Abstract
Here we investigate the promoter control of alternative splicing by studying two transcriptional activators on templates under replicating conditions. SV40 large T-antigen (T-Ag) activates template replication only 2-fold but transcription 25-fold. T-Ag-mediated replication, reported to inhibit RNA polymerase II elongation, provokes a 10- to 30-fold increase in the inclusion of the fibronectin EDI exon into mature mRNA. The T-Ag effect is exon specific, occurs in cis and depends strictly on DNA replication and not on cell transformation. VP16, an activator of transcriptional initiation and elongation, has a similar effect on transcription but the opposite effect on splicing: EDI inclusion is inhibited by 35-fold. VP16 completely reverts the T-Ag effect, but a VP16 mutant with reduced elongation ability provokes only partial reversion. Both T-Ag and VP16 promote conspicuous co-localization of mRNA with nuclear speckles that contain the SR protein SF2/ASF, a positive regulator of EDI inclusion. Therefore, we conclude that co-localization of transcripts and speckles is not sufficient to stimulate EDI inclusion.
Keywords: alternative splicing/SV40 T-Ag/VP16
Introduction
The production of mature, translatable mRNA in most eukaryotic cells involves transcription by RNA poly merase II (pol II) and three RNA processing mechanisms: capping, splicing and cleavage/polyadenylation. Among these, alternative splicing appears as a widespread means for producing polypeptide diversity from a single gene (Black, 2000). In human fibronectin (FN), for example, up to 20 different polypeptide variants arise from alternative splicing in three regions of a single gene (Gutman and Kornblihtt, 1987). However, this figure remains modest when compared with that of the Drosophila dscam gene, where an extremely complex array of alternative exons could potentially give rise to 38 016 DSCAM proteins, of which 49 mRNA species have already been identified (Schmucker et al., 2000). In spite of the estimation that 35% of human genes are expressed through alternative splicing (Croft et al., 2000), and the sophisticated functional, cell type and developmental specificities documented in many cases, the mechanisms of alternative splicing regulation are poorly understood. A key role in splice site choice regulation is played by members of the SR (Ser/Arg-rich) family of proteins. These proteins participate in constitutive and alternative splicing, by binding to exonic splicing enhancers (ESEs) and enhancing or repressing spliceosome assembly at adjacent splice sites. One model suggests that alternative splicing in different cell types or at different points in time is regulated by variation in relative abundance of SR proteins. However, SR proteins do not seem to have a highly specific tissue distribution, which suggests the existence of more complex regulatory mechanisms. SR proteins are also present in the nucleoplasm, move rapidly throughout the nucleus (Phair and Misteli, 2000) and some are able to shuttle between the nucleus and the cytoplasm (Cáceres et al., 1998).
Recent evidence indicates that transcription and pre-mRNA processing are not independent events. On the contrary, there is a high degree of co-ordination since all three processing reactions occur in intimate association with the elongating RNA pol II (for reviews see Bentley, 1999; Travers, 1999; Hirose and Manley, 2000; Proudfoot, 2000; Cramer et al., 2001). Transcriptional activation of pol II genes provokes association of SR proteins like SF2/ASF to sites of transcription (Misteli et al., 1997). This re-localization does not occur if RNA pol II has a truncated C-terminal domain (CTD) (Misteli and Spector, 1999), which is consistent with the fact that truncation of the CTD causes defects in cleavage/polyadenylation and splicing (McCracken et al., 1997). We have recently extended the concept of a functional coupling between transcription and splicing by demonstrating that differences in promoter structure lead to differences in alternative splicing of the transcript (Cramer et al., 1997). The system analyzed involved the EDI exon (also named EDA or EIIIA), which encodes a facultative repeat of FN (Kornblihtt et al., 1984). Alternative splicing of EDI varies during embryonic development, ageing, and in proliferative processes such as healing and cancer (Kornblihtt et al., 1996). EDI contains an ESE (Mardon et al., 1987; Lavigueur et al., 1993; Caputi et al., 1994) that is targeted by the SR proteins SF2/ASF and 9G8. Overexpression of SF2/ASF and 9G8 markedly stimulates EDI inclusion, but the effect of these proteins is modulated by the promoter (Cramer et al., 1999), demonstrating that cell-specific alternative splicing is not solely controlled by signals in the RNA molecule, but by a more complex process involving cell-specific promoter occupation.
In our search for a mechanism to explain the promoter control we studied the effects on alternative splicing of two activators with opposite effects on pol II elongation. SV40 large T-antigen (T-Ag) activates template replication by only 2-fold but transcription of replicated templates by 25-fold. Replication-mediated transcriptional activation provokes an important decrease in RNA pol II processivity, shown by the generation of shorter transcripts (Nahreini and Mathews, 1995; Williams et al., 1996). In these conditions, T-Ag provokes a 10- to 30-fold increase in the inclusion of the EDI exon. VP16, an activator of both transcriptional initiation and elongation, has the opposite effect on splicing: EDI inclusion is inhibited by 35-fold. Both T-Ag and VP16 promote nuclear co-localization of transcripts with SF2/ASF, which indicates that the association of SR proteins and transcripts detected by microscopy is not as relevant for the fate of alternative splicing as thought previously.
Results
Promoter modulation of alternative splicing is observed in stably transfected cell lines
Human hepatoma Hep3B cells were transiently transfected with the minigenes containing the α-globin, FN or CMV promoters (Figure 1), and alternative splicing of EDI was assessed by RT–PCR. As we previously demonstrated (Cramer et al., 1997, 1999), inclusion of EDI is lower when transcription is driven by the α-globin promoter compared with the FN or CMV promoters (Figure 2A, lanes 1, 3 and 5). To test whether the promoter modulation of alternative splicing was also observed in a physiological chromatin context, we generated Hep3B cell lines stably transfected with the α-globin and FN promoter constructs. EDI splicing patterns observed in the stable transfectants correlate with those of the transient transfectants: the α-globin promoter provokes lower EDI inclusion than the FN promoter regardless of the EDI inclusion levels of the endogenous FN mRNAs (Figure 2B). These results suggest that the physiological chromatin assembly of the integrated minigenes does not abolish the control of splice site selection by the promoter, originally found with episomal templates.
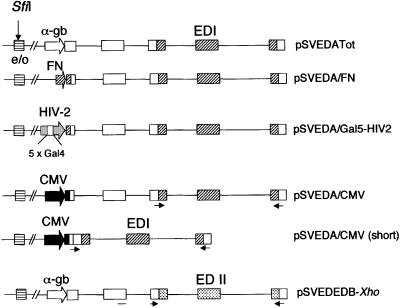
Fig. 1. Schemes of the minigenes carrying the different promoters. Codes for DNA sequences. White, human α-globin; dashed or dotted, human FN; black, CMV; horizontal hatching, SV40 enhancer/origin of replication (e/o); gray, HIV2. Thin arrows show the primers used to amplify the mRNA splicing variants by RT–PCR. The SfiI site used to disrupt the SV40 origin of replication is indicated.
Fig. 2. (A) Effect of T-Ag on EDI alternative splicing in the context of different promoters. Hep3B cells were transfected with 800 ng of the corresponding minigene plasmid and 800 ng of a plasmid expressing T-Ag (even lanes) or pBSKS+ (Stratagene) (odd lanes). (B) Endogenous (EDIe) and minigene-driven (EDIm) alternative splicing patterns of Hep3B cell lines stably transfected with different promoter minigenes. (C) Effect of the presence of the SV40 origin of replication on alternative splicing elicited by minigenes transfected in COS-7 cells. RNA splicing variants were detected by radioactive RT–PCR and analyzed in 6% native polyacrylamide gels. Ratios between radio activity in EDI+ bands and radioactivity in EDI– bands are shown under each lane.
SV40 T-Ag stimulates inclusion of the EDI alternative exon
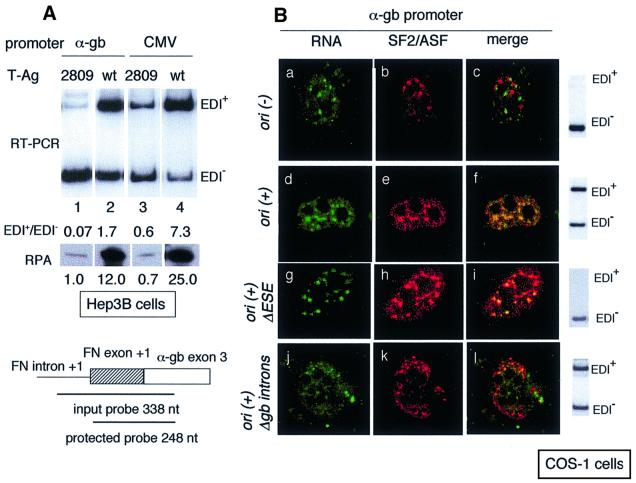
Plasmids maintained as episomes in mammalian cells are known to acquire nucleosomal organization. Replication of these plasmids provokes a more specific nucleosomal assembly (Cereghini and Yaniv, 1984; Gruss et al., 1990) that affects transcriptional regulation. To examine the role of template replication in alternative splicing, we co-transfected Hep3B cells with α-globin, FN or CMV promoter minigenes and a plasmid expressing SV40 T-Ag, taking advantage of the fact that the constructs in Figure 1 carry the SV40 origin of replication and therefore replicate only in the presence of T-Ag. Expression of T-Ag stimulates EDI inclusion with all three promoters by 10- to 30-fold (Figure 2A, lanes 2, 4 and 6), preserving the different promoter efficacies with respect to splicing. In fact, in the presence of T-Ag, FN and CMV promoters are respectively 2.2- and 7.7-fold better ‘includers’ than the α-globin promoter. The effect of T-Ag was corroborated in transfections of the minigenes into COS-7 cells that express SV40 T-antigen constitutively. Figure 2C shows that EDI inclusion is ∼20-fold higher in COS-7 cells than in Hep3B cells not supplemented with T-Ag (compare lanes 1 and 3 of Figure 2C with lanes 1 and 5 of Figure 2A). To prove that this difference was caused by template replication we disrupted the SV40 origin of replication by digestion with SfiI according to the method described by Proudfoot et al. (1992). This disruption affects only one of the two T-Ag-binding sites present in the SV40 enhancer/origin region of constructs in Figure 1 (binding site II; Fanning and Knippers, 1992). The high-affinity T-Ag-binding site I remains unaffected. The resulting ori(–) constructs transfected into COS-7 cells elicited much lower levels of EDI inclusion than the ori(+) constructs and similar to those of the ori(+) constructs transfected into Hep3B cells (Figure 2C, lanes 2 and 4).
The T-Ag effect on splicing is not due to cell transformation
T-Ag is a multifunctional phosphoprotein that not only participates in SV40 DNA replication but also promotes cell transformation. The latter is mainly due to T-Ag’s ability to associate with the retinoblastoma protein Rb and with p53. The Rb binding activity is restricted to a small region between amino acid (aa) residues 105 and 114, while the DNA replication activity is supported by different regions of the molecule including the DNA polymerase-α binding site (aa residues 1–83) and the ATPase and helicase activities located in the center of the molecule (Figure 3A). When transcription is driven by ori(+) templates in Hep3B cells, the T-Ag mutant lacking the Rb binding activity (K1 mutant; Zhu et al., 1991) is as potent as the wild type in stimulating EDI inclusion (Figure 3B, lanes 2 and 3). However, two T-Ag mutants that fail to support DNA replication (mutants 2809 and 2811; Zhu et al., 1991) are only 10% effective compared with the wild type (Figure 3B, lanes 4 and 5). Mutants 2809 and 2811 do not bind p53. The fact that the 10% remnant activity of these mutants on ori(+) constructs (lanes 1, 4 and 5) is comparable to the activity of the wt T-Ag on the ori(–) constructs (lanes 6 and 7) rules out p53 as a mediator of the effect on splicing. The behaviors of the K1, 2809 and 2811 T-Ag mutants in trans and of the ori(–) minigene mutants in cis allow us to conclude that the profound changes in splicing are due to some qualitative change in the minigene templates caused by replication and not to an indirect role of T-Ag through transformation.
Fig. 3. Effect of T-Ag mutants on EDI alternative splicing. (A) Scheme of the SV40 T-Ag molecule with the location of its binding and enzymatic activities and the positions of mutants K1, 2809 and 2811. (B) EDI alternative splicing in Hep3B cells co-transfected with pSVEDA/CMV having (lanes 1–5) or lacking (lanes 6–10) the SV40 origin of replication and plasmids expressing the wild type or different mutants of T-Ag (Zhu et al., 1991).
The T-Ag effect is exon specific
We have previously shown that SF2/ASF and 9G8 are the SR proteins involved in EDI alternative splicing in vivo, and that their effect is mediated through the recognition of the ESE present in the EDI exon (Cramer et al., 1999) (Figure 4A). Mutation of the ESE abolishes EDI inclusion into mature mRNA and prevents the stimulatory effect of SF2/ASF and 9G8 (Cramer et al., 1999). Figure 4B (lanes 1–4) shows that disruption of the ESE completely prevents the stimulation of EDI inclusion by T-Ag, indicating that SF2/ASF and/or 9G8 are needed in the mechanism that links replication to alternative splicing. To test whether the T-Ag effect on alternative splicing is specific to the EDI ESE or a more general phenomenon, we replaced the ESE with a binding site for a different SR protein, SRp-55. This replacement is functional since overexpression of SRp-55 stimulates inclusion of the alternative exon (Figure 4A). T-Ag stimulates the inclusion of this mutated exon in the same way as it affects the wild-type exon (Figure 4B, lanes 5 and 6). We also tested whether T-Ag influenced the inclusion of EDII (EDB or EIIIB; Figure 4A), another FN exon whose alternative splicing is controlled by SRp-40 and intronic elements (Lim and Sharp, 1998; lanes 7 and 8). When EDI and EDII minigenes are co-transfected in the same cells, T-Ag stimulates EDI but not EDII inclusion into mature mRNA (Figure 4A, lanes 9–12). As positive control, overexpression of SRp-40 stimulates inclusion of the EDII exon in our system (Figure 4A). We conclude that T-Ag specifically affects splice site choice at the EDI exon.
Fig. 4. The effect of T-Ag is exon specific. (A) Diagrams showing different alternative exons and their corresponding cis-controlling elements used to evaluate the effects of T-Ag on alternative splicing. ESE, exonic splicing enhancer for SF2/ASF (GAAGAAGAG) and for SRp-55 (GCACGGAC). Positive controls for the activation of exon inclusion by overexpression of SRp-55 and SRp-40 are shown. (B) Effect of T-Ag on alternative splicing of an EDI exon in which the natural ESE was disrupted (lanes 1–4) or replaced by the SRp-55 target site (lanes 5 and 6). Lanes 7–12, effect of T-Ag on EDII alternative splicing. Hep3B cells were co-transfected with pSVEDEDB-Xho (lanes 7–12) and either pBSKS+ (Stratagene) (lanes 7 and 8) or pSVEDATot (lanes 9–12). The plasmid expressing T-Ag (Zhu et al., 1991) was co-transfected in the odd lanes. RT–PCRs for EDI (lanes 1–6, 9 and 10) and for EDII (lanes 7, 8, 11 and 12) were as in Materials and methods.
The regulation of splicing by T-Ag-dependent replication is a cis effect
The splicing pattern elicited by a non-replicating [ori(–)] minigene is not affected by the introduction in the same cell of a replicating [ori(+)] minigene, and vice versa. We co-transfected COS-7 cells with two different constructs, pSVEDA/CMV and pSVEDA/CMV (short), both depicted in Figure 1, with or without an SV40 origin of replication. pSVEDA/CMV (short) is a variant of pSVEDA/CMV that lacks both globin introns and globin exon 2. Both constructs have the same promoter and give rise to the same EDI alternative splicing variants. However, the absence of globin exon 2 in pSVEDA/CMV (short) permitted the design of specific primers to distinguish their corresponding transcripts by RT–PCR. The splicing pattern of the transcripts arising from pSVEDA/CMV ori(–) alone is not modified by the presence in the same cell of either pSVEDA/CMV (short) ori(+) or pSVEDA/CMV (short) ori(–). Accordingly, the splicing patterns characteristic of pSVEDA/CMV (short) ori(+) and pSVEDA/CMV (short) ori(–) are not affected by the presence in the same cell of copies of pSVEDA/CMV ori(–) (data not shown). These experiments indicate that the replication effect is a property conferred exclusively to the replicated template itself, which can not be imposed in trans to other template molecules.
T-Ag stimulates the number of plasmid templates per nucleus only 2-fold
To determine whether the effect of T-Ag on alternative splicing correlated with a significant increase in the number of minigene templates, we estimated the total number of templates by recovering plasmid DNA from isolated nuclei of the transfected COS-7 cells (Hirt, 1967) and counting the number of colonies after transformation of Escherichia coli. Unlike plasmid molecules that have replicated in the mammalian cell, those that have entered the mammalian cell but have not replicated retain adenine methylation at the GATC sequences (dam-methylation) acquired in the bacterial cell, and are therefore insensitive to digestion with MboI. This property allowed us to determine the number of template copies that had replicated within the eukaryotic cell nucleus by treating the recovered plasmid DNA with MboI before transformation. Table I shows that, as previously reported in other SV40-based systems (Proudfoot et al., 1992; Nahreini and Mathews, 1995), T-Ag has a modest effect on the number of plasmid templates with only a 2.3-fold increase. Consistent with this result, the number of transcription foci detected by in situ hybridization probed with pSVEDATot labeled by nick translation shows little increase in the presence of T-Ag (1.63-fold) compared with the stimulation of EDI inclusion (23-fold). This points to qualitative changes in the DNA template caused by a few rounds of replication rather than a simple quantitative effect.
Table I. Effects of the presence of the SV40 origin of replication on the number and expression of plasmid constructs transfected in COS-1 cells.
| Transfected construct (pSVEDATot) |
Ratio | ||
|---|---|---|---|
| ori (–) | ori (+) | ori (+)/ori (–) | |
| Total plasmida (no MboI digestion) | 100 | 226 | 2.26 |
| Non-replicated plasmida (after MboI digestion) | 105 | 118 | 1.12 |
| Transcription foci per nucleus | 49.2 ± 10 | 80.4 ± 17.0 | 1.63 |
| % PML bodies associated to transcription foci | 13.1 (n = 290) | 60.8 (n = 268) | 4.64 |
| EDI+/EDI– | 0.6 | 14.0 | 23.33 |
aColony forming units obtained from plasmids recovered from 2.105 COS-1-transfected cells.
T-Ag-directed replication is restricted to specific nuclear domains named ND10 or PML bodies (promyelocytic leukemia nuclear bodies). The presence of a core origin and the expression of T-Ag are the only necessary conditions to target chimeric plasmids harboring the SV40 origin to PML bodies, when the increase in template copies is negligible as it happens in our experiments (Tang et al., 2000). In order to confirm the replicative nature of our effect we estimated the co-localization of PML bodies (detected by immunofluorescence) with transcription foci (detected by FISH) of COS-1 cells transfected either with replicative or non-replicative minigenes. Table I shows that when the transfected minigenes replicate, 60.8% of the PML bodies co-localize with transcription foci. In non-replicative conditions, only 13.1% of the PML bodies co-localize. We conclude that the effect of T-Ag on alternative splicing correlates with the localization of the template plasmids to PML bodies.
T-Ag increases mRNA levels and promotes co-localization of transcripts with speckles
The fact that T-Ag does not increase significantly the number of plasmid templates per cell suggests that its effect on alternative splicing might be the consequence of transcriptional activation of the replicated templates. Hep3B cells were transfected with α-globin or CMV promoter constructs [both ori(+)] and plasmids expressing the 2809 mutant or wild-type T-Ag. Since the 2809 mutant is unable to promote replication, its expression provides information on the transcriptional effect of T-Ag, while expression of wild-type T-Ag provides information on both replication and transcriptional effects. Figure 5A shows that compared with mutant 2809, wild-type T-Ag provokes 12- and 36-fold more transcription from the α-globin- [Figure 5A, RNase protection assays (RPAs) in lanes 1 and 2] and CMV-driven (RPAs in lanes 3 and 4) reporter genes, respectively. The magnitude of this promoter activation in the presence of wild-type T-Ag greatly exceeded the increase in template number. This result agrees with previous findings that promoter activity is greatly augmented on a replicative DNA template compared with a non-replicative one (Nahreini and Mathews, 1995).
Fig. 5. T-Ag elicits higher mRNA levels and co-localization of transcripts with speckles. (A) RPAs to measure RNA levels produced by T-Ag on Hep3B cells transfected with α-globin (lanes 1 and 2) and CMV (lanes 3 and 4) promoter constructs. Hep3B cells were co-transfected with the mentioned minigenes and plasmids expressing the 2809 mutant (lanes 1 and 3) or the wild-type version (lanes 2 and 4) of SV40 T-Ag. The design of the probe for the RPA is shown at the bottom. (B) In situ hybridization to minigene RNA (transcription foci, green) and immunofluorescence to SF2/ASF (speckles, red) of COS-1 cells transfected with different constructs. Panels a–c show a transfection with pSVEDATot ori(–). Panels d–f show a transfection with pSVEDATot ori(+). Panels g–i show a transfection with pSVEDA Δ2e (α-globin promoter, deleted ESE) ori(+). Panels j–l show a transfection minigene construct lacking the α-globin introns, similar in structure to pSVEDA/CMV (short) depicted in Figure 1, but under the control of the α-globin promoter. Co-localization (yellow) is conspicuous in panels f, i and l, but negligible in panel c. Gel electrophoreses of the RT–PCR products for EDI splicing of each transfection are shown to the right.
To investigate whether this increase correlates with a differential association of transcripts to speckles containing SR proteins we performed transfections of COS-1 cells with ori(–) and ori(+) constructs and double labeling of nuclei; transcription foci were detected by FISH in green, while speckles were detected by immunofluorescence in red, with an antibody against SF2/ASF. Ori(–) constructs display multiple transcription foci dispersed throughout the nucleus (Figure 5B, panel a), presenting no conspicuous association with speckles (panel c). On the contrary, a conspicuous co-localization of RNA and speckles is observed when ori(+) constructs are transfected, independently of whether they carry a wild-type (Figure 5B, panels d–f) or a disrupted (panels g–i) ESE. Transcripts produced by an ori (+) short minigene that lacks the globin introns also co-localize with speckles (Figure 5B, panels j–l).
Transcript association to speckles is not sufficient to stimulate EDI inclusion
To rule out the possibility that the effect of T-Ag on alternative splicing is the trivial consequence of an increase in transcriptional activity and a subsequent increase in the amounts of pre-mRNA that associate with speckles, we investigated the effects on splicing of VP16, a non-related transcriptional activator. This herpes virus protein activates transcription by stimulating both initiation and elongation (Blau et al., 1996). Since VP16 lacks a DNA-binding domain, we studied the effect of overexpressing a GAL4-VP16 fusion protein on a minigene carrying five Gal4-binding sites located upstream of the HIV2 minimal promoter (construct shown in Figure 1; Cramer et al., 1999), under non-replicating conditions. Although, like T-Ag-mediated replication, VP16 provokes a large increase in transcription (RPA in Figure 6A) and a subsequent increase in co-localization with speckles (Figure 6B, panels d and e), the effect on alternative splicing is opposite to that of T-Ag: VP16 provokes a 35-fold decrease in EDI inclusion (Figure 6A, RT–PCR of lanes 1 and 2). Most importantly, the effects of VP16 and T-Ag on splicing remain opposite even when, under conditions of similar transfection efficiencies and template concentrations, VP16 elicits RNA levels that are 3.2 times higher than those provoked by T-Ag (Figure 6A, compare lanes 2 and 4). The main conclusion of these experiments is that activation of transcription and enhancement of transcript association to speckles containing SF2/ASF do not necessarily result in augmented EDI inclusion. In the case of VP16, activation of transcription and co-localization with speckles correlate with a reduction of inclusion of similar magnitude to the augmentation caused by T-Ag. Therefore, other mechanisms must be operating during transcription that determine whether a transcriptional activator provokes inclusion or exclusion of EDI.
Fig. 6. Effects of VP16 on alternative splicing, mRNA levels and co-localization. (A) COS-1 cells were transfected with 600 ng of pSVEDA/Gal5-HIV2 either eo(+) (lane 4) or eo(–) (lanes 1–3) and 100 ng of plasmids expressing GAL4 (lanes 1 and 4), GAL4-VP16 (lane 2) or GAL4-SW6 (lane 3). Alternative splicing was assessed by RT–PCR and mRNA levels were quantified by RPA. (B) In situ hybridization to minigene RNA (transcription foci, green) and immunofluorescence to SF2/ASF (speckles, red) of COS-1 cells transfected with pSVEDA/Gal5-HIV2 eo(–). The cells were co-transfected with a plasmid expressing GAL4 (panels a–c) or GAL4-VP16 (panels d–f). Co-localization (yellow) is conspicuous in panel f, but negligible in panel c. (C) COS-1 cells were transfected with 600 ng of pSVEDA/Gal5-HIV2 eo(+) and 100 ng of plasmids expressing GAL4 (lane 1), GAL4-SW6 (lane 2) or GAL4-VP16 (lane 3). Alternative splicing was assessed by RT–PCR and mRNA levels were quantified by RPA. (D) In situ hybridization to minigene RNA (transcription foci, green) and immunofluorescence to SF2/ASF (speckles, red) of COS-1 cells transfected with pSVEDA/Gal5-HIV2 eo(+). The cells were co-transfected with a plasmid expressing GAL4 (panels a–c) or GAL4-VP16 (panels d–f). Co-localization (yellow) is conspicuous in panels c and f. The intensity of the green signal was much higher in panel d than in panel a.
The VP16 domain involved in RNA pol II elongation is important to revert the T-Ag effect on alternative splicing
It is known that the few rounds of plasmid replication elicited by T-Ag determine a more compact nucleosomal organization that is associated with lower RNA pol II processivity (Nahreini and Mathews, 1995). On the other hand, the ability of VP16 to stimulate transcriptional elongation (Blau et al., 1996) correlates with its effect in chromatin reconfiguration (Pazin and Kadonaga, 1998). In this context, it becomes plausible that the opposite effects of T-Ag and VP16 on splicing are the consequence of changes in RNA pol II elongation, with lower processivity associated with higher EDI inclusion and higher processivity associated with EDI exclusion. Overexpression of GAL4-VP16 activates transcription from a replicative Gal5-HIV2 promoter template and provokes a 5-fold reduction in EDI inclusion (Figure 6C, compare lanes 1 and 3). This indicates that the effect of T-Ag-mediated replication can be reverted by VP16 and suggests that these two regulators act anatgonistically on alternative splicing. A reciprocal effect of T-Ag on VP16 is evident from the comparison of Figure 6A, lane 2 with Figure 6C, lane 3: T-Ag increases by 20-fold the ratios already diminished by VP16.
A mutant of VP16, named SW6, which has point mutations of Phe residues at positions 442, 473, 475 and 479, stimulates transcriptional initiation but is defective in its ability to support RNA pol II elongation (Blau et al., 1996). Overexpression of GAL4-SW6 is less potent than GAL4-VP16 in reverting the T-Ag effect on splicing (Figure 6C, lane 2), consistent with a role for transcript elongation in the control of alternative splicing. GAL4-SW6 is also less potent than GAL4-VP16 in bringing down EDI inclusion under non-replicating conditions (Figure 6A, lane 3).
Discussion
It is estimated that the 35 000 genes in the human genome could produce several hundred thousand different proteins as a result of alternative splicing (Black, 2000). Therefore, the study of the molecular bases of alternative splicing regulation becomes especially relevant in the post-genome era. Our previous findings that promoter identity modulates alternative splicing were obtained in a transient expression system (Cramer et al., 1997, 1999). These observations are extended to more physiological conditions in this paper as we show that the promoter effect is also seen in cell lines stably transfected with the same minigenes used in transient experiments (Figure 2B), which indicates that the acquisition of a structured chromatin is compatible with mechanisms of coupling between transcription and splicing.
The effect of promoters on alternative splicing could have a widespread impact on the diversity of proteins expressed under different physiological conditions. In our search for the mechanism underlying the promoter effect we decided to investigate the effects of transcriptional regulators in transiently transfected cells where the plasmid templates were allowed to replicate and therefore mimic a more physiological chromatin structure. In this context, we found that overexpression of SV40 T-Ag stimulates the inclusion of the alternatively spliced FN EDI exon by 10- to 30-fold. We demonstrated that the T-Ag effect is due to transcriptional activation under replicating conditions and not to cell transformation (Figure 3), and that, as expected for a replicative effect, it is observed only in cis. It was also clear that the dramatic stimulation of alternative splicing by replication does not correlate with the modest increase in the number of templates (∼2-fold; Table I), which rules out a mechanism in which transcription or splicing factors were titrated out by an increase in template DNA molecules. We also showed that the number of transcription foci per nucleus increases modestly upon replication and that the nuclear domains (PML bodies), reported to be the sites for replication (Tang et al., 2000), co-localize preferentially with transcription foci derived from ori(+) minigenes (Table I). Taken together, these experiments strongly support that the T-Ag effect is due to qualitative rather than quantitative modifications of the template caused by replication.
T-Ag does not stimulate inclusion of a mutant EDI exon in which the ESE has been disrupted or of EDII, but it does activate the inclusion of an EDI exon in which the target site for SF2/ASF has been replaced with that of another SR protein (SRp-55) (Figure 4). In fact, unlike EDI splicing, EDII splicing is controlled by SRp-40 through regulatory elements located in the 3′ intron (Lim and Sharp, 1998) and in the exon (Du et al., 1997). In our system, overexpression of SRp-40 has little effect on EDI inclusion (Cramer et al., 1999), but activates EDII inclusion by ∼5-fold (Figure 4A). This evidence suggests that T-Ag plays a selective role in regulating alternative splicing, depending on the nature of the exon involved. This correlates with the differential in vivo behavior of FN EDI and EDII splicing in particular situations like liver regeneration, where EDI is upregulated but EDII remains unchanged (Caputi et al., 1995).
Figure 5 shows that T-Ag-mediated replication provokes both a several-fold increase in transcription and a conspicuous co-localization of transcripts with nuclear speckles containing SF2/ASF, a specific activator of EDI inclusion. Transcriptional activation was observed with two different promoters although to different extents: 12-fold for the α-globin promoter and 36-fold for the CMV promoter. Both these transcriptional effects are much greater than the elevation of DNA template number.
T-Ag acts simultaneously as a DNA replication factor and as a transcriptional activator. Nahreini and Mathews (1995) dissected both activities in an expression system using the HIV1 LTR. In agreement with our findings, these authors showed that replication only increases template plasmid numbers by 2- to 3-fold but enhances transcriptional activation by 40-fold. The disruption of the SV40 origin of replication by a mutation similar to the one used here (SfiI site) abrogates the transcriptional effect. The HIV1 LTR gives rise to two classes of transcripts, a class of short non-polyadenylated RNAs and a class of full length poly(A)+ transcripts. In the absence of replication, only the full-length transcript is produced, while upon replication an intrinsic property of the template is modified to augment the formation of short over long transcripts. It was hypothesized that the ‘intrinsic’ property is the chromatin structure that is modified by replication. Plasmids containing SV40 origins, when transfected into mammalian cells, are known to be assembled into chromatin structures that are restructured when the DNA is replicated (Cereghini and Yaniv, 1984).
Whether the change in chromatin structure is causal or not, there is no doubt that T-Ag-mediated replication provokes shorter transcripts (Nahreini and Mathews, 1995; Williams et al., 1996). In fact, Figure 7 shows that in our transfection system, even when steady state mRNA levels are assessed, T-Ag-mediated replication causes a 2-fold increase in the proportion of proximal over distal transcripts. This effect is observed with three minigenes with different promoters. These observations prompted us to investigate two alternative hypotheses to explain the elevation of EDI inclusion caused by T-Ag-dependent replication: (i) the effect is the trivial consequence of an increase in transcript levels and a subsequent increase in SF2/ASF recruitment to the sites of transcription; and (ii) the effect is the consequence of a decrease in RNA pol II elongation and/or processivity.
Fig. 7. T-Ag-dependent replication increases the proportion of shorter transcripts. RPAs of transcripts produced by minigenes with three different promoters transfected into COS-1 cells under replicative [ori(+)] and non-replicative [ori(–)] conditions. Transcripts were detected with a proximal (globin intron 1–exon 2 boundary) and a distal (FN intron +1–exon +1 boundary) probe, described in Materials and methods. Proximal/distal ratios of the protected probe band quantifications are shown.
A key tool to decide between the two hypotheses was the use of the herpes virus transcriptional activator VP16. Like T-Ag, VP16 provokes an important elevation in transcript levels and in their co-localization with speckles. However, it has the opposite effect on splicing: it inhibits EDI inclusion by 35-fold (Figure 6A). This is sufficient to rule out hypothesis 1, but the mechanism of action of VP16 provides the clue for the support of hypothesis 2. VP16 activates transcription by stimulating both initiation and elongation (Blau et al., 1996). If T-Ag-mediated replication inhibits but VP16 activates elongation and if EDI inclusion depends on elongation, VP16 should revert the effects of T-Ag on EDI splicing. This is in fact what we observed in the experiments of Figure 6, which become relevant in opting for the second hypothesis in two respects. On one hand, VP16 increases the RNA levels over those already elevated by T-Ag but reverts EDI inclusion completely: the EDI+/EDI– ratios for the Gal5-HIV2 promoter construct decrease from 10.5 upon replication to 1.9 in the presence of both replication and VP16. On the other hand, SW6, the VP16 mutant that diminishes elongation but preserves initiation, is less potent than VP16 in reverting the T-Ag effect. These findings agree with antagonistic transcriptional effects of VP16 and replication reported by Williams et al. (1996), who demonstrated that the ratio of full-length over aborted transcripts goes from 0.3 to 5.1 when VP16 is added to a replicating template. Further evidence for an involvement of pol II elongation comes from the study of the SV40 enhancer. The transcriptional effects of the presence of the SV40 enhancer in a template are equivalent to those of adding VP16, as both promote RNA pol II elongation (Yankulov et al., 1994; Krumm et al., 1995; Blau et al., 1996). Consistently, we found that deletion of the SV40 enhancer from constructs depicted in Figure 1 provokes higher inclusion of the EDI exon under non-replicative conditions (S.Kadener and A.R.Kornblihtt, unpublished results). The mechanism by which VP16 activates elongation seems to be related to the promotion of histone acetylation. Two yeast histone acetyl transferase complexes (SAGA and NuA4) were reported to interact with GAL4-VP16. While SAGA acetylates H3 histones restricted to the promoter region, targeting of the NuA4 complex led to a broader domain of H4 acetylation >3 kpb (Vignali et al., 2000). This acetylation of internal regions of the gene might facilitate elongation (Krumm et al., 1998), influencing processes like alternative splicing (see below).
An unexpected conclusion of our experiments is that association of speckles to transcripts detected by microscopy does not seem to be as critical as first thought for the fate of alternative splicing (Smith et al., 1999; Misteli, 2000). Both T-Ag (Figure 5B, panels d–f) and VP16 (Figure 6B, panels d–f) cause a striking co-localization of transcripts and SF2/ASF, which is independent from the presence of the globin introns flanking the constitutive exons of the minigenes (Figure 5B, panels j–l). However, T-Ag-dependent replication provokes inclusion while VP16 provokes exclusion of the EDI exon. Furthermore, co-localization is enhanced in the presence of both treatments in which VP16 reverts the elevation of EDI inclusion caused by T-Ag (Figure 6D, panels d–f). We think that high levels of co-localization are the consequence of high levels of transcription. High levels of co-localization are independent of whether the transcripts harbor the target site for the SR proteins accumulated in the speckles; Figure 5B (panels g–i) shows that even in conditions in which EDI inclusion is completely prevented by a disruptive mutation in its ESE, the transcripts co-localize with speckles that contain SF2/ASF.
In a previous work (Cramer et al., 1999) we hypothesized that the promoter effect on alternative splicing could be caused by differential recruitment of splicing factors to the elongating RNA pol II. In such a model, different promoters would provoke different pol II complexes with different abilities to carry splicing factors to the nascent transcript regions involved in alternative splicing. Results reported here favor an alternative model in which specific transcriptional regulation can be, in certain situations, more relevant than splicing factor recruitment, in agreement with the proposal of a kinetic role for transcription in splicing made by Eperon’s laboratory many years ago. Eperon et al. (1988) found critical differences in the role of a stem and loop pre-mRNA secondary structure in the usage of an alternative 5′ splice site, depending on whether the primary transcript was presented already made in a full-length version to the splicing machinery (in vitro) or whether splicing was taking place co-transcriptionally (in vivo). This led them to propose that pre-mRNA is free to fold only within a limited period after transcription, which points to a model in which the kinetics of pre-mRNA synthesis become relevant for the presentation of target sequences and/or secondary structures to the splicing apparatus. A similar mechanism involving a kinetic link between transcription and splicing was suggested from experiments in which RNA pol II pause sites affect alternative splicing by delaying the transcription of an essential splicing inhibitory element (DRE) required for regulation of tropomyosin exon 3 (Roberts et al., 1998).
How can a kinetic model explain why EDII splicing is not affected by T-Ag-mediated replication? The simplest explanation is that pol II processivity affects alternative splicing only of those exons with a particular internal or surrounding pausing architecture. If this is the case, some alternative exons like EDI might be subjected to the processivity control while others like EDII might not.
The discovery of the promoter modulation of alternative splicing allowed us to predict that factors which regulate alternative splicing could be acting through promoters and that cell-specific alternative splicing may not simply result from the differential abundance of ubiquitous SR proteins, but from a more complex process involving cell-specific promoter occupation. The use of a promoter swapping strategy (Cramer et al., 1997, 1999) proved to be very useful in defining the transcriptional control of alternative splicing. However, promoters are not swapped in nature. The present report confirms the physiological implications of our predictions by showing that different transcriptional activators acting on a single promoter have dramatic and opposite effects on alternative splicing that are independent from SR protein abundance.
Materials and methods
Plasmids
EDI splicing: pSVEDATot (α-globin promoter) (Caputi et al., 1994), pSVEDA/CMV (cytomegalovirus promoter) and pSVEDA/FN (FN promoter) (Cramer et al., 1997); pSVEDA Δ2e (α-globin promoter, mutated ESE), pSVEDA Δ2e/FN (FN promoter, mutated ESE) and pSVEDA/Gal5-HIV2 (Cramer et al., 1999). A variant of pSVEDA/CMV lacking globin introns 1 and 2 and exon 2, named pSVEDA/CMV (short), was constructed by excision of a 510 bp XbaI segment from pSVEDA/CMV. EDII splicing: pSVEDEDB-Xho (α-globin promoter) (Paolella et al., 1988). All previous constructs contain the SV40 origin of replication. Disruption of the SV40 origin of replication was carried out by digestion with SfiI (Proudfoot et al., 1992), followed by treatment with T4-DNA polymerase and re-ligation.
Transfections
Approximately 1.8 × 105 human hepatoma Hep3B cells or monkey COS-7 or COS-1 cells were transiently transfected with 6 µl of lipofectamine (Life Technologies) and 2 µg of total plasmid DNA in 35 mm tissue culture dishes. Cells were harvested 24 or 48 h post-transfection. Co-transfection with pCMVβgal allowed standardization of the RNA samples by transfection efficiencies; an aliquot of the cells was used to measure β-galactosidase activity and the rest was used to prepare total RNA by a single step procedure (Chomczynski and Sacchi, 1987). Stable cell lines were generated by co-transfection with a plasmid expressing neomycin phosphotransferase and selection with G418 (geneticin).
Radioactive RT–PCR amplification
Amounts of RNA corresponding to equal β-galactosidase activity OD readings at 415 nm were used as templates for M-MLV reverse transcriptase and subsequent PCR amplification in the presence of [α-32P]dCTP as previously described (Cramer et al., 1999). Specific primers for EDI (Caputi et al., 1994) or EDII (Paolella et al., 1988) splicing products were previously described. RT–PCR products were electrophoresed in 6% acrylamide native gels and detected by autoradiography. Radioactivity in the bands was measured in a scintillation counter (Cerenkov method).
RNase protection assay
The distal riboprobe template was obtained by subcloning into pBSKS+ (Stratagene), a 338-bp PCR fragment from pSVEDATot encompassing the minigene intron +1–exon +1 boundary of FN and α-globin exon (Figure 5). For the proximal riboprobe, a 268 bp XmaI–HindIII fragment from the same plasmid, encompassing the globin intron 1–exon 2 boundary, was used. Plasmids were linearized with HindIII and riboprobes were obtained by in vitro transcription from the T7 promoter in the presence of 60 µCi of [α-32P]UTP (3000 Ci/mmol). Approximately 10 µg of total RNA from transfected cells were co-precipitated with 105 c.p.m. of riboprobe, resuspended in 25 µl of hybridization buffer (40 mM PIPES pH 6.7, 400 mM NaCl, 1 mM EDTA, 80% v/v deionized formamide) and denatured for 5 min at 90°C. Subsequent annealing was carried out overnight at 45°C. RNase digestion was performed by addition of 300 µl of RNase solution (2 µg/ml RNase A, 40 U/ml RNase T1) and incubated for 30 min at 30°C. The reaction was stopped by addition of SDS and proteinase K to a final concentration of 0.6% (w/v) and 0.1 mg/ml, respectively, and incubation at 37°C for 15 min. Samples were extracted once with phenol:chloroform and ethanol-precipitated in the presence of yeast tRNA as carrier. After denaturing, samples were electrophoresed in 6% sequencing gels.
Immunohistochemistry and fluorescence in situ hybridization
For the simultaneous detection of speckles (splicing factor compartments) and specific RNA sequences we first carried out in situ hybridization with a biotinylated nick-translated probe corresponding to pSVEDATot, followed by labeling with fluorescein isothiocyanate (FITC)-avidin according to Misteli and Spector (1999). Secondly, fixed cells were incubated with Mab103, a specific antibody to SF2/ASF (provided by Adrian Krainer) and rhodamine-labeled secondary antibodies. Confocal images were obtained using a Zeiss Axiovert LSM 510 confocal laser scanning microscope.
Acknowledgments
Acknowledgements
We thank T.Misteli, T.Santa Coloma and M.Bianchini for help with confocal microscopy; D.Bentley for the VP16 and SW6 constructs and valuable criticisms; C.Cole for T-Ag mutant constructs; F.E.Baralle for the EDB (EDII) construct; A.Krainer for antibodies to SF2/ASF; G.Maul for antibodies to PML bodies; and D.Refojo, O.Coso and D.Paz for valuable help. This work was supported by grants from the Fundación Antorchas, the ICGEB, the Universidad de Buenos Aires, the ANPCyT and the CONICET. A.R.K. and A.S. are investigators of the CONICET.
References
- Bentley D. (1999) Coupling of RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol., 11, 347–351. [DOI] [PubMed] [Google Scholar]
- Black D.L. (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell, 103, 367–370. [DOI] [PubMed] [Google Scholar]
- Blau J., Xiao,H., McCracken,S., O’Hare,P., Greenblatt,J. and Bentley,D. (1996) Three functional classes of transcriptional activation domains. Mol. Cell. Biol., 16, 2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres J.F., Screaton,G.R. and Krainer,A.R. (1998) A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev., 12, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M., Casari,G., Guenzi,S., Tagliabue,R., Sidoli,A., Melo,C.A. and Baralle,F.E. (1994) A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res., 22, 1018–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M., Melo,C.A. and Baralle,F.E. (1995) Regulation of fibronectin expression in rat regenerating liver. Nucleic Acids Res., 23, 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghini S. and Yaniv,M. (1984) Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J., 3, 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Cramer P., Pesce,C.G., Baralle,F.E. and Kornblihtt,A.R. (1997) Functional association between promoter structure and transcript alternative splicing. Proc. Natl Acad. Sci. USA, 94, 11456–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Cáceres,J.F., Cazalla,D., Kadener,S., Muro,A.F., Baralle,F.E. and Kornblihtt,A.R. (1999) Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell, 4, 251–258. [DOI] [PubMed] [Google Scholar]
- Cramer P., Srebrow,A., Kadener,S., Werbajh,S., de la Mata,M., Melen,G., Nogués,G. and Kornblihtt,A.R. (2001) Coordination between transcription and pre-mRNA processing. FEBS Lett., 498, 179–182. [DOI] [PubMed] [Google Scholar]
- Croft L., Schandorff,S., Clark,F., Burrage,K., Arctander,P. and Mattick,J.S. (2000) ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nature Genet., 24, 340–341. [DOI] [PubMed] [Google Scholar]
- Du K., Peng,Y., Greenbaum,L.E., Haber,B.A. and Taub,R. (1997) HRS/SRp40-mediated inclusion of the fibronectin EIIIB exon, a possible cause of increased EIIIB expression in proliferating liver. Mol. Cell. Biol., 17, 4096–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L.P., Graham,I.R., Griffiths,A.D. and Eperon,I.C. (1988) Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell, 54, 393–401. [DOI] [PubMed] [Google Scholar]
- Fanning E. and Knippers,R. (1992) Structure and function of simian virus large T tumor antigen. Annu. Rev. Biochem., 61, 55–85. [DOI] [PubMed] [Google Scholar]
- Gruss C., Gutiérrez,C., Burhauns,W.C., De Pamphilis,M.L., Koller,T. and Sogo,J.M. (1990) Nucleosome assembly in mammalian cell extracts before and after DNA replication. EMBO J., 9, 2911–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman A. and Kornblihtt,A.R. (1987) Identification of a third region of cell-specific alternative splicing in human fibronectin mRNA. Proc. Natl Acad. Sci. USA, 84, 7179–7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y. and Manley,J.L. (2000) RNA polymerase II and the integration of nuclear events. Genes Dev., 14, 1415–1429. [PubMed] [Google Scholar]
- Hirt B. (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol., 26, 365–369. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A.R., Vibe-Pedersen,K. and Baralle,F.E. (1984) Human fibronectin: molecular cloning evidence for two mRNA species differing by an internal segment coding for a structural domain. EMBO J., 3, 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A.R., Pesce,C.G., Alonso,C.R., Cramer,P., Srebrow,A., Werbajh,S.E. and Muro,A.F. (1996) The fibronectin gene as a model for splicing and transcription studies. FASEB J., 10, 248–257. [DOI] [PubMed] [Google Scholar]
- Krumm A., Hickey,L.B. and Groudine,M. (1995) Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev., 9, 559–572. [DOI] [PubMed] [Google Scholar]
- Krumm A., Madisen,L., Yang,X.J., Goodman,R., Nakatani,Y. and Groudine,M. (1998) Long-distance transcriptional enhancement by the histone acetyltransferase PCAF. Proc. Natl Acad. Sci. USA, 95, 13501–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigueur A., La Branche,H., Kornblihtt,A.R. and Chabot,B. (1993) A splicing enhancer in the human fibronectin alternate exon EDI interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev., 7, 2405–2417. [DOI] [PubMed] [Google Scholar]
- Lim L.P. and Sharp,P.A. (1998) Alternative splicing of the EIIIB exon depends on specific TGCATG repeats. Mol. Cell. Biol., 18, 3900–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon H.J., Sebastio,G. and Baralle,F.E. (1987) A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res., 15, 7725–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Fong,N., Yankulov,K., Ballantyne,S., Pan,G., Greenblatt,J., Patterson,S.D., Wickens,M. and Bentley,D.L. (1997) The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature, 385, 357–361. [DOI] [PubMed] [Google Scholar]
- Misteli T. (2000) Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J. Cell Sci., 113, 1841–1849. [DOI] [PubMed] [Google Scholar]
- Misteli T. and Spector,D.L. (1999) RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell, 3, 697–705. [DOI] [PubMed] [Google Scholar]
- Misteli T., Cáceres,J.F. and Spector,D.L. (1997) The dynamics of a pre-mRNA splicing factor in living cells. Nature, 387, 523–527. [DOI] [PubMed] [Google Scholar]
- Nahreini P. and Mathews,M.B. (1995) Effect of simian virus 40 origin of replication on transcription from the human immunodeficiency virus type 1 promoter. J. Virol., 69, 1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolella G., Henchcliffe,C., Sebastio,G. and Baralle,F.E. (1988) Sequence analysis and in vivo expression show that alternative splicing of ED-B and ED-A regions of the human fibronectin gene are independent events. Nucleic Acids Res., 16, 3545–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin M.J. and Kadonaga,J.T. (1998) Transcriptional and structural analysis of chromatin assembled in vitro. In Gould,H. (ed.), Chromatin: A Practical Approach. Oxford University Press, Oxford, UK, pp. 173–194.
- Phair R.D. and Misteli,T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J. (2000) Connecting transcription to messenger RNA processing. Trends Biochem. Sci., 25, 290–293. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J., Lee,B.A. and Monks,J. (1992) Multiple Sp1 binding sites confer enhancer-independent, replication-activated transcription of HIV-1 and globin gene promoters. New Biol., 4, 369–381. [PubMed] [Google Scholar]
- Roberts G.C., Gooding,C., Mak,H.Y., Proudfoot,N.J. and Smith,C.W.J. (1998) C0-transcriptional commitment to alternative splice site selection. Nucleic Acids Res., 26, 5568–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D., Clemens,J.C., Shu,H., Worby,C.A., Xiao,J., Muda,M., Dixon,J.E. and Zipursky,S.L. (2000) Drosophila DSCAM is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell, 101, 671–684. [DOI] [PubMed] [Google Scholar]
- Smith K.P., Moen,P.T., Wydner,K.L., Coleman,J.R. and Bentley Lawrence,J. (1999) Processing of endogenous pre-mRNA in association with SC-35 domains is gene specific. J. Cell Biol., 144, 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Bell,P., Tegtmeyer,P. and Maul,G.G. (2000) Replication but not transcription of Simian Virus 40 DNA is dependent on nuclear domain 10. J. Virol., 74, 9694–9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. (1999) Chromatin modification by DNA tracking. Proc. Natl Acad. Sci. USA, 96, 13634–13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M., Steger,D.J., Neely,K.E. and Workman,J.L. (2000) Distribution of acetylated histones resulting from GAL4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J., 19, 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R., Lee,B., Jackson,S. and Proudfoot,N. (1996) Activation domains of transcription factors mediate replication dependent transcription from a minimal HIV-1 promoter. Nucleic Acids Res., 24, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankulov K., Blau,J., Purton,T., Roberts,S. and Bentley,D.L. (1994) Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell, 77, 749–759. [DOI] [PubMed] [Google Scholar]
- Zhu J., Abate,M., Rice,P.W. and Cole,C.N. (1991) The ability of Simian Virus 40 large T antigen to immortalize primary mouse embryo fibroblasts cosegregates with its ability to bind to p53. J. Virol., 65, 6872–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]