Abstract
We have analysed the role of topoisomerase II (topo II) in plasmid DNA replication in Xenopus egg extracts, using specific inhibitors and two-dimensional gel electrophoresis of replication products. Topo II is dispensable for nuclear assembly and complete replication of plasmid DNA but is required for plasmid unlinking. Extensive unlinking can occur in the absence of mitosis. Replication intermediates generated in the absence of topo II activity have an increased positive superhelical stress (+ΔLk), suggesting a deficiency in precatenane removal. The geometry of replication intermediates cut by poisoning topo II with etoposide and purified by virtue of their covalent attachment to topo II subunits demonstrates that topo II acts behind the forks at all stages of elongation. These results provide direct evidence for unlinking replicating DNA by precatenane removal and reveal a division of labour between topo I and topo II in this eukaryotic system. We discuss the role of chromatin structure in driving DNA unlinking during S phase.
Keywords: DNA replication/DNA topoisomerases/nuclear assembly/topoisomerase inhibitors/Xenopus
Introduction
Topoisomerases unlink DNA during and after replication (Ullsperger et al., 1995). Lk, the linking number of the parental DNA strands, must be reduced to zero for separation of daughter DNA molecules. ΔLk is the difference between Lk and Lk0, the value of Lk for the same DNA molecule in relaxed form. The unwinding of parental DNA by replicative helicases causes a compensatory (+)ΔLk that must be removed by topoisomerases. Champoux and Been (1980) suggested that the (+)ΔLk could take the form of (+) supercoils in the unreplicated region as well as windings of the two newly replicated regions around each other. Thus, topoisomerases could unlink replication intermediates (RIs) by acting either in front of or behind the fork. The form of (+)ΔLk in the replicated DNA was subsequently named precatenane, due to structural similarity to catenanes and to distinguish it from the (+)ΔLk in the unreplicated DNA (Ullsperger et al., 1995). Any (+)ΔLk not removed before termination would form catenanes, and these would be unlinked after replication.
This model did not achieve immediate acceptance because early electron microscopy (EM) observations of circular RIs showed supercoils but no precatenanes, suggesting that topoisomerases only act ahead of the fork. However, studies of plasmid replication with purified Escherichia coli enzymes suggested that precatenanes are important in unlinking during replication (Peng and Marians, 1993; Hiasa and Marians, 1994, 1996). Moreover, both (–) precatenanes and (–) supercoils were observed on purified RIs accumulated by replication of plasmids containing two termination sites in E.coli (Peter et al., 1998). An EM artefact apparently caused earlier studies to miss precatenanes. Finally, a topological analysis of knots trapped within arrested RIs suggested that (–) precatenanes exist in E.coli cells (Sogo et al., 1999).
However, removing (–) precatenanes would just increase Lk. Arrested RIs from E.coli cells have a (–)ΔLk, presumably due to gyrase activity after replication arrest. Direct evidence for precatenanes on (+)ΔLk RIs, as predicted by Champoux and Been, has been lacking. In fact, (+)ΔLk RIs prepared by adding intercalating agents to purified (–)ΔLk RIs contain neither supercoils nor precatenanes. Instead, the (+) topological stress is relieved by re-annealing of the parental strands and formation of a Holliday junction, a process called fork reversal (Postow et al., 2001; J.B.Schvartzman, personal communication). It remains unclear, at least in bacteria, whether transient (not arrested) RIs carry a (+) or a (–)ΔLk and whether protein binding inside the cell prevents fork reversal and/or spinning and (+) precatenane formation.
In bacteria, two type 2 topoisomerases can unlink replicating DNA. Gyrase introduces (–) supercoils in front of the forks and may suffice to overcome the (+)ΔLk generated by replication until late RI stages, while topoisomerase IV (topo IV) is responsible for decatenating complete replication products (reviewed in Levine et al., 1998). Studies with purified enzymes support a role for precatenane unlinking by topo IV (Peng and Marians, 1993; Hiasa and Marians, 1996). However, in topo IV mutants, newly synthesized plasmid DNA accumulates as catenanes with the same node number distribution as transient catenanes in wild-type cells (Zechiedrich and Cozzarelli, 1995). Thus, in vivo evidence for precatenane removal by topo IV is lacking. In fact, the recent discovery that topo IV relaxes (+) supercoils 20-fold faster than (–) supercoils suggests that it may unlink DNA in front of the fork as efficiently as gyrase (Crisona et al., 2000).
Eukaryotes lack unconstrained (–) supercoils and the (–) supercoiling activity of gyrase. Thus, free (+) supercoils and possibly (+) precatenanes are expected to form during elongation. Both topo I and topo II can remove (+) supercoils in vitro. Studies with yeast mutants showed that replication can initiate in the absence of both enzymes, but elongation stops after a couple of thousand base pairs (Kim and Wang, 1989). Either topo I or topo II (but not topo III) can support further elongation and completion of S phase (Uemura and Yanagida, 1986; Brill et al., 1987). Topo II is strictly required at mitosis for separation of sister chromatids (Holm et al., 1985; Uemura and Yanagida, 1986). Similar observations were made for the replication of naked SV40 DNA in cell-free extracts (Yang et al., 1987) or with purified proteins (Ishimi et al., 1992b). The usual interpretation is that either topo I or topo II can remove (+) supercoils to drive elongation, while topo II is required to remove catenane crossings persisting after replication. However, it is unclear whether topo II relaxes (+) supercoils in front of the forks, like topo I, or removes (+) precatenanes behind the forks. Studies of SV40 minichromosome replication in cellular extracts or in infected cells suggest that topo II inhibition in some cases blocks elongation at the late RI stage (Richter et al., 1987; Ishimi et al., 1992a,b), but in other cases allows synthesis of complete catenated dimers, even though late RIs accumulate (Sundin and Varshavsky, 1981; Ishimi et al., 1995 and references therein; reviewed in Snapka, 1996). Thus, the implication of topo II in the late stages of DNA synthesis in higher eukaryotes is unclear.
Another unresolved issue is what drives decatenation of daughter DNA molecules. In vitro, topo II catalyses both catenation and decatenation of DNA rings, and favours catenation at high DNA concentration (Krasnow and Cozzarelli, 1982). Given the high concentration of DNA in vivo, one expects the equilibrium to be toward catenation. Mechanical separation of sister chromatids during mitosis has been proposed to drive decatenation (Holm, 1994; Duplantier et al., 1995). Although experiments in yeast and Xenopus show that topo II is required for mitotic chromosome condensation and segregation (reviewed in Holm, 1994), it is not known whether decatenation is postponed entirely until mitosis or already starts in S or G2 phases.
To investigate these questions, we have studied the effect of topo II inhibition on DNA replication in Xenopus egg extracts. Because studying replication and topology of a long linear chromosome would be difficult, we focused on circular plasmid DNA. Any plasmid DNA incubated in Xenopus egg extracts is replicated under cell cycle control, but only after it has been assembled by the egg extract into chromatin and then into synthetic nuclei, in which replication occurs at discrete foci as in normal nuclei (Blow and Laskey, 1986; Blow and Sleeman, 1990; Cox and Laskey, 1991). Small plasmids (<15 kb) support a single, randomly located initiation event that closely mimics replication of chromosomal domains in early embryonic nuclei (Hyrien and Méchali, 1992, 1993; Mahbubani et al., 1992; Lucas et al., 2000). Although caution is required because plasmids may be free of some of the topological restraints of long linear chromosomes, extrapolation from this system to what happens inside cells seems reasonable.
We have analysed the effect of various topo II inhibitors on plasmid DNA replication using high-resolution two-dimensional gel electrophoresis of replication products. ICRF-193 traps the enzyme in the form of a closed protein clamp without introducing DNA breaks (Tanabe et al., 1991). Etoposide (VP-16) and teniposide (VM-26) poison topo II by stabilizing a covalent reaction intermediate, the cleavable complex. Subsequent treatment with protein denaturants reveals the DNA strand breaks and the covalent linking of a topoisomerase subunit to the 5′ end of the broken DNA (Chen et al., 1984). The covalent intermediates can be extracted selectively with phenol prior to deproteinization. These properties were exploited to map the sites of topo II action during DNA replication. Our results provide direct evidence for the Champoux and Been model in this eukaryotic system, and reveal a division of labour between topo I and topo II. We suggest a role for chromatin assembly in driving DNA unlinking behind the replication fork.
Results
Topo II is dispensable for nuclear assembly and replication of plasmid DNA
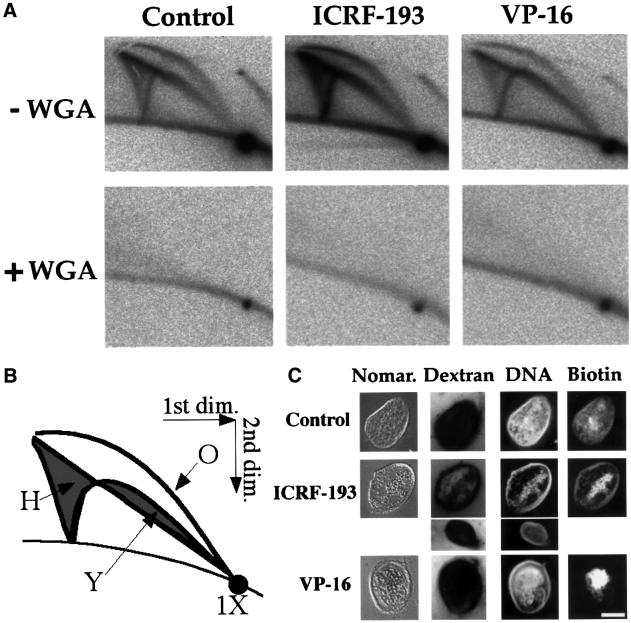
We first confirmed that 100 µM ICRF-193 or 50 µM VP-16 strongly (>98%) inhibits decatenation of kinetoplast DNA by crude Xenopus egg extracts (Shamu and Murray, 1992; Takasuga et al., 1995; data not shown). We then analysed the effect of these drugs on plasmid replication (Figure 1A, upper row). pBR322 DNA was incubated in egg extracts in the presence of [α-32P]dATP for 90 min, cut with EcoRI and analysed using the two-dimensional gel technique of Brewer and Fangman (1987). Since pBR322 DNA contains a single EcoRI site and supports a single, randomly located initiation event, RIs consisted of bubbles and double Ys. Replicated plasmids (1× spot) and complete RI arcs were detected in the control or when 100 µM ICRF-193 or 200 µM VP-16 was added at the start of the incubation (Figure 1A, upper row). Similar results were obtained with 50 µM VM-26, 50–200 µM VP-16 or 50–200 µM ICRF-193 (not shown).
Fig. 1. Effect of topo II inhibition on nuclear assembly and replication of plasmid DNA in Xenopus egg extracts. (A) pBR322 DNA was incubated for 90 min in an egg extract in the presence of [α-32P]dATP, and in the presence or absence of 100 µM ICRF-193, 200 µM VP-16 and 1 mg/ml WGA as indicated. The DNA was purified, cut with EcoRI and analysed by two-dimensional gel electrophoresis. (B) Schematic diagram of two-dimensional gel patterns. The linear 1× spot corresponds to fully replicated plasmids. The three classes of RIs (O, bubble arc; Y, simple Y arc; H, double Y smear) are shown. The arc above the 1× spot consists of heterogeneously sized open circles formed by end joining of a minor population of broken molecules in the plasmid preparation. The faint labelling of the 1× spot in the WGA samples is presumably due to repair synthesis. (C) Light microscopic appearance of pseudo-nuclei assembled from pMM-36, a 36 kb plasmid (Lucas et al., 2000). pMM-36 DNA was incubated in egg extract for 3 h with 20 µM biotin-16-dUTP, and with or without 100 µM ICRF-193 or 200 µM VP-16 as indicated. Samples were supplemented with1 mg/ml Hoechst 33258 to show DNA, 0.1 vol. of fluorescein-tagged streptavidin (Amersham) to show biotin, and 12 µM rhodamine-tagged dextran 70S (Sigma) to show exclusion by the nuclear membrane, and were then viewed unfixed under fluorescence (Dextran, DNA, Biotin) or Nomarski optics (Nomar.). Some nuclei in the ICRF-193 sample showed a faint dextran signal, probably due to increased fragility and artefactual damage. An unambiguous example of strong dextran exclusion is shown by a smaller nucleus. Bar, 5 µm.
This observation was surprising, since it was reported that replication of plasmid DNA depends on prior nuclear assembly (Blow and Sleeman, 1990) and that nuclear assembly is blocked by 40 µM VM-26 (Newport, 1987). We first verified that even with topo II inhibitors, plasmid DNA was assembled into pseudo-nuclei surrounded by a closed membrane that excludes fluorescent dextran and that replication occurred within these pseudo-nuclei (Figure 1C). Nevertheless, most of the DNA remained condensed at the centre of the pseudo-nuclei and detached from the nuclear envelope, and in some nuclei the DNA associated with the envelope replicated less efficiently. Secondly, we demonstrated that plasmid DNA replication in the presence or absence of topo II inhibitors was completely blocked by wheat germ agglutinin (WGA), an inhibitor of nuclear transport (Finlay et al., 1987) (Figure 1A, bottom row). WGA blocks sperm chromatin replication in egg extracts by preventing nuclear formation as well as by blocking protein import into pre-formed nuclei (Cox, 1992). In contrast, WGA has no effect on DNA synthesis on single-stranded templates (Cox, 1992), nor on sperm chromatin replication when the need for nuclear assembly is circumvented by sequential addition of cytosolic and nuclear extracts (Walter et al., 1998). Therefore, WGA has no effect on DNA replication other than by inhibition of nuclear assembly. We conclude that replication of plasmid DNA occurs exclusively in pseudo-nuclei and that topo II inhibition does not abolish nuclear assembly nor replication of plasmid DNA in egg extracts.
Nevertheless, phosphoimager quantification showed that the amount of RIs was increased and the 1× spot was decreased in the presence of topo II inhibitors. Thus, both the turnover of RIs and the completion of plasmid synthesis were slower. Forks accumulated at all stages of elongation and not specifically near termination (quantitations not shown). As demonstrated in other experiments (see Supplementary data available at The EMBO Journal Online), these differences are due to an ∼2-fold slowing of fork progression. However, replication forks can proceed at normal speed if topo II inhibitors are only added after nuclear assembly (see Supplementary data). Therefore, topo II is required to optimize nuclear assembly for replication but not to stimulate elongation per se.
Topo II rapidly decatenates daughter duplexes in S phase
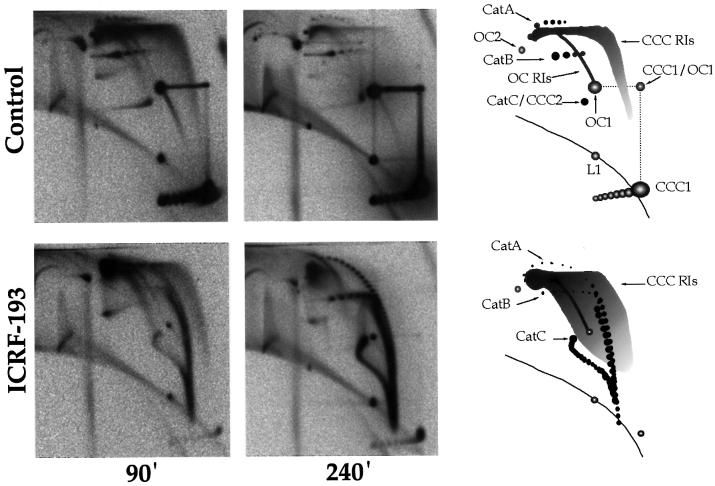
Next, we analysed the effects of topo II inhibition on the topology of DNA replication. Aliquots of pBR322 replication reactions were removed at 90 and 240 min and analysed on two-dimensional gels without restriction enzyme digestion (Figure 2). The migration behaviour of the various forms of plasmid monomers, RIs and catenated dimers is diagrammed in Figure 2. Evidence for these assignments is based on treatment with DNase I and topo I (Brewer and Fangman, 1987; Brewer et al., 1988; Martin-Parras et al., 1998). Further evidence based on topo II treatment (Figure 3) and partial digestion with EcoRI or S1 nuclease (Figure 5) is reported here.
Fig. 2. Topological analysis of plasmid replication. pBR322 DNA was incubated in an egg extract in the presence of [α-32P]dATP and in the presence or absence of 100 µM ICRF-193. Samples were removed at 90 and 240 min, and analysed by two-dimensional gel electrophoresis without restriction enzyme digestion. Interpretative diagrams are shown on the right. L, linear molecules; CCC, covalently closed circles; OC, open circles; the numbers refer to the multimeric state (1, monomers, 2, dimers). CCC1/OC1, molecules that migrated as CCC1 during the first dimension, were subsequently nicked, and migrated as OC1 during the second dimension. Cat A, Cat B, Cat C refer to OC1–OC1, OC1–CCC1 and CCC1–CCC1 catenated dimers, respectively. OC RIs and CCC RIs are open and covalently closed circular replication intermediates, respectively.
Fig. 3. Identification of CCC RIs and catenated dimers by resolution with topo II. pBR322 was replicated in an egg extract without ICRF-193 for 90 min or with 100 µM ICRF-193 for 240 min, in order to maximize the accumulation of RIs and catenated dimers, respectively. Purified replication products were analysed by two-dimensional gel electrophoresis either directly (left) or after treatment with D.melanogaster topo II (right). Arrows point to CCC RIs (top) and arrowheads to catenanes (bottom left) or to OC and relaxed CCC monomers (bottom right).
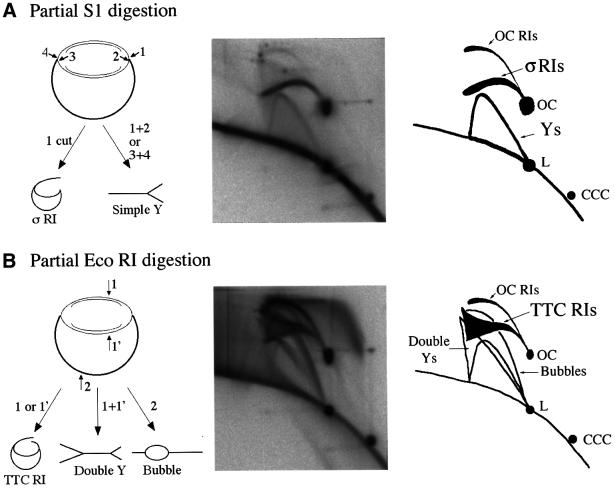
Fig. 5. Identification of σRIs and TTC RIs. pBR322 was replicated in an egg extract for 90 min. Purified replication products were partially digested with S1 nuclease (A) or with EcoRI (B) and analysed by two-dimensional gel electrophoresis. Predicted products (left), experiments (middle) and interpretative diagrams (right) are shown. For clarity, CCC RIs and catenanes are not shown.
In the controls (Figure 2, top row), most replication products were open circular (OC) and covalently closed circular (CCC) monomers. RIs were abundant at 90 min but scarce at 240 min, indicating that most nuclei had finished replicating at that time. The migration behaviour of RIs is discussed later. A small amount of catenated dimers was detected at both time points. A-type (OC–OC) and B-type (OC–CCC) catenanes were resolved as two series of spots of decreasing intensity, each corresponding to a certain node number. Only catenanes with 2 to <10 nodes (1 to <5 intertwinings) were observed. In many experiments (e.g. Figure 3, top left panel), the two-noded catenane was the only detectable species. C-type (CCC–CCC) catenanes were not resolved from each other, and co-migrated with supercoiled plasmid dimer (CCC2). These results show that in uninhibited extracts, precatenane and catenane nodes either seldom form or are removed very rapidly, during or shortly after plasmid replication. As the extracts were blocked in interphase by addition of cycloheximide, which prevents cyclin synthesis and entry in mitosis, we conclude that a nearly complete unlinking can occur prior to mitosis.
In the presence of 100 µM ICRF-193 (Figure 2, bottom), very few monomers (OC and CCC) were produced. The signal accumulated in three arcs, which we interpret as A-, B- and C-type catenated dimers with a broad node number distribution. This assignment was confirmed by treatment with calf thymus topo II (Figure 3, bottom): the three arcs disappeared and were replaced by OC and relaxed CCC monomers. In the 240 min sample (Figure 2, bottom, middle panel), A- and B-type catenanes were resolved as individual spots ranging from 2 to >40 nodes. The most intertwined catenanes probably had 70–80 nodes. The migration of C-type catenanes also differed from the control. The trail emanating downwards from the spot of lowly intertwined C-type catenanes to merge with the bottom ends of A- and B-type catenanes arcs presumably consisted of highly intertwined C-type catenanes. A similar electrophoretic behaviour has been reported for C-type catenanes produced by exposure of replicating SV40 to ICRF-193, and has been attributed to an inverse relationship between catenation and superhelicity (Permana et al., 1994). Although this arc seemed continuous, we do not think it consisted of RIs because it accumulated with time (compare 90 and 240 min) and was converted to OC and CCC upon topo II treatment (Figure 3, bottom). In the 90 min sample (Figure 2, bottom left panel), only catenanes with >16 nodes were clearly detected. Thus, upon topo II inhibition, newly synthesized DNA essentially appeared in the form of highly intertwined catenated dimers. These results confirmed, as expected, that topo II is required for optimum unlinking of daughter duplexes in egg extracts.
The topology of CCC RIs was analysed by treatment with topo II. Untreated CCC RIs migrated as a broad smear rising upwards from the CCC1 ladder and extending mainly to the right of OC RIs (Figures 2, top row and 3, top left). Topo II-treated, relaxed CCC RIs migrated to the left and very close to OC RIs (Figure 3, top right), as a thinner arc along which RIs were resolved essentially according to mass. Similar results were obtained with calf thymus topo I (Martin-Parras et al., 1998). Since (+)ΔLk RIs have no net writhe and migrate as relaxed RIs due to fork reversal (Postow et al., 2001; J.B.Schvartzman, personal communication), the faster migration of untreated RIs in the first dimension must be due to compaction by (–) precatenanes and (–) supercoils. In the second dimension, the high (+)ΔLk introduced by ethidium bromide caused all RIs to migrate as relaxed RIs. The ΔLk of untreated RIs had two potential sources: the (+)ΔLk generated by replication and the (–)ΔLk constrained by nucleosomes in unreplicated DNA. The electrophoretic behaviour of untreated RIs shows that when both topo I and topo II are active, the resulting ΔLk remains negative until the very last helix turns are replicated.
The migration of CCC RIs from ICRF-193-treated extracts was different from that of control RIs. The signal distribution across the smear was shifted to the left, especially in the upper part of the gel, suggesting a globally less negative ΔLk (Figure 2). Since catenanes with up to 80 nodes were seen, the ΔLk of the latest RIs (>90% replicated) should range from 0 to +40. According to Postow et al. (2001) and J.B.Schvartzman (personal communication), these (+)ΔLk RIs will migrate as relaxed RIs of the same mass. Indeed, most of the signal from late RIs concentrated at the upper left corner of the smear, at the position of relaxed late RIs (compare with Figure 3, top right). The changes in the electrophoretic behaviour of CCC RIs upon topo II inhibition reveal that topo II actively unlinks DNA during elongation in a way that cannot be taken over fully by topo I. This less complete unlinking of RIs provides a simple explanation for the increase in catenane complexity observed upon topo II inhibition. The increase in ΔLk appears stronger for late RIs, suggesting that the unlinking action of topo II that cannot be taken over by topo I increases as replication progresses. This is suggestive of precatenane rather than supercoil removal because the larger the replicated region, the more the ΔLk should be expressed as precatenanes (Peter et al., 1998; reviewed in Snapka, 1996), and because topo I, which is abundant in these extracts, can remove supercoils but not precatenanes.
Topo II acts behind the forks during replication elongation
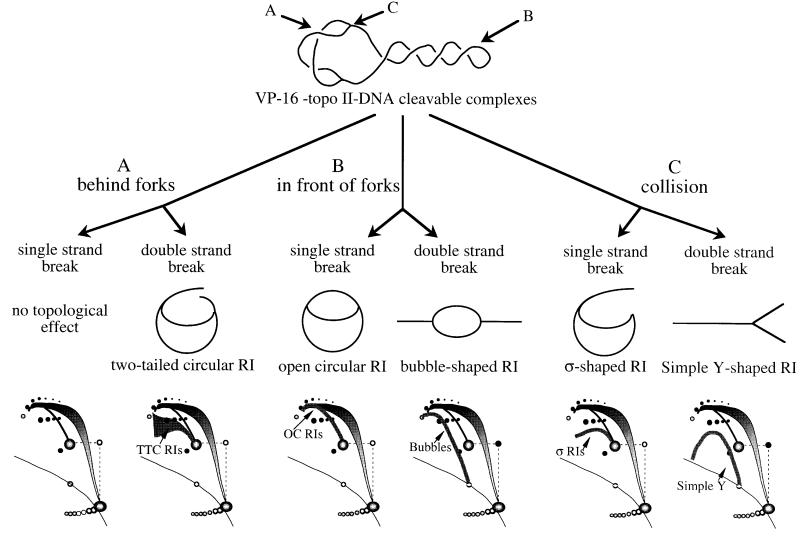
In order to map where topo II acts during replication, the location of VP-16-induced breaks on RIs was assessed by two-dimensional-gel electrophoresis. VP-16 induces both double- and single-stranded breaks (DSBs and SSBs) in plasmid DNA in egg extracts (see Supplementary data). If topo II acts in the replicated region, an SSB will not change the topology of RIs, and a DSB will convert CCC and OC RIs into two-tailed circular RIs (TTC RIs; Figure 4A). On the other hand, if topo II acts in the unreplicated region, an SSB will convert CCC RIs into OC RIs and a DSB will convert CCC and OC RIs into bubble-shaped fragments (Figure 4B). Finally, a collision between a replication fork and a cleavable complex in unreplicated DNA will result in sigma-shaped RIs (σRIs; in the case of an SSB) and simple Y RIs (in the case of a DSB) (Figure 4C). The resulting two-dimensional gel patterns should allow discrimination between these three scenarios.
Fig. 4. Mapping the sites of topo II action on plasmid RIs by trapping topo II–DNA cleavable complexes with VP-16. Products expected after a single or a double-strand break behind (A) or in front of (B) the fork or after collision of the fork with the cleavable complex (C) are shown. The expected changes in two-dimensional gel patterns are shown at the bottom.
The migration behaviour of bubbles, simple Ys and OC RIs is well documented (Brewer and Fangman, 1987). To identify σRIs, purified RIs were partially digested with S1 nuclease and the products were analysed on two-dimensional gels (Figure 5A). S1 cut parental DNA at the single-stranded regions of the forks, converting OC RIs and CCC RIs into σRIs (one cut), simple Y RIs (two cuts at one fork) and linear plus OC fragments (cuts at both forks). The resulting gel allowed the unambiguous identification of σRIs as the eyebrow-shaped arc starting and extending leftwards from the OC spot, as previously suggested (Belanger et al., 1996; Martin-Parras et al., 1998). To identify TTC RIs, purified RIs were partially digested with EcoRI (Figure 5B). As pBR322 replication initiates randomly in egg extracts, EcoRI cut at random in the unreplicated region (bubbles) or in one (TTC RIs) or both (double Ys) replicated arms. The arc of TTC RIs started from the OC spot much like σRIs, but broadened in a triangular smear above the position where σRIs would migrate. The migration of TTC RIs with respect to σRIs was reminiscent of the migration of double Ys with respect to simple Ys. Presumably, TTC RIs of identical masses but different lengths of the two tails migrate similarly in the first dimension but differently in the second dimension, with asymmetric forms migrating faster (closer to σRIs).
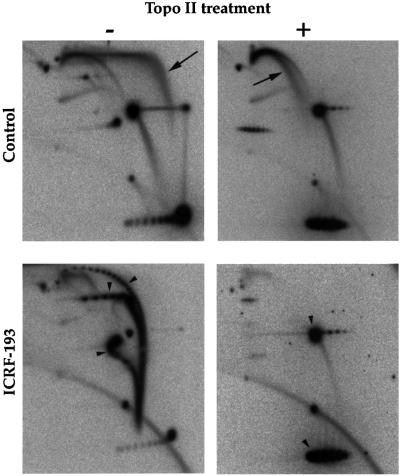
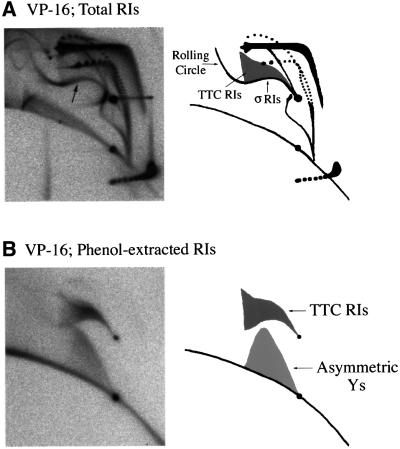
Figure 6A shows the two-dimensional gel patterns generated when 50 µM VP-16 was present in the extract. Control (no drug) gels are shown in Figures 2 and 3, top left panels. Topo II inhibition by VP-16 resulted in the accumulation of highly intertwined catenated dimers, as with ICRF-193. However, the inhibition of topo II by 50 µM VP-16 was less complete than with 100 µM ICRF-193, as judged by the distribution of catenane node number, the amount of OC and CCC monomers and the migration of CCC RIs. Importantly, we observed in the VP-16-treated samples a new, composite arc (arrow) consisting of material either absent or less abundant in control or ICRF-193-treated samples. The rightmost part of the signal consisted of a strong eyebrow of σRIs surmounted by a faint triangular smear of TTC RIs. This signal was consistent with a mixture of DSBs behind the forks and SSBs at the fork (pathways A/DSB + C/SSB in Figure 4). Alternatively, this signal could result from DSBs exclusively behind but predominantly close to the forks (pathway A/DSB only). The leftmost part of the signal prolonged the eyebrow, but not the triangular smear, upwards and leftwards beyond the 2× position. This arc of >2× σRIs probably arose by ligation of a nascent strand to the broken parental strand of a 1–2× σRI to create a rolling circle intermediate, as seen when an SSB is produced by DNase activity (Gourlie and Pigiet, 1983) or by topo I poisoning (Snapka, 1986). Alternatively, rolling circles could result from repair of a TTC RI by the action of a flap endonuclease, gap filling and ligation. Neither a bubble arc, a simple Y arc nor a convincing increase of OC RIs was detected, failing to support pathways B/SSB, B/DSB and C/DSB. Overall, these results suggest that topo II poisoning by VP-16 results in DSBs behind the forks and possibly SSBs only a short distance ahead of the forks.
Fig. 6. Topo II acts behind replication forks. pBR322 DNA was replicated for 90 min in the presence of 50 µM VP-16. The total replication products (A) or the phenol-extracted covalent topo II–DNA complexes (B) were analysed by two-dimensional gel electrophoresis. The composite arc of σRIs, TTC RIs and rolling circle RIs induced by VP-16 is shown by an arrow on the top left panel. Interpretative diagrams are shown on the right.
To investigate further the patterns of topo II cleavage on RIs, replication reactions were extracted with phenol prior to proteinase K digestion to enrich for topo II–RI covalent complexes. DNA molecules in the phenol phase and the aqueous phase were purified, digested with proteinase K and analysed separately by two-dimensional gel electrophoresis. In the control (no drug) reactions, no material was found in the phenol phase (data not shown). In the presence of 50 µM VP-16, a small but significant fraction of the labelled DNA was extracted in the phenol phase because of its covalent association with topo II (Figure 6B). This extraction was abolished by proteinase K treatment prior to phenol addition (data not shown). The phenol-extracted material consisted of: (i) a small amount of OC and linear molecules; (ii) a complete arc of TTC RIs; and (iii) a smear of molecules migrating between simple Ys and linears, as described for asymmetric Ys (i.e. molecules with three branches of unequal length) (Martin-Parras et al., 1992). The presence of TTC RIs in the phenol phase confirms that DSBs behind the forks are mediated by topo II. Asymmetric Ys were not detected in many experiments (e.g. not in Figure 6A). They may result from a nick at one fork of a TTC RI, or from degradation of a symmetric Y generated by pathway C/DSB (see Discussion). In contrast to TTC RIs and asymmetric Ys, neither the 1–2× σRIs nor the >2× rolling circle RIs were found in the phenol phase, although both were induced by VP-16 treatment. σRIs may be created by some nuclease induced by VP-16, or topo II may be removed from the broken forks (see Discussion). Finally, the lack of OC RIs and CCC RIs in the phenol phase shows that topo II-bound SSBs are found neither in unreplicated DNA nor in replicated arms.
We noted that only a small fraction of replicating DNA was cleaved upon VP-16 treatment, as observed for SV40 RIs extracted from VM-26-treated cells (Avemann et al., 1988). Furthermore, only a small fraction of the cleaved products was extracted with phenol. In contrast, non-replicating DNA was cleaved and phenol extracted efficiently (see Supplementary data). These results may be due to reversal or processing of cleavable complexes on replicating DNA (see Discussion).
In summary, topo II-bound DSBs induced by 50 µM VP-16 are located mainly if not exclusively behind the forks, as predicted if topo II is to remove precatenanes during replication. The lack of detectable DSBs in front of the forks suggests that such cleavage is infrequent or reversed very efficiently. The completeness of the arc of TTC RIs suggests that topo II unlinks precatenanes at all stages of elongation and not specifically near termination, and acts at various positions behind the forks. Further evidence for a covalent association of topo II with pulse-labelled nascent strands is presented in the Supplementary data.
Discussion
In this work, we provide evidence that: (i) topo II is somewhat dispensable for nuclear assembly and replication of plasmid DNA in Xenopus egg extracts, but is required for optimal unlinking of replicating DNA; (ii) precatenanes form at all stages of elongation but are unlinked rapidly by topo II behind the fork; and (iii) extensive unlinking of daughter plasmids does not require mitosis.
Role of topo II in assembly of replication-competent nuclei
We demonstrate that topo II inhibition by VP-16, VM-26 or ICRF-193 does not abolish nuclear assembly nor replication of plasmid DNA, although it perturbs the intranuclear distribution of chromatin and slows down its replication. This result contrasts with the reported inhibition of nuclear assembly from bacteriophage λ DNA by VM-26 (Newport, 1987). As bacteriophage λ DNA was ligated into large multimers before addition to the egg extract, topo II may be needed to resolve tangles that form during this ligation step but not when plasmid DNA is used. Our results are more consistent with the effects of ICRF-193 on sperm chromatin in egg extracts (Takasuga et al., 1995). ICRF-193 prevents decondensation of sperm heads. The nuclear membrane forms and grows but makes very few connections with the condensed chromatin. DNA replication nevertheless occurs, although initiation is retarded and the rate of DNA synthesis per nucleus is decreased. Here, we show that in the presence of VP-16 or ICRF-193, most of the DNA remains condensed in the nuclear interior, and plasmid replication is slower due to a longer pre-replication lag and a slower fork progression. However, replication forks progress at a normal rate if topo II inhibitors are added after nuclear assembly. Therefore, topo II is required to optimize nuclear assembly for replication but not for elongation per se.
Topo II is not required specifically for elongation of late RIs
Many reports have suggested that topo II inhibition specifically slows down the late stage of SV40 minichromosome replication (Sundin and Varshavsky, 1981; Ishimi et al., 1995 and references therein; reviewed in Snapka, 1996). This conclusion was based largely on the accumulation of undigested SV40 RIs at a specific position in one-dimensional agarose gels (although some neutral chloroquine two-dimensional gels failed to confirm this; see Permana et al., 1994). We do observe an analogous accumulation of CCC RIs (in our two-dimensional gel system, at the upper left of the CCC RI smear) upon topo II inhibition. However, the analysis of EcoRI-digested RIs unambiguously demonstrates that the ratio of late RIs to early RIs is not increased. We reinterpret the accumulated signal as follows. In the presence of topo II, late RIs form a series of (–)ΔLk topoisomers that are spread through the gel. In the absence of topo II, late RIs have a high (+)ΔLk, causing them to undergo fork reversal after deproteinization (Postow et al., 2001; J.B.Schvarztman, personal communication) and to accumulate at the position of relaxed RIs during electrophoresis. These results imply that a high (+)ΔLk can build up without impeding the unwinding of the terminus region, presumably because the (+)ΔLk of late RIs is expressed as precatenanes rather than supercoils.
Evidence for the Champoux and Been model
Two lines of evidence strongly argue that precatenanes form on replicating Xenopus chromatin and are removed rapidly by topo II. First, the geometry of RIs cut by poisoning topo II with VP-16 shows that topo II acts behind the forks. Secondly, the selective extraction of topo II–RI covalent complexes shows a covalent association of topo II behind but not in front of the forks and a covalent association of topo II with pulse-labelled nascent strands.
Furthermore, RIs generated in the presence of topo II inhibitors have an increased ΔLk, as shown by two-dimensional gel electrophoresis. This implies that topo II contributes to replicative unlinking in a way that cannot be taken over fully by topo I. While plasmid replication proceeds at ∼8 nucleotides per second, it is estimated that a single topo I molecule can remove from 20 to 200 supercoils per second (Stivers et al., 1997; T.Strick, personal communication). Topo I is abundant in egg extracts and should not be limiting for removing supercoils unless its access to unreplicated DNA becomes limited by its decreasing size or by proteins or kinetic factors. As topo II–DNA cleavable complexes are detected behind but not in front of the forks at all elongation stages, the topo II-mediated unlinking for which topo I cannot substitute most probably consists of removing precatenanes rather than supercoils.
Our evidence for precatenane removal by topo II is consistent with the inference of precatenane removal by topo IV, a bacterial type II topoisomerase, during plasmid replication with purified proteins (Peng and Marians, 1993; Hiasa and Marians, 1996). However, the latter studies neither directly demonstrated precatenane production nor showed that it was a substrate.
One limitation of our study is that it is restricted to small plasmids replicating in a crude extract. In contrast to soluble (e.g. SV40) replication systems, plasmids in frog egg extracts are assembled into and replicate exclusively within synthetic nuclei that closely resemble normal embryonic nuclei. Nevertheless, the formation of precatenane implies that replication forks are free to rotate and distribute the ΔLk either transiently or permanently, in apparent contrast to their attachment to nuclear substructure (reviewed in Hyrien et al., 1997). We cannot exclude the possibility that fork spinning is easier for such plasmids than for large chromosomes in vivo.
Lack of detectable topo II–DNA cleavable complexes in front of the forks
It is widely believed that topo II can unlink replicating DNA by removing (+) supercoils in front of the forks, just as topo I. There is good evidence that cleavable complexes trapped by topo I poisons are not themselves toxic but can be converted into DSBs, the cytotoxic lesion, by encounter with a moving replication fork (reviewed in Fortune and Osheroff, 2000). This idea has been extended to topo II poisons without much evidence.
Our analysis of phenol-purified topo II–RI covalent complexes provides clear evidence for cleavable complexes behind the forks (TTC RIs) but not in front of the forks (bubbles or OC RIs). VP-16 does induce breakage at the forks (σRIs), but these breaks are not covalently joined to topo II and we cannot exclude the possibility that they are mediated by some nuclease rather than by topo II. Asymmetric Ys are joined to topo II and may result from collision with a DSB (pathway C/DSB in Figure 4) followed by degradation of the resulting symmetric Ys. However, asymmetric Ys were not observed in many experiments, and DSBs prior to collision (bubbles) were never detected. Asymmetric Ys may also result from an SSB at one fork of a TTC RI, perhaps during sample manipulation. In short, we did not find unequivocal evidence for a destructive interaction of replication forks with drug-stabilized topo II cleavable complexes.
Why should topo II fail to work in front of the forks? It is unlikely that chromatin masks supercoil nodes or favours twist over writhe, because topo II does work on non-replicating plasmids. Supercoils may be suppressed during replication because helicases/polymerases generate tension in unreplicated DNA or because topo I relaxes twist before DNA can writhe. Sequestration of topo II behind the fork by DNA substrate affinity appears unlikely because the rates of decatenation of singly linked catenanes (∼7/s; Baird et al., 1999) and relaxation of (+) or (–) supercoils (∼3/s; Strick et al., 2000) are similar. Yet, multiply linked catenanes were not studied, and enzyme processivity may matter. Differences in chromatin structure may also alter the equilibrium distribution or the relative accessibility of supercoil and precatenane nodes (see below). Interaction with replication proteins might sequester topo II behind the fork.
Another possibility is that cleavable complexes in front of the forks are reversed (repaired) specifically in egg extracts. Topo II association with σRIs may be reversed upon conversion of σRIs to rolling circle templates. A replication-coupled removal of cleavable complexes may explain the low efficiency of VP-16-induced cleavage and topo II trapping on replicating DNA. Cleavable complexes located in front of the forks represent a more immediate threat than those located behind the forks and may constitute preferred repair substrates. Concordantly, we found that camptothecin, a topo I poison reported to induce fork breakage in SV40 (Snapka, 1986; Avemann et al., 1988), induces surprisingly few σRIs in egg extracts (data not shown).
Various responses to topo I- and topo II-mediated DNA damage have been reported. A conserved enzyme specifically hydrolyses the topo I–DNA bond (Pouliot et al., 1999). Topo I and topo II poisons trigger SUMO-1 conjugation to the respective enzyme (Mao et al., 2000a,b). Bacteriophage T4 replication forks blocked in vivo by drug-induced topo II cleavable complexes remain intact, presumably due to topoisomerase resealing (Hong and Kreuzer, 2000). It would not be surprising to find that Xenopus eggs primed for accelerated replication during early development are able to remove efficiently the topo II–DNA complexes located in front of the forks.
A model for rapid unlinking of sister chromatids during S phase
In frog egg extracts that assemble mitotic spindles, adding topo II inhibitors prior to anaphase induction prevents sister chromatid separation. Therefore, some catenation persists until metaphase (Shamu and Murray, 1992). Mechanical separation of sister chromatids at anaphase was proposed to drive complete decatenation (Holm, 1994; Duplantier et al., 1995). However, the progress of decatenation prior to metaphase has not been assessed. Here, we used egg extracts blocked in interphase to show that replicated plasmids are decatenated extensively, although the high DNA concentration and aggregation conditions found in synthetic nuclei should favour catenation. We suggest that a mechanism for driving decatenation already exists in S phase.
Type 2 topoisomerases use ATP hydrolysis to untangle DNA molecules below thermal equilibrium (Rybenkov et al., 1997a), but this may not fully explain our observations. We suggest that chromatin assembly at the replication fork drives decatenation. Nucleosomes are normally assembled on daughter strands at ∼260 bp of the elongation point (Gasser et al., 1996). If nucleosome fibres are more refractory to catenation than naked DNA, newly formed precatenanes may hinder nucleosome assembly. This could in turn facilitate decatenation by topo II, allowing assembly of nucleosomes (and higher order structures) to proceed and prevent recatenation. In this way, the two sister chromatids would be decatenated as they are replicated. Bacteria do not have nucleosomes, but unconstrained (–) supercoils due to gyrase action. Negative supercoiling greatly reduces the equilibrium constant for catenation of small plasmids (Rybenkov et al., 1997b). Thus, supercoiling in bacteria and chromatin assembly in eukaryotes may play a comparable role in driving decatenation of newly replicated DNA.
Materials and methods
Stock solutions of ICRF-193 (17.7 mM; a gift from Zenyaku Kogyo), VP-16 (10 mM; Sigma) and VM-26 (10 mM; a gift from S.Gasser) were prepared in dimethyl sulfoxide (DMSO; Sigma). ICRF-193 was used at 100–200 µM, VP-16 at 50–200 µM and VM-26 at 25–100 µM in the extract. WGA (Sigma) was made up in distilled water at 10 mg/ml and added to the extract to 1 mg/ml. pBR322 DNA was prepared from the Rec+ BL21 strain by standard lysis and caesium chloride gradient centrifugation.
Plasmid DNA was added at 5 ng/µl to Xenopus egg extracts prepared as described (Hyrien and Méchali, 1992). [α-32P]dATP was added to label replication products directly. ICRF-193, VP-16, VM-26 or an equivalent amount of DMSO without drug (control) was added as indicated. The reactions were stopped at the indicated times with an equal volume of 1% sarkosyl, 80 mM EDTA, 600 mM NaCl, and incubated for 1 h at 37°C with 150 µg/ml RNase A. Proteinase K was added at 400 µg/ml and incubation continued overnight. DNA was extracted three times with phenol–chloroform, precipitated with ethanol and resuspended in TE. In an attempt to minimize putative resealing of VP-16- or VM-26-cleavable complexes during DNA extraction, we checked differents parameters of the protocol. SDS (2%) was substituted for 1% sarkosyl, and EDTA was added at 0, 1 or 10 min. We also tried immediate addition of higher amounts (2 mg/ml) of proteinase K. These modifications did not change the recovery of cleavage products.
The selective extraction of cleavable complexes with phenol is described in the Supplementary data.
For the topo II relaxation experiment shown in Figure 3, plasmid DNA isolated from egg extracts (150 ng) was resuspended in 85 µl of topo II buffer, incubated with 20 U of purified Drosophila melanogaster topo II for 2.5 h at 30°C, purified by phenol–chloroform extraction and ethanol precipitation, and resuspended in TE.
For two-dimensional gel analysis, 150 ng of purified DNA were loaded on each gel either directly or after digestion with EcoRI as described (Hyrien and Méchali, 1992). The two-dimensional gel method in Tris–borate–EDTA described in Brewer and Fangman (1987) was used with modifications [first dimension, 0.4% (w/v) agarose gel, 1 V/cm, 18 h room temperature; second dimension, 1.1% agarose gel, 0.3 µg/ml ethidium bromide, 5 V/cm, 7–8 h, 4°C]. The gels were blotted in 0.4 M NaOH onto Hybond-N+ and analysed on a Fuji BAS 1000 phosphoimager as described (Lucas et al., 2000).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank J.-L.Sikorav and T.Strick for reading the manuscript and for discussions, and the referees for insightful criticism. We thank Zekyaku Kogyo for the gift of ICRF-193 and S.Gasser for the gift of VM-26. This work was supported by the ATIPE-CNRS, the Ligue Nationale Française Contre le Cancer (Comité de Paris) and the Association pour la Recherche sur le Cancer. I.L. was supported by fellowships from the MENESR and the Association pour la Recherche sur le Cancer.
References
- Avemann K., Knippers,R., Koller,T. and Sogo,J.M. (1988) Camptothecin, a specific inhibitor of type I DNA topoisomerase, induces DNA breakage at replication forks. Mol. Cell. Biol., 8, 3026–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird C.L., Harkins,T.T., Morris,S.K. and Lindsley,J.E. (1999) Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl Acad. Sci. USA, 96, 13685–13690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger K.G., Mirzayan,C., Kreuzer,H.E., Alberts,B.M. and Kreuzer, K.N. (1996) Two-dimensional gel analysis of rolling circle replication in the presence and absence of bacteriophage T4 primase. Nucleic Acids Res., 24, 2166–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J. and Laskey,R.A. (1986) Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell, 47, 577–587. [DOI] [PubMed] [Google Scholar]
- Blow J.J. and Sleeman,A.M. (1990) Replication of purified DNA in Xenopus egg extract is dependent on nuclear assembly. J. Cell Sci., 95, 383–391. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman,W.L. (1987) The localization of replication origins on ARS plasmids in S.cerevisiae. Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- Brewer B., Sena,E. and Fangman,W. (1988) Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. In Kelly,T. and Stillman,B. (eds), Cancer Cells 6. Eukaryotic DNA Replication. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 229–234.
- Brill S.J., DiNardo,S., Voelkel-Meiman,K. and Sternglanz,R. (1987) Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature, 326, 414–416. [DOI] [PubMed] [Google Scholar]
- Champoux J. and Been,M. (1980) Topoisomerases and the swivel problem. In Alberts,B. (ed.), Mechanistic Studies of DNA Replication and Genetic Recombination: ICN-UCLA Symposia on Molecular and Cellular Biology. Academic Press, New York, NY, pp. 809–815.
- Chen G.L., Yang,L., Rowe,T.C., Halligan,B.D., Tewey,K.M. and Liu,L.F. (1984) Nonintercalative antitumor drugs interfere with the breakage–reunion reaction of mammalian DNA topoisomerase II. J. Biol. Chem., 259, 13560–13566. [PubMed] [Google Scholar]
- Cox L.S. (1992) DNA replication in cell-free extracts from Xenopus eggs is prevented by disrupting nuclear envelope function. J. Cell Sci., 101, 43–53. [DOI] [PubMed] [Google Scholar]
- Cox L.S. and Laskey,R.A. (1991) DNA replication occurs at discrete sites in pseudonuclei assembled from purified DNA in vitro. Cell, 66, 271–275. [DOI] [PubMed] [Google Scholar]
- Crisona N.J., Strick,T.R., Bensimon,D., Croquette,V. and Cozzarelli, N.R. (2000) Preferential relaxation of positively supercoiled DNA by E.coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev., 14, 2881–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplantier B., Jannink,G. and Sikorav,J.L. (1995) Anaphase chromatid motion: involvement of type II DNA topoisomerases. Biophys. J., 69, 1596–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay D.R., Newmeyer,D.D., Price,T.M. and Forbes,D.J. (1987) Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol., 104, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune J.M. and Osheroff,N. (2000) Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol., 64, 221–253. [DOI] [PubMed] [Google Scholar]
- Gasser R., Koller,T. and Sogo,J.M. (1996) The stability of nucleosomes at the replication fork. J. Mol. Biol., 258, 224–239. [DOI] [PubMed] [Google Scholar]
- Gourlie B.B. and Pigiet,V.P. (1983) Polyoma virus minichromosomes: characterization of the products of in vitro DNA synthesis. J. Virol., 45, 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiasa H. and Marians,K.J. (1994) Topoisomerase III, but not topoisomerase I, can support nascent chain elongation during Θ-type DNA replication. J. Biol. Chem., 269, 32655–32659. [PubMed] [Google Scholar]
- Hiasa H. and Marians,K.J. (1996) Two distinct modes of strand unlinking during Θ-type DNA replication. J. Biol. Chem., 271, 21529–21535. [DOI] [PubMed] [Google Scholar]
- Holm C. (1994) Coming undone: how to untangle a chromosome. Cell, 77, 955–957. [DOI] [PubMed] [Google Scholar]
- Holm C., Goto,T., Wang,J.C. and Botstein,D. (1985) DNA topoisomerase II is required at the time of mitosis in yeast. Cell, 41, 553–563. [DOI] [PubMed] [Google Scholar]
- Hong G. and Kreuzer,K.N. (2000) An antitumor drug-induced topoisomerase cleavage complex blocks a bacteriophage T4 replication fork in vivo. Mol. Cell. Biol., 20, 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. and Méchali,M. (1992) Plasmid replication in Xenopus eggs and egg extracts: a 2D gel electrophoretic analysis. Nucleic Acids Res., 20, 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. and Méchali,M. (1993) Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J., 12, 4511–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Maric,C. and Lucas,I. (1997) Role of nuclear architecture in the initiation of eukaryotic DNA replication. Biochimie, 79, 541–548. [DOI] [PubMed] [Google Scholar]
- Ishimi Y., Ishida,R. and Andoh,T. (1992a) Effect of ICRF-193, a novel DNA topoisomerase II inhibitor, on simian virus 40 DNA and chromosome replication in vitro. Mol. Cell. Biol., 12, 4007–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y., Sugasawa,K., Hanaoka,F., Eki,T. and Hurwitz,J. (1992b) Topoisomerase II plays an essential role as a swivelase in the late stage of SV40 chromosome replication in vitro. J. Biol. Chem., 267, 462–466. [PubMed] [Google Scholar]
- Ishimi Y., Ishida,R. and Andoh,T. (1995) Synthesis of simian virus 40 C-family catenated dimers in vivo in the presence of ICRF-193. J. Mol. Biol., 247, 835–839. [DOI] [PubMed] [Google Scholar]
- Kim R.A. and Wang,J.C. (1989) Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J. Mol. Biol., 208, 257–267. [DOI] [PubMed] [Google Scholar]
- Krasnow M.A. and Cozzarelli,N.R. (1982) Catenation of DNA rings by topoisomerases. Mechanism of control by spermidine. J. Biol. Chem., 257, 2687–2693. [PubMed] [Google Scholar]
- Levine C., Hiasa,H. and Marians,K.J. (1998) DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication and drug sensitivities. Biochim. Biophys. Acta, 1400, 29–43. [DOI] [PubMed] [Google Scholar]
- Lucas I., Chevrier-Miller,M., Sogo,J.M. and Hyrien,O. (2000) Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J. Mol. Biol., 296, 769–786. [DOI] [PubMed] [Google Scholar]
- Mahbubani H.M., Paull,T., Elder,J.K. and Blow,J.J. (1992) DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res., 20, 1457–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Desai,S.D. and Liu,L.F. (2000a) SUMO-1 conjugation to human DNA topoisomerase II isozymes. J. Biol. Chem., 275, 26066–26073. [DOI] [PubMed] [Google Scholar]
- Mao Y., Sun,M., Desai,S.D. and Liu,L.F. (2000b) SUMO-1 conjugation to topoisomerase I: a possible repair response to topoisomerase-mediated DNA damage. Proc. Natl Acad. Sci. USA, 97, 4046–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Parras L., Hernandez,P., Martinez-Robles,M.L. and Schvartzman, J.B. (1992) Initiation of DNA replication in ColE1 plasmids containing multiple potential origins of replication. J. Biol. Chem., 267, 22496–22505. [PubMed] [Google Scholar]
- Martin-Parras L., Lucas,I., Martinez-Robles,M.L., Hernandez,P., Krimer,D.B., Hyrien,O. and Schvartzman,J.B. (1998) Topological complexity of different populations of pBR322 as visualized by two-dimensional agarose gel electrophoresis. Nucleic Acids Res., 26, 3424–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J. (1987) Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell, 48, 205–217. [DOI] [PubMed] [Google Scholar]
- Peng H. and Marians,K.J. (1993) Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Natl Acad. Sci. USA, 90, 8571–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permana P.A., Ferrer,C.A. and Snapka,R.M. (1994) Inverse relationship between catenation and superhelicity in newly replicated simian virus 40 daughter chromosomes. Biochem. Biophys. Res. Commun., 201, 1510–1517. [DOI] [PubMed] [Google Scholar]
- Peter B.J., Ullsperger,C., Hiasa,H., Marians,K.J. and Cozzarelli,N.R. (1998) The structure of supercoiled intermediates in DNA replication. Cell, 94, 819–827. [DOI] [PubMed] [Google Scholar]
- Postow L., Ullsperger,C., Keller,R.W., Bustamante,C., Vologodskii,A.V. and Cozzarelli,N.R. (2001) Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem., 276, 2790–2796. [DOI] [PubMed] [Google Scholar]
- Pouliot J.J., Yao,K.C., Robertson,C.A. and Nash,H.A. (1999) Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science, 286, 552–555. [DOI] [PubMed] [Google Scholar]
- Richter A., Strausfeld,U. and Knippers,R. (1987) Effects of VM26 (teniposide), a specific inhibitor of type VI DNA topoisomerase, on SV40 DNA replication in vivo. Nucleic Acids Res., 15, 3455–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybenkov V.V., Ullsperger,C., Vologodskii,A.V. and Cozzarelli,N.R. (1997a) Simplification of DNA topology below equilibrium values by type II topoisomerases. Science, 277, 690–693. [DOI] [PubMed] [Google Scholar]
- Rybenkov V.V., Vologodskii,A.V. and Cozzarelli,N.R. (1997b) The effect of ionic conditions on the conformations of supercoiled DNA. II. Equilibrium catenation. J. Mol. Biol., 267, 312–323. [DOI] [PubMed] [Google Scholar]
- Shamu C.E. and Murray,A.W. (1992) Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J. Cell Biol., 117, 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapka R.M. (1986) Topoisomerase inhibitors can selectively interfere with different stages of simian virus 40 DNA replication. Mol. Cell. Biol., 6, 4221–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapka R.M. (1996) The SV40 Replicon Model for Analysis of Anticancer Drugs. R.G.Landes Co., Austin, TX.
- Sogo J.M., Stasiak,A., Martinez-Robles,M.L., Krimer,D.B., Hernandez, P. and Schvartzman,J.B. (1999) Formation of knots in partially replicated DNA molecules. J. Mol. Biol., 286, 637–643. [DOI] [PubMed] [Google Scholar]
- Stivers J.T., Harris,T.K. and Mildvan,A.S. (1997) Vaccinia DNA topoisomerase I: evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry, 36, 5212–5222. [DOI] [PubMed] [Google Scholar]
- Strick T.R., Croquette,V. and Bensimon,D. (2000) Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature, 404, 901–904. [DOI] [PubMed] [Google Scholar]
- Sundin O. and Varshavsky,A. (1981) Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell, 25, 659–669. [DOI] [PubMed] [Google Scholar]
- Takasuga Y., Andoh,T., Yamashita,J. and Yagura,T. (1995) ICRF-193, an inhibitor of topoisomerase II, demonstrates that DNA replication in sperm nuclei reconstituted in Xenopus egg extracts does not require chromatin decondensation. Exp. Cell Res., 217, 378–384. [DOI] [PubMed] [Google Scholar]
- Tanabe K., Ikegami,Y., Ishida,R. and Andoh,T. (1991) Inhibition of topoisomerase II by antitumor agents bis(2,6-dioxopiperazine) derivatives. Cancer Res., 51, 4903–4908. [PubMed] [Google Scholar]
- Uemura T. and Yanagida,M. (1986) Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated mitosis. EMBO J., 5, 1013–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullsperger C.J., Vologodskii,A.V. and Cozzarelli,N.R. (1995) Unlinking of DNA by topoisomerases during DNA replication. In Lilley,D.M.J. and Eckstein,F. (eds), Nucleic Acids and Molecular Biology. Vol. 9. Springer-Verlag, Berlin, Germany, pp. 115–142.
- Walter J., Sun,L. and Newport,J. (1998) Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell, 1, 519–529. [DOI] [PubMed] [Google Scholar]
- Yang L., Wold,M.S., Li,J.J., Kelly,T.J. and Liu,L.F. (1987) Roles of DNA topoisomerases in simian virus 40 DNA replication in vitro. Proc. Natl Acad. Sci. USA, 84, 950–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich E.L. and Cozzarelli,N.R. (1995) Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev., 9, 2859–2869. [DOI] [PubMed] [Google Scholar]