in bp are indicated on the left). (C) Western blots detecting Lsm10 and Sm proteins associated with 28-WT or 28-OPT RNAs in transfected 293T cells. Nuclear extracts were prepared and processed as described for (A), except that precipitations were performed with magnetic streptavidin beads with (+) or without (–) biotinylated anti-28 oligonucleotide. The top two panels are blots of material precipitated from the same nuclear extracts used in (B). Alternatively, 293T cells were co-transfected with 28-WT or 28-OPT and plasmids for either HA-tagged Sm D2 (two middle panels) or D3 (two bottom panels). Anti-HA and Y12 anti-Sm antibodies were used to detect HA-tagged proteins and Sm B/B′, respectively. Input, sample of original 28-WT extract from each experiment.
For the critical experiment, the stably transfected 293T cells expressing HA-tagged Lsm10 were transfected with plasmids 28-WT and 28-OPT. These encoded murine U7 Sm WT and U7 Sm OPT snRNAs with the first 20 nt (complementary to the histone DSE) replaced by an unrelated sequence of 28 nt. After transient transfection into the cells, the expression of the snRNAs was analysed by RNase protection (Figure 5B). Quantitation of the band intensities revealed that 28-OPT RNA was expressed ∼3 times more efficiently than 28-WT RNA, consistent with earlier findings (Grimm et al., 1993). Next, nuclear extracts from the transfected cells were precipitated with a biotinylated oligonucleotide complementary to the 28 nt sequence and the precipitates were analysed by western blotting with either anti-HA antibody to detect the HA-tagged Lsm10 or with anti-Sm antibodies to detect Sm B/B′ (Figure 5C, top panels). The B/B′ polypeptides were found in the pellets of 28-WT- and 28-OPT-transfected cells in approximately the same ratio as the corresponding U7 snRNAs, indicating that both 28-WT and 28-OPT RNAs had assembled into snRNPs and that they were precipitated with similar efficiency. In contrast, HA-tagged Lsm10 was present in much higher amounts in the affinity-enriched 28-WT snRNPs than in the corresponding sample from cells transfected with 28-OPT. Control experiments in which plasmids encoding HA-tagged Sm D2 or D3 were transfected into 293T cells along with the 28-WT and 28-OPT constructs indicated that HA-tagged Sm D2 associated exclusively with 28-OPT RNA (Figure 5C, middle panels), whereas HA-tagged Sm D3 associated with both RNAs in similar ratios as the endogenous Sm B/B′ polypeptides (Figure 5C, bottom panels). These experiments demonstrated that the specific presence of Lsm10 and lack of Sm D2 in the U7 snRNP were at least in part dictated by the special Sm binding site of U7 snRNA.
Discussion
Lsm10 is a new member of the Sm-like protein family
In this paper, we have adapted a large-scale snRNP purification strategy (Bach et al., 1990) to obtain fractions enriched for human U7 snRNPs (Figure 1A and B). A 14 kDa polypeptide isolated from such a fraction was identified as a new Sm-like protein (Figure 2). In vivo, this protein, called Lsm10, is stably associated with the U7 snRNP (Figure 3) and has a similar subcellular distribution as was reported earlier for U7 snRNA (Frey and Matera, 1995; Figure 4). On the basis of these results, Lsm10 is an integral component of U7 snRNPs and the first U7-specific protein to be characterized molecularly.
Sequence similarities, and in particular the presence of the conserved Sm motifs 1 and 2, identify Lsm10 as a new member of the Sm/Lsm protein family (Figure 2). The highest similarity is found with Sm D1 and D3 as well as with Lsm2 and Lsm4, which show 30, 24, 30 and 28% identity, respectively. These similarities are mostly confined to the Sm motifs, but there is significant divergence at the C-termini, where Lsm10 has a single GR dipeptide compared with the GR repeats found in Sm D1 and D3. In Sm D1, this repeat region was shown to be an epitope for anti-Sm antibodies (Hirakata et al., 1993).
Concerning the Sm motifs, the Lsm10 sequence is identical to at least one of the four comparison sequences in 16 of 32 amino acids of motif 1 and in 9 of 14 positions of motif 2 (Figure 2B). Moreover, there are only two deviations from the Sm motif consensus sequences: a conserved glycine at position 8 of motif 1 is replaced by glutamic acid in Lsm10; however, as Lsm2 has aspartic acid at this position, this change may be conservative. The second change affects a conserved arginine at position 7 of motif 2, which is replaced by threonine; this loss of positive charge may be compensated by the arginine at position 9 of Lsm10 (the sequence RGX in the other Sm/Lsm proteins is occupied by TGR in Lsm10).
The fact that we failed to find any non-vertebrate Lsm10 sequences by database searches raises the interesting question whether the polypeptide composition of invertebrate U7 snRNPs is different or whether the Lsm10 sequence has diverged below the detection limits of the search algorithms. Another less likely possibility is that histone mRNAs could be processed in these organisms by a pathway that does not involve a U7 snRNP. The only certified non-vertebrate U7 snRNA sequence known to date is that from the sea urchin Psammechinus miliaris (Strub et al., 1984).
U7 snRNPs have a novel Sm protein composition
The spliceosomal snRNPs can be regarded as containing two distinct protein modules. First, during the cytoplasmic assembly phase, the seven Sm proteins associate with the single-stranded RNA Sm binding site to form the doughnut-like Sm core (Figure 6). In addition to this structure, which seems to be identical for all spliceosomal snRNPs, most snRNPs contain snRNP-specific proteins. Where analysed, these have been shown to bind, after import of the Sm core assembly product into the nucleus, either to regions of the snRNA other than the Sm binding site, or, via protein–protein interactions, to the Sm core and/or other snRNP-specific proteins already bound to the RNA (reviewed in Lührmann et al., 1990).
Fig. 6. Tentative model for the structure of a U7 snRNP-specific Sm core. A conventional Sm core complex containing all seven Sm proteins as proposed by Kambach et al. (1999) is shown schematically on the left. In the U7 snRNP (shown on the right), the missing D1 and D2 proteins may be replaced by the Lsm10 protein characterized in this paper and by U7-p50, which can be UV-cross-linked to the U7 Sm binding site (Mital et al., 1993; Stefanovic et al., 1995; see text).
Our characterization of the polypeptide composition of highly purified U7 snRNPs (Figure 1C and D) indicates that the U7 snRNP does not conform to this pattern. Rather, it appears to have a non-canonical Sm core structure composed of five standard Sm proteins together with one or more U7-specific proteins (Figure 6). Therefore, at least some of the U7 snRNP-specific proteins, unlike the particle-specific proteins of spliceosomal snRNPs, may be part of a U7 snRNP-specific Sm core and may not show the behaviour of the spliceosomal snRNP-specific proteins described above. The evidence for this is as follows. (i) Our most highly purified U7 snRNP preparation contains only five Sm proteins, with Sm D1 and D2 being conspicuously absent (Figure 1C and D). A previous analysis of purified U7 snRNPs had revealed polypeptides co-migrating on SDS gels with all of the common Sm proteins (Smith et al., 1991). However, these proteins were not analysed in detail and the gel system used in that study did not allow for the separation of the Sm D proteins; nevertheless, it was noted that the D proteins might be under-represented.
(ii) In a study of the assembly of U7 snRNPs in Xenopus oocytes (Stefanovic et al., 1995), we have previously shown that the Sm B and G proteins could be cross-linked to the Sm binding site of U7 snRNA to similar positions as have recently been characterized for Sm proteins assembled onto a consensus Sm binding site oligonucleotide (Urlaub et al., 2001). Thus, at least the Sm B and G proteins seem to interact with U7 snRNA in a structure that is similar to an Sm core, raising the question which proteins might replace the missing D1 and D2 proteins in this structure.
(iii) As we have characterized Lsm10 as a new Sm-like protein and as an integral component of the U7 snRNP, it is a likely candidate to replace one of the two missing proteins in the Sm core of U7 snRNPs. Because of the high similarity to Sm D1, we have tentatively placed Lsm10 in the position of D1 in Figure 6.
(iv) Our most highly purified U7 snRNP preparation contained additional polypeptides of 50 and 70 kDa (Figure 1C). We hope to characterize these proteins in the near future. A 50 kDa polypeptide had already been detected in the purified U7 snRNP preparation of Smith et al. (1991), but a 70 kDa band was not identified. Moreover, we previously showed that in addition to the Sm proteins B and G mentioned above, a protein of 40 kDa could be UV-cross-linked to U7 snRNA in Xenopus oocytes (Stefanovic et al., 1995). Three similar UV adducts of U7 snRNA could also be formed in nuclear extracts from mouse mastocytoma cells; however, the largest adduct seemed to be ∼10 kDa larger than that seen in Xenopus oocytes, i.e. it appeared to correspond to a protein of ∼50 kDa (Mital et al., 1993). It is likely, although not proven, that the 40 kDa protein seen to interact with the U7 Sm binding site in the oocytes is the Xenopus homologue of the 50 kDa protein found in affinity-purified U7 snRNPs (Smith et al., 1991 and Figure 1C). Our previous findings that the 40-kDa protein cross-link could be mapped to the Sm binding site, within 2 and 4 nt of the B and G cross-links, respectively, and that this photoadduct could already be detected in the cytoplasm, prior to import of the assembled snRNPs into the nucleus (Stefanovic et al., 1995), make it a good candidate for the missing member of a heptameric U7-specific Sm core (Figure 6). To fulfil this function, the 50 kDa U7-specific protein presumably should also contain Sm motifs or at least assume a similar fold as the Sm/Lsm proteins.
(v) However, the argument for an involvement of the 50 kDa protein is speculative and alternative explanations exist. For example, the U7 snRNP might have a hexameric, rather than heptameric, Sm core. This would be compatible with the U7 snRNP’s low stability during biochemical fractionation as well as with its inefficient assembly in vivo.
The replacement of individual members of the Sm core by one or more U7-specific proteins is reminiscent of the two Lsm complexes containing Lsm2–7. There, the replacement of a single subunit changes the nature of the particle, its subcellular localization, as well as its function. In combination with Lsm8, the complex binds to U6 snRNA in the nucleus and is important for U6 snRNP metabolism and pre-mRNA splicing (Cooper et al., 1995; Seraphin, 1995; Achsel et al., 1999; Gottschalk et al., 1999; Mayes et al., 1999; Salgado-Garrido et al., 1999). In contrast, when Lsm8 is replaced by Lsm1, the Lsm structure forms part of a cytoplasmic complex involved in mRNA decapping and degradation (Boeck et al., 1998; Bouveret et al., 2000; Tharun et al., 2000).
Implications for the assembly and function of U7 snRNPs
Although many aspects of the above model remain to be proven, it has interesting implications for both the assembly and function of U7 snRNPs. It is interesting to note that the Sm D1 and D2 proteins that are missing in the U7 snRNP form heterodimers in vivo, i.e. they can be regarded as parts of the same functional module. All together, three subcomplexes of the Sm core have been described, containing the B–D3, D1–D2 and F–E–G proteins, respectively (Lehmeier et al., 1994; Raker et al., 1996). As an intermediate leading to the formation of an Sm core, the D1–D2 and F–E–G subcomplexes are first thought to bind to the snRNA, followed by binding of B–D3 to complete the structure (Raker et al., 1996). Thus, it is possible that a distinct U7-specific heterodimer recognizes the special Sm binding site and combines with the conserved F–E–G and B–D3 subcomplexes to form the U7 snRNP. Our finding that the association of Lsm10 with U7 snRNA as well as the absence of Sm D2 from the particle are to a large extent specified by the sequence of the Sm binding site (Figure 5) is fully consistent with this possibility. Nevertheless, the fact that low amounts of HA-tagged Lsm10 also associated with 28-OPT RNA (Figure 5C) suggests that other features of the U7 snRNA may also contribute to the specificity of assembly.
Interestingly, U7 snRNPs formed with U7 Sm OPT RNA are non-functional in histone RNA 3′-end processing (Grimm et al., 1993; Stefanovic et al., 1995). So far, the only structural correlate for this functional difference had been the inability of U7 Sm OPT snRNA to form the UV adduct with the 40/50 kDa U7-specific protein (Stefanovic et al., 1995). The unusual polypeptide composition of U7 snRNPs (Figure 1) and the preferential association of Lsm10 with 28-WT compared with 28-OPT snRNA (Figure 5) now provide a rational explanation for this functional difference. In this context, Lsm10 [presumably helped by the other U7-specific protein(s)] is likely to be functionally important for histone RNA 3′ processing.
Furthermore, the U7-specific ribonucleoprotein structure may also govern the subcellular distribution of the particle. A U7 snRNA derivative similar to U7 Sm OPT, but containing the Sm binding site sequence of U2 snRNA, when injected into X.laevis oocytes, failed to become concentrated in spheres, the Xenopus equivalents of CBs, in contrast to WT U7 snRNA (Wu et al., 1996).
Materials and methods
U7 snRNP purification
Total snRNPs were prepared from HeLa cell nuclear extracts by anti-m3G immunoaffinity chromatography followed by separation on a 10–30% glycerol gradient (Bach et al., 1990). The peak containing 12S U1 and U2 snRNPs was fractionated on a Resource Q column using a 0–1 M KCl gradient.
For affinity selections, 30 pmol of biotinylated antisense oligonucleotide were incubated with 1.5 ml of enriched Resource Q fraction (Figure 1) or with nuclear extract from one 10 cm dish of transfected 293T cells (Figure 5). Magnesphere streptavidin beads (Promega), pre-blocked with 100 µg each of bovine serum albumin, yeast tRNA and glycogen (Figure 1) or with K21 nuclear extract (Stauber et al., 1990; Figure 5), were used to precipitate the oligonucleotide-bound snRNPs. The precipitate was washed three times with 20 mM HEPES pH 7.9, 100 mM KCl, 20% glycerol, 0.25 mM EDTA, 0.5 mM dithiothreitol (DTT) and 0.1% NP-40. The beads were transferred to a new tube and boiled in loading buffer prior to SDS–PAGE.
The antisense 2-O-methyl-oligoribonucleotides used were: U7 snRNA, 5′- AGAGCUGUAA (3′ biotin); U1 snRNA, 5′-GCCAGGUAGUAU dC*dC*dC*dC*dT (where dC* is biotinylated 2′-deoxycytidine); 28-WT or 28-OPT snRNAs, 5′-ACAGCCCCACCAACT (3′-biotin).
Protein analysis
Proteins were resolved on 12 or 15% high-N,N,N′,N′-tetramethylethylene diamine (TEMED) SDS–polyacrylamide gels (Will et al., 1994). Western blots were probed with Y12 anti-Sm (Lerner et al., 1981), anti-D2, anti-D3, anti-F, anti-G (Raker et al., 1996) and anti-HA (BAbCo) antibodies, and developed by the enhanced chemiluminescence method (Amersham).
Relevant polypeptide bands were excised and analysed at the Harvard Microchemistry Facility by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (µLC/MS/MS) on a Finnigan LCQ quadrupole ion trap mass spectrometer. The spectra were correlated with sequences from the non-redundant DDBJ/EMBL/GenBank (nr) and EST databases (dbEST) using the Sequest algorithm (Chittum et al., 1998). Alternatively, MALDI-TOF analysis was performed at University of Aberdeen Protein Laboratory.
Plasmids
IMAGE EST clones were obtained from the UK Human Genome Mapping Project (HGMP) Resource centre. The Lsm10 ORF was amplified by PCR from EST AA043070 and subcloned into appropriate vectors. For expression in Escherichia coli as an N-terminal–GST fusion, it was inserted into Gex4T-2 (Pharmacia). A PstI deletion removed the central part encoding the Sm motifs and fused the first 22 to the last 24 codons. The pcDNA3-HA mammalian cell expression vector with an N-terminal HA tag was made by inserting annealed DNA oligonucleotides encoding the HA epitope (MAYPYDVPDYASLE) into the HindIII and XhoI sites of pcDNA3 (Invitrogen). For U7 snRNAs with modified base-pairing region, the first 20 nt of U7 snRNA in plasmids p49 (U7 Sm WT) and p74 (U7 Sm OPT; Grimm et al., 1993) were replaced by the 28 nt sequence 5′-AAGUUGGUGGGGCUGUUGGCUCGAGUGC-3′. Further details are available on request.
RNA analytical techniques
Methods for 3′-end labelling of RNAs (Lamond and Sproat, 1994), native gel analysis of U7 snRNPs (Melin et al., 1992) and RNase mapping (Grimm et al., 1993) have been described. Primer extensions were performed with oligodeoxynucleotides complementary to nt 18–33 and 10–25 of U7 (Soldati and Schümperli, 1988) and U1 snRNAs, respectively.
Cell cultures and extracts
293T cells were grown to 50–60% confluency in 10 cm dishes and transfected with 10 µg of DNA using Lipofectamine (Life Technologies). For stable expression of HA-Lsm10, cells were co-transfected with pcDNA3-HA-Lsm10 and a hygromycin B resistance plasmid, followed by selection with hygromycin B. Transfections with 28-U7 WT or 28-U7 OPT plasmids were carried out on two consecutive days. Cells were harvested 48 h after the last transfection.
For preparation of nuclear extracts, cells were lysed by incubation in 10 mM HEPES pH 7.9, 100 mM KCl, 1 mM EDTA, 1 mM DTT, 0.5% NP-40 for 5 min at 4°C, mixed vigorously and centrifuged at 5000 r.p.m. at 4°C. The nuclear pellets were washed with lysis buffer lacking NP-40, resuspended in 250 mM Tris–HCl pH 7.9, 100 mM KCl, 1 mM EDTA, 1 mM DTT, 0.5% NP-40, 20% glycerol and shaken at 4°C for 1 h. After centrifugation at 10 000 r.p.m. for 30 min at 4°C, the extract was collected and used directly for further experiments.
Immunological techniques
The insoluble GST–Lsm10 was recovered from the pellet fraction of E.coli BL21 (Pharmacia), resolved by SDS–PAGE, and a gel fragment mixed with adjuvants was used for immunization of rabbits. Antibodies were affinity purified by overnight incubation with recombinant proteins blotted on nitrocellulose membrane. A depletion with strips containing GST alone was followed by selection with GST–Lsm10 (for Lsm10full antibody) or GST–Lsm10ΔPstI (for Lsm10ΔPstI antibody). The last filter was washed three times with 20 mM Tris pH 7.6, 137 mM NaCl for 5 min each. Bound antibodies were eluted with 0.2 M glycine pH 2.8, neutralized, and stored at 4°C.
Immunoprecipitations were performed with HeLa nuclear extracts (Will et al., 1994). Immunoprecipitated RNAs were recovered by proteinase K treatment and phenol extraction.
For immunofluorescence microscopy, HeLa cells grown on cover slips were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min. After permeabilization with 0.1% Triton X-100 in PBS for 2 min, the cells were incubated for 1 h with Lsm10ΔPstI antibody (diluted 1:100 in PBS with 1% goat serum) and/or monoclonal p80 coilin antibody SP10 (Sleeman et al., 1998; 1:100 000). Cells were then washed in PBS and incubated with goat anti-rabbit (1:500; coupled to Oregon Green) and/or anti-mouse (1:500; Texas Red) antibodies before being mounted with Fluoprep (bioMérieux SA, France). All incubations were carried out at room temperature.
Acknowledgments
Acknowledgements
The p80-coilin antibody was a gift of C.Smythe and had been produced in the laboratory of A.I.Lamond. We thank O.Mühlemann for critical comments, M.Ku for assistance with immunolocalization, V.Metzler for the 28-WT and 28-OPT plasmids, and J.Gygax, A.Badoiun and G.Heyne for technical help. This work was supported by the State of Bern and by Swiss National Science Foundation grant 31-52619.97 to D.S. and B.M. Work in B.M.’s laboratory at Aberdeen University was supported by the Wellcome Trust and that in R.L.’s laboratory by a grant from the DFG (SFB 523/A8).
References
- Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achsel T., Stark,H. and Lührmann,R. (2001) The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl Acad. Sci. USA, 98, 3685–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bach M., Bringmann,P. and Lührmann,R. (1990) Purification of small nuclear ribonucleoprotein particles with antibodies against modified nucleosides of small nuclear RNAs. Methods Enzymol., 181, 232–257. [DOI] [PubMed] [Google Scholar]
- Boeck R., Lapeyre,B., Brown,C.E. and Sachs,A.B. (1998) Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol., 18, 5062–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond U.M., Yario,T.A. and Steitz,J.A. (1991) Multiple processing-defective mutations in a mammalian histone premessenger RNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev., 5, 1709–1722. [DOI] [PubMed] [Google Scholar]
- Bouveret E., Rigaut,G., Shevchenko,A., Wilm,M. and Seraphin,B. (2000) A Sm-like protein complex that participates in mRNA degradation. EMBO J., 19, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H., Raymackers,J., Union,A., de Keyser,F., Meheus,L. and Lührmann,R. (2000) The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem., 275, 17122–17129. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol,A., Ebel,J.P., Lazar,E., Haendler,B. and Jacob,M. (1982) U2 RNA shares a structural domain with U1, U4 and U5 RNAs. EMBO J., 1, 1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C.B., Padgett,R.A. and Sharp,P.A. (1998) Evolutionary fates and origins of U12-type introns. Mol. Cell, 2, 773–785. [DOI] [PubMed] [Google Scholar]
- Chittum H.S., Lane,W.S., Carlson,B.A., Roller,P.P., Lung,F.D., Lee,B.J. and Hatfield,D.L. (1998) Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry, 37, 10866–10870. [DOI] [PubMed] [Google Scholar]
- Collins B.M., Harrop,S.J., Kornfeld,G.D., Dawes,I.W., Curmi,P.M. and Mabbutt,B.C. (2001) Crystal structure of a heptameric Sm-like protein complex from Archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol., 309, 915–923. [DOI] [PubMed] [Google Scholar]
- Cooper M., Johnston,L.H. and Beggs,J.D. (1995) Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J., 14, 2066–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z. and Marzluff,W.F. (1999) Formation of the 3′ end of histone mRNA. Gene, 239, 1–14. [DOI] [PubMed] [Google Scholar]
- Frey M.R. and Matera,A.G. (1995) Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl Acad. Sci. USA, 92, 5915–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A., Neubauer,G., Banroques,J., Mann,M., Lührmann,R. and Fabrizio,P. (1999) Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6⋅U5] tri-snRNP. EMBO J., 18, 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Stefanovic,B. and Schümperli,D. (1993) The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J., 12, 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann H., Fabrizio,P., Raker,V.A., Foulaki,K., Hornig,H., Brahms,H. and Lührmann,R. (1995) snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J., 14, 2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakata M., Craft,J. and Hardin,J.A. (1993) Autoantigenic epitopes of the B and D polypeptides of the U1 snRNP. Analysis of domains recognized by the Y12 monoclonal anti-Sm antibody and by patient sera. J. Immunol., 150, 3592–3601. [PubMed] [Google Scholar]
- Hoch S.O., Eisenberg,R.A. and Sharp,G.C. (1999) Diverse antibody recognition patterns of the multiple Sm-D antigen polypeptides. Clin. Immunol., 92, 203–208. [DOI] [PubMed] [Google Scholar]
- Kambach C., Walke,S., Young,R., Avis,J.M., de la Fortelle,E., Raker,V.A., Lührmann,R., Li,J. and Nagai,K. (1999) Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell, 96, 375–387. [DOI] [PubMed] [Google Scholar]
- Lamond A.I. and Sproat,B.S. (1994) Isolation and characterization of ribonucleoprotein complexes. In Higgins,S.J. and Hames,B.D. (eds), RNA Processing. Vol. 1. Oxford University Press, Oxford, UK, pp. 103–140.
- Lehmeier T., Raker,V., Hermann,H. and Lührmann,R. (1994) cDNA cloning of the Sm proteins D2 and D3 from human small nuclear ribonucleoproteins: evidence for a direct D1–D2 interaction. Proc. Natl Acad. Sci. USA, 91, 12317–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E.A., Lerner,M.R., Janeway,C.A. and Steitz,J.A. (1981) Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl Acad. Sci. USA, 78, 2737–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liautard J.P., Sri-Widada,J., Brunel,C. and Jeanteur,P. (1982) Structural organization of ribonucleoproteins containing small nuclear RNAs from HeLa cells. Proteins interact closely with a similar structural domain of U1, U2, U4 and U5 small nuclear RNAs. J. Mol. Biol., 162, 623–643. [DOI] [PubMed] [Google Scholar]
- Lührmann R., Kastner,B. and Bach,M. (1990) Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta, 1087, 265–292. [DOI] [PubMed] [Google Scholar]
- Mayes A.E., Verdone,L., Legrain,P. and Beggs,J.D. (1999) Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J., 18, 4321–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin L., Soldati,D., Mital,R., Streit,A. and Schümperli,D. (1992) Biochemical demonstration of complex formation of histone pre-mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J., 11, 691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mital R., Albrecht,U. and Schümperli,D. (1993) Detection of UV-induced RNA:protein crosslinks in snRNPs by oligonucleotides complementary to the snRNA. Nucleic Acids Res., 21, 1049–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Query,C.C. and Sharp,P.A. (1993) Splicing of precursors to mRNA by the spliceosome. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 303–357.
- Müller B. and Schümperli,D. (1997) The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin. Cell Dev. Biol., 8, 567–576. [DOI] [PubMed] [Google Scholar]
- Mura C., Cascio,D., Sawaya,M.R. and Eisenberg,D.S. (2001) The crystal structure of a heptameric archaeal Sm protein: implications for the eukaryotic snRNP core. Proc. Natl Acad. Sci. USA, 98, 5532–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker V.A., Plessel,G. and Lührmann,R. (1996) The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J., 15, 2256–2269. [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J., Bragado-Nilsson,E., Kandels-Lewis,S. and Seraphin,B. (1999) Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J., 18, 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F., Gilmartin,G.M., Bannwarth,W. and Birnstiel,M.L. (1986) Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature, 323, 777–781. [DOI] [PubMed] [Google Scholar]
- Seraphin B. (1995) Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J., 14, 2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J., Lyon,C.E., Platani,M., Kreivi,J.P. and Lamond,A.I. (1998) Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp. Cell Res., 243, 290–304. [DOI] [PubMed] [Google Scholar]
- Smith H.O., Tabiti,K., Schaffner,G., Soldati,D., Albrecht,U. and Birnstiel,M.L. (1991) Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc. Natl Acad. Sci. USA, 88, 9784–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati D. and Schümperli,D. (1988) Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol. Cell. Biol., 8, 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H., Dube,P., Lührmann,R. and Kastner,B. (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature, 409, 539–543. [DOI] [PubMed] [Google Scholar]
- Stauber C., Soldati,D., Lüscher,B. and Schümperli,D. (1990) Histone-specific RNA 3′ processing in nuclear extracts from mammalian cells. Methods Enzymol., 181, 74–89. [DOI] [PubMed] [Google Scholar]
- Stefanovic B., Hackl,W., Lührmann,R. and Schümperli,D. (1995) Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res., 23, 3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Galli,G., Busslinger,M. and Birnstiel,M.L. (1984) The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J., 3, 2801–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., He,W., Mayes,A.E., Lennertz,P., Beggs,J.D. and Parker,R. (2000) Yeast Sm-like proteins function in mRNA decapping and decay. Nature, 404, 515–518. [DOI] [PubMed] [Google Scholar]
- Toro I., Thore,S., Mayer,C., Basquin,J., Seraphin,B. and Suck,D. (2001) RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J., 20, 2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub H., Raker,V.A., Kostka,S. and Lührmann,R. (2001) Sm protein–Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J., 20, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walke S., Bragado-Nilsson,E., Seraphin,B. and Nagai,K. (2001) Stoichiometry of the Sm proteins in yeast spliceosomal snRNPs supports the heptamer ring model of the core domain. J. Mol. Biol., 308, 49–58. [DOI] [PubMed] [Google Scholar]
- Will C.L., Kastner,B. and Lührmann,R. (1994) Analysis of ribonucleoprotein interactions. In Higgins,S.J. and Hames,B.D. (eds), RNA Processing. Vol. 1. Oxford University Press, Oxford, UK, pp. 141–177.
- Will C.L., Schneider,C., Reed,R. and Lührmann,R. (1999) Identification of both shared and distinct proteins in the major and minor spliceosomes. Science, 284, 2003–2005. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Murphy,C. and Gall,J.G. (1996) The Sm binding site targets U7 snRNA to coiled bodies (spheres) of amphibian oocytes. RNA, 2, 811–823. [PMC free article] [PubMed] [Google Scholar]