Abstract
Human MutLα, a heterodimer of hMLH1 and hPMS2, is essential for DNA mismatch repair. Inactivation of the hmlh1 or hpms2 genes by mutation or epigenesis causes genomic instability and a predisposition to hereditary non-polyposis cancer. We report here the X-ray crystal structures of the conserved N-terminal 40 kDa fragment of hPMS2, NhPMS2, and its complexes with ATPγS and ADP at 1.95, 2.7 and 2.7 Å resolution, respectively. The NhPMS2 structures closely resemble the ATPase fragment of Escherichia coli MutL, which coordinates protein–protein interactions in mismatch repair by undergoing structural transformation upon binding of ATP. Unlike the E.coli MutL, whose ATPase activity requires protein dimerization, the monomeric form of NhPMS2 is active both in ATP hydrolysis and DNA binding. NhPMS2 is the first example of a GHL ATPase active as a monomer, suggesting that its activity may be modulated by hMLH1 in MutLα, and vice versa. The potential heterodimer interface revealed by crystallography provides a mutagenesis target for functional studies of MutLα.
Keywords: DNA binding/GHL ATPase/HNPCC/mismatch repair/PMS2
Introduction
DNA mismatch repair (MMR) corrects replication errors and is thus essential for maintaining genome integrity. Mutation of the genes encoding MMR proteins, MutL and MutS, or alteration of the expression level of MutL results in a mutator phenotype which is often manifested in micro-satellite instability (Kolodner, 1996; Modrich and Lahue, 1996; Buermeyer et al., 1999). In addition, these MMR proteins have also been implicated in mitotic and meiotic recombination, drug resistance and transcription-coupled repair (Anthoney et al., 1996; Datta et al., 1996; Mellon and Champe, 1996; Denamur et al., 2000). Interest in the MMR system was heightened when the direct link between mutations in the mismatch repair genes and hereditary nonpolyposis colon cancer (HNPCC) was established (Fishel and Kolodner, 1995; HNPCC Mutation Database, 1997; Peltomaki and Vasen, 1997).
MutL and its homologs are found in organisms ranging from Escherichia coli to humans. MutL plays an essential role in MMR by coordinating various protein–protein interactions, thereby earning the pseudonyms ‘molecular matchmaker’ and ‘molecular switch’ (Sancar and Hearst, 1993; Ban et al., 1999). MutL proteins are either homo- or heterodimers with subunits of 70–100 kDa. The N-terminal ∼350 residues of MutL homologs share extensive sequence homology; the remaining C-terminal region shows little sequence similarity, but has been shown to be responsible for protein dimerization in E.coli, yeast and humans (Li and Modrich, 1995; Pang et al., 1997; Drotschmann et al., 1998; Raschle et al., 1999).
The structure of the conserved N-terminal 40 kDa fragment of E.coli MutL, LN40, has been determined, and this domain has been shown to possess both an ATPase and DNA binding activity (Ban and Yang, 1998). The apo-LN40 comprises 331 residues and is monomeric, with about one-fifth of the protein disordered in the crystal. In the subsequently determined structure of an LN40–ADPnP complex, the entire polypeptide chain of LN40 becomes ordered and the protein dimerizes (Ban and Yang, 1998; Ban et al., 1999). Several residues that undergo dramatic conformational changes are involved in nucleotide binding, but many contribute to the dimer interface. The ATPase activity of E.coli MutL depends on the association of the N-terminal conserved region and appears to be regulated by DNA, in particular by single-stranded (ss)DNA (Ban et al., 1999). The structural transformation induced by ATP binding and hydrolysis allows MutL to switch interactions with various proteins and coordinate the MMR process (Ban and Yang, 1998; Junop et al., 2001).
LN40 shares sequence and structural similarity with the ATPase fragment of DNA gyrase B (NgyrB) and the ATP binding domain of Hsp90 (Wigley et al., 1991; Prodromou et al., 1997; Stebbins et al., 1997; Ban et al., 1999). Members of these three protein families lack the conventional ATPase signature motif, the Walker A motif, but share four uniquely conserved sequence motifs and a common tertiary structure. They are collectively called the GHL ATPases (Ban and Yang, 1998). These four sequence motifs have also been found to form an ATP binding site in bacterial histidine kinases similar to that of the GHL ATPases (Mushegian et al., 1997; Bilwes et al., 2001).
Protein dimerization appears to be essential for the enzymatic activity among the GHL ATPases and histidine kinases studied thus far. For instance, histidine kinases catalyze the phosphotransfer reaction between two protein subunits in trans. The kinase core domain of one subunit binds ATP and phosphorylates a conserved histidine residue of the second subunit (Robinson et al., 2000). In DNA gyrase, a single ATP binding site comprises residues from both protein subunits (Wigley et al., 1991). Similarly, dimerization of LN40 and its ATPase activity are interdependent. Mutations that weaken the association of the LN40 region also reduce the ATPase activity of MutL (Ban et al., 1999). In addition, dimerization in Hsp90 upon ATP binding has also been observed (Prodromou et al., 2000). The emerging scheme is that dimerization may be a common theme among these ATPases and kinases, and yet it serves a unique function in each protein family. All of these proteins form homodimers, however, and all interactions between subunits are reciprocal.
Humans possess several mutL-like genes, including hpms1, hpms2, hmlh1 and hmlh3, all of which form heterodimers (Li and Modrich, 1995; Raschle et al., 1999; Lipkin et al., 2000). hMutLα, in which hMLH1 is paired with hPMS2, is essential for MMR in humans (Li and Modrich, 1995; Prolla et al., 1998). Functions of the heterodimer are apparently asymmetric in MMR since mutations of the equivalent residues in the two subunits based on sequence conservation result in different mutator phenotypes (Tran and Liskay, 2000). Many missense mutations in hmlh1 and a few in hpms2 segregate with HNPCC in genetic screens (HNPCC Mutation Database, 1997; Peltomaki and Vasen, 1997). Several of these mutations have been mapped to the potential ATP binding site (Ban et al., 1999). Results from two-hybrid analyses suggested that yeast MutLα can bind ATP, and binding of ATP promotes the association of the N-terminal region (Tran and Liskay, 2000). However, no ATPase activity has been reported with purified eukaryotic MutL homologs in vitro.
To uncover the potential ATPase activity of MutLα, and to elucidate the relationship between the two protein subunits and their functional roles in mismatch repair as well as in cancer biology, we have cloned, expressed and purified the conserved 40 kDa N-terminal fragment of hPMS2 (NhPMS2), and found both an ATPase and DNA binding activity intrinsic to NhPMS2. Here, we present the crystal structures of the apo-NhPMS2, NhPMS2–ATPγS and NhPMS2–ADP complexes, and discuss the structural and functional implications of the ATPase activity associated with the monomeric NhPMS2, an exception in the GHL ATPase superfamily.
Results
NhPMS2 hydrolyzes ATP and binds DNA
We cloned the N-terminal conserved regions of human MLH1, PMS1 and PMS2 according to the sequence alignment with E.coli MutL (Ban and Yang, 1998), and expressed each with removable N-terminal histidine affinity tags in E.coli. Only the N-terminal hPMS2, NhPMS2, expressed in a soluble form. Attempts to refold the N-terminal fragments of hMLH1 and hPMS1 from inclusion bodies or to generate soluble protein by co-expression of both subunits of each heterodimer, either truncated or full length, were unsuccessful. The purified NhPMS2, which contains residues 1–365 of hPMS2, was examined for DNA binding and ATPase activity.
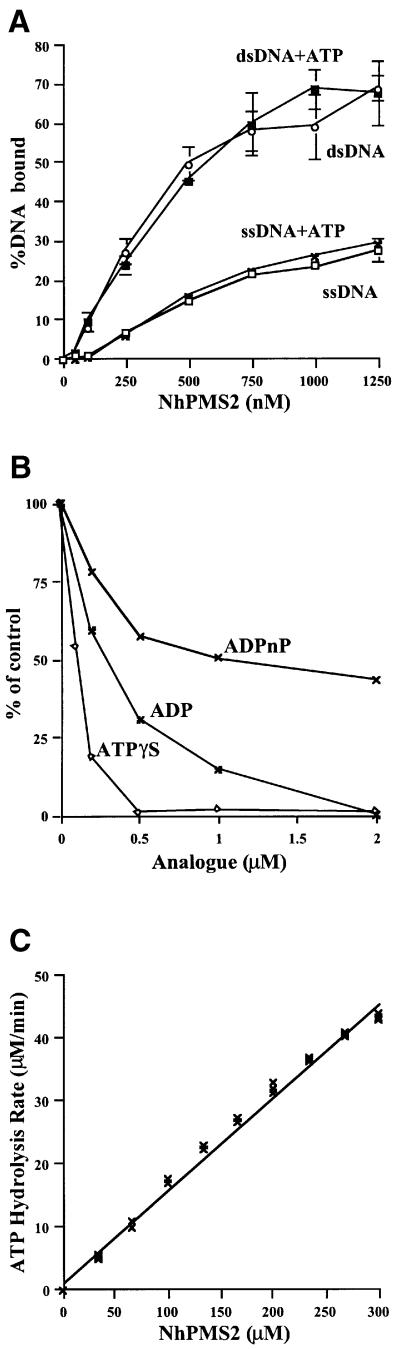
DNA gel mobility shift assays showed a discrete band corresponding to NhPMS2–DNA complexes after incubating NhPMS2 with either double-stranded (ds) or ssDNA of 110 bases in length. In contrast to E.coli MutL (Ban et al., 1999), NhPMS2 preferentially binds to dsDNA and its affinity for DNA does not increase with the addition of ATP (Figure 1A).
Fig. 1. DNA binding and ATPase activity of NhPMS2. (A) A plot of the percentage of either single stranded or homoduplex of 110 bp DNA (3 ng) bound to NhPMS2 versus increasing amount of NhPMS2, in the presence or absence of ATP. (B) Inhibition of the NhPMS2 ATPase activity by increasing amount of ATPγS, ADP or ADPnP. (C) Linear correlation between the NhPMS2 ATPase activity and protein concentrations.
Incubation of NhPMS2 with α-32P-labeled ATP resulted in the hydrolysis of ATP to ADP and Pi. The measured Km of NhPMS2 for ATP is ∼100 µM and kcat is 0.2 min–1 at pH 8 and room temperature. The NhPMS2 ATPase activity is not affected by the presence of DNA. The kcat of NhPMS2 is lower than that of the intact E.coli MutL (0.4 min–1), but much higher than the equivalent LN40 of E.coli MutL (0.04 min–1), and the Km is essentially identical for all three proteins (Ban and Yang, 1998; Ban et al., 1999). This ATPase activity is most effectively inhibited by ATPγS, and less effectively by ADP and ADPnP (Figure 1B). In order to verify that this ATPase activity is intrinsic to NhPMS2, we mutated residue Glu41, the equivalent to Glu29 of E.coli MutL, which acts as the general base in ATP hydrolysis (Ban et al., 1999). The NhPMS2-E41A mutant shows no detectable ATPase activity, thus confirming that NhPMS2 is a genuine ATPase.
It came as a surprise that NhPMS2 can hydrolyze ATP at a level close to that of the MutL full-length protein. As a GHL ATPase, hPMS2 is expected to require dimerization with hMLH1 for ATPase activity. We reasoned that it might be possible that the N-terminal fragment of hPMS2, like LN40, could form a homodimer in the presence of nucleotide. Dimeric NhPMS2, however, was not detectable by gel filtration, protein cross-linking or equilibrium ultracentrifugation after incubation with non-hydrolyzable ATP analogs (data not shown). It is possible that NhPMS2 forms a transient dimer to allow ATP hydrolysis, but dissociates too quickly to be detected even when ATPγS is present. To check whether dimerization, however transient, is required for the ATPase activity of NhPMS2, we measured the ATPase activity of NhPMS2 as a function of protein concentration. The ATPase activity of NhPMS2 was linear with increasing protein concentration and did not show a cooperative effect (Figure 1C), suggesting that the active form of NhPMS2 is monomeric.
Crystal structure of NhPMS2
We were able to crystallize the soluble NhPMS2 fragment, which diffracted X-rays to better than 2 Å Bragg spacings. Despite the conservation of amino acid sequence and the ATPase activity, efforts to solve the NhPMS2 structure by molecular replacement using either the LN40 or LN40–ADPnP structure as a search model were unsuccessful. The structure of NhPMS2 was eventually solved using the multiwavelength anomalous dispersion (MAD) method on the selenomethionine-substituted protein. The final model contains residues 29–365 with two internal loops disordered and is refined to 1.95 Å resolution (Table I). There are two NhPMS2 molecules in each asymmetric unit related by a non-crystallographic dyad axis. However, this dyad relationship does not appear to be functionally relevant.
Table I. Data collection and refinement statistics.
| Data collection | Low energy | Se edge | Se peak | High energy | NhPMS2–ATPγS | NhPMS2–ADP |
|---|---|---|---|---|---|---|
| Wavelength (Å) | 0.9879 | 0.9793 | 0.9788 | 0.9686 | 1.5418 | 1.5418 |
| Resolution (Å)a | 30–1.95 (1.98–1.95) | 30–2.1 (2.14–2.1) | 30–2.1 (2.14–2.1) | 30–2.0 (2.05–2.0) | 25–2.7 (2.8–2.7) | 25–2.7 (2.8–2.7) |
| Completeness (%)a | 97.7 (99.3) | 96.6 (95.3) | 95.8 (78.6) | 95.4 (73.9) | 95.1 (89.3) | 92.3 (91.7) |
| Rmerge (%)a,b | 6.2 (31.8) | 6.1 (24.2) | 6.5 (23.4) | 6.6 (34.6) | 6.1 (36.2) | 6.4 (30.4) |
|
I/σ(I)b |
18.5 (1.6) |
21.6 (3.2) |
20.8 (3.1) |
19.9 (2.4) |
14.5 (1.8) |
12.5 (2.1) |
| Refinement |
|
|
|
|
|
|
| Resolution (Å) | 20–1.95 | 25–2.7 | 25–2.7 | |||
| Reflections work set | 48 335 | 18 884 | 18 569 | |||
| Reflections test set | 5470 | 1385 | 1371 | |||
| Protein atoms | 4822 | 4713 | 4707 | |||
| Solvent atoms | 292 | 33 | 50 | |||
| R (Rfree) (%)c | 21.8 (24.3) | 23.7 (27.0) | 23.0 (26.2) | |||
| R.m.s.d. in bonds (Å) | 0.007 | 0.010 | 0.010 | |||
| R.m.s.d. in angles (°) | 1.3 | 1.8 | 1.7 | |||
| Mean B values (Å2) | 35.9 | 55.9 | 51.2 |
aNumber in parentheses correspond to the last resolution shell.
bRmerge = ΣhΣi|Ihi – <Ih>|/Σ<Ih>.
cR = Σ||Fobs| – |Fcalc||/Σ|Fobs|.
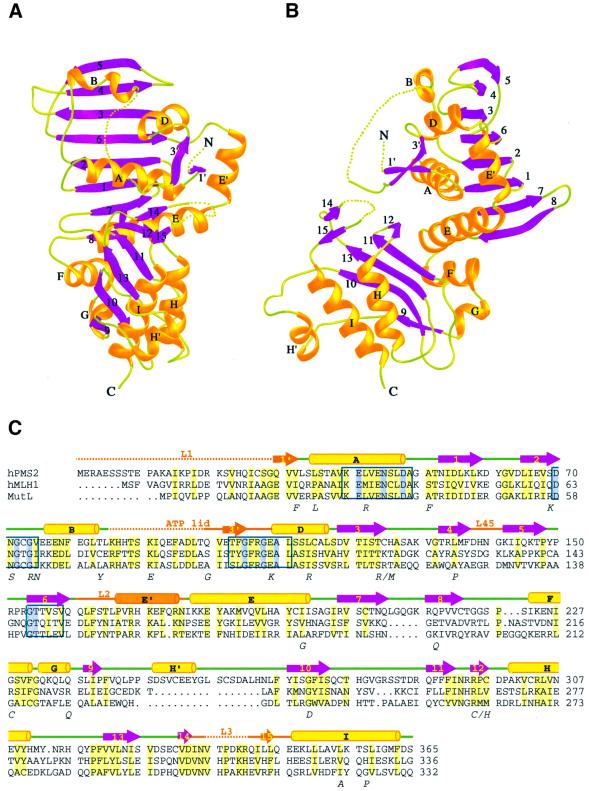
The structure of NhPMS2 consists of two α/β domains connected by two short helices, αF and αG (residues 222–239) (Figure 2). The N-terminal domain (residues 29–221) comprises a mixed β-sheet of eight strands (β1–8) packed against a layer of five helices (αA–E). In addition, this domain contains a pair of short parallel β-strands, β1′ and β3′, held together by four pairs of hydrogen bonds. Residues 86–95 are disordered. The second domain (residues 239–365) consists of a five-stranded mixed β-sheet (β9–13) and three helices (αH′, αH and αI) forming an α/β-barrel and two short β-strands, β14 and β15, covering one end of the barrel. Residues 336–341, which connect β14 and β15, are disordered (Figure 2).
Fig. 2. Structure of NhPMS2. (A and B) Orthogonal views of NhPMS2 structure in ribbon diagram. β-strands are shown as purple arrows, α and 310 helices as gold ribbons and connecting loops as light green coils. Disordered loops are shown as green dotted lines. Secondary structures are labeled using the same convention as for the LN40 structure. New structural elements in NhPMS2 are labeled with the same name as the following structural element with a prime (E′ helix lies between D and E helices). N- and C-termini are marked for clarity. (C) Sequence alignment of hPMS2, hMLH1 and MutL. Loops undergoing structural transformation in LN40 are labeled and shown in orange. ATP binding motifs (I–IV) are boxed in blue. The residues conserved in the GHL superfamily are shaded in blue, the rest of the conserved residues are shaded in light yellow.
Structural comparison of NhPMS2 with LN40
Superimposition of the NhPMS2 and LN40 structures confirms an overall structural similarity between the two homologs, as predicted from the sequence alignment (Ban and Yang, 1998) (Figure 3). NhPMS2 contains several new structural elements due to an addition of 34 amino acid residues in comparison with LN40 (Figure 2C). The N-terminal L1 loop alone contains an addition of 12 residues. Although mostly disordered in the crystal structure, residues 30–32 form a β-strand, β1′, which is absent in all three structural forms of LN40. A two-residue insertion results in the shift of the β8-strand (Figure 2C) (Ban and Yang, 1998). The other major addition in the NhPMS2 occurs in the second structural domain comprising residues 248–265, which results in a helix αH′ inserted between strands β9 and 10 (Figure 2).
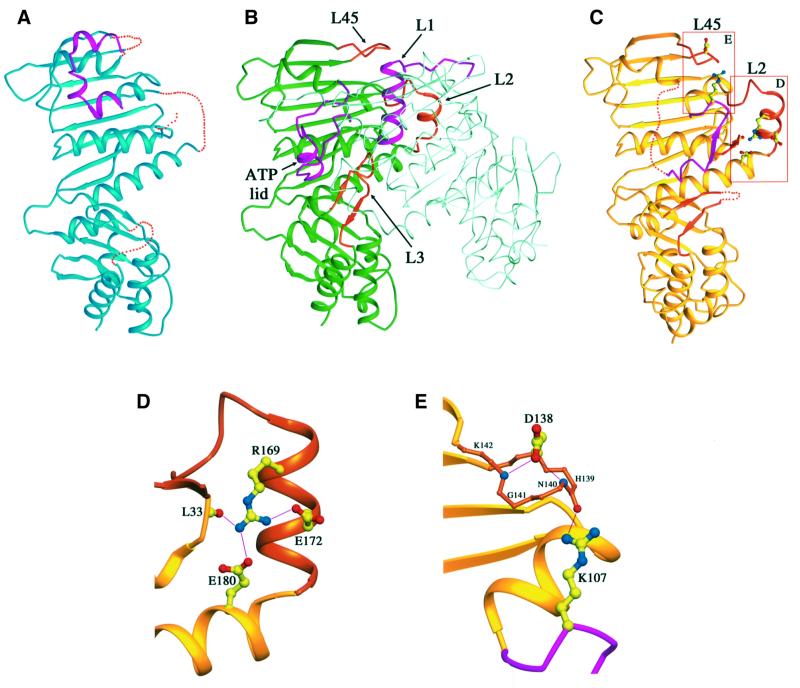
Fig. 3. Comparison of LN40 and NhPMS2 structures. Ribbon diagrams of (A) apo-LN40 in blue, (B) dimeric LN40 in complex with ADPnP, one monomer shown in dark green and the other as a Cα trace in light green, and (C) NhPMS2 in yellow. The ATP lid and L1 loop are shown in magenta, and other loops undergoing structural transformation in orange. Dotted lines represent disordered loops. (D) An enlarged view of L2 of NhPMS2. Side chains of Arg169, Glu172 and Glu180 and main-chain carbonyl group of residue Leu33 are shown as ball-and-sticks. Carbon atoms are shown in yellow, oxygen in red, and nitrogen in blue. Hydrogen bonds are shown as thin lines. (E) An enlarged view of L45. Cα trace of L45 with main-chain nitrogen atoms of residues Asn140 and Lys142 marked as blue spheres. Side chains of Asp138 and Arg107 are central for stabilizing the loop.
ATP binding induces a structural transformation of LN40, which includes reorienting the N- and C-terminal domains by 20° and the ordering of five flexible loops upon dimerization (Figures 2B, 3A and B) (Ban and Yang, 1998; Ban et al., 1999). Interestingly, half of the loops that undergo structural changes in LN40 are already ordered in the monomeric NhPMS2 structure in the absence of a ligand (Figure 3C) and are stabilized by the hydrogen bond networks within a single subunit (Figure 3D and E), instead of between subunits as observed in LN40. The relative orientation between the two domains is also more similar to that of the LN40–ADPnP complex. The overall NhPMS2 structure resembles that of a single LN40 subunit in the ADPnP complex more closely than the LN40 apo-protein structure (Figure 3).
Three loops, L2, L3 and L45, are either fully or partially ordered in the apo-protein of NhPMS2. The L2 loop (residues 162–176) adopts a helical conformation (αE′) much like the structure of the L2 loop in the LN40–ADPnP complex (Figure 3B and C). The presence of one additional residue in L2 of NhPMS2 allows helix αE′ to gain an extra turn. In NhPMS2, αE′ is stabilized by an intramolecular hydrogen bond network (Figure 3D). Similarly, L45 (residues 137–142) is stabilized by interactions within the loop and further anchored into the protein core structure by a hydrogen bond from a main-chain carbonyl oxygen to the guanidinium group of Arg107 (Figure 3E). The αH′ helix and the following loop, unique in NhPMS2, buttress β14 and β15 of the L3 loop to contact the N-terminal domain (residues 96–101), as in the LN40–ADPnP complex. The L3 hairpin loop is thus mostly ordered, except residues 336–341 near the tip of the β-turn.
The ATP binding site
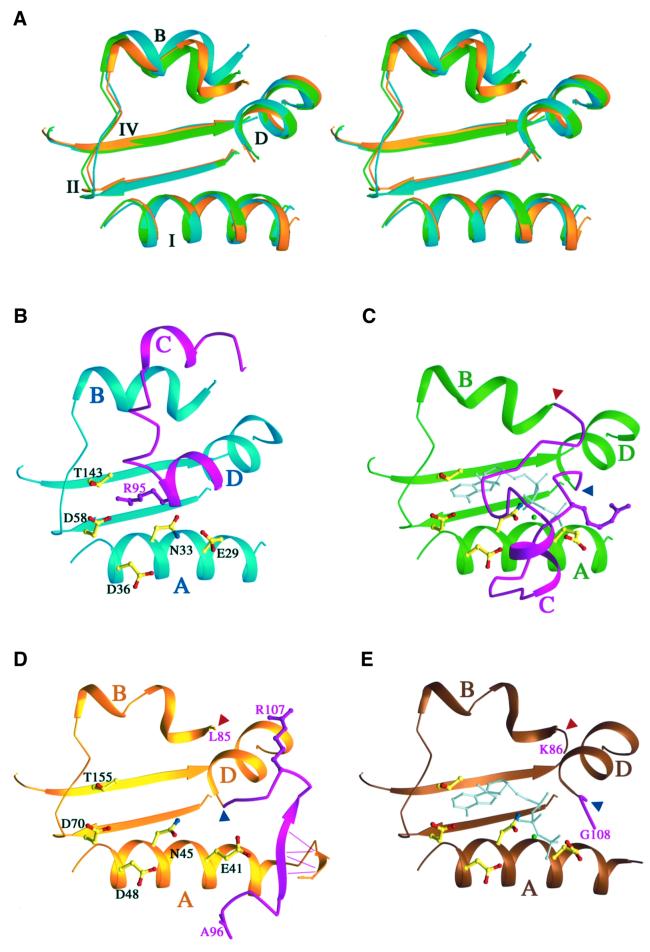
The structures of motifs I, II and IV, which form a platform for ATP binding in the GHL ATPases and histidine kinases, are conserved in all structures determined to date, including NhPMS2, regardless of the presence or absence of ATP (Figures 3 and 4A) (Wigley et al., 1991; Prodromou et al., 1997; Stebbins et al., 1997; Ban and Yang, 1998; Ban et al., 1999; Bilwes et al., 1999, 2001). The ATP lid (residues 85–109), which encompasses Motif III (Figure 2C) and can form a cover to enclose the nucleotide, is very flexible and adopts different conformations in different proteins depending on the presence of ligands (Figure 4) (Prodromou et al., 2000). In the LN40 structure without ligand, parts of the ATP lid are disordered, while the ordered portion, Arg95 in particular, occupies the ATP binding site. In the LN40–ADPnP structure, helix D, which contains Arg95, unfolds by one turn and unblocks the ATP binding site. In addition, helix αC shifts by over 10 Å towards the dimer interface, and the disordered part of the ATP lid becomes fully ordered and wraps around the nucleotide (Figure 4B and C).
Fig. 4. Comparison of the active site of LN40 and NhPMS2. (A) Ribbon diagram of the constant part of the ATP binding site. Superimposition of NhPMS2 (gold), LN40–ADPnP (green) and apo-LN40 (blue) encompassing motifs I, II and IV, and helices B and D is shown in stereo. (B–E) Ribbon diagrams of the complete ATP binding site of apo-LN40 (B), LN40 complexed with ADPnP (C), apo-NhPMS2 (D), and NhPMS2 complexed with ATPγS (E). The ATP lids are shown in solid magenta when traceable. The start and end residues of the ATP lid of NhPMS2 structures are labeled and marked with red and blue arrowheads, respectively. Side chains of conserved residues involved in nucleotide binding are shown as ball-and-sticks. Arg95 of LN40 and its equivalent, Arg107 of hPMS2, are shown as magenta ball-and-sticks. The bound nucleotide is shown as a gray stick model.
In NhPMS2, residues 86–95 of the ATP lid are disordered and the ordered portion, residues 96–109 encompassing the motif III of the GHL ATP superfamily, assumes different structures from those found in the LN40 and LN40–ADPnP complex (Figure 4B–D). In NhPMS2, the interactions with β1′ of L1 and β14 of the second domain hold the ATP lid such that the ATP binding site is wide open (Figure 4D). Arg107 of NhPMS2, the equivalent to Arg95 of MutL, is far removed from the ATP binding site and interacts with the L45 loop instead (Figure 3E). Helix αD in the structure of NhPMS2 is as short as, and oriented similarly to, αD in the LN40–ADPnP complex. In addition, helix αB, which leads into the ATP lid, and L3 in the second domain, which contains a catalytically important Lys residue, are oriented towards the active site in a manner very similar to those found in the LN40–ADPnP structure (Figures 3C and 4D). The NhPMS2 active site is therefore engaged to receive ATP and lacks only the ordered ATP lid in comparison with the fully active LN40–ADPnP structure.
NhPMS2–ATPγS structure: monomeric NhPMS2 binds ATP
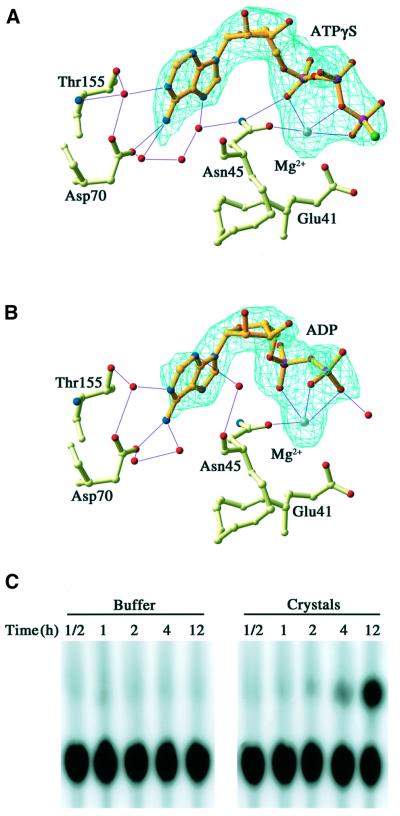
Based on the efficient inhibition of NhPMS2 ATPase activity by the non-hydrolyzable ATP analog ATPγS (Figure 1B), we set out to examine an activated state of NhPMS2 by growing crystals of the NhPMS2–ATPγS complex. The crystals of NhPMS2–ATPγS complex were obtained only under conditions containing >2.0 M phosphate salt and were isomorphous to that of the apo-protein. The electron density map obtained from the co-crystal of NhPMS2 and ATPγS suggested low occupancy of ATPγS in the ATP binding site and increased movement of the L1 loop and the ATP lid.
To alleviate the problem of low occupancy, which is likely to be due to high salt concentrations in the crystallization buffer, we transferred apo-NhPMS2 crystals grown at high salt concentrations to a stabilization buffer containing 25% PEG4000 and low concentrations of salt, and soaked ATPγS into the protein crystal (see Materials and methods). The resulting difference Fourier map unambiguously showed the ATPγS in the active site of both NhPMS2 molecules in the asymmetric unit, which was refined at full occupancy (Figure 5A).
Fig. 5. NhPMS2 binds ATPγS and hydrolyzes ATP. (A and B) Omit-map of the ATP binding pocket in the structures of NhPMS2 complexed with ATPγS (A), and ADP (B). Fo – Fc maps were calculated after omitting the nucleotide and Mg2+ ion, contoured at 2.5 σ in green, and superimposed on the final refined ATPγS and ADP. Side chains involved in nucleotide binding, Glu41 and Asn45 (motif I), Asp70 (II) and Thr155 (IV) are also shown. Nucleotides are shown in gold ball-and-stick models; nitrogen, oxygen, phosphorus, sulfur and Mg2+ ion are shown in blue, red, purple, green and light gray, respectively. (C) [α-32]P-labeled ATP hydrolysis by NhPMS2 crystals analyzed on a TLC plate. The left five lanes are of the buffer solution and the right five lanes are of the NhPMS2 crystals at various time points as indicated.
The structure of the NhPMS2–ATPγS complex has the same overall shape as the apo-NhPMS2. The side-chain rotamer conformation of the general base, Glu41 on motif I, is switched to point towards, instead of away from, the γ-phosphate (Figure 4D and E). The hydrogen bonds holding the β1′ of L1 and β3′ of the ATP lid together are no longer present, thus releasing the sequestered ATP lid (Figure 4D and E). Residues 31–33 of the L1 loop form a loop unrelated to its conformation in the apo-NhPMS2 structure. However, crystal-packing contacts do not allow the ATP lid of NhPMS2 to form, as is found in the LN40–ADPnP structure. The extended portion of the ATP lid can easily form, but the αC helix would clash with the neighboring molecule at the β-turn that connects strands seven and eight. As a result, the ATP lid is disordered from residues 87 to 107 (Figure 4E). Interestingly, the traceable ends that lead into the ATP lid suggest that the projection of the lid is towards the bound ATPγS, like that in the LN40–ADPnP complex (Figure 4C–E).
All the interactions between ATPγS and motifs I, II and IV of NhPMS2, including those mediated by water molecules, are very similar to those observed in the LN40–ADPnP complex (Ban et al., 1999). In particular, hydrogen bonds that define the specificity for the adenine base are conserved, and Asn45, which is absolutely conserved, coordinates the Mg2+ ion, which in turn chelates the oxygen atoms of α, β and γ phosphates. Even though the ATP lid does not adopt a defined structure, it probably makes transient interactions with the phosphate moiety of the bound ATPγS. The structure of the NhPMS2–ATPγS complex clearly shows that a monomer of NhPMS2 has all the required elements for binding ATP.
Monomeric NhPMS2 hydrolyzes ATP
Based on the ability of NhPMS2 to bind ATPγS in the crystal lattice, we designed the following two experiments to confirm that the monomeric NhPMS2 possesses ATPase activity. As an initial approach, we checked the ATPase activity of crystallized NhPMS2 since it was trapped as monomers. We washed crystals of the apo-NhPMS2 in a low salt stabilizing solution to remove any free protein molecules and detected hydrolysis of ATP by addition of α-32P-labeled ATP to the stabilizing mother liquor (Figure 5). To confirm that ATP is hydrolyzed to ADP by NhPMS2 in the crystal, we then soaked a native crystal in the low salt stabilizing solution with 1 mM ATP and Mg2+ for 12 h, and collected the X-ray diffraction data of the ‘live’ crystal. The resulting 2Fo – Fc and Fo – Fc maps showed electron density of ADP with no sign of the γ-phosphate (Figure 5B). The ATP binding site is similar overall to that of the NhPMS2–ATPγS structure, including the disordered ATP lid (residues 86–109). Thus, the enzymatic studies carried out in solution as well as in the crystal lattice confirm that the monomeric NhPMS2 has all the required elements for binding and hydrolyzing ATP.
Discussion
Implications of the NhPMS2 ATPase activity
Our observation of the ATPase activity of the monomeric NhPMS2 establishes a new paradigm for GHL ATPases and histidine kinases, which in all previous cases had required dimerization for measurable ATPase activity. A unique feature of the MutL subfamily of GHL ATPases is that all the residues that coordinate ATP and catalyze hydrolysis belong to a single polypeptide chain. In E.coli MutL, ATP binding precedes dimerization. The N302A MutL mutant, which is defective in dimerization, binds ATP similarly to the wild type (Ban et al., 1999). Dimerization of E.coli MutL concurs with the ordering of five loops, which presumably reinforces the formation of the catalytic center. In the monomeric NhPMS2, two of these five loops are fully ordered. The L3 loop, which contains the catalytically important Lys340, an equivalent of K307 in E.coli MutL, is mostly ordered. Lys340 remains disordered in the nucleotide complex, but was restricted enough in conformation to perform its catalytic role in ATP hydrolysis. An ordered ATP lid is apparently not essential for ATP hydrolysis by NhPMS2.
hPMS2 depends on an association with hMLH1 to form MutLα in vivo for the MMR function, despite being independently active in ATP binding and hydrolysis. Truncated hPMS2 lacking the C-terminal region responsible for dimerization with MLH1 is often associated with a mutator phenotype and HNPCC, indicating that the ATPase activity of PMS2 alone is insufficient for normal MMR (HNPCC Mutation Database, 1997; Peltomaki and Vasen, 1997). hPMS2 probably does not form a homodimer, and dimers of NhPMS2 are not observed in solution. Superimposition of NhPMS2 onto the dimeric LN40 results in clashes at L1, L2, L3 and the ATP lid, despite the fact that large portions of L1 and the ATP lid are disordered (Figure 2C). Interestingly, the N-terminal region of hMLH1 contains three residues fewer than LN40, instead of 34 residues more as in the case of NhPMS2 (Figure 2C). The resulting minimal structure of hMLH1 may provide space either to homodimerize or accommodate the extra structural elements of NhPMS2. By evolving into heterodimers, in which each subunit may become optimized to specialized functions, eukaryotic proteins perhaps become more efficient in carrying out MMR. Indeed, even though prokaryotic MutS and MutL are homodimers, structural analyses indicate that at least E.coli and Taq MutS are functionally asymmetric (Lamers et al., 2000; Obmolova et al., 2000).
Functional dissection of MutLα is hampered by the lack of an in vitro reconstituted eukaryotic mismatch repair assay and the lack of identification of proteins other than MutS homologs, which interact with the MLH1–PMS2 heterodimer to complete MMR. The asymmetry of the heterodimeric hMutLα certainly suggests many modes of interaction between the MLH1 and PMS2, between MutLα and its effector proteins, and between hMutLα and DNA. The ATPase activity of NhPMS2 may either be enhanced or inhibited by hMLH1. The lack of measurable ATPase activity of the isolated intact hMutLα may be due to inactivation during the purification process, but it may result from genuine inhibition by intersubunit interactions. If so, future research is needed to elucidate where the inhibition occurs, such as at the stage of ATP binding, hydrolysis, or product release, and also the significance of the MutLα ATPase activity (or the lack of it) in the process of MMR.
DNA binding versus ATPase activity
A DNA binding groove is formed between the two N-terminal ATPase fragments in E.coli MutL that is far away from the ATP binding site (Ban et al., 1999). The presence of ATP or non-hydrolyzable ATP analogs enhances DNA binding by MutL and the presence of DNA, most effectively ssDNA, stimulates the MutL ATPase activity. The interdependence of DNA binding and the ATPase activity of MutL is likely mediated by the dimerization of the N-terminal domains (Ban et al., 1999). Since NhPMS2 does not form a homodimer upon association with ATP, it is not surprising that its DNA binding property is not affected by the presence of ATP.
Binding of DNA by MutL is essential in E.coli MMR. Mutations that eliminate the DNA binding by MutL also abolish the MMR in vivo (M.S.Junop, W.Yang and J.H.Miller, in preparation). The DNA binding cleft is probably retained in the eukaryotic MutL homologs upon dimerization of the N-terminal region, judging from the structure of NhPMS2. Although residues involved in DNA binding by E.coli MutL are not conserved in human MutL homologs, positive charges on helix αI in the center of the DNA binding cleft are preserved (Ban et al., 1999). The additional helix αH′ adjacent to αI in NhPMS2 potentially increases the DNA–protein interface (Ban et al., 1999). Besides helices αH′ and αI, residues on helix αG and the loop connecting β-strands 10 and 11 also contribute to the overall positive charge in NhPMS2. The variability of size, shape and charge distribution of the DNA binding cleft indicates that the mode of DNA recognition may be different in each MutL homolog, such as the preference for ss or dsDNA and the exact location of the DNA binding surface. Although the ATPase and DNA binding activities of NhPMS2 appear to be independent, it is possible that in the MLH1–PMS2 heterodimer the two activities influence one another.
HMutLα and HNPCC
The roles of PMS2 and MLH1 in MMR are apparently not equivalent, as indicated by previous studies (Prolla et al., 1998; Tran and Liskay, 2000). Point mutations of yeast MutLα showed that defective MLH1 is more deleterious to MMR than defective PMS2 (Tran and Liskay, 2000). Many missense mutations isolated from HNPCC kindreds are located in hMLH1, and only nonsense mutations are found in NhPMS2 (Peltomaki and Vasen, 1997). The limited conformational changes observed in the structures of NhPMS2 and its complexes with ATPγS and ADP suggest that PMS2 may provide a relatively constant structural platform for MLH1 to be the ‘molecular switch’ and communicate with other molecules.
We examined the 25 currently known HNPCC missense mutations within the N-terminal conserved region of hMLH1 (Peltomaki and Vasen, 1997), 10 of which were not known when the LN40 structures were determined (Ban and Yang, 1998; Ban et al., 1999). According to homologous sequence-based structural modeling (Figure 2C), six of them (I25F, N64S, C77Y, K84E, S93G and E102K) are predicted to be located on or adjacent to the conserved motifs I, II and III, thereby affecting the ATPase activity of hMLH1. H329P is located in the middle of helix αI, which forms the DNA binding groove. Mutation to proline is likely to disrupt the helix formation and hence the function of hMLH1. Interestingly, the remaining three mutations, Q62K, E199Q and R217C, seem to be located on the exposed surface on the side of the molecule opposite the ATPase active site. Whether this exposed surface is involved in molecular interactions significant for MMR awaits future determination.
Conclusion
The ATPase activity found in the monomeric NhPMS2 indicates that dimerization is not essential for every member of the GHL family to hydrolyze ATP. The correlation between dimerization and ATPase activity is perhaps pertinent to the homodimeric members, but not to the heterodimeric ones. Internal stabilization of critical regions in eukaryotic MutL homologs, which are mobile in E.coli MutL, affords them ATPase activity in the absence of dimerization. The independent ATPase activity of each protein subunit in hMutLα is probably functionally significant. The structures of NhPMS2 illuminate the regions where hPMS2 is likely to interact with hMLH1, such as the disordered L1 loop and the αC helix on the ATP lid. Future mutagenesis studies can thus target residues in these key areas. A cluster of genes homologous to the conserved N-terminal region of hPMS2 has been found on human chromosome 7 (Horii et al., 1994; Nicolaides et al., 1995; Kondo et al., 1999). These truncated pms2-like gene products do not interact with hMLH1, but some may contain the ATPase activity and play important biological roles as yet undefined. The structural transformation, the versatility of the enzymatic activity and the multitude of inter- as well as intramolecular regulation revealed in MutL, and now in hPMS2, provide a foundation for the dissection of the multiple functions of MutL and its homologs.
Materials and methods
Protein purification and crystallization
NhPMS2 of residues 1–365 was subcloned into the pET15b vector (pWY1161). The His-tagged NhPMS2 was expressed in E.coli BL21(DE3) (Novagen) after induction by isopropyl-β-d-thiogalactopyranoside at 30°C for 4 h. Protein was purified on a Ni affinity column (Pharmacia) using the standard protocol and followed by a Mono-Q column pre-equilibrated with 150 mM KCl (Pharmacia). NhPMS2 protein flowed through the Mono-Q column while most of the contaminants were retained. The N-terminal His tag was removed by thrombin (Novagen) digestion at 12 U per mg of protein for 1 h at room temperature. The NhPMS2 protein with His tag removed was finally purifed using a Mono-S column (Pharmacia) pre-equilibrated with 20 mM HEPES pH 6.3, 150 mM KCl, 1 mM EDTA, 5 mM dithiothreitol (DTT) and 5% glycerol, and eluted with a linear gradient from 150 to 500 mM KCl. NhPMS2 was concentrated to 10 mg/ml in 20 mM Tris buffer pH 8.5, 200 mM KCl, 0.1 mM EDTA, 5 mM DTT and 5% glycerol. Six liters of E.coli culture yielded ∼0.5–1 mg of pure NhPMS2. Selenomethionyl (SeMet) NhPMS2 was overexpressed in B834(DE3) using minimal media as described (Hendrickson et al., 1990), and purified as for the native protein.
Crystals of NhPMS2 were grown using the hanging drop vapor diffusion method. The crystallization buffer contained 2–2.4 M Na/K phosphate pH 6.2 and 0.2 M LiCl. Crystals were flash-frozen in liquid propane in the mother liquor with an addition of 20% glycerol as a cryo-protectant. NhPMS2–ATPγS complex was formed by soaking native crystals in a solution containing 25% PEG 4K, 0.1 M Na/K phosphate pH 6.2, 0.1 M LiCl, 5 mM MgCl2 and 20 mM ATPγS at room temperature for 30 min. NhPMS2–ADP complex was formed by soaking crystals in the same solution containing 1 mM ATP instead of ATPγS at room temperature overnight. Subsequently, crystals were frozen using the same mother liquor with 10% ethylene glycol as a cryo-protectant.
Data collection and structure determination
A native data set at 2.4 Å resolution was collected using an R-AXIS IV image plate detector mounted on an RU200 rotating anode X-ray generator (Rigaku). The space group was determined to be P212121 with unit cell dimensions a = 74.06 Å, b = 74.15 Å, c = 134.1 Å. There are two monomers in the asymmetric unit. The native crystal was determined to contain pseudo-merohedral twinning (α = 0.05) due to the similarity of the a and b axes. Twinning was overcome by using SeMet-labeled protein and replacing Li2SO4 with LiCl in the crystallization buffer; the unit cell dimensions became a = 74.16 Å, b = 74.85 Å, c = 135.34 Å.
The MAD data from a SeMet NhPMS2 crystal were collected at low-energy remote, absorption edge, absorption peak and high-energy remote up to 1.95 Å resolution at the X9B beamline at the National Synchrotron Light Source, Brookhaven National Laboratory. Data sets were indexed and scaled using DENZO and SCALEPACK (Table I) (Otwinowski and Minor, 1997). Eight of 10 possible selenium sites were found and refined at 2.3 Å resolution using SOLVE (Terwilliger and Berendzen, 1999), which produced a mean figure of merit of 0.52 and an overall score of 68. After solvent flattening with DM (CCP4, 1994), all of the traceable residues of the NhPMS2 molecule were easily modeled into the experimental map. After refinement using standard protocols in CNS (Brunger et al., 1998), the final model of NhPMS2 contains residues 29–85, 96–335 and 342–365 of molecule A, residues 33–85, 108–333 and 343–365 of molecule B, and 292 water molecules (Table I). Over 91% of the residues are within the most favored regions in the Ramachandran plot and the rest are in the additional allowed regions.
The complete diffraction data sets of the NhPMS2–ATPγS and NhPMS2–ADP crystals were collected at home to 2.7 Å each. These crystals were nearly isomorphous to those of the apo-protein (Table I). After rigid body refinement, both 2Fo – Fc and Fo – Fc maps showed clear density for the nucleotide moiety. Final models include residues 31–86, 108–335 and 342–364 for the ATPγS complex and residues 27–85, 110–333 and 344–364 for the ADP complex, a nucleotide molecule and a Mg2+ ion for each molecule in the asymmetric unit. NhPMS2–ATPγS has 91% and NhPMS2–ADP 90% of the residues within the most favored regions of the Ramachandran plot and the rest are in the additional allowed regions.
NhPMS2 mutants
The E41A mutant of NhPMS2 (pWY1192) was derived from plasmid pWY1161 using the QuickChange site-directed mutagenesis kit (Stratagene). The sequence of the mutant was verified using DNA sequencer PRISM-310 (ABI). NhPMS2-E41A was expressed and purified as for the native protein.
ATPase activity assay
The ATPase activity of NhPMS2 and NhPMS2-E41A was assayed by incubating the protein with α-32P-labeled ATP in 20 mM Tris pH 8, 1 mM DTT, 90 mM KCl and 5 mM MgCl2. Reactions were initiated by addition of α-32P-labeled ATP, incubated at room temperature for 0.5–4 h, and terminated by the addition of an equal volume of 50 mM EDTA. Km and kcat were obtained with 1 µM NhPMS2 and 25 µM–1 mM ATP over a 2 h reaction time course. ATP and ADP were separated on TLC plates developed in 0.75 M KH2PO4 and quantified using a TYPHOON 8600 phosphorImaging plate system (Molecular Dynamics).
ATPase activity of the NhPMS2 crystals was assayed in 25% PEG 4K, 0.1 M Na/K phosphate pH 6.2, 0.1 M LiCl, 5 mM MgCl2, containing 0.2 mM α-32P-labeled ATP. Crystals were washed repeatedly in the stabilization buffer to remove all soluble protein carried in the crystallization mother liquor. Aliquots of the supernatant were extracted at various time points and mixed with an equal volume of 50 mM EDTA. Reaction products were separated on TLC plates as described above.
DNA binding assay
DNA binding of NhPMS2 was determined by the gel electrophoretic mobility shift assay using a similar protocol to that of Bende and Grafstrom (1991). Three nanograms of 5′ 32P end-labeled ss or dsDNA 110 bp in length were incubated with increasing concentrations of NhPMS2, as indicated in Figure 1, in 20 mM Tris pH 8, 50 mM KCl, 1 mM DTT, 5 mM MgCl2 and 0.1 mg/ml bovine serum albumin on ice for 1 h. The reaction samples (10 µl) were analyzed on a 6% polyacrylamide gel and quantified using a TYPHOON 8600 phosphorImaging plate system. The effect of ATP on DNA binding was checked by addition of 1 mM ATP to the reaction mixtures.
Figures 2A–5B were generated using RIBBONS (Carson, 1987).
The coordinates of NhPMS2, NhPMS2–ATPγS and NhPMS2–ADP have been deposited with the Protein Data Bank (PDB). The accession codes are 1h7s, 1h7u and 1ea6.
Acknowledgments
Acknowledgements
We thank Drs Z.Dauter and C.Ogata for synchrotron beamline support, Dr H.Ling for assistance in X-ray data collection, B.Vogelstein and K.Kinzler for the hMLH1, hPMS1 and hPMS2 clones, E.Deye for constructing the original NhPMS2 plasmid, and R.Craigie and D.Leahy for comments on the manuscript. A.G. and M.S.J. are supported by post-doctoral fellowships from the Human Frontier Science Program Organization and Medical Research Council of Canada, respectively.
References
- Anthoney D.A., McIlwrath,A.J., Gallagher,W.M., Edlin,A.R. and Brown,R. (1996) Microsatellite instability, apoptosis, and loss of p53 function in drug-resistant tumor cells. Cancer Res., 56, 1374–1381. [PubMed] [Google Scholar]
- Ban C. and Yang,W. (1998) Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell, 95, 541–552. [DOI] [PubMed] [Google Scholar]
- Ban C., Junop,M. and Yang,W. (1999) Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell, 97, 85–97. [DOI] [PubMed] [Google Scholar]
- Bende S.M. and Grafstrom,R.H. (1991) The DNA binding properties of the MutL protein isolated from Escherichia coli. Nucleic Acids Res., 19, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilwes A.M., Alex,L.A., Crane,B.R. and Simon,M.I. (1999) Structure of CheA, a signal-transducing histidine kinase. Cell, 96, 131–141. [DOI] [PubMed] [Google Scholar]
- Bilwes A.M., Quezada,C.M., Croal,L.R., Crane,B.R. and Simon,M.I. (2001) Nucleotide binding by the histidine kinase CheA. Nature Struct. Biol., 8, 353–360. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Buermeyer A.B., Deschenes,S.M., Baker,S.M. and Liskay,R.M. (1999) Mammalian DNA mismatch repair. Annu. Rev. Genet., 33, 533–564. [DOI] [PubMed] [Google Scholar]
- Carson M. (1987) Ribbon models of macromolecules. J. Mol. Graph., 5, 103–106. [Google Scholar]
- CCP4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Datta A., Adjiri,A., New,L., Crouse,G.F. and Jinks Robertson,S. (1996) Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur E. et al. (2000) Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell, 103, 711–721. [DOI] [PubMed] [Google Scholar]
- Drotschmann K., Aronshtam,A., Fritz,H.J. and Marinus,M.G. (1998) The Escherichia coli MutL protein stimulates binding of Vsr and MutS to heteroduplex DNA. Nucleic Acids Res., 26, 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R. and Kolodner,R.D. (1995) Identification of mismatch repair genes and their role in the development of cancer. Curr. Opin. Genet. Dev., 5, 382–395. [DOI] [PubMed] [Google Scholar]
- Hendrickson W.A., Horton,J.R. and LeMaster,D.M. (1990) Seleno methionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J., 9, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HNPCC mutation database (1997) (cited April 1, 2001) http://www.nfdht.nl/databasemlh1.htm
- Horii A., Han,H.J., Sasaki,S., Shimada,M. and Nakamura,Y. (1994) Cloning, characterization and chromosomal assignment of the human genes homologous to yeast PMS1, a member of mismatch repair genes. Biochem. Biophys. Res. Commun., 204, 1257–1264. [DOI] [PubMed] [Google Scholar]
- Junop M.S., Obmolova,G., Rausch,K., Hsieh,P. and Yang,W. (2001) Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell, 7, 1–12. [DOI] [PubMed] [Google Scholar]
- Kolodner R. (1996) Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev., 10, 1433–1442. [DOI] [PubMed] [Google Scholar]
- Kondo E., Horii,A. and Fukushige,S. (1999) The human PMS2L proteins do not interact with hMLH1, a major DNA mismatch repair protein. J. Biochem. (Tokyo), 125, 818–825. [DOI] [PubMed] [Google Scholar]
- Lamers M.H., Perrakis,A., Enzlin,J.H., Winterwerp,H.H., de Wind,N. and Sixma,T.K. (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature, 407, 711–717. [DOI] [PubMed] [Google Scholar]
- Li G.M. and Modrich,P. (1995) Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc. Natl Acad. Sci. USA, 92, 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin S.M., Wang,V., Jacoby,R., Banerjee-Basu,S., Baxevanis,A.D., Lynch,H.T., Elliott,R.M. and Collins,F.S. (2000) MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nature Genet., 24, 27–35. [DOI] [PubMed] [Google Scholar]
- Mellon I. and Champe,G.N. (1996) Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. and Lahue,R. (1996) Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem., 65, 101–133. [DOI] [PubMed] [Google Scholar]
- Mushegian A.R., Bassett,D.E.,Jr, Boguski,M.S., Bork,P. and Koonin,E.V. (1997) Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Proc. Natl Acad. Sci. USA, 94, 5831–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaides N.C., Carter,K.C., Shell,B.K., Papadopoulos,N., Vogelstein,B. and Kinzler,K.W. (1995) Genomic organization of the human PMS2 gene family. Genomics, 30, 195–206. [DOI] [PubMed] [Google Scholar]
- Obmolova G., Ban,C., Hsieh,P. and Yang,W. (2000) Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature, 407, 703–710. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pang Q., Prolla,T.A. and Liskay,R.M. (1997) Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol. Cell. Biol., 17, 4465–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltomaki P. and Vasen,H.F. (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The international collaborative group on hereditary nonpolyposis colorectal cancer. Gastroenterology, 113, 1146–1158. [DOI] [PubMed] [Google Scholar]
- Prodromou C., Roe,S.M., O’Brien,R., Ladbury,J.E., Piper,P.W. and Pearl,L.H. (1997) Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell, 90, 65–75. [DOI] [PubMed] [Google Scholar]
- Prodromou C., Panaretou,B., Chohan,S., Siligardi,G., O’Brien,R., Ladbury,J.E., Roe,S.M., Piper,P.W. and Pearl,L.H. (2000) The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J., 19, 4383–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla T.A. et al. (1998) Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nature Genet., 18, 276–279. [DOI] [PubMed] [Google Scholar]
- Raschle M., Marra,G., Nystrom-Lahti,M., Schar,P. and Jiricny,J. (1999) Identification of hMutLβ, a heterodimer of hMLH1 and hPMS1. J. Biol. Chem., 274, 32368–32375. [DOI] [PubMed] [Google Scholar]
- Robinson V.L., Buckler,D.R. and Stock,A.M. (2000) A tale of two components: a novel kinase and a regulatory switch. Nature Struct. Biol., 7, 626–633. [DOI] [PubMed] [Google Scholar]
- Sancar A. and Hearst,J.E. (1993) Molecular matchmakers. Science, 259, 1415–1420. [DOI] [PubMed] [Google Scholar]
- Stebbins C.E., Russo,A.A., Schneider,C., Rosen,N., Hartl,F.U. and Pavletich,N.P. (1997) Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell, 89, 239–250. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P.T. and Liskay,R.M. (2000) Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLα. Mol. Cell. Biol., 20, 6390–6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley D.B., Davies,G.J., Dodson,E.J., Maxwell,A. and Dodson,G. (1991) Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature, 351, 624–629. [DOI] [PubMed] [Google Scholar]