Abstract
Several factors have been biochemically characterized based on their ability to increase the overall rate of transcription elongation catalyzed by the multiprotein complex RNA polymerase II (Pol II). Among these, the ELL family of elongation factors has been shown to increase the catalytic rate of transcription elongation in vitro by suppressing transient pausing. Several fundamental biological aspects of this class of elongation factors are not known. We have cloned the Drosophila homolog (dELL) in order to test whether ELL family proteins are actually associated with the elongating Pol II in vivo. Here we report that dELL is a nuclear protein, which, like its mammalian homologs, can increase the catalytic rate of transcription elongation by Pol II in vitro. Interestingly, we find that dELL co-localizes extensively with the phosphorylated, actively elongating form of Pol II at transcriptionally active sites on Drosophila polytene chromosomes. Furthermore, dELL is relocalized from a widespread distribution pattern on polytenes under normal conditions to very few transcriptionally active puff sites upon heat shock. This observation indicates a dynamic pattern of localization of dELL in cells, which is a predicted characteristic of a Pol II general elongation factor. We also demonstrate that dELL physically interacts with Pol II. Our results strongly suggest that dELL functions with elongating RNA polymerase II in vivo.
Keywords: ELL/elongation/heat shock genes/RNA polymerase II/transcription
Introduction
The regulation of gene expression by the eukaryotic RNA polymerase II (Pol II) is controlled by the action of a variety of transcription factors. Transcriptional regulation by Pol II proceeds through a set of multiple stages denoted as pre-initiation, initiation, promoter clearance, elongation and termination. For more than 20 years, research from many laboratories has centered around understanding the pre-initiation and initiation stages of eukaryotic mRNA synthesis. These studies have resulted in defining biochemical roles for the components of the basal transcription machinery and its interacting proteins, and have firmly established the cellular roles of these factors.
A requirement for Pol II elongation factors was predicted from early biochemical studies on the catalytic properties of transcription elongation. These early studies demonstrated that purified Pol II lacked the ability to catalyze mRNA chain elongation in vitro at rates sufficient to account for the observed rates of mRNA synthesis in vivo (Ucker and Yamamoto, 1984; Izban and Luse, 1992; Tennyson et al., 1995). Transcription elongation by purified Pol II in vitro results in transient and/or stable pausing, which sometimes ends with arrest (Reines et al., 1996; Uptain and Chamberlain, 1997; Uptain et al., 1997). Therefore, it has been hypothesized that transcription elongation factors are required to increase the overall rate of transcription and/or prevent premature pausing and arrest by Pol II during the expression of many eukaryotic genes (Uptain and Chamberlain, 1997; Uptain et al., 1997). Based on this observation, to date several Pol II elongation factors have been characterized biochemically using in vitro transcription assays. The Pol II elongation factors fall into at least three functional classes. One class includes P-TEFb, DSIF (Spt4, Spt5), Spt6 and SII. P-TEFb prevents elongation arrest by ATP analog DRB, DSIF is required for DRB to inhibit transcript elongation, and SII prevents DNA sequence-dependent arrest (Reines and Mote, 1993; Rudd et al., 1994; Marshall and Price, 1995; Marshall et al., 1996; Reines et al., 1996; Mancebo et al., 1997; Zhu et al., 1997; Peng et al., 1998; Hartzog et al., 1998; Shilatifard, 1998a,b; Wada et al., 1998; Yamaguchi et al., 1999; Price, 2000). The elongation rate of transcrip tion by Pol II in vivo is also controlled by the presence of nucleosomal DNA. This has led to the discovery of the second class factors, which regulate the rate of transcription elongation through nucleosomes. A member of this class of factors is FACT (facilitates chromatin transcription) (Orphanides et al., 1998). The third class of Pol II elongation factors operates to increase the catalytic rate of transcription elongation by altering the Km and/or the Vmax of the elongating polymerase, and is comprised of TFIIF, Elongin and the ELL family (ELL, ELL2 and ELL3) (Price et al., 1989; Bradsher et al., 1993; Garrett et al., 1994; Duan et al., 1995; Reines et al., 1996; Shilatifard et al., 1996, 1997a,b; Miller et al., 2000). It is not clear whether all of the above factors function at the same time with the elongating polymerase; however, the observed rate of transcription in vivo suggests that several of these factors may work together to regulate the elongation stage of transcription catalyzed by Pol II.
During the past few years, biochemical studies have demonstrated the importance of the promoter clearance and the elongation stages of transcription (Dvir et al., 1996, 1997; Kugal and Goodrich, 1998; Kumar et al., 1998; Moreland et al., 1999; Bradsher et al., 2000; Tremeau-Bravard et al., 2001). Some Pol II elongation factors have been biochemically implicated in processes such as transcriptional regulation of the human immunodeficiency virus genome (Mancebo et al., 1997; Zhu et al., 1997; Parada and Roeder, 1999), heat shock and stress response (Andrulis et al., 2000; Kaplan et al., 2000), neuronal development (Guo et al., 2000) and pathogenesis of human cancer (DiMartino et al., 2000).
As mentioned above, TFIIF, Elongin and the ELLs are all capable of stimulating the overall elongation rate catalyzed by Pol II by suppressing transient pausing by the enzyme in vitro. Previous studies have shown that bacterial and eukaryotic RNA polymerases are susceptible to pausing at each step of nucleotide addition (Reines et al., 1996). Because the duration of pausing is often greater than the rate of phosphodiester bond formation (kcat), it has been proposed that elongating Pol II cycles between active and inactive conformations at each step of nucleotide addition in vivo. Although we know a great deal about the biochemical properties of Pol II elongation factors (TFIIF, Elongin and ELLs), we do not know whether any of these factors actually function with the actively elongating polymerase in vivo. Also, little is known about the extent of their role and association with transcription complexes in the context of chromatin.
To determine whether ELL can function with the elongating Pol II in vivo, we have studied biological properties of the Drosophila melanogaster homolog of ELL (dELL). We show here that dELL is a nuclear protein and, like its mammalian homolog, it can increase the catalytic rate of transcription elongation by Pol II in vitro. To define whether dELL is associated with the elongating Pol II in vivo, we have generated polyclonal antibodies to dELL and have also generated transgenic flies that express epitope-tagged dELL. We found that both the endogenous dELL and transgenic tagged dELL co-localized extensively with the phosphorylated, actively elongating form of Pol II at transcriptionally active sites on Drosophila polytene chromosomes. Upon heat shock, dELL dramatically relocalized to a very few transcriptionally active heat shock puff sites. This observation indicates a dynamic localization pattern of dELL. We also found that dELL is biochemically associated with Pol II in whole fly extract. This study provides strong evidence for a general role for the ELL family of proteins in transcription elongation by Pol II in vivo.
Results
Identification and sequence analysis of the Drosophila homolog of ELL
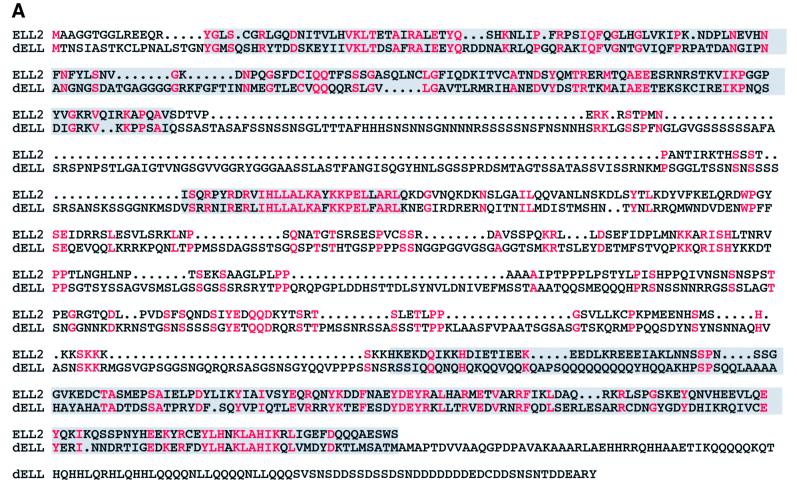
To define the biochemical and cellular properties of the Pol II elongation factor ELL, we have cloned the gene encoding the unique Drosophila ELL protein dELL. dELL is most similar to human ELL2, based on its primary sequence (Figure 1A), and it is the only ELL-related protein in Drosophila. There are three distinct domains of extensive homology. First is the region from amino acids 15 to 175 of ELL2, which also has a very high degree of homology to the ELL and ELL3 human homologs. This domain has been characterized by deletion mutagenesis of ELL, and has been shown to be required for in vitro elongation activity (Shilatifard et al., 1997a,b). A second region of nearly identical sequence in the central portion of the two proteins is unique to ELL2 and dELL; it is not seen in either ELL or ELL3, and it has not been characterized. The C-terminal domain is the most highly conserved domain in the ELL family (Miller et al., 2000) and a homologous domain is also found in the C-terminal half of dELL. Interestingly, we have demonstrated that the conserved C-terminal domain of ELL is necessary and sufficient for the immortalization activity of an mixed lineage leukemia (MLL)–ELL fusion protein in myeloid progenitor cells (DiMartino et al., 2000).
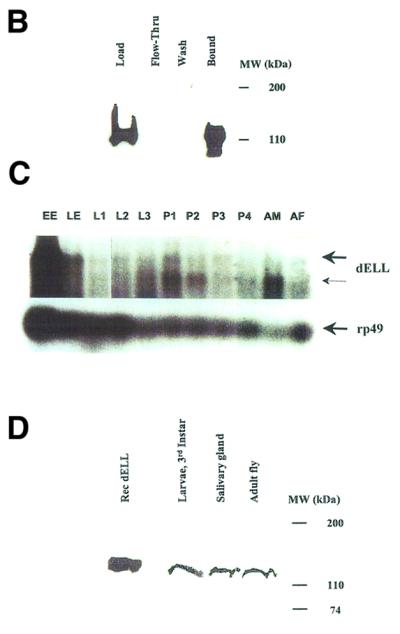
Fig. 1. Domain structure and conservation and expression pattern of D.melanogaster ELL. (A) Sequence alignment of dELL with human ELL2. Amino acid identities are highlighted in red. Gray boxes indicate the sequences homologous to the N-terminal elongation activation domain and the C-terminal immortalization domain of ELL, and a central region of high homology between dELL and ELL2. (B) Affinity chromatography and western analysis of recombinant dELL. His6-tagged protein was expressed and one-step purified by nickel chromatography as described in Materials and methods. Chromatographic fractions were analyzed by western blotting with anti-Express monoclonal antibody (Invitrogen) to visualize recombinant dELL. As expected, a single band of ∼120 kDa appears in both the load and bound fractions. (C) Expression of dELL transcripts during development. (Top) An autoradiograph of a northern blot showing the two dELL mRNAs. (Bottom) An autoradiograph from the same northern blot probed for rp49 transcript as loading control. Each lane contained 5 µg of total RNA isolated from Oregon R wild-type flies at the developmental stage indicated: EE, 0–30 min after egg laying (AEL); LE, 30 min–16 h AEL; L1–L3, first to third instar larvae; P1–P4, first to fourth day of pupation; AM, 0- to 1-day-old male adults; AF, 0- to 1-day-old adult females. (D) Western analysis of dELL protein expression at different stages of fly development. Protein extracts were prepared from third instar larvae, salivary gland tissue from third instar larvae and adult Drosophila as described in Materials and methods. Extracts were subjected to western analysis with purified anti-dELL polyclonal antisera to examine relative levels of protein expression at the selected developmental stages. A single polypeptide band (∼120 kDa) was detected in the extracts from each stage, and this polypeptide co-migrates with recombinant dELL. Protein concentrations in each extract were determined prior to loading to ensure that total protein in each lane was similar.
dELL can stimulate elongation by Pol II
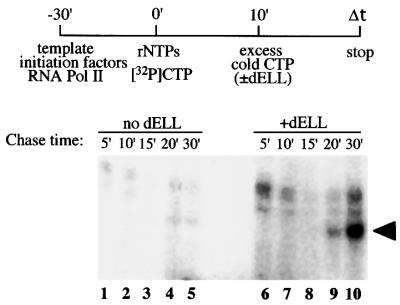
Each of the three human ELL homologs (ELL, ELL2 and ELL3) exhibits the ability to increase the catalytic rate of transcription elongation catalyzed by Pol II from adenovirus major late (AdML) promoter constructs in vitro (Shilatifard et al., 1996, 1997a; Miller et al., 2000). To test the in vitro elongation activity of dELL, we expressed ELL in Escherichia coli as a His6-tagged protein. Recombinant dELL was purified by affinity chromatography and tested in a pulse–chase transcription assay. Accumulation of runoff transcripts from the AdML promoter was determined by autoradiography. Runoff transcripts became readily visible within 20 min in the presence of dELL after the chase was initiated, and a large amount of transcript was observed after 30 min (Figure 2, lanes 6–10). Under conditions where no dELL was added, transcript levels were virtually undetectable after 30 min (Figure 2, lanes 1–5). These results demonstrate that dELL can increase the catalytic rate of transcription elongation catalyzed by Pol II in vitro.
Fig. 2. Drosophila ELL can increase the catalytic rate of transcription elongation catalyzed by RNA polymerase II in vitro. Pre-initiation complexes were formed in the presence of AdMLP DNA, TBP, TFIIB, TFIIF, TFIIE, TFIIH and RNA Pol II. Transcription reactions were initiated by the addition of rNTPs as described in Materials and methods. Nascent transcripts were chased after the addition of cold CTP in the presence or absence of recombinant dELL for the times indicated. Runoff transcripts were analyzed in a 6% acrylamide gel containing 7 M urea and 0.51× TBE, and visualized by PhosphorImager analysis.
Expression of dELL during development
To characterize the expression of dELL during different stages of development in Drosophila, we have analyzed the mRNA and protein levels at several stages. Northern blot analysis of total RNA isolated from 10 developmental stages was performed to identify the dELL transcript and its relative level of expression at various times throughout development. Using a radiolabeled antisense RNA generated from the cloned cDNA, we detected two transcripts ranging from ∼5.0 to 6.2 kb in size (Figure 1C). The upper (thicker arrow) indicates the position of the bands corresponding to full-length dELL transcript. The lower (thinner arrow) indicates the position of an alternatively spliced form of dELL transcript. This form of dELL appears to be higher in the adult male. It is possible that this spliced version of the dELL transcript may be a replacement of human ELL3 (which is testes specific) in the adult male fly. The full-length transcripts for dELL appear to be maternally loaded, as seen by their abundance in 30′ old embryos, before zygotic transcription begins in the developing organism. It must be noted here that we have also seen alternatively spliced forms of mammalian ELLs. Following the peak of the mRNA level in the early embryogenesis, levels appear to be quite similar.
To confirm dELL protein expression, polyclonal antiserum to recombinant dELL was raised and affinity purified for use in detecting endogenous dELL. Extracts from Drosophila larvae or adult flies were subjected to western blot analysis. dELL is detectable in larvae and adults, corresponding to the northern data, suggesting that a steady-state level of dELL protein expression is maintained throughout development. In extracts of larval salivary glands, the purified dELL polyclonal antibody specifically recognizes a single band of ∼120 kDa co-migrating with the recombinant dELL expressed in bacteria (Figure 1D). This demonstrates the presence of dELL in salivary glands and that the purified dELL polyclonal antibody does not cross-react with other polypeptides in the salivary gland.
dELL is localized to the nuclei of Drosophila embryos
To determine the distribution of dELL in embryos, dELL was localized by immunofluorescence. Like its mammalian homolog, dELL is a nuclear protein with ubiquitous distribution in early and late embryos (Figure 3). Co-localization of dELL with the pattern of DAPI staining and with Pol II staining occurred in all cells, with no indication of tissue specificity at this stage of development.
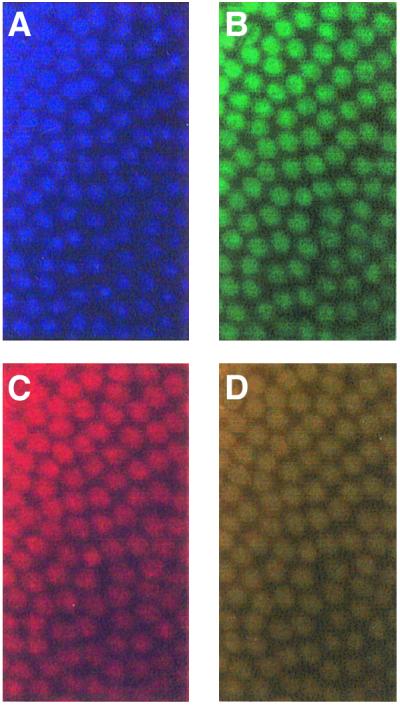
Fig. 3. Whole-mount immunofluorescence detection of ELL in D.melanogaster embryo. Pre-cellularization Drosophila embryos were fixed as described in Materials and methods and probed either with (A) DAPI to visualize DNA, (B) monoclonal antibody directed against phosphorylated Pol II or (C) purified polyclonal anti-dELL serum. (D) Overlay of (B) and (C).
Immunolocalization of dELL in polytene chromosomes
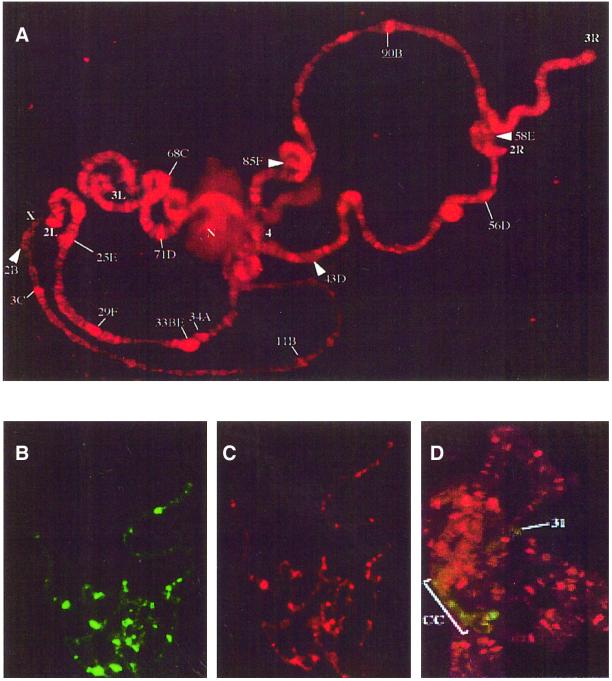
To determine whether dELL is associated with the elongating polymerase in vivo and in the presence of chromatin components, we examined the distribution of dELL on polytene chromosomes from the salivary glands of third instar larvae. dELL is found at many sites along all the polytene chromosome arms (Figure 4A). To confirm the specificity of our purified polyclonal antibody, we also examined the distribution of epitope-tagged dELL expressed in transgenic Drosophila. Figure 4B and C shows that the immunofluorescence for tagged dELL (using anti-His monoclonal antibody) (Figure 4B) co-localizes with immunofluorescence for dELL (using dELL purified polyclonal antibody) (Figure 4C). These results demonstrate that endogenous dELL (Figure 4A) is widely dispersed on developmental puff sites, which are generally recognized as sites of active transcription (Pelling, 1964; Plagens et al., 1976). Among the prominent dELL binding sites are the loci encoding the salivary gland glue proteins sgs 3, 4, 5, 7 and 8, which are highly transcribed in mid-third instar larval salivary glands (Figure 4A; Korge, 1975; Guild and Shore, 1984; Crowley et al., 1983).
Fig. 4. Drosophila ELL is associated with euchromatin on transcriptionally active sites. (A) Immunofluorescence localization of dELL on polytene chromosomes. Wild-type polytene chromosomes were prepared from third instar larvae salivary glands and probed with purified polyclonal antibodies raised against dELL. (B and C) To ensure that the purified dELL polyclonal antibodies were specific for dELL, we also generated a tagged dELL transgenic fly. Polytene chromosomes isolated from the transgenic dELL third instar larvae salivary glands were probed either with (B) monoclonal antibody directed against the His6-tag on dELL or (C) purified polyclonal antibodies directed against dELL. (D) Polytene chromosomes were probed for dELL (red) and the heterochromatin-associated protein HP1 (green). The bracket indicates the heterochromatic chromocenter (cc), while a prominent region of euchromatic staining by HP1 in region 31 is indicated by a line (31).
Unexpectedly, we found nucleolar staining using the purified anti-dELL polyclonal serum, although Pol II is absent from the nucleolus. The significance of this observation is unclear, but may indicate a previously unanticipated role for the ELL family of proteins in RNA Pol I activity. Nucleolar staining was not observed when anti-His6 antibody was used to immunolocalize transgenic His6-tagged dELL. The possible role of dELL in RNA Pol I activity is not clear at this point.
While dELL is distributed widely throughout the euchromatic chromosome arms, double staining of the polytene chromosome with antibodies directed against heterochromatin binding protein 1 (HP1) and dELL showed no detectable overlap (Figure 4D), indicating that dELL is largely excluded from the heterochromatin.
dELL co-localizes extensively with Pol IIo on polytene chromosomes
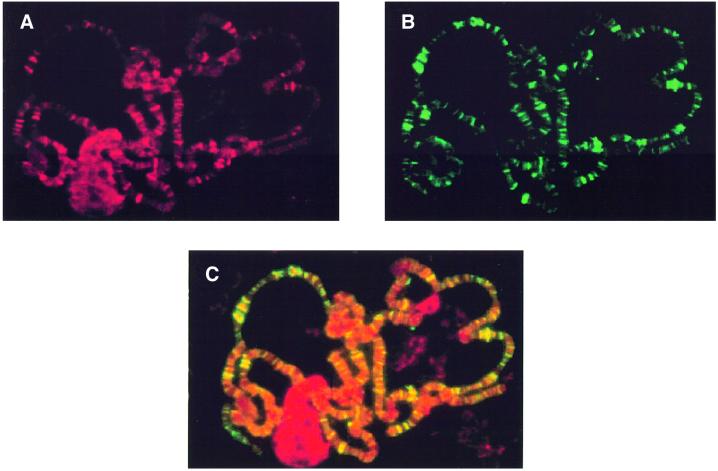
It is well established that the elongating form of Pol IIo is hyperphosphorylated at its C-terminal domain (Dahmus, 1996; Myers et al., 1998; Wada et al., 1998; Cho et al., 1999; Otero et al., 1999; Yamaguchi et al., 1999). We have examined whether dELL co-localizes with the hyperphosphorylated Pol II on polytene chromosomes from the salivary glands of third instar larvae. Chromosome squashes were stained simultaneously with antibodies against dELL (purified polyclonal) (Figure 5A) and Pol IIo (monoclonal H14; Covance) (Figure 5B), and the appropriate secondary antibodies and fluorescence images were generated for each fluorophore. dELL co-localizes extensively with Pol IIo at a number of transcriptionally active sites on polytene chromosomes (Figure 5C), consistent with a role for dELL as a Pol II elongation factor in vivo.
Fig. 5. Co-localization of dELL with hyperphosphorylated RNA polymerase II (Pol IIo) on polytene chromosomes. (A) Immunofluorescence detection of dELL. (B) Immunolocalization of Pol IIo. (C) Overlay of (A) and (B). Co-localization of red and green signals appears yellow.
Heat shock induces relocalization of dELL to activated heat shock genes
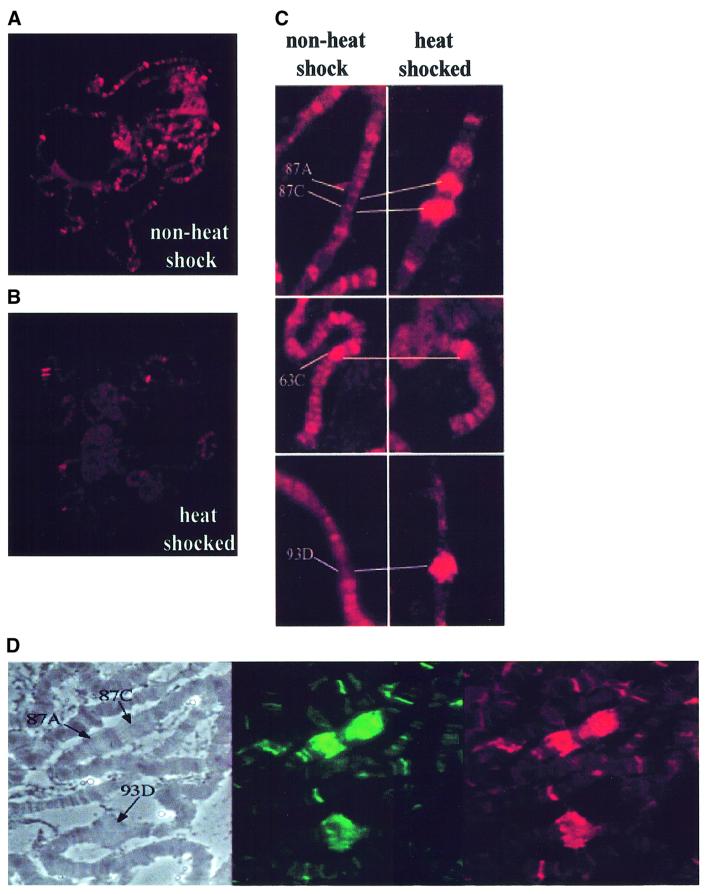
Heat shock in Drosophila results in a dramatic relocalization of RNA Pol II from sites of developmentally regulated genes to a small number of vigorously transcribed heat shock genes (Greenleaf et al., 1978). To determine whether dELL accompanies RNA Pol II mobilization upon heat shock, we compared the distribution of dELL on wild-type polytene chromosomes under normal and heat shock conditions (Figure 6). As shown in Figure 6A, there is a wide distribution of dELL on polytene chromosomes before heat shock. Upon heat shock, there is a dramatic relocalization of dELL to a small number of puffs (Figure 6B). Detailed analysis indicated that a significant amount of dELL is relocalized to selected heat shock puffs upon heat shock (Figure 6C). Most prominent are the 87A and 87C sites and the puff site at 93D. We have also analyzed these heat shock puff sites for the presence of hyperphosphorylated RNA Pol II and dELL (Figure 6D). These data demonstrate that dELL is mobilized together with Pol IIo during heat shock, consistent with the hypothesis that dELL functions as a transcription factor in vivo.
Fig. 6. Heat shock induces a dramatic relocalization of dELL to heat shock loci. Immunofluorescence detection of wild-type third instar larvae salivary glands polytene chromosomes probed with polyclonal antibodies directed against dELL (A) before and (B) after heat shock induction. (C) Presence of dELL on heat shock loci 87A, 87C, 63C and 93D (left panel) before and (right panel) after heat shock induction. (D) Co-localization of Pol IIo and dELL at heat shock puffs 87A, 87C, and 93D in heat-shocked polytene chromosomes. Phase contrast (left panel), staining for Pol IIo (middle panel) and staining for dELL (right panel) on a heat-shocked polytene chromosome.
RNA Pol II and dELL are part of a common complex in vivo
The co-localization of dELL and RNA Pol II suggests that they are part of a complex at sites of active transcription elongation. To provide evidence for the presence of a common biochemical complex containing dELL and RNA Pol II in vivo, we prepared total fly extracts from equal masses of control flies and transgenic flies expressing His6-tagged dELL. The polyhistidine tag permits a one-step chromatographic enrichment of dELL-associated polypeptides using a nickel chelating resin to anchor dELL and its associated polypeptides. Western blot analysis of fractionated extracts is shown in Figure 7. As expected, His-tagged dELL is retained by the resin (data not shown). Aliquots from nickel chelating resin were examined for the presence of Pol II using a Pol II-specific monoclonal antibody. Pol II is specifically retained in extracts containing His-tagged dELL, demonstrating that the two proteins interact directly or indirectly in vivo. While only a fraction of the total Pol II is retained by His6-dELL, this is expected since the tagged protein presumably competes with endogenous dELL for RNA polymerase binding.
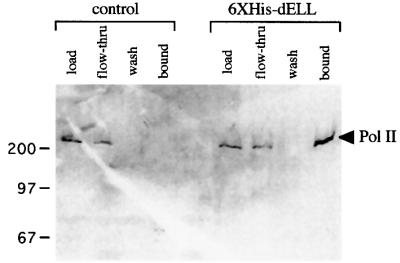
Fig. 7. RNA polymerase II and dELL can physically interact. To determine biochemical interaction between dELL and Pol II, wild-type and His6-tagged dELL transgenic fly extracts were fractionated by nickel chromatography. Load, flow-through, wash and bound fractions were subjected to 12% SDS–PAGE, and fractions were analyzed for the presence of Pol II by western blot analysis using a Pol II-specific monoclonal antibody as described in Materials and methods.
Discussion
Transcriptional elongation represents a key regulated step in the expression of eukaryotic genes (reviewed in Uptain et al., 1997; Shilatifard, 1998a; Reines et al., 1999; Conaway and Conaway, 2000). Several biochemical factors have been identified that enhance Pol II elongation activity in vitro, suggesting multiple layers of cellular control. The specific roles of these diverse elongation factors and the interplay and/or functional redundancy between them are poorly understood. Conceptually, one can consider the elongation mechanism as two steps: promoter escape and processive elongation. The ELL family of proteins behave in vitro as enhancers of Pol II processive elongation by suppressing transient pausing by polymerase (Shilatifard et al., 1997a,b; Elmendorf et al., 2001). These biochemical studies implicate the ELL family of proteins in promoting Pol II catalysis. Previous studies demonstrated that both human and murine ELLs are nuclear proteins (Thirman et al., 1997; DiMartino et al., 2000), consistent with a role in transcription in vivo.
Here, we present the first direct evidence placing an ELL family of proteins at sites of active transcription elongation in vivo. dELL is a nuclear protein in all cell types examined. We find that dELL is broadly distributed on polytene chromosomes. Upon heat shock, dELL undergoes a massive redistribution that accompanies the redistribution of Pol II and the switch to heat shock gene transcription. We show that dELL is complexed with Pol II in vivo, providing the first evidence that the ELL family of elongation factors is directly associated with the catalytic machinery of transcription in the cell.
dELL is a structurally and functionally conserved member of the ELL family of Pol II elongation factors
Three mammalian ELL family proteins have been identified, all with biochemical elongation activity but differing in expression pattern. ELL and ELL2 are widely expressed (Thirman et al., 1994; Shilatifard et al., 1997), while ELL3 is testis specific (Miller et al., 2000). dELL shows significant sequence homology to all three mammalian ELL family proteins, but is most closely related to ELL2. Importantly, the conserved sequences in dELL include N- and C-terminal domains with previously defined properties in ELL: the N-terminal domain is required for transcription elongation activity in vitro (Shilatifard et al., 1997a,b), and the conserved C-terminal domain is required for immortalization activity as part of an MLL–ELL chimeric protein (DiMartino et al., 2000). Based on the annotated Drosophila genome sequence, dELL appears to be the unique ELL family protein in Drosophila. Its ubiquitous expression in development suggests that dELL has a broad role in transcription, perhaps reflecting an ancestral role for ELL family proteins from which specialized or redundant mammalian isoforms evolved.
ELL-like proteins and related sequences have been reported in all mammals tested, as well as in chickens and fish (Thirman et al., 1994, 1997), but are apparently absent in Saccharomyces cerevisiae and Caenorhabditis elegans. Thus, the ELL family of elongation factors may represent an adaptation of higher metazoan development. In metazoans, many developmentally important genes have large transcription units owing to the presence of introns, which can sometimes be of immense size. ELL family proteins may have evolved to support the efficient transcription of large genes and/or the high level expression of particular tissue-specific genes during tissue differentiation.
In addition to its structural homology to mammalian ELL family proteins, dELL behaves like a functional homolog of mammalian ELL in that it enhances the Pol II catalytic rate in vitro. The fact that the Drosophila protein can stimulate activity of the mammalian Pol II demonstrates a remarkable conservation of function. Thus, the properties of dELL in Drosophila are likely to reflect conserved properties shared with the mammalian ELL family of proteins.
dELL is a Pol II-associated factor in vivo
Previous studies have demonstrated that the ELL family of proteins can increase Pol II catalytic rates in vitro (Shilatifard et al., 1996, 1997a,b; Miller et al., 2000). Our immunofluorescence data clearly show that dELL is associated with elongating Pol II in vivo. dELL is concentrated at chromosomal sites of active transcription, notably at sites of intense developmental gene transcription such as the salivary gland glue protein loci. The distribution of dELL on polytene chromosomes is nearly co-extensive with that of phosphorylated Pol II, suggesting a very general role in transcription. This inference is further supported by the dramatic redistribution of dELL that accompanies a brief heat shock. Previous studies (Andrulis et al., 2000; Kaplan et al., 2000; Lis et al., 2000) show a similar broad distribution and heat-shock-dependent redistribution for certain other elongation factors (pTEFb, Spt5 and Spt6). Thus, dELL appears to act in concert with Pol II and a suite of elongation factors at a large number of genes.
Our data provide independent support for a dELL–Pol II interaction by demonstrating a chromatographic co-enrichment of His6-tagged dELL and Pol II. Taken together with the in vitro elongation activity of dELL in a purified system, this suggests that dELL contacts Pol II in vivo. While further biochemical and structural studies are required to demonstrate the nature of dELL–Pol II binding, dELL has emerged as a strong candidate for a global transcriptional regulator in Drosophila. Together with the evolutionary conservation of the ELL family of proteins from flies to humans, this suggests an important role for this family of factors in metazoan development.
Roles for dELL in transcriptional elongation
Heat shock treatment results in a striking mobilization of dELL from a large number of euchromatic sites to a small number of loci corresponding to heat shock genes. In Drosophila polytene chromosomes, the heat shock response involves regression of the developmental puffs, corresponding to a dramatic reduction in transcription of developmental genes, concomitant with the appearance of a small set of heat shock puffs corresponding to the intensely transcribed heat shock genes (Ashburner, 1970). The widespread chromosomal binding of dELL under non-heat shock conditions and its absence from heterochromatic sites, its mobilization from developmental genes to heat shock genes upon heat shock, and its general co-localization with Pol II are all consistent with a general role for dELL in gene expression. Taken together with its ability to enhance Pol II processivity in vitro, dELL is likely to have a general role in transcriptional elongation in vivo. It must be noted here that the sum of dELL signals associated with the heat shock loci appears to correspond to a small fraction of the sum of dELL signal observed before the heat shock. This can be explained either by a very short half-life for the dELL protein and/or message, or the dELL being saturated at the heat shock loci and, therefore, the rest of the dELL is released from the polytene chromosomes.
Detailed analysis of dELL distribution patterns on polytene chromosomes reveals a few exceptional cases of Pol II binding with little or no detectable dELL. A potential explanation for this would be if these exceptional cases represent genes with short transcription units and/or genes that are transcribed at low levels. By this model, the amount of dELL present at such loci would be low, and the immunostaining inapparent, while dELL accumulates to high levels at larger, more highly transcribed genes. Alternatively, dELL may form distinct complexes at different loci, in some cases masking the protein from interaction with antibody. Indeed, mammalian ELL has been isolated as part of a larger protein complex (Shilatifard, 1998c). Such complexes may provide a mechanism for regulating ELL–Pol II interactions at specific loci. Clearly, the identification of a unique ELL family protein in Drosophila opens the door to genetic and biological analyses that, together with biochemistry, can define the mechanism of ELL and its interactions with other Pol II general elongation factors in vivo.
Materials and methods
Plasmid construction
A small expressed sequence tag (EST) clone containing a portion of dELL cDNA was used for probing a Drosophila cDNA library (Brown and Kaftos, 1988). The full-length dELL cDNA was cloned into a pRSET vector containing dELL cDNA with an upstream in-frame N-terminal His6 epitope tag and an Express epitope tag. The dELL cDNA and accompanying epitope tags were excised from the vector as an NdeI–EcoRI restriction fragment, with the NdeI end blunted by the Klenow fragment. This restriction fragment was then ligated to the EcoRI–SalI fragment of the HIC-L vector (Kraus et al., 1988), in which the SalI end was also blunted by the Klenow fragment. The resulting construct placed the dELL cDNA and accompanying epitope tags downstream of the Hsp70 promoter. Subsequently, the entire cDNA, tags and promoter sequence were excised from HIC-L as a NotI restriction fragment, which was then ligated into a NotI-restricted pYC1.8 vector (Fridell and Searles, 1991). Transgenic flies carrying this construct were used for polytene chromosome staining and co-immunoprecipitation experiments.
To generate radiolabeled RNA from the dELL cDNA for the developmental northern analysis of dELL expression, the dELL cDNA was excised from the pRSETb construct in the same manner as described above, containing an EcoRI-restricted end at its C-terminus and a blunted NdeI end at its N-terminus. This fragment was then ligated to the SmaI–EcoRI fragment of pGEM-3 (Promega), where it was placed under a T7 promoter at its C-terminal end. This was then used to transcribe antisense RNA in the reaction described below.
Northern blot analysis
Five micrograms of total RNA isolated from D.melanogaster at each of the 10 developmental stages (gift of Dale Dorsett) were denatured in MOPS/formaldehyde/formamide solution and separated by electrophoresis on a 1% formaldehyde gel at low voltage. Resolved RNAs were transferred to a Biotrans (ICN) membrane and cross-linked to the membrane by UV irradiation. The membrane was incubated in hybridization solution (50% formamide, 10% PEG 8000, 3.5% SDS, 150 mM sodium pyrophosphate, 250 mM NaCl, 1 mM EDTA, 2× Denhardts) for at least 1 h at 65°C before the addition of radiolabeled probe. The antisense RNA probe was prepared using the pGEM vector containing dELL cDNA under the control of the T7 promoter at the C-terminus. Antisense RNA transcription was performed using T7 RNA polymerase (Promega) under the following reaction conditions: 40 mM Tris–HCl pH 7.9, 10 mM NaCl, 6 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine, 0.05% Tween-20, 0.5 mM each ATP, GTP and UTP, 0.5 µCi of [32P]CTP (ICN), 0.1 U RNAsin (Promega) and 2 µg of template DNA. Reactions were carried out at 37°C for 60 min, and then treated with RQ1 DNase (Promega) for 15 min to remove plasmid template. Antisense RNA was then purified using the RNeasy RNA purification kit (Qiagen), added to the hybridization mixture and incubated overnight at 65°C. The blot was then washed successively in buffers containing decreasing concentrations of SSC and SDS until background radiation levels reached a minimum. Transcripts were visualized by autoradiography or Phosphor Imager. To visualize the loading control, pGEM containing Drosophila RP49 cDNA (gift of Dale Dorsett) under T7 was used to generate the antisense RNA radiolabeled probe.
Expression and purification of recombinant dELL
dELL in pRSETb was used to transform BL21 (DE3) pLysE E.coli. Positive transformants were selected by double selection with ampicillin (100 µg/ml) and chloramphenicol (34 µg/ml), and were then used to produce overnight cultures in liquid broth containing each antibiotic. Cultures were diluted 1:50 and allowed to grow to an OD600 of ∼0.4, and then induced with isopropyl-β-d-thiogalactopyranoside (Roche) at 0.5–1 mM concentrations for ∼4 h. Cells were harvested by centrifugation and lysed by repeated freeze–thaw cycles in 1× phosphate-buffered saline (PBS). Inclusion bodies were separated from soluble proteins by ultracentrifugation, and then homogenized in buffer A (6 M guanidine– HCl, 40 mM Tris) to solubilize the recombinant protein. Homogenates were then ultracentrifuged again to separate soluble protein from cell debris. His-tagged recombinant protein was purified using Probond nickel chelating resin (Invitrogen). Briefly, proteins were bound to resin in buffer A. Resin–protein conjugates were washed with buffer B (buffer A + 40 mM imidazole) three times to remove non-specifically bound contaminants. Protein was eluted from resin with buffer C (buffer A + 300 mM imidazole) and vortexing. This was followed by centrifugation to separate the resin from the supernatant containing the eluted protein. Load, flow-through, wash and bound fractions were analyzed by western blotting with anti-Express and anti-His6 antibodies (Invitrogen) to examine the efficiency of purification. Using this same strategy, ∼5 mg of recombinant protein were expressed and purified (the purity and identity of the expressed dELL were determined by MALDI mass spectroscopy analysis) for the purpose of raising polyclonal antisera. Protein samples were sent to Research Genetics (Huntsville, AL) for antibody production. For use in the in vitro transcription assay, purified recombinant dELL was renatured by dialysis in buffer containing 250 mM KCl. The polyclonal antisera from animals injected with dELL were purified over a dELL affinity column and the purified polyclonal serum was tested for specificity towards dELL by western blot analysis. Also, extracts prepared from salivary glands isolated from transgenic larvae expressing dELL demonstrated a significant increase in a polypeptide of the same size as recombinant dELL in western analysis, demonstrating the specificity of the purified polyclonal antibodies towards dELL.
In vitro transcription elongation assay
Vector containing an AdML promoter was cut with EcoRI and NdeI. The linear fragment was purified by gel electrophoresis and used in in vitro transcription assays. Pre-initiation complexes were assembled at the AdML promoter with recombinant TBP, TFIIB, TFIIE, TFIIF and purified rat TFIIH and RNA polymerase II as described (Shilatifard et al., 1996). Transcription was initiated by the addition of 50 µM ATP, 50 µM GTP, 2 µM UTP, 10 µCi of [32P]CTP (ICN) (specific activity 3000 Ci/mmol) and 7 mM MgCl2. After 10 min at 28°C, 100 µM non-radioactive CTP was added to the reaction mixture, and short transcripts were chased in the presence or absence of purified and renatured recombinant dELL for the times indicated. Transcripts were analyzed by electrophoresis through a 6% acrylamide–7 M urea–0.5× TBE gel and developed using a Molecular Dynamics (Sunnyvale, CA) PhosphorImager. Mock purified and renatured protein was used in the absence of dELL as a control for the experiment.
Germ line transformation and fly culture
v36f; ry506 embryos were injected with the Hsp70–dELL cDNA construct, together with the helper plasmid pπ25.7wc (Karess and Rubin, 1984), essentially as described (Spradling, 1986). G0 survivors were crossed to v36f; ry506 flies and F1 adults were screened for rescue of the vermilion eye color. Transgenic lines used in this study had inserts on the third chromosome, and were carried over a TM2, ry Ubx balancer.
Immunofluorescence staining
Embryos were dechorionated and fixed in 4% formaldehyde and stained with purified dELL polyclonal or phospho-Pol II-specific monoclonal antibody (H14). Appropriate secondary antibodies (Cy3 for dELL and Cy2 for Pol II at 1:1000 dilution; Jackson Laboratory) were used and images were recorded with a SPOT CCD camera (Diagnostic Instruments, Inc.) using PAX-it™ imaging software (Midwest Information Systems, Inc.).
Salivary glands were dissected in gland buffer (Cohen and Gotchel, 1971), fixed for 30 s in 2% formaldehyde in gland buffer, then in 45% acetic acid for 3 min, before storage in 67% glycerol/33% PBS at –20°C. We found that varying the fixation between 10 and 60 s made no difference to the staining. Anti-His6G antibody (Invitrogen) was diluted 1:100 with incubation for 1 h. For optimal staining with the polyclonal dELL antibody, purified dELL polyclonal (dilution was 1:25) was incubated with polytene chromosomes overnight. Appropriate secondary antibodies (Jackson Laboratory) as indicated in the figure legends were used at 1:1000 dilution. Fluorescence detection was by epifluorescence using an Olympus BX60 fluorescence microscope with an NB barrier filter for fluorescein and Cy2 detection, and a WG barrier filter for rhodamine and Cy3. Images were recorded with a SPOT CCD camera (Diagnostic Instruments, Inc.) using PAX-it™ imaging software (Midwest Information Systems, Inc.).
Co-precipitation of dELL and RNA Pol II
Equal masses of transgenic flies expressing His-tagged dELL under heat shock conditions or non-transgenic flies of similar genetic background were homogenized in ice-cold NUN buffer (1 M urea, 300 mM NaCl, 25 mM HEPES pH 7.9) (Lavery and Schibler, 1993). Extracts were cleared by ultracentrifugation and supernatants were mixed with Probond resin that was equilibrated with cold NUN buffer. Binding was carried out at 4°C for 1 h with gentle agitation. Resin–protein conjugates were gently washed 3–4 times with three volumes of NUN buffer, and bound proteins were eluted with solution containing 300 mM imidazole. Load, flow-through, wash and bound fractions from each sample set (transgenic or non-transgenic) were analyzed by western blotting for the presence of dELL (using anti-Express monoclonal antibody) and RNA pol II (using Pol II-specific monoclonal antibody as described above). Proteins of interest were visualized using the Renaissance ECL kit (Amersham).
Accession number
The DDBJ/EMBL/GenBank accession No. for Drosophila ELL is AF416770.
Acknowledgments
Acknowledgements
We thank Nikolous Spoerel for sharing sequence data before the publication of the Drosophila genome. We are also grateful to Drs Nathalie Aulner and Dale Dorsett for the generous gift of Drosophila RNA and fruitful discussions and conversations. We thank Drs Ueli Schibler, John Lis and Abdul Whaheed for helpful conversations, comments and encouragements. This work was supported by NIH grant 1R55GM057005 to J.C.E. and ACS grant RP69921801, NIH grant 1R01CA089455 and Mallinckrodt Foundation award to A.S. A.S. is Scholar of the Leukemia and Lymphoma Society.
References
- Andrulis E.D., Guzmán,E., Döring,P., Werner,J. and Lis,J.T. (2000) High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev., 14, 2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. (1970) The genetic analysis of puffing in polytene chromosomes of Drosophila. Proc. R. Soc. Lond. B Biol. Sci., 176, 319–327. [DOI] [PubMed] [Google Scholar]
- Bradsher J.N., Jackson,K.W., Conaway,R.C. and Conaway,J.W. (1993) An RNA polymerase II transcription factor SIII.I. Identification, purification and properties. J. Biol. Chem., 268, 25587–25593. [PubMed] [Google Scholar]
- Bradsher J., Coin,F. and Egly,J.M. (2000) Distinct roles for the helicases of TFIIH in transcript initiation and promoter escape. J. Biol. Chem., 275, 2532–2538. [DOI] [PubMed] [Google Scholar]
- Brown N.H. and Kafatos,F.C. (1988) Functional cDNA libraries from Drosophila embryos. J. Mol. Biol., 203, 425–437. [DOI] [PubMed] [Google Scholar]
- Cho H., Kim,T.K., Mancebo,H., Lane,W.S., Flores,O. and Reinberg,D. (1999) A protein phosphatase functions to recycle RNA polymerase II. Genes Dev., 13, 1540–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L.H. and Gotchel,B.V. (1971) Histones of polytene and nonpolytene nuclei of Drosophila melanogaster. J. Biol. Chem., 246, 1841–1848. [PubMed] [Google Scholar]
- Conaway J.W. and Conaway,R.C. (2000) Transcription elongation and human disease. Annu. Rev. Biochem., 68, 301–319. [DOI] [PubMed] [Google Scholar]
- Crowley T.E., Bond,M.W. and Meyerowitz,E.M. (1983) The structural genes for three Drosophila glue proteins reside at a single polytene chromosome puff locus. Mol. Cell. Biol., 3, 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus M.E. (1996) Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem., 271, 19009–19012. [DOI] [PubMed] [Google Scholar]
- DiMartino J.F., Miller,T., Ayton,P.M., Landewe,T., Hess,J.L., Cleary,M.L. and Shilatifard,A. (2000) A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood, 96, 3887–3893. [PubMed] [Google Scholar]
- Duan D.R. et al. (1995) Inhibition of transcription elongation by the VHL tumor suppressor protein. Science, 269, 1402–1406. [DOI] [PubMed] [Google Scholar]
- Dvir A., Conaway,R.C. and Conaway,J.W. (1996) Promoter escape by RNA polymerase II. A role for an ATP cofactor in suppression of arrest by polymerase at promoter-proximal sites. J. Biol. Chem., 271, 23352–23356. [DOI] [PubMed] [Google Scholar]
- Dvir A., Tan,S., Conaway,J.W. and Conaway,R.C. (1997) Promoter escape by RNA polymerase II. Formation of an escape-competent transcriptional intermediate is a prerequisite for exit of polymerase from the promoter. J. Biol. Chem., 272, 28175–28178. [DOI] [PubMed] [Google Scholar]
- Elmendorf B.J., Shilatifard,A., Yan,Q., Conaway,J.W. and Conaway,R.C. (2001) Transcription factors TFIIF, ELL and Elongin negatively regulate SII-induced nascent transcript cleavage by non-arrested RNA polymerase II elongation intermediates. J. Biol. Chem., 276, 23109–23114. [DOI] [PubMed] [Google Scholar]
- Fridell Y.W. and Searles,L.L. (1991) Vermilion as a small selectable marker gene for Drosophila transformation. Nucleic Acids Res., 19, 5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett K.P., Tan,S., Bradsher,J.N., Lane,W.S., Conaway,J.W. and Conaway,R.C. (1994) Molecular cloning of an essential subunit of an RNA polymerase II elongation factor SIII. Proc. Natl Acad. Sci. USA, 91, 5237–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf A.L., Plagens,U., Jamrich,M. and Bautz,E.K.F. (1978) RNA polymerase B (or II) in heat induced puffs of Drosophila polytene chromosomes. Chromosoma, 65, 127–136. [DOI] [PubMed] [Google Scholar]
- Guild G.M. and Shore,E.M. (1984) Larval salivary gland secretion proteins in Drosophila. Identification and characterization of the Sgs-5 structural gene. J. Mol. Biol., 179, 289–314. [DOI] [PubMed] [Google Scholar]
- Guo S., Yamaguchi,Y., Schilbach,S., Wada,T., Lee,J., Goddard,A., French,D., Handa,H. and Rosenthal,A. (2000) A regulator of transcriptional elongation controls vertebrate neuronal development. Nature, 408, 366–369. [DOI] [PubMed] [Google Scholar]
- Hartzog G.A., Wada,T., Handa,H. and Winston,F. (1998) Evidence that Spt4, Spt5 and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae.Genes Dev., 12, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban M.G. and Luse,D.S. (1992) RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J. Biol. Chem., 267, 13647–13655. [PubMed] [Google Scholar]
- Kaplan C.D., Morris,J.R., Wu,C. and Winston,F. (2000) Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D.melanogaster. Genes Dev., 14, 2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R.E. and Rubin,G.M. (1984) Analysis of P transposable element functions in Drosophila. Cell, 38, 135–146. [DOI] [PubMed] [Google Scholar]
- Korge G. (1975) Chromosome puff activity and protein synthesis in larval salivary glands of Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 72, 4550–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus K.W., Lee,Y.H., Lis,J.T. and Wolfner,M.F. (1988) Sex-specific control of Drosophila melanogaster yolk protein 1 gene expression is limited to transcription. Mol. Cell. Biol., 8, 4756–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel J.F. and Goodrich,J.A. (1998) Promoter escape limits the rate of RNA polymerase II transcription and is enhanced by TFIIE, TFIIH and ATP on negatively supercoiled DNA. Proc. Natl Acad. Sci. USA, 95, 9232–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K.P., Akoulitchev,S. and Reinberg,D. (1998) Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proc. Natl Acad. Sci. USA, 95, 9767–9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery D.J. and Schibler,U. (1993) Circadian transcription of the cholesterol 7 α hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev., 7, 1871–1884. [DOI] [PubMed] [Google Scholar]
- Lis J.T., Mason,P., Peng,J., Price,D.H. and Werner,J. (2000) P-TEFb kinase recruitment and function at heat shock loci. Genes Dev., 14, 792–803. [PMC free article] [PubMed] [Google Scholar]
- Mancebo H.S. et al. (1997) P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev., 11, 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall N.F. and Price,D.H. (1995) Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem., 270, 12335–12338. [DOI] [PubMed] [Google Scholar]
- Marshall N.F., Peng,J., Xie,Z. and Price,D.H. (1996) Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem., 271, 27176–27183. [DOI] [PubMed] [Google Scholar]
- Miller T., Williams,K., Johnstone,R.W. and Shilatifard,A. (2000) Identification, cloning, expression and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J. Biol. Chem., 275, 32052–32056. [DOI] [PubMed] [Google Scholar]
- Moreland R.J., Tirode,F., Yan,Q., Conaway,J.W., Egly,J.M. and Conaway,R.C. (1999) A role for the TFIIH XPB DNA helicase in promoter escape by RNA polymerase II. J. Biol. Chem., 274, 22127–22130. [DOI] [PubMed] [Google Scholar]
- Myers L.C., Gustafsson,C.M., Bushnell,D.A., Lui,M., Erdjument-Bromage,H., Tempst,P. and Kornberg,R.D. (1998) The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev., 12, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., LeRoy,G., Chang,C.H., Luse,D.S. and Reinberg,D. (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell, 92, 105–116. [DOI] [PubMed] [Google Scholar]
- Otero G., Fellows,J., Li,Y., de Bizemont,T., Dirac,A.M., Gustafsson,C.M., Erdjument-Bromage,H., Tempst,P. and Svejstrup,J.Q. (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell, 3, 109–118. [DOI] [PubMed] [Google Scholar]
- Parada C.A. and Roeder,R.G. (1999) A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J., 18, 3688–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelling C. (1964) Ribonukein saure synthese der reisenchromosomen. Chromosoma, 15, 71–122. [DOI] [PubMed] [Google Scholar]
- Peng J., Marshall,N.F. and Price,D.H. (1998) Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem., 273, 13855–13860. [DOI] [PubMed] [Google Scholar]
- Plagens U., Greenleaf,A.L. and Bautz,E.K.F. (1976) Distribution of RNA polymerase on Drosophila polytene chromosomes as studied by indirect immunofluorescence. Chromosoma, 59, 157–165. [DOI] [PubMed] [Google Scholar]
- Price D.H. (2000) P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol., 20, 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.H., Sluder,A.E. and Greenleaf,A.L. (1989) Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol. Cell. Biol., 9, 1465–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D. and Mote,J. (1993) Elongation factor SII-dependent transcription by an RNA polymerase II through a sequence-specific DNA-binding protein. Proc. Natl Acad. Sci. USA, 90, 1917–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D., Conaway,J.W. and Conaway,R.C. (1996) The RNA polymerase II general elongation factors. Trends Biochem. Sci., 21, 351–355. [PMC free article] [PubMed] [Google Scholar]
- Reines D., Conaway,R.C. and Conaway,J.W. (1999) Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr. Opin. Cell Biol., 11, 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd M.D., Izban,M. and Luse,D.S. (1994) The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc. Natl Acad. Sci. USA, 91, 8057–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. (1998a) Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J., 12, 1437–1446. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. (1998b) The RNA polymerase II general elongation complex. Biol. Chem., 379, 27–31. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. (1998c) Identification and purification of Holo-ELL complex: evidence for the presence of ELL-associated protein. J. Biol. Chem., 273, 11212–11217. [DOI] [PubMed] [Google Scholar]
- Shilatifard A., Lane,W.S., Jackson,K.W., Conaway,R.C. and Conaway,J.W. (1996) An RNA polymerase II elongation factor encoded by the human ELL gene. Science, 271, 1873–1876. [DOI] [PubMed] [Google Scholar]
- Shilatifard A., Duan,D.R., Haque,D., Florence,C., Schubach,W.H., Conaway,J.W. and Conaway,R.C. (1997a) ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc. Natl Acad. Sci. USA, 94, 3639–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A., Haque,D., Conaway,R.C. and Conaway,J.W. (1997b) Structure and function of RNA polymerase II elongation factor ELL. Identification of two overlapping ELL functional domains that govern its interaction with polymerase and the ternary elongation complex. J. Biol. Chem., 272, 22355–22363. [DOI] [PubMed] [Google Scholar]
- Spradling A.C. (1986) P element mediated transformation. In Roberts, D.B. (ed.), Drosophila, A Practical Approach. IRL Press, Oxford, UK. pp 175–197. [Google Scholar]
- Tennyson C.N., Klamut,H.J. and Worton,R.G. (1995) The human dystrophin gene requires 16 h to be transcribed and is cotranscriptionally spliced. Nature Genet., 9, 184–190. [DOI] [PubMed] [Google Scholar]
- Thirman M.J., Levitan,D.A., Kobayashi,H., Simon,M.C. and Rowley,J.D. (1994) Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc. Natl Acad. Sci. USA, 91, 12110–12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirman M.J., Diskin,E.B., Bin,S.S., Ip,H.S., Miller,J.M. and Simon,M.C. (1997) Developmental analysis and subcellular localization of the murine homologue of ELL. Proc. Natl Acad. Sci. USA, 94, 1408–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremeau-Bravard A., Perez,C. and Egly,J.M. (2001) A role of the C-terminal part of p44 in the promoter escape activity of transcription factor IIH. J. Biol. Chem., 276, 27693–27697. [DOI] [PubMed] [Google Scholar]
- Ucker D.S. and Yamamoto,K.R. (1984) Early events in the stimulation of mammary tumor virus RNA synthesis by glucocorticoids. Novel assays of transcription rates. J. Biol. Chem., 259, 7416–7420. [PubMed] [Google Scholar]
- Uptain S.M. and Chamberlain,M.J. (1997) Escherichia coli RNA polymerase terminates transcription efficiently at rho-independent terminators on single-stranded DNA templates. Proc. Natl Acad. Sci. USA, 94, 13548–13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain S.M., Kane,C.M. and Chamberlain,M.J. (1997) Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem., 66, 117–172. [DOI] [PubMed] [Google Scholar]
- Wada T., Takagi,T., Yamaguchi,Y., Watanabe,D. and Handa,H. (1998) Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J., 17, 7395–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Wada,T., Watanabe,D., Takagi,T., Hasegawa,J. and Handa,H. (1999) Structure and function of human transcription elongation factor DSIF. J. Biol. Chem., 274, 8085–8092. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Peery,T., Peng,J., Rananathan,Y., Marshall,N.F., Marshall,T., Amendt,B., Mathews,M.B. and Price,D.H. (1997) Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev., 11, 2622–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]