Abstract
Agrobacterium tumefaciens causes crown gall disease in dicotyledonous plants by introducing a segment of DNA (T-DNA), derived from its tumour-inducing (Ti) plasmid, into plant cells at infection sites. Besides these natural hosts, Agrobacterium can deliver the T-DNA also to monocotyledonous plants, yeasts and fungi. The T-DNA integrates randomly into one of the chromosomes of the eukaryotic host by an unknown process. Here, we have used the yeast Saccharomyces cerevisiae as a T-DNA recipient to demonstrate that the non-homologous end-joining (NHEJ) proteins Yku70, Rad50, Mre11, Xrs2, Lig4 and Sir4 are required for the integration of T-DNA into the host genome. We discovered a minor pathway for T-DNA integration at the telomeric regions, which is still operational in the absence of Rad50, Mre11 or Xrs2, but not in the absence of Yku70. T-DNA integration at the telomeric regions in the rad50, mre11 and xrs2 mutants was accompanied by gross chromosomal rearrangements.
Keywords: Agrobacterium/genomic instability/non-homologous end-joining /T-DNA integration/telomeres
Introduction
Agrobacterium tumefaciens causes crown gall disease in plants by transferring an oncogenic segment of DNA, the transferred or T-DNA, to plant cells at wound sites (Chilton et al., 1977; Tinland and Hohn, 1995). The T-DNA is derived from an ∼200 kb tumour-inducing (Ti) plasmid, which is present in the bacterium. Plant phenolic compounds, produced by the wounded plant cells, induce expression of virulence genes located elsewhere on the Ti plasmid. The virulence protein VirD2 introduces nicks at the 24 bp border repeats, which flank the T-region (Wang et al., 1987; Pansegrau, 1993). This leads to the formation of a single-stranded DNA copy from the T-region, which is called the T-strand (Stachel et al., 1986). The T-strand is bound at the 5′ end by the VirD2 protein and it is this single stranded nucleoprotein complex that is delivered into plant cells (Ward and Barnes, 1988; Chaudhury et al., 1994; Tinland et al., 1994; Yusibov et al., 1994). There it is co-operatively bound by the VirE2 protein (Citovsky et al., 1989), which is delivered separately into plant cells by the bacterium (Vergunst et al., 2000), and targeted to the nucleus by the presence of a nuclear localization signal in the VirE2 and VirD2 proteins (Tinland et al., 1992; Rossi et al., 1993; Tzfira et al., 2000). T-DNA integration occurs at random positions in the genome by a process of non-homologous recombination (NHR) (Offringa et al., 1990; Mayerhofer et al., 1991; Tinland and Hohn, 1995; Gelvin, 2000). Although the processing and transfer of T-DNA to plants is reasonably well understood, (host) factors involved in T-DNA integration are just beginning to be identified (Gelvin, 2000; Mysore et al., 2000).
The T-DNA itself does not encode enzymes that are involved in integration. As the Agrobacterium proteins VirD2 and VirE2 accompany the T-DNA to the plant nucleus, it is reasonable to propose that they may be involved in T-DNA integration. The VirD2 protein indeed is important for the accuracy of the integration of T-DNA (Tinland et al., 1995). The VirE2 protein probably protects the T-DNA from nucleolytic degradation and eases its translocation into the nucleus (Rossi et al., 1996; Zupan et al., 1996). Therefore, VirD2 and VirE2 are important for T-DNA transfer and nuclear targeting, but do not seem to play an essential role in the integration process per se. In accordance with this, in vitro studies showed that a T-DNA ligation-integration reaction is mediated by plant enzymes, which implies a role for host factors in T-DNA integration (Ziemienovicz et al., 2000).
T-DNA transfer can also occur, at least under laboratory conditions, to yeasts and fungi (Bundock et al., 1995; Bundock and Hooykaas, 1996; De Groot et al., 1998; Gouka et al., 1999) where, in the absence of DNA homology, integration occurs by a similar process of NHR as in plants. In contrast, integration occurs by homologous recombination (HR) when the T-DNA carries homology with the yeast genome. This was not found in plants where T-DNA sharing extensive homology with the plant genome still integrates mainly by NHR (Offringa et al., 1990). These important findings indicate that the process of T-DNA integration into the host genome is predominantly determined by host factors. Recently, Salomon and Puchta (1998) showed that T-DNA could be captured during DNA double-strand break (DSB) repair in plants. This suggests that DSB repair provides a pathway for T-DNA integration. However, as the right ends of the T-DNAs that had integrated into the DSB were all truncated, it is possible that this does not represent the most common form of T-DNA integration.
Studies on the repair of DNA DSBs in the yeast Saccharomyces cerevisiae revealed that there are two general recombination mechanisms: one that requires homology between the two recombining DNA molecules (HR) and one that is independent of such homology [non-homologous end-joining (NHEJ)] (reviewed by Critchlow and Jackson, 1998; Haber, 2000). Several mechanisms have been described for the repair of DSBs by HR, most of which rely on the action of genes of the RAD52 epistasis group (RAD50-RAD59, MRE11 and XRS2) (reviewed by Sung et al., 2000). Studies on the repair of DSBs under conditions where HR was impossible revealed that at least 10 genes are required for repair by NHEJ (YKU70, YKU80, LIG4, LIF1, SIR2, SIR3, SIR4, RAD50, MRE11 and XRS2) (reviewed by Tsukamoto and Ikeda, 1998; Lewis and Resnick, 2000). Most of these NHEJ genes have additional functions in telomere length maintenance (RAD50, MRE11, XRS, YKU70, YKU80 and SIR2-SIR4; Porter et al., 1996; Boulton and Jackson, 1998; Chamankkah and Xiao, 1999; Gallego and White, 2001) and/or transcriptional silencing at the telomeres (YKU70, YKU80 and SIR2-SIR4; Aparicio et al., 1991; Boulton and Jackson, 1998).
We have now studied the role of host proteins in the integration of T-DNA by NHR. As the results obtained so far on this topic with plants are controversial (Gelvin, 2000), we have now employed the yeast S.cerevisiae as a model organism to investigate which of the genes encoding for recombination enzymes are necessary for T-DNA integration. To this end T-DNA integration in wild-type was compared with that in isogenic strains carrying disruptions of these genes. The results show for the first time that the NHEJ proteins Yku70, Rad50, Mre11, Xrs2, Lig4 and Sir4 are required for the integration of T-DNA into the host genome. We discovered a minor pathway for T-DNA integration at the telomeric regions, which is still operational in the absence of Rad50, Mre11 and Xrs2, but not in the absence of Yku70. T-DNA integration at the telomeric regions in the rad50, mre11 and xrs2 mutants was accompanied by gross chromosomal rearrangements.
Results
A versatile T-DNA to study integration by NHR in yeast
T-DNA, which lacks homology with the yeast genome, has been described to integrate by NHR (Bundock and Hooykaas, 1996). The T-DNA that was used in this study carried the URA3 gene and to prevent homology between the T-DNA and the yeast genome the URA3 gene was removed from the genome of the T-DNA recipient. In order to be able to study the integration of T-DNA by NHR in various yeast strains, independent of the genetic background, a novel T-DNA vector (pSDM8000) was constructed with the KanMX selectable marker between the T-DNA border repeats (Figure 1; Wach et al., 1994). This marker, which allows selection of transgenic yeasts resistant to G418, consists of heterologous DNA and thus the T-DNA of pSDM8000 lacks any homology with the yeast genome. Integration of the T-DNA from pSDM8000 into the yeast genome can therefore only occur by NHR. The T-DNA from pSDM8000 integrated in the wild-type yeast strains YPH250 and JKM115 at frequencies of 1.6 × 10–7 and 1.2 × 10–5, respectively (Table I). For 10 T-DNA insertions the integration site was established after retrieval of the linked genomic sequences by the Vectorette PCR (data not shown). This confirmed that integration of the T-DNA from pSDM8000 had occurred by NHR as was described previously (Bundock and Hooykaas, 1996).
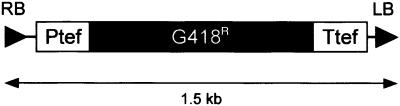
Fig. 1. Schematic representation of the T-DNA from pSDM8000. The T-DNA from pSDM8000 was used in co-cultivation experiments to study T-DNA integration by NHR in recombination defective S.cerevisiae strains. The T-DNA contains the KanMX cassette, which consists of the kan resistance gene of the Escherichia coli transposon Tn903 under control of transcriptional and translational sequences of the filamentous fungus Ashbya gossypii TEF gene. This module allows selection of S.cerevisiae transformants resistant against the antibiotic G418 (Wach et al., 1994).
Table I. Frequencies of T-DNA integration by NHR in recombination defective yeast strains.
| Strain | Genotype | Frequency of G418-resistant colonies ± SEM (×10–8)a | Relative frequency of G418-resistant colonies (%)b |
|---|---|---|---|
| YPH250 | WT | 16 ± 9.6 | 100 |
| YPH250rad51 | rad51Δ | 14 ± 7.8 | 88 |
| YPH250rad52 | rad52Δ | 38 ± 8.4c | 238 |
| YPH250yku70 | yku70Δ | <0.0075c | <0.05 |
| YPH250rad50 | rad50Δ | 0.80 ± 0.40c | 5.0 |
| YPH250lig4 | lig4Δ | 0.37 ± 0.36c | 2.3 |
| JKM115 | WT | 1150 ± 0.50 | 100 |
| JKM129 | xrs2Δ | 27 ± 6.0c | 2.3 |
| JKM138 | mre11Δ | 29 ± 3.0c | 2.5 |
| JKM120sir4 | sir4Δ | 15 ± 1.9c | 1.3 |
aAll yeast strains were co-cultivated with LBA1119(pSDM8000). Averages of two or more independent experiments are shown. Frequencies are depicted as the number of G418-resistant colonies divided by the output number of yeast cells (cells/ml). SEM= standard error of the mean.
bThe relative frequency of T-DNA integration by NHR is (frequency in the mutant/frequency in the wild-type (WT)) × 100%.
cThe means of the frequency of G418-resistant colonies seen in the wild-type (WT) and the mutant were tested significantly different in a Student’s t-test (p <0.05).
NHEJ proteins are required for T-DNA integration by NHR
Many proteins involved in various forms of DNA recombination and DNA repair have been identified in yeast. In order to determine their possible role in T-DNA integration, we performed T-DNA transfer experiments and compared T-DNA integration in wild-type yeasts with that in isogenic strains carrying disruptions of the recombination genes RAD51, RAD52, YKU70, LIG4, MRE11, RAD50, XRS2 and SIR4. The results are summarized in Table I. In the rad51 and rad52 mutants that are defective in HR, the frequencies of T-DNA integration by NHR were, respectively, similar and slightly higher (∼2- to 3-fold) than observed for the wild-type yeast strain. The Rad51 and Rad52 proteins apparently do not play a role in T-DNA integration by NHR and possibly Rad52 even suppresses T-DNA integration by NHR. In contrast, in the lig4, mre11, rad50, xrs2 and sir4 mutants the frequency of T-DNA integration by NHR was reduced dramatically and T-DNA integration seems to be abolished in the yku70 mutant, as no G418-resistant colonies were obtained in experiments performed with this mutant. The genes impaired in these mutants have been described as being involved in the repair of genomic DSBs by NHEJ (Tsukamoto and Ikeda, 1998; Lewis and Resnick, 2000). We conclude that the proteins Yku70, Lig4, Mre11, Rad50, Xrs2 and Sir4, are also required for the integration of T-DNA by NHR into the host genome. In addition, Yku70 seems to be essential for T-DNA integration as in its absence integration of T-DNA was never observed.
T-DNA integrates preferentially at (sub)telomeric regions in rad50, mre11 and xrs2 mutants and forms repeat structures in the genome of a lig4 mutant
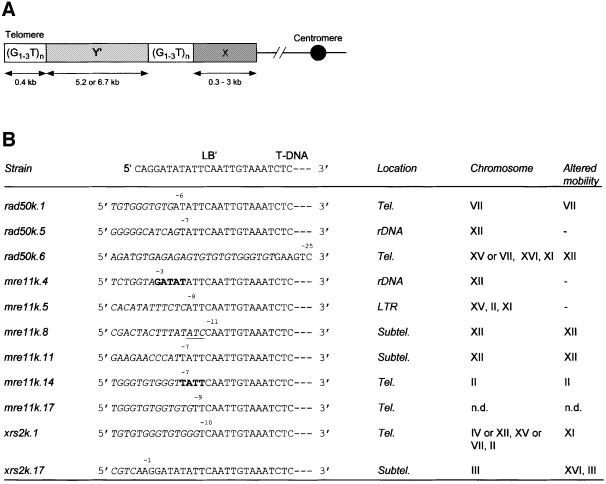
From co-cultivation experiments with the rad50, mre11, xrs2, lig4 and sir4 mutants, we obtained a small number of G418-resistant colonies. The features of T-DNA integration were determined for a number of these lines. The yeast genomic sequences linked to the T-DNA were isolated by Vectorette PCR or thermal asymmetric interlaced–PCR (TAIL–PCR), sequenced and used in a BLAST search in the yeast genome database to determine the T-DNA integration sites. Strikingly, analysis of the genomic DNA junctions to the left end of the T-DNA revealed that in two out of three rad50, four out of six mre11 and two out of two xrs2 mutants analysed, T-DNAs had integrated into telomeres or subtelomeric regions (Table II; Figure 2). Saccharomyces cerevisiae telomeres generally consist of one or more copies of Y′ and X elements followed by telomerase-generated C(1–3)A/TG(1–3) repeats (Figure 2; Zakian, 1996). In the rad50k.1, rad50k.6, mre11k.14, mre11k.17 and xrs2k.1 mutants, the left end of the T-DNA was fused to this typical telomerase-generated C(1–3)A/TG(1–3) repeat (Figure 2). Other than these telomeric insertions we found one T-DNA insertion in the long terminal repeat (LTR) of the Ty transposable element (mre11k.5; Table II and Figure 2) and two insertions in the rDNA region, present in chromosome XII (mre11k.4 and rad50k.5; Table II and Figure 2).
Table II. Genomic distribution of T-DNA integrated by NHR in rad50, mre11, xrs2 and sir4 mutants in comparison with the wild-type after T-DNA transfer from pSDM8000.
| Yeast strain (genotype) | (Sub)telomeric region | LTR | rDNA | Elsewhere |
|---|---|---|---|---|
| rad50Δ | 2 | 0 | 1 | 0 |
| mre11Δ | 4 | 1 | 1 | 0 |
| xrs2Δ | 2 | 0 | 0 | 0 |
| sir4Δ | 0 | 0 | 0 | 6 |
| Wild-typea | 1 | 2 | 1 | 50 |
aPreviously, target sites for T-DNA integration in the genome of S.cerevisiae strain RSY12 were determined in 44 T-DNA transformed lines (Bundock and Hooykaas, 1996; Bundock, 1999). In addition, we determined the position of T-DNA integration in 10 T-DNA transformed S.cerevisiae YPH250 lines.
Fig. 2. T-DNA integrates preferentially at (sub)telomeric regions in rad50, mre11 and xrs2 mutants. (A) Schematic representation of a yeast chromosome end, showing the position of the telomeric (G1–3T) repeats, the Y′ and X elements and the centromere. Y′ and X elements are present at only a subset of all chromosomes and are often found in association with internal tracts of (G1–3T) repeats. (B) Junction sequences of T-DNA left end and genomic DNA of the S.cerevisiae rad50, mre11 and xrs2 mutants. Genomic DNA sequences are shown in italics, T-DNA sequences in normal capitals. Bold sequences represent microhomology of the T-DNA left end with the integration site. Filler DNA sequences are underlined and depicted in italics. The numbers above the sequences represent the number of base pairs deleted from the T-DNA left end. Tel. = telomeric region; Subtel. = subtelomeric region; rDNA = ribosomal DNA region; LTR = long terminal repeat of Ty element; – = none of the 16 chromosomes showed an altered mobility; n.d. = not determined; LB′ = remnant of T-DNA left border repeat.
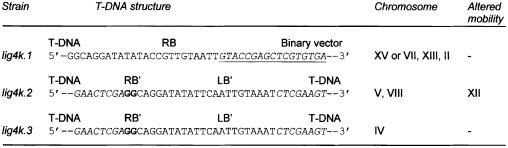
We obtained only three colonies from co-cultivations with the lig4 mutant. Unfortunately, the location of the T-DNA insertion could not be established in these three lig4 mutants. In one mutant, the right end of the T-DNA was found to be fused to sequences of the binary vector (lig4k.1; Figure 3). Most likely only the left border repeat (LB) was processed during T-strand formation in Agrobacterium, resulting in the transfer and integration of the binary vector. In the two other lig4 mutants a perfect fusion between the right and left end of two T-DNAs was found (lig4k.2 and lig4k.3; Figure 3). This showed that two T-DNA copies had integrated at the same position in the yeast genome, resulting in the formation of a direct repeat structure. T-DNA repeat structures were only found in ∼3% of T-DNA transformed wild-type (data not shown), suggesting that the absence of Lig4 favours the formation of complex T-DNA structures in the yeast genome.
Fig. 3. T-DNA forms repeat structures in the genome of a lig4 mutant. Sequences of T-DNA structures as found in the genome of the S.cerevisiae lig4 lines are depicted. Sequences of the binary vector are underlined and depicted in italics. T-DNA sequences are shown in italics, except for the border sequences. The right border repeat (RB) is presented in bold for lig4k.2 and lig4k.3 to distinguish between RB and LB sequences. RB′ and LB′ = fused remnants of the RB and LB, respectively.
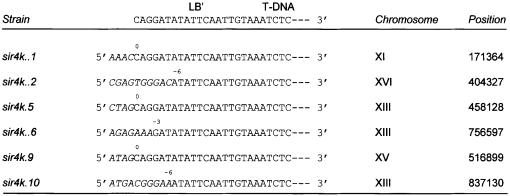
Analysis of six T-DNA insertions in the sir4 mutant revealed no preference for integration at the telomeres as found for the wild-type strain (Table II and Figure 4). T-DNA integrations in the wild-type are characterized by truncation of the T-DNA left end and the presence of microhomology between the left end of the T-DNA and the target site (Offringa et al., 1990; Mayerhofer et al., 1991; Tinland and Hohn, 1995; Bundock and Hooykaas, 1996). Similar features were seen for T-DNA integrations in the rad50, mre11, xrs2 and sir4 mutants (Figures 2 and 4).
Fig. 4. The position of T-DNA integration by NHR in the genome of a sir4 mutant is not biased. Junction sequences of the T-DNA left end and genomic DNA of the sir4 mutant are depicted. Genomic DNA sequences are shown in italics, T-DNA sequences in normal capitals. The numbers above the sequences represent the number of base pairs deleted from the left T-DNA end.
We conclude that in the rad50, mre11 and xrs2 mutants the T-DNA, if integrated at all, becomes preferentially inserted in (sub)telomeric regions. Yku70 may play an important role in this novel pathway for T-DNA integration at the telomeres as in its absence no such insertions were found. In contrast, disruption of SIR4 did affect the efficiency of T-DNA integration by NHR, but not the position of T-DNA integration. In the lig4 mutant T-DNA integrated rarely and formed repeat structures at so far unknown positions in the genome.
T-DNA integration in rad50, mre11, xrs2 and lig4 mutants is accompanied by chromosomal instability
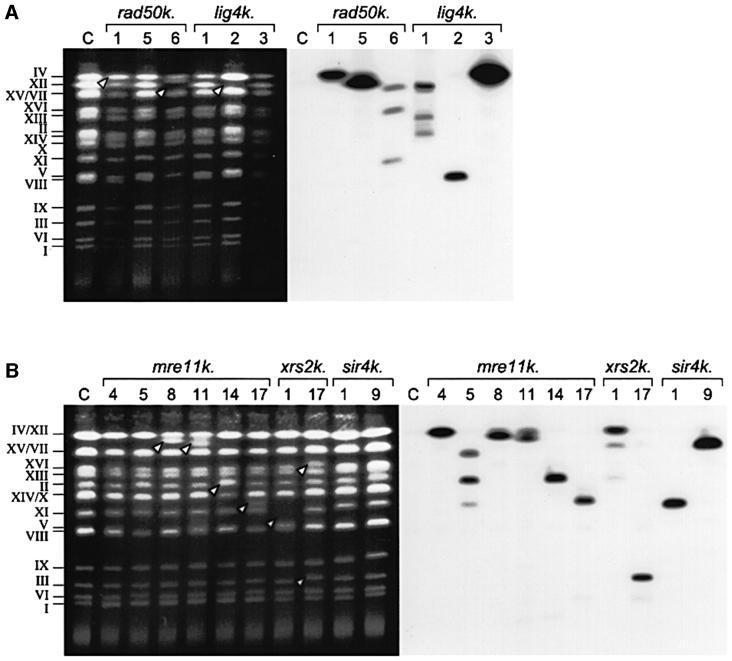
Telomeres consist of highly conserved and homologous sequences (Figure 2; Zakian, 1996). When the telomere sequences flanking the T-DNA inserts in the NHEJ mutants were used in a BLAST search, several yeast chromosomes were found to be the possible target for T-DNA integration. In order to determine in which chromosome the T-DNAs had integrated in the rad50, mre11, xrs2 and lig4 mutants, intact chromosomes were isolated, separated in a CHEF gel, blotted on a membrane and hybridized with the KanMX probe which anneals to the T-DNA (Figure 5).
Fig. 5. T-DNA integration is accompanied by chromosomal rearrangements in rad50, mre11, xrs2 and lig4 mutants. CHEF-gel analysis of T-DNA integration events in the genome of S.cerevisiae rad50, mre11, xrs2, lig4 and sir4 mutants was studied. Chromosomes from G418-resistant S.cerevisiae colonies obtained after co-cultivation with A.tumefaciens carrying pSDM8000 were isolated, separated on a CHEF gel (left panels in A and B) and blotted on a membrane. The membrane was hybridized with a labelled KanMX probe that anneals to the T-DNA and an autoradiograph was made (right panels in A and B). (A) T-DNA transformed rad50 and lig4 mutants and their isogenic and untransformed wild-type control (C). (B) T-DNA transformed mre11, xrs2 and sir4 mutants and their isogenic and untransformed wild-type control (C). White arrowheads indicate chromosomes with an altered mobility.
As a control, two sir4 strains and the two rad50 and mre11 strains in which the T-DNA had inserted outside the telomeres were analysed. In the sir4k.1 and sir4k.9 mutants the T-DNA was present in chromosome XI and XV, respectively (Table II; Figure 5), confirming the sequence data. For the rad50k.5 and mre11k.4 mutants the sequence data indicated that T-DNA insertions were located in the rDNA region, present in chromosome XII (Figure 5). In line with this for both mutants a band on the blot corresponding to chromosome XII was seen, indicating that the T-DNA indeed had inserted in chromosome XII. In the mre11k.5 mutant the T-DNA insertion was present in an LTR element according to the sequence analysis (Figure 5). Unexpectedly, for this mutant three bands were seen on the blot, indicating the presence of T-DNA insertions in chromosomes XV or VII (which were not visually separated on the gel), II and XI. It is conceivable that one T-DNA had integrated in a LTR element on one of these chromosomes and was subsequently duplicated and translocated to the other chromosomes (Kim et al., 1998). Similarly, the chromosomes into which the T-DNA had inserted in the other rad50, mre11 and xrs2 mutants were assigned (Figure 2).
The rad50 and mre11 mutants, that had T-DNA insertions in rDNA or LTR elements, did not show any changes in chromosome mobility when compared with the wild-type. In contrast, in all rad50, mre11 and xrs2 mutants that contained a T-DNA inserted at the (sub)telomeric region, changes in the mobility of one or more of the chromosomes were seen. In most of these mutants, rad50k.1, mre11k.8, mre11k.11, mre11k.14, mre11k.17 and xrs2k.17, the chromosome into which the T-DNA had inserted showed an altered mobility. This was confirmed by the use of chromosome specific probes (data not shown). In this way it became apparent, for instance, that the T-DNA insertion in the rad50k.1 mutant was present on chromosome VII, which now co-migrated with chromosome IV instead of XV (Figure 5). In a minority of the mutants, rad50k.6 and xrs2k.1, an altered mobility was seen of a chromosome that did not contain a T-DNA insertion. For instance, in the rad50k.6 mutant T-DNA insertions were found in chromosome XV or VII, XVI and XI (Figures 2 and 5). None of these chromosomes showed a change in mobility. However, with a probe specific for chromosome XII a change in the mobility of this chromosome was observed, although a T-DNA insertion was not present in this chromosome (Figure 5).
The position of the T-DNA insertions in the lig4 mutant could not be determined as sequence analysis only revealed T-DNA repeat structures. CHEF gel analysis of the lig4k.1, lig4k.2 and lig4k.3 mutants revealed that T-DNA had integrated in chromosome XV or VII, XIII and II (lig4k.1), V and II (lig4k.2) and IV (lig4k.3) (Figures 3 and 5). In the lig4k.2 mutant chromosome XII had an altered mobility, although a T-DNA had not integrated into this chromosome.
In summary, by combining sequence and CHEF-gel data we can draw several conclusions. In the NHEJ mutants rad50, mre11 and xrs2 the T-DNA integrates preferentially into telomeric regions, but these events are associated with the formation of gross chromosomal rearrangements as detected by altered chromosome mobility. Strikingly, the chromosomes that show rearrangements are not always the chromosomes that contain the T-DNA insertion. As we did not observe chromosomal rearrangements in the wild-type strain transformed by A.tumefaciens or in untransformed rad50, mre11, xrs2 and lig4 mutants (data not shown) we can only conclude that introduction and/or integration of T-DNA in these NHEJ mutants is accompanied by genetic instability. Whether this instability is a consequence of T-DNA integration at the telomeres or that T-DNA is captured at the telomeres during repair of already existing telomeric instability in the NHEJ mutants needs to be investigated.
Discussion
Agrobacterium tumefaciens delivers T-DNA into cells of its natural host, dicotyledonous plants, monocotyledonous plants, yeasts and fungi. In the absence of homology with the host genome the T-DNA integrates randomly into any of the chromosomes of these eukaryotic hosts by NHR. Here, we have used the yeast S.cerevisiae as a T-DNA recipient to investigate which host proteins might play a role in T-DNA integration. We demonstrate that the NHEJ proteins Yku70, Rad50, Mre11, Xrs2, Lig4 and Sir4 are required for the integration of T-DNA into the host genome. It was described before that these proteins have distinct functions in the repair of genomic DSBs by NHEJ (Tsukamoto and Ikeda, 1998; Lewis and Resnick, 2000). We infer from this that DNA DSB repair by NHEJ provides a pathway for T-DNA integration. A recent study showed that T-DNAs can indeed be captured during DSB repair in plants (Salomon and Puchta, 1998) However, as the right ends of the T-DNAs that had integrated into the DSB were all truncated, in contrast to the right ends of the most commonly found T-DNAs, it is probable that T-DNA integration occurs usually at other sites than DSBs. The present work shows that such T-DNA integrations are nevertheless dependent on NHEJ enzymes.
It would be interesting to know whether the dependence on the NHEJ enzymes for integration is typical for the Agrobacterium T-DNA or more general for introduced non-homologous DNA molecules. Schiestl et al. (1994) showed that a mutation in RAD50 reduces the frequency of integration of non-homologous, BamHI-treated DNA into the yeast genome by restriction enzyme-mediated integration (REMI) after yeast transformation. The role of RAD50 in such integration can be explained by the fact that the accompanying restriction enzyme induces DSBs in the genome where integration can occur by NHEJ.
The Rad51 and Rad52 proteins play important roles in HR in yeast. The frequency of T-DNA integration by NHR was comparable with wild-type levels in the rad51 mutant, but >2-fold higher in the rad52 mutant. This latter observation may be explained by the idea that Rad52 and Yku70 are competing agents, channelling recombination into either HR or NHR, respectively (Haber, 1999). Yku70-mediated integration of T-DNA by NHR may be more efficient as the competing pathway of HR is not operative in the absence of Rad52.
T-DNA integrates preferentially at (sub)telomeric regions in rad50, mre11 and xrs2 mutants. The telomeres comprise ∼2% of the yeast genome (Zakian, 1996). Therefore, 2% of the T-DNA insertions would be expected at the telomeres. In fact in the wild-type we have found one telomeric T-DNA insertion in 54 analysed wild-type lines (Table II). In contrast, in the rad50, xrs2 and mre11 mutants eight out of 11 T-DNAs had integrated in this area. The Rad50, Mre11 and Xrs2 proteins play a minor role in telomeric silencing (Boulton and Jackson, 1998). Thus, an explanation might be that reduced silencing at the telomeric region makes this part of the chromosome more accessible for T-DNA, thereby facilitating T-DNA integration at (sub)telomeric regions. The Sir4 protein plays an important role in transcriptional silencing at the telomeres (Aparicio et al., 1991). However, we did not find a bias for T-DNA integration at these regions in the sir4 mutant. Therefore, we conclude that the absence of telomeric silencing in the rad50, mre11 and xrs2 mutants is not responsible for the integration of T-DNA at the telomeres. We speculate that an alternative T-DNA integration pathway, which leads to specific integration at the telomeres or ribosomal DNA repeat, can replace the normal integration pathway in the absence of an active Rad50–Mre11–Xrs2 complex. One possibility is that this pathway may operate in vivo to repair DNA aberrations occurring during DNA replication of repeated DNA. Alternatively, it may be used for the restoration of the telomeric structure. The Rad50–Mre11–Xrs2 complex and the Yku70 protein are involved in telomere length maintenance and rad50, mre11, xrs2 and yku70 mutants show shortened telomeres (Porter et al., 1996; Boulton and Jackson, 1998). The telomeric structure has to be restored in order to survive and this process may lead to the incorporation of T-DNA at the telomeres. As we did not obtain such T-DNA insertions in the yku70 mutant, it may be that YKU70 plays an important role in this pathway.
It has been found that the lack of Sir genes induces the loss of silencing of cryptic mating-type genes. This leads to changes in expression of mating-type specific genes, which in turn leads to a reduced efficiency of NHEJ (Lee et al., 1999). The Sir4 protein may also play a minor role in DSB repair by NHEJ by mediating an altered local chromatin structure at the break site, thereby making the break site more accessible for the repair enzymes (Boulton and Jackson, 1998; Critchlow and Jackson, 1998). Both options can explain why the T-DNA integration frequency is reduced in the sir4 mutant and the pattern of integration remains as in the wild-type.
In the absence of Rad50, Mre11 and Xrs2, and to a lesser extent Lig4, gross chromosomal rearrangements have been observed (Chen and Kolodner, 1999). In our studies we did not detect such chromosomal rearrangements, neither in untransformed wild-type nor in the untransformed rad50, mre11, xrs2 and lig4 mutants (data not shown). However, chromosomal rearrangements were often seen in the yeast mutants after transformation by Agrobacterium and more specifically in the rad50, mre11 and xrs2 mutants after the T-DNA had integrated at (sub)telomeric regions. We infer that either the integration of T-DNA or the presence of a T-DNA insertion at (sub)telomeric regions is specifically accompanied by chromosomal rearrangements. It has been observed before that Ty elements can induce chromosomal rearrangements by inserting at telomeric regions (Kim et al., 1998). Alternatively, T-DNAs may be captured specifically by cells with chromosomal instability. Translocations were the principal class of rearrangements seen in rad50, mre11 and xrs2 mutants (Myung et al., 2001) and might reflect the rearrangements seen in the rad50, mre11 and xrs2 mutants in which the T-DNA had integrated at the (sub)telomeric region. However, further experiments should elucidate the type of rearrangements observed in these mutants.
We observed that in the absence of Lig4 T-DNA integration is strongly reduced. It was remarkable that the few lines that were obtained after transformation by Agrobacterium had T-DNA repeat structures. We may assume that due to the absence of Lig4 chromosomal breaks are sealed less efficiently and that the T-DNAs that are captured by such breaks have more time to recombine or fuse to extrachromosomal copies of T-DNA, resulting in the accumulation of T-DNAs in repeat structures. The repeats consist of fused left and right borders and it is therefore likely that the VirD2 strand transferase mediated formation of these repeat structures (Pansegrau et al., 1993).
The structure of the T-DNA as seen after integration by NHR into the genome of plants, fungi and yeasts is similar (Mayerhofer et al., 1991; Bundock and Hooykaas, 1996; De Groot et al., 1998). This has been taken as evidence that T-DNA integration in these diverse organisms occurs by a conserved mechanism. In agreement with this expectation the NHR machinery, which we found to be used for random T-DNA integration in yeast, has been well conserved during eukaryotic evolution. Further experimentation will have to prove that plant and fungal orthologues of the yeast YKU70, RAD50, MRE11, XRS2, LIG4 and SIR4 genes also play an important role in T-DNA integration in these organisms.
Materials and methods
Yeast strains
The yeast strains that were used are listed in Table III. Yeast mutants isogenic to the haploid YPH250 strain were constructed using the one-step disruption method (Rothstein, 1991) after lithium acetate transformation (Gietz et al., 1992). Disruption of YKU70, LIG4, RAD50, RAD51 and RAD52 was confirmed by PCR and Southern blot analysis.
Table III. Yeast strains.
| Strain | Genotype | Reference |
|---|---|---|
| YPH250 | MATα, ura3-52, lys2–801, ade2-101, trp1-Δ1, his3-Δ200, leu2-Δ1 | Sikorski and Hieter (1989) |
| YPH250rad51 | MATα, ura3-52, lys2–801, ade2-101, trp1-Δ1, his3-Δ200, leu2-Δ1, rad51::LEU2 | this study |
| YPH250rad52 | MATα, ura3-52, lys2–801, ade2-101, trp1-Δ1, his3-Δ200, leu2-Δ1, rad52::LEU2 | this study |
| YPH250yku70 | MATα, ura3-52, lys2–801, ade2-101, trp1-Δ1, his3-Δ200, leu2-Δ1, yku70::LEU2 | this study |
| YPH250rad50 | MATα, ura3-52, lys2–801, ade2-101, trp1-Δ1, his3-Δ200, leu2-Δ1, rad50::hisG | this study |
| YPH250lig4 | MATα, ura3-52, lys2–801, ade2-101, trp1-Δ1, his3-Δ200, leu2-Δ1, lig4::HIS3 | this study |
| JKM115 | MATα, Δho, Δhml::ADE1, Δhmr::ADE1, ade1, leu2-3,112, lys5, trp1::hisG, ura3-52 | Moore and Haber (1996) |
| JKM129 | MATα, Δho, Δhml::ADE1, Δhmr::ADE1, ade1, leu2-3,112, lys5, trp1::hisG, ura3-52, xrs2::LEU2 | Moore and Haber (1996) |
| JKM138 | MATα, Δho, Δhml::ADE1Δhmr::ADE1, ade1, leu2-3,112, lys5, trp1::hisG, ura3-52, mre11::hisG | Moore and Haber (1996) |
| JKM120sir4 | MATα, Δho, HMLα, HMRα, ade1, leu2-3,112, lys5, trp1::hisG, ura3-52, his3::hisG′-URA3-hisG′, sir4::HIS3 | Lee et al. (1999) |
Construction of the binary vector pSD8000
To construct pSDM8000, a 1513 bp PvuII–EcoRV fragment carrying the KanMX marker was obtained from pFA6a (Wach et al., 1994) and was ligated into the unique HpaI site of pSDM14 (Offringa, 1992). The binary vector pSDM8000 was introduced into A.tumefaciens by electroporation (Den Dulk-Ras and Hooykaas, 1995).
Co-cultivations/T-DNA transfer experiments
Co-cultivations were performed as described earlier with slight modifications (Bundock et al., 1995). Agrobacterium strain LBA1119 (also known as EHA105; Hood et al., 1993) was used in all experiments and was grown overnight in LC medium prior to co-cultivation. The mix of Agrobacterium and S.cerevisiae cells was incubated for 9 days at 20°C. T-DNA containing S.cerevisiae strains were selected at 30°C on YPAD medium containing G418 (200 µg/ml) (Life Technologies/Gibco-BRL).
Vectorette PCR and TAIL–PCR
Chromosomal DNA was isolated using Qiagen’s Genomic Tips G/20 as per manufacturer’s protocol. An amount (1–2 µg) of genomic DNA was digested with EcoRI, ClaI, PstI or HindIII and run on a 1% TBE gel. Non-radioactive Southern analysis was performed. The membrane was hybridized with a digoxigenine-labelled KanMX probe to determine the size of T-DNA/genomic junction fragments (EcoRI and ClaI for right end containing fragments and PstI and HindIII for left end containing fragments). The KanMX probe, a 792 bp internal fragment of the KanMX marker, was made by PCR using primers kanmxp1 (5′-AGACTCACGTTTCGAGGCC-3′) and kanmxp2 (5′-TCACCGAGGCAGTTCCATAG-3′) and a Non-Radioactive DNA Labelling and Detection kit (Boehringer Mannheim). The restriction enzyme producing the smallest junction fragment was used for Vectorette PCR as described (http://genome-www.stanford.edu/group/botlab/protocols/vectorette.html). The Expand™ High Fidelity System (Boehringer Mannheim) was used to amplify fragments >2.5 kb, whereas sTaq DNA polymerase (SphaeroQ) was used for the amplification of fragments <2.5 kb. Primer kanmxp3 (5′-TCGCAGGTCTGCAGCGAGGAGC-3′) and kanmxp4 (5′-TCGCT CGACATCATCTGCCCAG-3′) were used to amplify right border junction and left border junction fragments, respectively.
TAIL–PCR was used to isolate right border junction fragments and was performed as described previously (Liu et al., 1995), except that KanMXp3, KanMXp5 (5′-TCACATCATGCCCCTGAGCTGC-3′) and KanMXp7 (5′-GGGTATTCTGGGCCTCCATG-3′) were used as right border specific primers.
The yeast genomic sequences that were isolated by Vectorette and TAIL–PCR were used in a BLAST search in the yeast genome database at http://www-genome.stanford.edu/SGD in order to determine the T-DNA integration sites.
T7 DNA polymerase sequencing
Vectorette PCR and TAIL–PCR products were cloned in pGEMTEasy (Promega) and sequenced using the T7 polymerase sequencing kit (Pharmacia) according to the manufacturer’s protocol.
CHEF gels
Complete and intact yeast chromosomes were isolated in agarose blocks as described previously (Schwartz and Cantor, 1984; De Jonge et al., 1986). Blocks were placed in the wells of a 0.25× TBE–agarose gel. A CHEF apparatus was used to separate the chromosomes (Bio-Rad). Electrophoresis was performed in 0.25× TBE at 14°C with an initial switch time of 40 s and a final switch time of 120 s at 200 V for 30 h. The LKB2016 VacuGene vacuum blotting apparatus was used with 10× standard saline citrate (SSC) to transfer the chromosomes to a positively charged Nylon membrane (Boehringer Mannheim). The membrane was probed with the KanMX probe (see Vectorette PCR) and autoradiography was done for 1 day using Fuji SuperRX film. Subsequently, the membrane was stripped in 0.5 M NaOH at 42°C, hybridized with a probe specific for chromosome XII, stripped again and hybridized with a probe specific for chromosome VII. Probes specific for chromosomes XII and VII, were made by PCR. A 1045 bp region, located downstream of the RAD5 gene, and a 1246 bp internal fragment of the LIF1 gene was amplified from genomic DNA isolated from the wild-type strain YPH250, respectively, using primers Lif1p1 (5′-AGCTGACGGAGTTCATTAGCT-3′) and Lif1p2 (5′-TACCGTTTCCGATTCTGTCT-3′) and Rad5p5 (5′-GGATTCGACAACAAGGGGTC-3′) and Rad5p6 (5′-GTGTGGTAAGAGCACCTGCC-3′).
Acknowledgments
Acknowledgements
We thank Professor J.Haber (Rosenstiel Center and Biology Department, Brandeis University, Waltham, MA) for kindly providing a number of yeast mutants, Dr H.Y.Steensma for technical assistance with the CHEF gel and Dr E.J.van der Zaal and Dr R.Offringa for critical reading of the manuscript. This work was supported by the EU RECOMBIO Biotech program BIO4-CT97-2028.
References
- Aparicio O.M., Billington,B.L. and Gottschling,D.E. (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci. Cell, 66, 1279–1287. [DOI] [PubMed] [Google Scholar]
- Boulton S.J. and Jackson,S.P. (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J., 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P. (1999) Agrobacterium tumefaciens-mediated transformation of yeasts and fungi. PhD Thesis, Leiden University, The Netherlands.
- Bundock P. and Hooykaas,P.J.J. (1996) Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc. Natl Acad. Sci. USA, 93, 15272–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P., Den Dulk-Ras,A., Beijersbergen,A. and Hooykaas,P.J.J. (1995) Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J., 14, 3206–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamankkah M. and Xiao,W. (1999) Formation of the yeast Rad50–Mre11–Xrs2 complex is correlated with DNA repair and telomere maintenance. Nucleic Acids Res., 27, 2072–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A.M., Dennis,E.S. and Brettell,R.I.S. (1994) Gene-expression following T-DNA transfer into plant cells is aphidicolin-sensitive. Aust. J. Plant Physiol., 21, 125–131. [Google Scholar]
- Chen C. and Kolodner,R.D. (1999) Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nature Genet., 23, 81–85. [DOI] [PubMed] [Google Scholar]
- Chilton M.D., Drummond,M.H., Merio,D.J., Sciaky,D., Montoya,A.L., Gordon,M.P. and Nester,E.W. (1977) Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell, 11, 263–271. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Wong,M.L. and Zambryski,P. (1989) Co-operative interaction of Agrobacterium VirE2 protein with single-stranded DNA: implications for the transfer process. Proc. Natl Acad. Sci. USA, 86, 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow S.E. and Jackson,S.P. (1998) DNA-end-joining: from yeast to man. Trends Biochem. Sci., 23, 394–398. [DOI] [PubMed] [Google Scholar]
- De Groot M.J.A., Bundock,P., Hooykaas,P.J.J. and Beijersbergen,A.G.M. (1998) Agrobacterium-mediated transformation of filamentous fungi. Nature Biotech., 16, 839–842. [DOI] [PubMed] [Google Scholar]
- De Jonge P., De Jongh,F.C.M., Meijers,R., Steensma,H.Y. and Scheffers,W.A. (1986) Orthogonal-field-alteration gel electrophoresis banding patterns of DNA from yeasts. Yeast, 2, 193–204. [DOI] [PubMed] [Google Scholar]
- Den Dulk-Ras A. and Hooykaas,P.J.J. (1995) Electroporation of Agrobacterium tumefaciens. In Nickoloff,J.A. (ed.), Plant Cell Electroporation and Electrofusion Protocols (4). Humana Press Inc., Totowa, NJ, pp. 63–72.
- Gallego M.E. and White,C.I. (2001) RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl Acad. Sci. USA, 98, 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S.B. (2000) Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol., 51, 223–256. [DOI] [PubMed] [Google Scholar]
- Gietz D., St. Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouka R.J., Gerk,C., Hooykaas,P.J.J., Bundock,P., Musters,W., Verrips,C.T. and De Groot,M.J.A. (1999) Transformation of Aspergillus awamori by Agrobacterium tumefaciens-mediated homologous recombination. Nature Biotech., 17, 598–601. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (1999) Gatekeepers of recombination. Nature, 398, 665–667. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (2000) Partners and pathways repairing a double-strand break. Trends Genet., 16, 259–264. [DOI] [PubMed] [Google Scholar]
- Hood E.E., Gelvin,S.B., Melchers,L.S. and Hoekema,A. (1993) New Agrobacterium helper plasmids for gene transfer to plants. Trangenic Res., 2, 208–218. [Google Scholar]
- Kim J.M., Vanguri,S., Boeke,J.D., Babriel,A. and Voytas,D.F. (1998) Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res., 8, 464–478. [DOI] [PubMed] [Google Scholar]
- Lee S.E., Paques,F., Sylvan,J. and Haber,J.E. (1999) Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol., 15, 767–770. [DOI] [PubMed] [Google Scholar]
- Lewis L.K. and Resnick,M.A. (2000) Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutagen. Res., 451, 71–89. [DOI] [PubMed] [Google Scholar]
- Liu Y.-G., Mitsukawa,N., Oosumi,T. and Whitier,R.F. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Mayerhofer R. et al. (1991) T-DNA integration: a mode of illegitimate recombination in plants. EMBO J., 10, 687–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K. and Haber,J.E. (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore K.S., Nam,J. and Gelvin,S.B. (2000) An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl Acad. Sci. USA, 97, 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Chen,C. and Kolodner,R.D. (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature, 411, 1073–1076. [DOI] [PubMed] [Google Scholar]
- Offringa R. (1992) Gene targeting in plants using the Agrobacterium vector system. PhD Thesis, Leiden University, The Netherlands.
- Offringa R., De Groot,M.J.A., Haagsman,H.J., Does,M.P., van den Elzen,P.J.M. and Hooykaas,P.J.J. (1990) Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. EMBO J., 9, 3077–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Schoumacher,F., Hohn,B. and Lanka,E. (1993) Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc. Natl Acad. Sci. USA, 90, 11538–11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter S.E., Greenwell,P.W., Ritchie,K.B. and Petes,T.D. (1996) The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res., 24, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L., Hohn,B. and Tinland,B. (1993) The VirD2 protein of Agrobacterium tumefaciens carries nuclear localization signals important for transfer of T-DNA to plants. Mol. Gen. Genet., 239, 345–353. [DOI] [PubMed] [Google Scholar]
- Rossi L., Hohn,B. and Tinland,B. (1996) Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl Acad. Sci. USA, 93, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R.J. (1991) One-step gene disruption in yeast. Methods Enzymol., 101, 202–211. [DOI] [PubMed] [Google Scholar]
- Salomon S. and Puchta,H. (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J., 17, 6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R.H., Zhu,J. and Petes,T.D. (1994) Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 4493–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D.C. and Cantor,C.R. (1984) Seperation of yeast chromosome-sized DNA’s by pulsed field gradient electrophoresis. Cell, 37, 67–75. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S.E., Timmerman,B. and Zambryski,P. (1986) Generation of single-stranded T-DNA molecules during the initial stages of T-DNA transfer from Agrobacterium tumefaciens to plant cells. Nature, 322, 706–712. [Google Scholar]
- Sung P., Trujillo,K.M. and Komen,S.V. (2000) Recombination factors of Saccharomyces cerevisiae. Mutagen. Res., 451, 257–275. [DOI] [PubMed] [Google Scholar]
- Tinland B. and Hohn,B. (1995) Recombination between prokaryotic and eukaryotic DNA: integration of Agrobacterium tumefaciens T-DNA into the plant genome. In Setlow,J.K. (ed.) Genetic Engineering. Plenum Press, NY, USA, pp. 209–229. [PubMed]
- Tinland B., Koukolßková-Nicola,Z., Hall,M.N. and Hohn,B. (1992) The T-DNA linked VirD2 protein contains two distinct functional nuclear localization signals. Proc. Natl Acad. Sci. USA, 89, 7442–7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B., Hohn,B. and Puchta,H. (1994) Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc. Natl Acad. Sci. USA, 91, 8000–8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B., Schoumacher,F., Gloeckler,V., Bravo-Angel,A.M. and Hohn,B. (1995) The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J., 14, 3585–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y. and Ikeda,H. (1998) Double-strand break repair mediated by DNA end-joining. Genes Cells, 3, 135–144. [DOI] [PubMed] [Google Scholar]
- Tzfira T., Ree,Y., Chen,M-H., Kunik,T. and Citovsky,V. (2000) Nucleic acid transport in plant-microbe interactions: the molecules that walk through the walls. Annu. Rev. Microbiol., 54, 187–219. [DOI] [PubMed] [Google Scholar]
- Vergunst A.C., Schrammmeijer,B. den Dulk-Ras,A., de Vlaam,C.M., Regensburg-Tuink,T.J. and Hooykaas,P.J.J. (2000) VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science, 290, 979–982. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Pöhlmann,R. and Philippsen,P (1994) New heterologous modules for classical gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wang K., Stachel,S.E., Timmerman,B. van Montagu,M. and Zambryski,P. (1987) Site-specific nick in the T-DNA border sequence as a result of Agrobacterium tumefaciens vir gene expression. Science, 235, 587–591. [DOI] [PubMed] [Google Scholar]
- Ward E.R. and Barnes,W.M. (1988) VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science, 242, 927–930. [Google Scholar]
- Yusibov V.M., Steck,T.R., Gupta,V and Gelvin,S.B. (1994) Association of single stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl Acad. Sci. USA, 91, 2994–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian V.A. (1996) Structure, function and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet., 30, 141–172. [DOI] [PubMed] [Google Scholar]
- Ziemienovicz A., Tinland,B., Bryant,J., Gloeckler,V. and Hohn,B. (2000) Plant enzymes but not Agrobacterium VirD2 mediate T-DNA ligation in vitro. Mol. Cell. Biol., 20, 6317–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan J.R., Citovsky,V. and Zambryski,P. (1996) Agrobacterium VirE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc. Natl Acad. Sci. USA, 93, 2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]