Abstract
B-cell differentiation is accompanied by a dramatic increase in cytoplasmic accumulation and stability of the IgM heavy chain (µ) secretory mRNA. Despite considerable effort, the mechanism is unknown. We have identified three short motifs upstream of the secretory poly(A) site, which, when mutated in the µ heavy chain gene, significantly increase the accumulation of the secretory form of poly(A)+ mRNA relative to the membrane form and regulate the expression of the secretory poly(A) site in a developmental manner. We show that these motifs bind U1A and inhibit polyadenylation in vitro and in vivo. Overexpression of U1A in vivo results in the selective inhibition of the secretory form. Thus, this novel mechanism selectively controls post-cleavage expression of the µ secretory mRNA. We present evidence that this mechanism is used to regulate alternative expression of other genes.
Keywords: IgM heavy chain secretory mRNA/novel U1A-binding motifs/poly(A) addition
Introduction
The poly(A) tail plays an important role in post-transcriptional control of gene expression in regulating mRNA transport, stability and translation (Ross, 1995; Gallie, 1998; Preiss and Hentze, 1999). Poly(A) tails are formed in the nucleus in conjunction with RNA splicing and 3′ end cleavage and, with the exception of histone mRNA, mRNAs that are not polyadenylated correctly are not transported to the cytoplasm and are degraded rapidly (Bousquet-Antonelli et al., 2000; Le Hir et al., 2000; Shatkin and Manley, 2000; Tollervey and Caceres, 2000; reviewed in Minvielle-Sebastia and Keller, 1999; Wahle and Ruegsegger, 1999; Zhao et al., 1999).
The IgM heavy chain (µ) pre-mRNA is processed alternatively to either a membrane or a secretory mRNA by alternative poly(A) site usage, which is regulated during B-cell development (Early et al., 1980; Galli et al., 1988; Perry et al., 1988; Peterson and Perry, 1989) (Figure 1A). This is accompanied by dramatic changes in total µ mRNA accumulation, which is not accounted for by an increase in transcription rate as the heavy chain enhancer is fully active even in pre-B cells (Gerster et al., 1986). These changes can be explained by changes in µ mRNA stability and nuclear to cytoplasmic transport (Darnell, 1982; Berberich and Schimpl, 1990). It has been shown that the half-life of µ mRNA roughly doubles from 3–8 to 14–16 h during differentiation (Mason et al., 1988; Cox and Emtage, 1989; Berberich and Schimpl, 1990; Reed et al., 1994). Perry and Kelley (1979) found a 6-fold change in total µ mRNA nuclear levels compared with a 150-fold change in cytoplasmic levels between 70Z/3 (a pre-B cell line) and MPC11 (a myeloma).
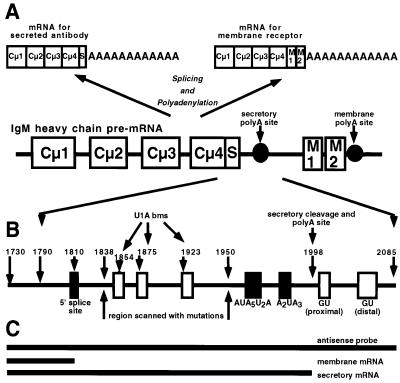
Fig. 1. Schematic model of the immunoglobulin secretory poly(A) site. (A) The genetic organization of the IgM heavy chain and its alternative processing to a secretory or a membrane form of mRNA. (B) The location of the secretory poly(A) site and relative location of the 5′ splice site, the U1A-binding motifs, the hexanucleotide sequence and the downstream GU-rich regions. Numbers indicate the positions of the ends of the RNA substrates and plasmid inserts referred to in the text. (C) The length and position of the antisense probe used for RNase protection assays and the protected fragments for the µ secretory and µ membrane mRNA, respectively, referred to in Figures 6 and 7.
Using lipopolysaccharide-stimulated primary B cells, Berberich and Schimpl (1990) distinguished µ secretory mRNA and µ membrane mRNA and found that the doubling of the half-life was due to a change in secretory µ mRNA stability as the half-life of the membrane form did not change. They found no change in the ratio of secretory to membrane mRNA accumulation in the nucleus but a drastic shift to µ secretory mRNA in the cytoplasm. These results suggest that mechanisms responsible for the increased stability and nuclear to cytoplasmic transport make a greater contribution to the increased expression of the µ secretory mRNA during B-cell differentiation than an up-regulated usage of the µ secretory poly(A) cleavage site.
In recent years, much work has focused on the regulation of cleavage efficiency at the secretory poly(A) site (Peterson and Perry, 1989; Peterson, 1992; Edwalds-Gilbert and Milcarek, 1995; Phillips et al., 1996, 1999; Phillips and Virtanen, 1997; Martincic et al., 1998; Takagaki and Manley, 1998; Veraldi et al., 2001). However, the molecular mechanism for the increased stability and nuclear to cytoplasmic transport of µ secretory mRNA has been unexplored.
Using a systematic mutational analysis, we have identified three motifs upstream of the µ secretory poly(A) site that inhibit accumulation of poly(A) µ secretory mRNA in vivo and regulate expression of the secretory poly(A) site during B-cell differentiation. All three motifs resemble the U1A-binding site on U1snRNP. It has been shown that U1A, independently of U1snRNP, binds the same sequence in its own 3′-untranslated region (UTR) and regulates its own production by inhibition of addition of a poly(A) tail to its own mRNA (Boelens et al., 1993; Gunderson et al., 1994, 1997). We therefore investigated the possibility that U1A has a similar effect on this heterologous mRNA. We show that recombinant U1A binds the three inhibitory motifs in vitro and that endogenous U1A binds these motifs in conjunction with the cleavage/polyadenylation-specific complex in nuclear extracts. U1A inhibits poly(A) tail addition in vitro and in vivo. U1A inhibits poly(A) addition to the µ secretory mRNA to a lesser extent than it does to its own mRNA, which is more appropriate for regulation of a heterologous mRNA. Furthermore, overexpression of U1A in vivo results in the selective inhibition of the µ secretory mRNA relative to the µ membrane mRNA, demonstrating selective post-cleavage control of the expression of the µ secretory mRNA. We scanned the sequences upstream of the poly(A) sites of the other immunoglobulin isotypes and found evidence that they also use this mechanism.
These results are the first demonstration of the physiological importance of the regulation of post-cleavage nuclear poly(A) addition in the regulation of alternative gene expression during development and may be used to regulate alternative expression of other genes, in particular the other immunoglobulin isotypes.
Results
Identification of multiple sites upstream of the µ secretory poly(A) site that inhibit expression in vivo
We have shown previously that the core sequence of the secretory poly(A) site (positions 1951–2085) consists of an extended AU-rich region, consisting of the consensus A2UA3 hexanucleotide sequence and an adjacent upstream AUA5U2A motif that sustains residual activity, and two downstream GU-rich regions (Phillips and Virtanen, 1997; Phillips et al., 1999) (Figure 1B). These sequences contain all the elements necessary and sufficient for cleavage/polyadenylation activity in vivo and to form a specific polyadenylation complex on this poly(A) site in vitro. However, the flanking sequences may be involved in the regulation of poly(A) site expression.
To look for sequences involved in the regulation of the accumulation of µ secretory mRNA, we carried out a systematic mutational analysis of the region between the extended AU-rich region and the upstream 5′ splice site (Figure 2; for location of motifs see Figure 1B). For this we used a dual luciferase reporter assay that we have used successfully previously (Phillips et al., 1996, 1999; Phillips and Virtanen, 1997) in which the secretory poly(A) site (1790–2085) is inserted downstream of the firefly luciferase gene in place of the poly(A) site. We systematically scanned the region upstream of the secretory poly(A) site with mutations by replacing eight nucleotides at a time with eight adenosines in sequential order, with three unchanged nucleotides between each sequence of eight, to make a series of 10 separate constructs (see Figure 2B). These constructs (numbered mut1–mut10) all contained a mutated 5′ splice site at position 1810–1815 (GTAAA to CAAAC) in order to exclude the effect of the inhibition by the 5′ splice site on this poly(A) site (Peterson and Perry, 1989; Gunderson et al., 1998). We transfected the constructs in triplicate into HeLa cells and into the plasmacytoma, J558L, along with a reference plasmid expressing Renilla luciferase, and harvested the cells 22 h later. The firefly luciferase activity was corrected for transfection efficiency and the results were expressed as a percentage of the wild-type secretory poly(A) site, and the results for J588L cells are presented in Figure 2A.
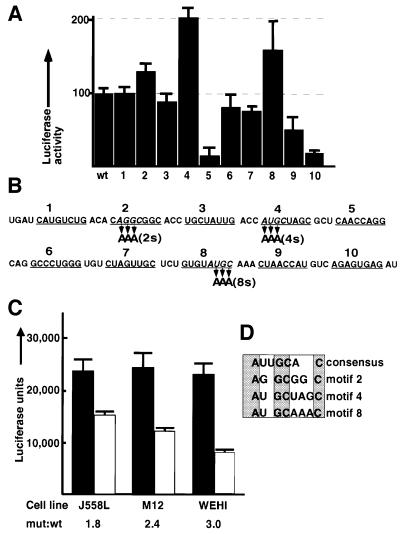
Fig. 2. Identification of motifs upstream of the secretory poly(A) site that inhibit polyadenylation in vivo in a developmentally regulated manner. Luciferase constructs containing the wild-type or mutant secretory poly(A) site from position 1790 to 2085 were transfected in triplicate into J558L, M12.4.1 and WEHI231 cells. (A) Luciferase activity in J558L cells of 10 constructs containing adenosine replacements of eight As in sequential order scanning the 113 nucleotide sequence from position 1838 to 1950 as indicated by bars and numbers in (B). Bars represent the mean of triplicates ± SE in both (A) and (C). (B) The sequence scanned by the adenosine replacements. The individual adenosine replacements are indicated by horizontal bars and the numbers 1–10. The ‘short’ mutations used in (C) are indicated via arrows and are designated 2s, 4s and 8s, respectively. (C) Luciferase activity of constructs containing the ‘short’ mutations 2s, 4s and 8s in combination in J558L, M12.4.1 and WEHI231 cells. (D) A sequence alignment of the consensus U1A-binding motif on U1snRNP with motifs 2, 4 and 8 (as indicated).
Mut4 gives the largest release of inhibition, with a 100% increase in luciferase activity, whereas mut8 results in a 50% increase (Figure 2A). Both these mutated sequences include the sequence AUGC (see Figure 2B). Mut2 also results in a small increase in luciferase expression and includes the sequence AGGC. Similar results were obtained in HeLa cells except that mut2 resulted in an increase of 75%, which was greater than that of mut8 in these cells (data not shown). Mut5 and mut10 result in significant decreases in luciferase activity. To confirm that the reduction in luciferase activity of mut5 was not an artifact, we tested several independent isolates of the mut5 plasmid. These produced the same result. Thus regions 5 and 10 upstream of the secretory poly(A) site have a positive effect on expression and will be the subject of a future investigation.
We refined the mutational analysis by replacing only three nucleotides in the inhibitory sequences with As. We mutated the sequence motif AUGC that was common to mut4 and mut8 and AGGC that was the closest match in mut 2 (see Figure 2B). We designated these shorter mutations with the suffix ‘s’ for ‘short’. The shorter mutations mut4s and mut8s reproduced the effects of the longer mutations mut4 and mut8 (data not shown). These mutants were combined to produce double mutations and a triple mutation. The triple mutant contains only nine (3 × 3) single nucleotide changes in a 212 nucleotide RNA.
To examine whether the inhibitory effect of these sequences on expression of the µ secretory poly(A) site is regulated during B-cell differentiation, we compared the ratio of expression from the luciferase constructs containing the 2s4s8s mutant poly(A) site to the wild type, in cells representing different stages of B-cell differentiation. J558L cells are plasmacytomas that have lost their endogenous immunoglobulin heavy chain and produce only the secretory form of µ mRNA from a transfected IgM heavy chain gene construct, WEHI231 is an immature B-cell line that produces equal quantities of endogenous secretory and membrane µ mRNA, while M12.4.1 cells are intermediate cells that produce endogenous IgG2a and twice as much secretory as membrane µ mRNA from a transfected construct.
As can be seen in Figure 2C, the plasmacytomas (J558L) show the least inhibitory effect of the sequences (lowest mutant to wild-type ratio) while the immature B cells (WEHI) show the highest ratio. The luciferase activity from constructs bearing the mutated motifs is similar in all three cells lines; the difference lies in those bearing wild-type sequences (Figure 2C). This shows that these sequences inhibit expression in a developmentally regulated manner. Furthermore, the ratio of the ratio of the J558L cells to the WEHI cells is similar to the ratio previously reported of the half-lives between these two cell lines (Mason et al., 1988).
An alignment of the motifs responsible for this inhibition and their flanking sequences revealed that all three sequences bear a resemblance to the U1A-binding site on U1snRNP (see Figure 2D). As U1A, independently of U1snRNP, has been shown to regulate its own production via inhibiting poly(A) addition to its own mRNA, we investigated the possibility that U1A binding of these motifs inhibits addition of a poly(A) tail to this heterologous mRNA.
Multiple (AU/GGC) motifs affect U1A binding upstream of the µ secretory poly(A) site
We first tested if the inhibitory motifs bind recombinant U1A in vitro. For these binding assays, we used substrates spanning nucleotides 1790–2001 representing the pre-cleaved RNA forms of the poly(A) sequences. Substrate 2001 was used instead of 1998, which is the actual site of cleavage, as inclusion of the additional bases produced cleaner PCR products.
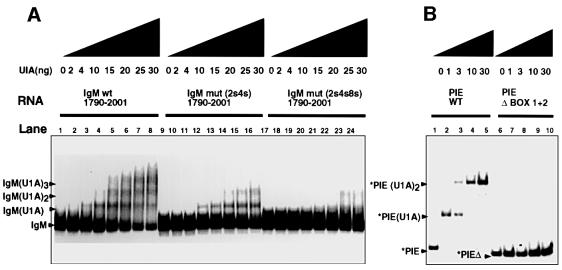
Figure 3A shows the in vitro binding pattern as increasing concentrations of recombinant U1A are added to the substrate and the effects of combinations of the short mutations. With the wild-type substrate, three U1A–RNA complexes are discernible, representing the binding of one, two or three U1A proteins, respectively, as the concentration of U1A increases (Figure 3A, lanes 1–8, indicated with arrows). Double mutant combinations of mut2s, mut 4s and mut8s resulted in a significant but not complete loss of U1A binding; the double mutant 2s4s is shown as an example in Figure 3A (lanes 9–16). When all three mutants are combined, U1A binding is abolished (lanes 17–24). Inclusion of mut3s in combinations with mut4s and mut8s had no effect on binding activity, showing that the random introduction of three As alone did not affect binding (data not shown). This analysis demonstrates that U1A binds all three inhibitory motifs in vitro and that each substrate can bind three molecules of U1A simultaneously. We included the pattern of binding of U1A to the inhibitory element in its own 3′-UTR for comparison with that of the µ secretory poly(A) site (Figure 3B). It can be seen that strength of binding of U1A to the µ secretory poly(A) site is lower than that of the binding of U1A to the inhibitory element in its own 3′-UTR. Compare the binding of 10 ng of U1A to the PIE element (Figure 3B, lane 4) with that of 10 ng of U1A to the µ secretory poly(A) site (Figure 3A, lane 4).
Fig. 3. The mutations that inhibit polyadenylation in vivo affect U1A binding in vitro. Electrophoretic mobility shift assays showing the binding of increasing amounts of recombinant U1A to (A) pre-cleaved immunoglobulin substrates containing either the wild-type sequence or multiple mutations as indicated; and (B) the wild-type or mutated PIE element in the 3′-UTR of U1A mRNA.
Endogenous U1A binds the inhibitory motifs in conjunction with the cleavage/polyadenylation-specific complex
We next wanted to test whether endogenous U1A binds the inhibitory motifs in vivo and is in a position to affect the function of the cleavage/polyadenylation apparatus. For this, we examined the effect of anti-U1A antibodies on the native cleavage/polyadenylation complex in nuclear extracts formed on the µ secretory poly(A) site in the presence and absence of the inhibitory motifs.
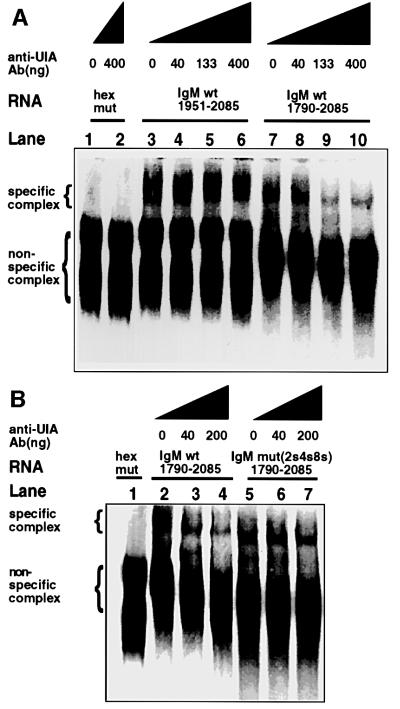
The cleavage/polyadenylation-specific complex is identifiable on native gels by its ability to form only on polyadenylation substrates that contain an intact AAU AAA hexanucleotide sequence (Gilmartin and Nevins, 1989) and previously shown for the µ secretory poly(A) site, in particular, by us (Phillips and Virtanen, 1997). We used the same substrates in lanes 1 and 2 of Figure 4A that we used previously to validate the identity of the complex, which is indicated with a bracket. These substrates, extending from position 1951 to 2085 (see Figure 1B), span the sequences we previously have shown to be necessary to bind the core polyadenylation complex (Phillips and Virtanen, 1997). However, they do not contain the inhibitory motifs and are used as a deletion control for the anti-U1A antibody experiments. We compared them with the longer substrates, extending upstream to position 1790, which do span the inhibitory motifs.
Fig. 4. Endogenous U1A binds RNA in conjunction with the cleavage/polyadenylation-specific complex that forms on the µ secretory poly(A) site in vitro. Electrophoretic mobility shift assays of HeLa cell nuclear extracts. (A) The formation of the polyadenylation-specific complex on immunoglobulin substrates with (lanes 7–10) or without (lanes 1–6) the region spanning the inhibitory motifs. Lanes 1 and 2 contain the substrate IgM(1951–2085) with four U to G mutations in the extended hexanucleotide sequence (Phillips and Virtanen, 1997). (B) The formation of the polyadenylation-specific complex on wild-type (lanes 2–4) or triple mutant (2s4s8s) immunoglobulin substrates (lanes 5–7). Lane 1 contains the substrate IgM(1951–2085) with four U to G mutations in the extended hexanucleotide sequence (Phillips and Virtanen, 1997). Anti-U1A antibodies were added in increasing concentrations as indicated. The positions of the specific and non-specific complexes are indicated.
Anti-U1A antiserum 856 did not affect the complex formed on the shorter substrate, IgM(1951–2085) (Figure 4A, lanes 3–6). However, when the substrate is extended upstream to include the inhibitory motif, IgM(1790–2085), introduction of increasing concentrations of anti-U1A antibody disrupts the complex (lanes 7–10). This shows that endogenous U1A binds RNA in conjunction with the native cleavage/polyadenylation-specific complex on the µ secretory poly(A) site when the region containing the U1A-binding sites is included. However, U1A is not part of the specific complex per se formed on the core sequences. Mutation of the inhibitory motifs abolished the ability of anti-U1A antibodies to disrupt the complex in the context of the longer substrate (Figure 4B, compare lanes 5–7 with 2–4). From these results, we conclude that endogenous U1A binds the inhibitory motifs in nuclear extracts and its binding sites are accessible in conjunction with the cleavage/polyadenylation-specific complex.
U1A inhibits poly(A) addition at the secretory poly(A) site in vitro
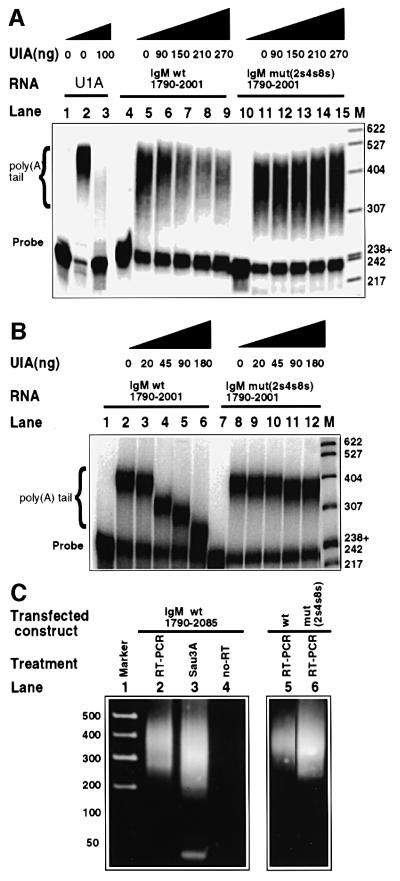
As U1A has been shown to inhibit poly(A) addition to its own mRNA (Gunderson et al., 1994), we next investigated whether U1A bound to the novel inhibitory motifs also affects poly(A) addition to this heterologous mRNA in vitro. We measured specific cleavage/polyadenylation specificity factor (CPSF)-dependent poly(A) addition at the µ secretory poly(A) site in nuclear extracts using the same pre-cleaved substrates that were shown to bind recombinant U1A in Figure 3A. Increasing concentrations of recombinant U1A reduced poly(A) addition to the wild-type pre-cleaved substrate but had no effect on the triple mutant pre-cleaved substrate (Figure 5A, lanes 5–9 and 11–15, respectively). This shows that U1A can inhibit poly(A) addition at the µ secretory poly(A) site and that U1A binding to the inhibitory motifs is responsible for this effect. U1A inhibits poly(A) addition to its own mRNA much more strongly than to the µ secretory poly(A) site (Figure 5A, compare lanes 2 and 3 with lanes 5–9). Nevertheless, U1A inhibits poly(A) addition of the µ secretory mRNA by the same mechanism that has been reported previously, namely via an inhibition of poly(A) polymerase (PAP) activity (Gunderson et al., 1994). This is shown in Figure 5B. We performed reconstituted CPSF-independent poly(A) addition assays (Bienroth et al., 1991) using only recombinant PAP, recombinant U1A and either pre-cleaved wild-type or pre-cleaved mut2s4s8s RNAs as substrates (Figure 5B). Increasing concentrations of U1A inhibited PAP activity on the wild-type substrate (lanes 2–6) but had no effect on the triple mutant RNA substrate (lanes 7–12). Thus, U1A bound upstream of the µ secretory poly(A) site has a direct inhibitory effect on PAP similar to its effect in the U1A autoregulatory system (Gunderson et al., 1994).
Fig. 5. U1A inhibits poly(A) polymerase activity at the secretory poly(A) site. (A) The effect of increasing concentrations of recombinant U1A with specific poly(A) addition in HeLa cell nuclear extracts on wild-type and triple mutant (2s4s8s) substrates. All lanes contain 2 µl of nuclear extracts, total protein content 15 µg, except lanes 1, 4 and 10, which contained no nuclear extract. (B) The effect of increasing concentrations of recombinant U1A with non-specific poly(A) addition on recombinant PAP activity on wild-type and triple mutant (2s4s8s) substrates. Increasing concentrations of recombinant U1A were added as indicated. All lanes contain 50 ng of recombinant PAP, except lanes 1 and 7, which contained no PAP. All substrates (including wild type) contained a mutated 5′ splice site at position 1810–1815. (C) The lengths of poly(A) tails formed in vivo from transfected luciferase constructs containing wild-type or triple mutant (2s4s8s) µ secretory poly(A) sites from Figure 2C (J558L cells) were measured by the LM-PAT method (Salles and Strickland, 1999). Lane 1, a 1 kb ladder (NEN); lane 2, the PCR products, which migrate as smears, for the wild type; lane 3, the products from lane 2 digested with Sau3A; lane 4, the same as lane 2 except that the reverse transcriptase was omitted during the reverse transcription step; lanes 5 and 6, results for the wild type and mutant as indicated.
Interestingly, the pattern of poly(A) tail inhibition was different in the two systems. In the recombinant system, increased U1A resulted in shorter poly(A) tails. In contrast, in nuclear extracts, poly(A) tails were not shorter but U1A inhibition resulted in fewer polyadenylated substrates (compare Figure 5B lanes 3–6 with A, lanes 6–9).
U1A causes a decreased abundance of normally polyadenylated µ secretory RNA rather than an overall shortening of poly(A) tails in vivo
To examine whether the inhibition via the inhibitory motifs resulted in an overall shortening of poly(A) tails or decreased the abundance of normally polyadenylated mRNA tails in vivo, we measured the length of the poly(A) tails formed on the wild-type and 2s4s8s mutated sequences of the µ secretory poly(A) sites placed downstream of the reporter constructs of Figure 2C, J558L cells. For this, we used a LM-PAT (ligation method, poly(A) tail measurement) assay (Salles and Strickland, 1999). We purified poly(A)+ mRNA from 107 transfected cells, which were found to have equivalent transfection efficiency as measured by Renilla activity. To amplify specifically the polyadenylated µ secretory sequences, we used two primers: (i) an oligo(dT) ‘anchor’ that anneals to poly(A) and (ii) a primer that hybridizes specifically with the µ secretory sequences present (positions 1790–1807). The primers were designed to span a Sau3A site at position 1838, which allowed verification of the PCR product by enzyme digestion (see Figure 5C, lane 3). We included a control with no reverse transcriptase to verify that genomic DNA was not amplified (Figure 5C, lane 4). The PCR products appear as smears that correspond to poly(A) tail length (see Figure 5C).
Figure 5C shows that mRNA from the wild-type and the triple mutant constructs form poly(A) tails of the same length (Figure 5C, compare lanes 5 and 6). The peak of intensity for both smears appears at ∼400, consistent with poly(A) tails of 200 residues for both, although the PCR product smear from the triple mutant appears more spread out in both directions than that from the wild type. The triple mutant is more intense than the wild type, consistent with an increased abundance of the poly(A)+ mutant mRNA. We conclude that poly(A) tails, if formed on the respective mRNAs, do not differ in ultimate length. Rather, these results are consistent with a reduction in the abundance of normally polyadenylated µ secretory sequences as a result of U1A inhibition.
The U1A-binding motifs inhibit accumulation of polyadenylated µ secretory mRNA in vivo
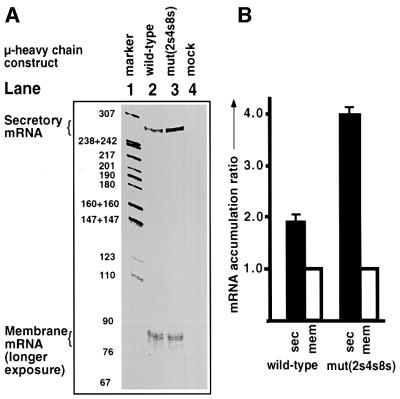
We next directly determined whether U1A decreases the abundance of normally polyadenylated µ secretory mRNA in vivo, in the context of the µ heavy chain gene, using quantitative RNase protection analysis (Figure 6). For this we used M12.4.1 cells, which produce a detectable amount of µ membrane mRNA from the transfected µ heavy chain-expressing plasmid, pµΔ3 (Grosschedl and Baltimore, 1985), providing a convenient internal reference for measurement of the accumulation of the secretory form of µ mRNA. These cells endogenously express IgG2a (Glimcher et al., 1982), which does not hybridize with the µ antisense probe used in the RNase protection assays (Figure 6A, lane 4).
Fig. 6. U1A-binding motifs inhibit accumulation of polyadenylated µ secretory mRNA in vivo. RNase protection assays. Wild-type or mutant plasmids containing the IgM heavy chain gene were transfected into M12.4.2 cells in triplicate. (A) RNase protection assay visualized by phosphorimagery. Lane 1 is the 32P-radiolabeled pBR322 MspI digest marker. Lanes 2–4 are mock, wild-type and mutant transfected poly(A)-selected mRNA, respectively. The secretory and membrane-protected fragments are indicated with brackets. (B) Phosphorimagery analysis of (A). The secretory measurements were corrected for the higher level of [32P]UTP due to a longer protected fragment and all the results were expressed as a ratio of the wild-type membrane-protected fragment. Bars represent the mean of triplicates ± SE.
At 17 h after transfection of wild-type- or triple mutant-containing pµΔ3 plasmids, poly(A)+ mRNA was extracted and subjected to RNase protection analysis using wild-type or mutant antisense RNA spanning the 5′ splice site and the secretory poly(A) site (see Figure 1C for location of the antisense probe and expected protected fragments). The expected sizes of the protected secretory and membrane mRNA fragments are 268 and 80 nucleotides, respectively. Triplicate analyses were quantitated and a representative of each of the wild type and mutant are presented in Figure 6A.
As can be seen in Figure 6A, introduction of mutations into the U1A-binding sites in the context of the whole µ heavy chain gene substantially increases the amount of polyadenylated µ secretory mRNA (compare the mutant with the wild type in lanes 3 and 2, respectively). The phosphorimagery values for the secretory product were corrected for the higher level of [32P]UTP due to a longer protected fragment, and all results were expressed as a ratio of the wild-type membrane protected fragment (see Figure 6B). For the wild type, the ratio of the secretory to the membrane form of mRNA was 1.9 ± 0.2 SE, which is comparable with what has been reported previously for this cell line (Peterson et al., 1991). Upon introduction of the mutations in the U1A-binding sites, this ratio increases to 4.0 ± 0.2. Thus, the U1A-binding motifs upstream of the secretory poly(A) site significantly inhibit the accumulation of polyadenylated µ secretory mRNA.
Overexpression of U1A in vivo selectively inhibits the secretory form of mRNA
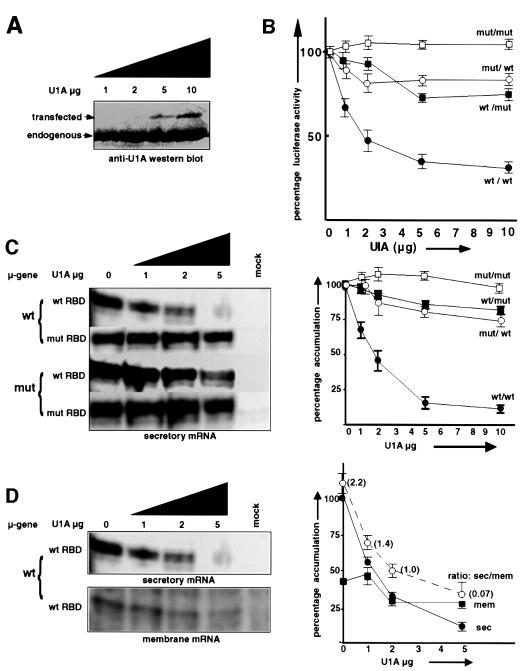
We tested the effect of overexpression of U1A on expression of the µ secretory form in vivo in M12.4.1 cells. We transfected sets of plasmids containing U1A in which the polyadenylation inhibitory element (PIE) had been deleted to avoid the regulatory feedback mechanism (Boelens et al., 1993). To control for the non-specific effects of overexpression, we used a U1A which contained Arg/Gly to Gly/Ser mutations at amino acids 52 and 53 within the RNA-binding domain (RBD) (Boelens et al., 1991). We tested another set of constructs in which we had inserted a flag tag in-frame between amino acids 275 and 276 near the C-terminal end, which allowed us to monitor expression of the transfected U1A (see Materials and methods). Using 10 and 5 µg of transfected plasmid resulted in a level of expression that was 22.4 and 11.0%, respectively, that of the endogenous U1A by quantitation by phosphorimagery after western blotting (see Figure 7A). Transfected U1A did not regulate the level of endogenous U1A detectably at 24 h after transfection.
Fig. 7. Overexpression of U1A in vivo selectively inhibits µ secretory mRNA via binding to the novel motifs. Increasing amounts of U1A (PIE deleted) with a wild-type (circles) or mutant (squares) RBD transfected into M12.4.1 cells. (A) The effect on expression of the flag-tagged transfected U1A relative to the endogenous U1A (anti-U1A antibody western blot). (B) The effect on luciferase expression from a construct containing either a wild-type (black) or a 2s4s8s mutant (white) µ secretory poly(A) site. (C) The effect on µ secretory mRNA relative to µ membrane mRNA from the wild-type or 2s4s8s mutated µ heavy chain gene (RNase protection assays, same symbols as in B). (D) The effect on µ secretory mRNA and µ membrane mRNA, separately. The dotted line represents the ratio of the secretory to membrane form as indicated. Values obtained were expressed as separate percentages of the wild-type or 2s4s8s mutant poly(A) site-containing constructs at zero transfected U1A, respectively. The mutant was 204 ± 15% of the wild type for the luciferase experiments and 195 ± 15% for the RNase protection experiments. All values are the mean of triplicates ± SE. Representative phosphorimages are shown in (C) and (D).
We first tested the effect of overexpression of U1A in the luciferase reporter assay (Figure 7B). Transfection of U1A containing a wild-type RBD significantly reduced luciferase expression from the construct containing the wild-type µ secretory poly(A) site (Figure 7B, black circles). Introduction of a mutation into either the RBD of U1A or into its binding site upstream of the µ secretory poly(A) site significantly reduced the ability of transfected U1A to inhibit luciferase activity (black squares and white circles, respectively). Combination of mutations in both the RBD of U1A and its binding motifs upstream of the µ secretory poly(A) site completely abolished inhibition even at higher levels of transfected U1A. The values were expressed as separate percentages of the wild-type or 2s4s8s mutant poly(A) site-containing constructs, at zero transfected U1A, respectively. In this way, the two sets of data were superimposed to allow a convenient comparison of the shapes of the curves. When compared with each other, the mutant was 204 ± 15% of the wild type. The flag-tagged set of constructs gave similar results to those without a tag.
A similar pattern was seen with the effect of overexpression of U1A on the expression of polyadenylated µ secretory mRNA in the context of the whole IgM heavy chain gene (Figure 7C). These experiments were performed with different ranges of transfected U1A plasmid, and both µ secretory and µ membrane fragments were quantitated by phosphorimagery and µ secretory mRNA was calculated relative to the µ membrane form for each sample. Once again, results for U1A inhibition of relative expression were expressed separately as a percentage of wild-type or mutant µ secretory poly(A) site values at zero U1A, respectively, for convenient comparison (mutant 195 ± 15% of wild type). Triplicates were pooled and are presented as curves in Figure 7C along with phosphorimages of the secretory µ mRNA protected fragment of representative experiments. Taken together, these results show that U1A specifically inhibits expression of the µ secretory poly(A) site via the novel U1A-binding sites in vivo.
We also quantitated whether U1A overexpression has a specific effect on the µ secretory mRNA relative to the µ membrane mRNA. Figure 7D shows that whereas overexpression of U1A causes a sharp decline in the accumulation of the secretory µ mRNA even at lower concentration, µ membrane mRNA is affected only moderately at the highest concentrations. Overexpression of U1A causes the ratio of secretory to membrane µ mRNA to drop from 2.2 ± 0.15 (with no transfected U1A) to 0.7 ± 0.3 (with 5 g of transfected U1A) (Figure 7D). Thus the control of U1A is exerted specifically on the secretory form and regulates its expression relative to the membrane form.
Discussion
We have elucidated an important mechanism regulating post-cleavage expression of µ secretory mRNA during B-cell differentiation. We identified three sites with the motif A(U/G)GC(N1–3)C which regulate expression of the µ secretory poly(A) site in a developmental manner. We showed that U1A binds these motifs in conjunction with the cleavage/polyadenylation-specific complex and inhibits poly(A) addition in vitro. Mutations in these motifs in the transfected µ heavy chain gene significantly increase polyadenylated µ secretory mRNA in vivo, and overexpression of U1A selectively inhibits the µ secretory mRNA relative to the membrane form only when these sites are intact. Taken together, these results show that U1A binds upstream of the µ secretory poly(A) site and selectively regulates poly(A) addition to the µ secretory mRNA.
U1A inhibits poly(A) addition at the µ secretory poly(A) site at a lower level than its own mRNA, although it regulates poly(A) addition to the µ secretory poly(A) site by the same mechanism via a direct inhibition of PAP activity. This less stringent effect is probably due to the lower binding strength of the novel binding sites and is particularly suited to a regulatory role for U1A at this poly(A) site, which is fine-tuned by competing weak interactions (Galli et al., 1988; Peterson, 1992; Peterson et al., 1994). In vivo, U1A appears to inhibit an early stage of poly(A) addition, decreasing the abundance of normally polyadenylated mRNA rather than an overall shortening.
Much work in recent years has focused on the regulation of the cleavage event at the secretory poly(A) site. However, the change in stability and increased nuclear to cytoplasmic transport of the secretory mRNA represents an important additional level of regulation. The finding that regulation of polyadenylation of the µ secretory mRNA can have a 2.2-fold effect on its accumulation raises the question of the relative contribution of the increased usage of the secretory poly(A) site and the increased polyadenylation of the mRNA to the increased expression of the µ secretory mRNA during B-cell differentiation. Peterson et al. (1991) measured the change in polyadenylation efficiency of the secretory poly(A) site between lymphomas and plasmacytomas using ratios of usage of tandem poly(A) sites in the two types of cells. The polyadenylation efficiency was found to change 2-fold. However, the native sequence between the secretory poly(A) site and Cµ4, which spans the U1A-binding sites demonstrated here, was included in their constructs. Therefore, the developmental changes in poly(A) tail acquisition, which we demonstrate here, were not excluded in those experiments. Lamson and Koshland (1984) examined the kinetics of the formation of the membrane and the secretory form during the stimulation of primary resting B cells. Their results clearly show a switch in production to that of the secretory form before the massive change in mRNA accumulation. However, as both increased processing to the secretory form and increased polyadenylation of the mRNA probably occur simultaneously, the relative contributions of the two processes are hard to dissect.
Is this mechanism used to regulate other Ig isotypes?
Our findings raise the possibility that expression of other genes might be regulated in the same way. The immunoglobulin gene locus undergoes switching to other immunoglobulin isotypes during the course of an immune response. The heavy chains of the other isotypes, IgG1, IgG2a, IgG2b, IgG3, IgA and IgE (γ1, γ2a, γ2b, γ3, α and ε, respectively) all undergo alternative processing of a common pre-mRNA in a manner similar to that of µ pre-mRNA during differentiation. The stability and/or cytoplasmic accumulation of the secretory mRNAs of γ2a, α and ε has also been reported to change during B-cell differentiation (Saxon et al., 1991; Eckmann et al., 1994; Milcarek et al., 1996). In addition, the stability of the κ light chain mRNA has been shown to change in co-ordination with the stability of γ2a secretory mRNA and α secretory mRNA, suggesting co-ordinate targeting by inducible factors (Eckmann et al., 1994; Milcarek et al., 1996).
We scanned the sequences of heavy chain genes of the above isotypes to look for sequences that match the novel U1A-binding sites, A(U/G)GC(N1–3)C. We found a frequency of one motif in 81 nucleotides upstream of the poly(A) sites between the hexanucleotide sequence and the 5′ splice site, representing an average of at least one motif per region, compared with an expected frequency of 1 in 170. There are two AUGC motifs upstream of the κ poly(A) site. This is in contrast to the frequency of the motif in the rest of the constant region genes, which was 1 in 280. Thus, the frequency of these motifs is skewed away from random in both directions. This suggests that these motifs are targeted to the region upstream of the secretory poly(A) site among immunoglobulin isotypes.
Conclusion
These results are the first demonstration of the physiological importance of the inhibition of nuclear poly(A) addition in the regulation of alternative gene expression during development. The more subtle effect of U1A on poly(A) addition at the µ secretory mRNA is more appropriate for a heterologous mRNA. Regulation of nuclear poly(A) addition via these novel U1A-binding sites may be used by other genes, in particular the other immunoglobulin isotypes.
Materials and methods
Plasmid constructs
For in vitro transcription, the PCR products from the secretory poly(A) site sequences, containing mutations and a 5′ EcoRI site and a 3′ XbaI site, introduced as part of the synthetic primers, were cloned into pGem 3Zf (containing a T7 promoter in the forward direction) between the EcoRI and XbaI sites. For transfection, a 5′ BglII site replaced the EcoRI site and PCR products were cloned into pPKLT55 (Phillips et al., 1996) containing the firefly luciferase gene between the BglII and XbaI sites replacing the poly(A) site. The forward primers 5′-GACTCTAGA(or AGATCT)GGACCGTGGACAAGTCC-3′ (1790 wild type) and 5′-GACTCTAGA (or AGATCT)GGACCGTCCACAAGTCCACTGCAAA CCCCACACTGTACAATG-3′ (1970 5′ splice site mutation) were combined with the reverse primers 5′-GCGTCTAGATAGGGTGGAGGCAAGTATGC-3′ (2085), to amplify from position 1790 to 2085, or 5′-AGTGACGTTTGAATGGATTTTTTTTATTTC-3′ (2001), to amplify from position 1970 to 2001 (nucleotide positions are numbered according to the mouse IgM sequence with accession No. V00818). PIE RNA templates were obtained by digestion of the plasmid Agwt at the HindIII site to give a substrate of 125 nucleotides (Boelens et al., 1993).
The adenosine replacement mutations were incorporated using crossover PCR as previously described (Phillips et al., 1999). The outside primers were the 1790 5′ splice mutation and 2085 as described above incorporating either EcoRI or BglII and XbaI restriction sites, respectively, depending on the cloning procedure. These were combined in sequence with pairs of oligonucleotides designed to introduce the respective mutations. For mut1, the pair of oligos used was the forward oligo 3′-CAATGTCTCCCTGATAAAAAAAAACACAGGCGGCACCTG-5′ combined with the exact reverse sequence. For mut2–mut10, similar pairs of oligos were used for which each subsequent 3′ and 5′ end as well as the region changed to eight As was shifted 11 bases downstream. For the single short mutation, the following forward oligos were used: mut4s, 3′-CACCTGCTATTGACCAAAATAGCGCTCAACCAGGCAGG-5′; mut8s, 3′-GTGTCCAGTTGCTCTGTGTAAAAAAACTAACCATGTCAGAGTGAG-5′; and mut24s, 3′-CTGATCATGTCT GACACAGAAAAAACCTGCTATTGACCAAAATAGCGCTCAACCAGGCAGG-5′. These were each combined with the exact reverse sequence as above. Subsequent mutations were incorporated by a similar procedure using single mutants as the PCR template.
For cloning of the mutant sequences into the µ heavy chain gene, the BglI–XhoI fragment of pµΔ3 (Grosschedl and Baltimore, 1985) was subcloned into pGM10 (Martin and Keller, 1996) to make pET300. PCR inserts were made between the ApaI and KpnI sites and mutations were introduced by crossover PCR as above. These were inserted into the BglII–XhoI (302–4897) fragment in pET300 in place of the wild-type ApaI–KpnI (1670–3170) fragment. The resulting mutant BglII–XhoI fragment was recloned back into pµΔ3 in place of the wild-type fragment.
For overexpression of U1A in vivo, we used the same set of two plasmids used for overexpression in Boelens et al. (1993). The PIE has been deleted in both constructs and the mutant contains an Arg/Gly to Gly/Ser mutation at amino acids 52 and 53, in RBD 1 (Boelens et al., 1991). To monitor the transfected protein more easily, we constructed a second set of two plasmids in which a flag tag sequence was inserted in-frame at the PstI site near the end of the coding sequence, which resulted in a flag tag between amino acids 275 and 276 near the C-terminal end of the U1A protein.
Recombinant proteins, nuclear extracts and RNA substrates
Untagged recombinant U1A was purified from Escherichia coli as described (Boelens et al., 1993). Recombinant bovine PAP, tagged at the C-terminus with six histidines, was purified from E.coli on Ni2+ NTA. The His-tagged C-terminus ensured that all of the C-terminal residues were present after purification (Gunderson et al., 1997). HeLa cell nuclear extracts were prepared as described (Dignam et al., 1983). RNA substrates were prepared by in vitro transcription as previously described (Phillips and Virtanen, 1997).
Electrophoretic mobility shift assays
Cleavage/polyadenylation-specific shifts were obtained as described (Phillips and Virtanen, 1997) using 100 000 c.p.m. of 32P-labeled RNA substrate and 11 µl of HeLa cell nuclear extract (10 µg/µl total protein). Anti-U1A antibodies were polyclonal rabbit antiserum 856 directed against a section of U1A spanning amino acids Ile93–Ser202 as described in Kambach and Mattaj (1992), kindly provided by I.Mattaj. These were added to the nuclear extracts before the RNA substrates. U1A binding assays were performed as previously described (vanGelder et al., 1993) using 10 000 c.p.m. of 32P-labeled RNA substrate per lane and recombinant U1A as indicated in the text.
Cell culture and transfection
J558L, HeLa and WEH321 cells were obtained from the European Collection of Animal Cell Cultures (Salisbury, UK). M12.4.1 cells were the kind gift of K.J.Kim (Kim et al., 1979). Plasmids were transfected into cells in log phase using Superfect (Qiagen) 20 µl/106 cells. Transfection efficiency was measured by co-transfection of Renilla SV40. Firefly and Renilla luciferase activity was measured using the Dual Luciferase Kit from Promega.
Poly(A) addition assays
Non-specific poly(A) addition assays were performed as previously described (Gunderson et al., 1994) with 50 ng of recombinant PAP and incubated for 30 min at 30°C. Poly(A) addition assays in nuclear extracts were performed as previously described (Phillips and Virtanen, 1997) using 15 µg of total protein HeLa cell nuclear extract and 20 000 c.p.m. of 32P-labeled RNA substrate.
Poly(A) tail measurement
This was performed by LM-PAT according to Salles and Strickland (1999). For the PCR step, the 5′ µ secretory poly(A) site-specific primer was 5′-GACTCTAGAGGACCGTCCACAAGTCCACTGCAAACCCCACACTGTACAATG-3′ and the 3′ primer was the oligo(dT) anchor as described (Salles and Strickland, 1999) to amplify from position 1790 to the poly(A) tail (to position 1998 plus tail) (Phillips et al., 1996), which incorporates a Sau3A restriction site at position 1838.
RNase protection assays
Wild-type or mutant plasmids containing the IgM heavy chain gene were transfected into M12.4.1 cells in triplicate. Poly(A) mRNA was extracted 17 h later using a Quickprep micro mRNA preparation kit from Pharmacia. mRNA levels were measured by RNase protection analysis according to Melton et al. (1984). Poly(A)+ RNA was hybridized overnight at 45°C with 100 000 c.p.m. uniformly 32P-labeled wild-type or mutant antisense RNA spanning position 1730–2085, which includes the 5′ splice site (1810) and the secretory poly(A) site (1998), in the presence of 5 µg of tRNA. Single-stranded RNA was digested using 50 U of RNase T1 and 0.5 µg of RNase A for 30 min at 37°C. Products were quantitated by phosphorimagery.
Acknowledgments
Acknowledgements
We would like to thank I.Mattaj for anti-U1A antiserum 856, J.Kim for the M12.4.1 cells and R.Grosschedl for the pµΔ3 plasmid. We would also like to thank Carol Wilusz and Jeff Wilusz for their helpful comments on the manuscript. The work was supported by the New Jersey Commission on Cancer Research 98-0717, the American Heart Foundation 98-0071T and NIH GM57286.
References
- Berberich I. and Schimpl,A. (1990) Regulation of Ig gene expression in normal lymphocytes: I. The half-life of the secreted µ chain mRNA differs from that of membrane µ chain mRNA in resting and activated B cells. Eur. J. Immunol., 20, 445–448. [DOI] [PubMed] [Google Scholar]
- Bienroth S., Wahle,E., Suter-Crazzolara,C. and Keller,W. (1991) Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J. Biol. Chem., 266, 19768–19776. [PubMed] [Google Scholar]
- Boelens W., Scherly,D., Jansen,E.J., Kolen,K., Mattaj,I.W. and van Venrooij,W.J. (1991) Analysis of in vitro binding of U1-A protein mutants to U1 snRNA. Nucleic Acids Res., 19, 4611–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens W.C., Jansen,E.J.R., van Venrooij,W.J., Stripecke,R., Mattaj,I.W. and Gunderson,S.I. (1993) The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell, 72, 881–892. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti,C. and Tollervey,D. (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell, 102, 765–775. [DOI] [PubMed] [Google Scholar]
- Cox A. and Emtage,J.S. (1989) A 6-fold difference in the half-life of immunoglobulin µ heavy chain mRNA in cell lines representing two stages of B cell differentiation. Nucleic Acids Res., 17, 10439–10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E. Jr (1982) Variety in the level of gene control in eukaryotic cells. Nature, 297, 365–371. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebowitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers,J., Davis,M., Calame,K., Bond,M., Wall,R. and Hood,L. (1980) Two mRNAs can be produced from a single immunoglobulin µ gene by alternative RNA processing pathways. Cell, 20, 313–319. [DOI] [PubMed] [Google Scholar]
- Eckmann L., Huang,G.T., Smith,J.R., Morzycka-Wroblewska,E. and Kagnoff,M.F. (1994) Increased transcription and coordinate stabilization of mRNAs for secreted immunoglobulin α heavy chain and κ light chain following stimulation of immunoglobulin A expressing B cells. J. Biol. Chem., 269, 33102–33108. [PubMed] [Google Scholar]
- Edwalds-Gilbert G. and Milcarek,C. (1995) Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol. Cell. Biol., 15, 6420–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie D.R. (1998) A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene, 216, 1–11. [DOI] [PubMed] [Google Scholar]
- Galli G., Guise,J., Tucker,P.W. and Nevins,J.R. (1988) Poly(A) site choice rather than splice site choice governs the regulated production of IgM heavy-chain RNAs. Proc. Natl Acad. Sci. USA, 85, 2439–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerster T., Picard,D. and Schaffner,W. (1986) During B cell differentiation enhancer activity and transcription rate of immunoglobulin heavy chain genes are high before mRNA accumulation. Cell, 45, 45–52. [DOI] [PubMed] [Google Scholar]
- Gilmartin G.M. and Nevins,J.R. (1989) An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev., 3, 2180–2189. [DOI] [PubMed] [Google Scholar]
- Glimcher L.H., Hamano,T., Asofsky,R., Herber-Katz,E., Hedrick,S., Schwartz,R.H. and Paul,W.E. (1982) I region-restricted antigen presentation by B cell–B lymphoma hybridomas. Nature, 298, 283–284. [DOI] [PubMed] [Google Scholar]
- Grosschedl R. and Baltimore,D. (1985) Cell-type specificity of immunoglobulin gene expression is regulation by at least three DNA sequence elements. Cell, 41, 885–897. [DOI] [PubMed] [Google Scholar]
- Gunderson S.I., Beyer,K., Martin,G., Keller,W., Boelens,W.C. and Mattaj,I.W. (1994) The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell, 76, 531–541. [DOI] [PubMed] [Google Scholar]
- Gunderson S.I., Vagner,S., Polycarpou-Schwarz,M. and Mattaj,I.W. (1997) Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev., 11, 761–773. [DOI] [PubMed] [Google Scholar]
- Gunderson S.I., Polycarpou-Schwarz,M. and Mattaj,I.W. (1998) U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell, 1, 255–264. [DOI] [PubMed] [Google Scholar]
- Kambach C. and Mattaj,I.W. (1992) Intracellular distribution of the U1A protein depends on active transport and nuclear binding to U1 snRNA. J. Cell Biol., 118, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.J., Kanellopoulos-Lanevin,C., Merwin,R.M., Sachs,D.H. and Asofsky,R. (1979) Establishment and characterisation of Balb/c lymphoma lines with B cell properties. J. Immunol., 122, 549–554. [PubMed] [Google Scholar]
- Lamson G. and Koshland,M.E. (1984) Changes in J-chain and µ-chain RNA expression as a function of B-cell differentiation. J. Exp. Med., 160, 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Moore,M.J. and Maquat,L.E. (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Martin G. and Keller,W. (1996) Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases and to other nucleotidyltransferases. EMBO J., 15, 2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Martincic K., Campbell,R., Edwalds-Gilbert,G., Souan,L., Lotze,M.T. and Milcarek,C. (1998) Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc. Natl Acad. Sci. USA, 95, 11095–11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J.O., Williams,G.T. and Neuberger,M.S. (1988) The half-life of immunoglobulin mRNA increases during B-cell differentiation: a possible role for targeting to membrane-bound polysomes. Genes Dev., 2, 1003–1010. [DOI] [PubMed] [Google Scholar]
- Melton D.A., Krieg,P.A., Rebagliati,M.R., Maniatis,T., Zinn,K. and Green,M.R. (1984) Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res., 12, 7035–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milcarek C., Suda-Hartman,M. and Croll,S.C. (1996) Changes in abundance of IgG 2a mRNA in the nucleus and cytoplasm of a murine B-lymphoma before and after fusion to a myeloma cell. Mol. Immunol., 33, 691–701. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L. and Keller,W. (1999) mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol., 11, 352–357. [DOI] [PubMed] [Google Scholar]
- Perry R.P. and Kelley,D.E. (1979) Immunoglobulin messenger RNAs in murine cell lines that have characteristics of immature B cells. Cell, 18, 1333–1339. [DOI] [PubMed] [Google Scholar]
- Perry R.P., Atchison,M.L., Kelley,D.E. and Peterson,M.L. (1988) Transcriptional and processing level control of immunoglobulin gene expression. Ann. N. Y. Acad. Sci., 546, 25–33. [DOI] [PubMed] [Google Scholar]
- Peterson M.L. (1992) Balanced efficiencies of splicing and cleavage–polyadenylation are required for µs and µm mRNA regulation. Gene Expr., 2, 319–327. [PMC free article] [PubMed] [Google Scholar]
- Peterson M.L. and Perry,R.P. (1989) The regulated production of µm and µs mRNA is dependent on relative efficiencies of µs poly(A) site usage and to the Cµ4-to-M1 splice. Mol. Cell. Biol., 9, 726–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M.L., Gimmi,E.R. and Perry,R.P. (1991) The developmentally regulated shift from membrane to secreted µ mRNA production is accompanied by an increase in cleavage–polyadenylation efficiency but no measurable change in splicing efficiency. Mol. Cell. Biol., 11, 2324–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M.L., Bryman,M.B., Peiter,M. and Cowan,C. (1994) Exon size affects competition between splicing and cleavage–polyadenylation in the immunoglobulin µ gene. Mol. Cell. Biol., 14, 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. and Virtanen,A. (1997) The murine IgM secretory poly(A) site contains dual upstream and downstream elements which affect polyadenylation. Nucleic Acids Res., 25, 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C., Schimpl,A., Dietrich-Goetz,W., Clements,J.B. and Virtanen,A. (1996) Inducible nuclear factors binding the IgM heavy chain pre-mRNA secretory polyA site. Eur. J. Immunol., 26, 3144–3152. [DOI] [PubMed] [Google Scholar]
- Phillips C., Kyriakopoulou,C.B. and Virtanen,A. (1999) Identification of a stem–loop structure important for polyadenylation at the murine IgM secretory poly(A) site. Nucleic Acids Res., 27, 429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T. and Hentze,M.W. (1999) From factors to mechanisms: translation and translational control in eukaryotes. Curr. Opin. Genet. Dev., 9, 515–521. [DOI] [PubMed] [Google Scholar]
- Reed D.J., Hawley,J., Dang,T. and Yuan,D. (1994) Role of differential mRNA stability in the regulated expression of IgM and IgD. J. Immunol., 152, 5330–5336. [PubMed] [Google Scholar]
- Ross J. (1995) mRNA stability in mammalian cells. Microbiol. Rev., 59, 423–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles F.J. and Strickland,S. (1999) Analysis of poly(A) tail lengths by PCR: the PAT assay. Methods Mol. Biol., 118, 441–448. [DOI] [PubMed] [Google Scholar]
- Saxon A., Kurbe-Leamer,M., Behle,K., Max,E.E. and Zhang,K. (1991) Inhibition of human IgE production via FcεR-II stimulation results from a decrease in the mRNA for secreted but not membrane εH chains. J. Immunol., 147, 4000–4006. [PubMed] [Google Scholar]
- Shatkin A.J. and Manley,J.L. (2000) The ends of the affair: capping and polyadenylation. Nature Struct. Biol., 7, 838–842. [DOI] [PubMed] [Google Scholar]
- Takagaki Y. and Manley,J.L. (1998) Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell, 2, 761–771. [DOI] [PubMed] [Google Scholar]
- Tollervey D. and Caceres,J.F. (2000) RNA processing marches on. Cell, 103, 703–709. [DOI] [PubMed] [Google Scholar]
- vanGelder C.W.G., Gunderson,S.I., Jansen,E.J.R., Boelens,W.C., Polycarpou-Schwarz,M., Mattaj,I.W. and van Venrooij,W.J. (1993) A complex secondary structure in U1A pre-mRNA that binds two molecules of U1A protein is required for regulation of polyadenylation. EMBO J., 12, 5191–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraldi K.L., Arhin,G.K., Martincic,K., Chung-Ganster,L.H., Wilusz,J. and Milcarek,C. (2001) hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol., 21, 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E. and Ruegsegger,U. (1999) 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]