Abstract
The repair of oxidative base lesions in DNA is a coordinated chain of reactions that includes removal of the damaged base, incision of the phosphodiester backbone at the abasic sugar residue, incorporation of an undamaged nucleotide and sealing of the DNA strand break. Although removal of a damaged base in mammalian cells is initiated primarily by a damage-specific DNA glycosylase, several lyases and DNA polymerases may contribute to the later stages of repair. DNA polymerase β (Pol β) was implicated recently as the major polymerase involved in repair of oxidative base lesions; however, the identity of the lyase participating in the repair of oxidative lesions is unclear. We studied the mechanism by which mammalian cell extracts process DNA substrates containing a single 8-oxoguanine or 5,6-dihydrouracil at a defined position. We find that, when repair synthesis proceeds through a Pol β-dependent single nucleotide replacement mechanism, the 5′-deoxyribosephosphate lyase activity of Pol β is essential for repair of both lesions.
Keywords: base excision repair/cell extracts/DNA polymerase β/NTH1/OGG1
Introduction
Reactive oxygen species are formed continuously as a consequence of normal cellular metabolism and are also generated by a number of external factors including UV and ionizing radiation (reviewed in Lindahl, 1993; Demple and Harrison, 1994; Wallace, 1997). The reaction of active oxygen species with DNA results in numerous forms of base damage. An increased level of oxidized DNA bases has been observed after treatment of cells with UV, ionizing radiation or chemical mutagens that generate oxygen radicals (reviewed in Dizdaroglu, 1992; Demple and Harrison, 1994). Formation of oxidative lesions can have profound consequences for genomic stability due to the effects that these lesions can have on polymerase fidelity and processivity. Some DNA oxidation products are mutagenic, promoting misincorporation of nucleotides by polymerases. For example, 8-oxoguanine, one of the most commonly occurring oxidative lesions, can form a stable Hoogsteen base pair with adenine and thereby induce transversion mutations (Shibutani et al., 1991). Other oxidized DNA bases, such as thymine glycol, effectively terminate DNA replication and transcription and are cytotoxic (Ide et al., 1985; Clark and Beardsley, 1987). It is therefore essential that oxidized bases be removed from DNA efficiently in order to maintain genome integrity.
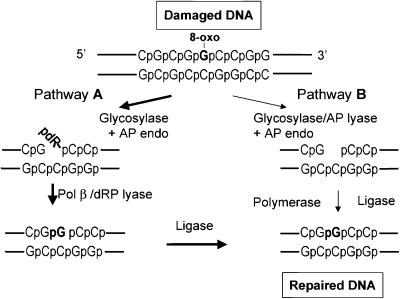
Base excision repair (BER) is the major repair system that contributes to the processing of oxidized purines and pyrimidines in mammalian cell extracts (Dianov et al., 1998, 2000). In mammalian cells, the two major DNA glycosylases implicated in BER of oxidative DNA damage are 8-oxoguanine-DNA glycosylase 1 (OGG1), which excises 8-oxoguanine and imidazole-ring opened purines, and NTH1 protein (the mammalian homologue of bacterial endonuclease III), which removes oxidized and fragmented pyrimidines (Wood et al., 2001). Glycosylases remove a damaged base through hydrolysis of the glycosylic bond linking the damaged base to the sugar, generating an abasic (AP) site (Lindahl and Wood, 1999). Normally, repair of an AP site proceeds through hydrolysis by AP endonuclease of the phosphodiester bond 5′ to the abasic site (Demple et al., 1991; Robson and Hickson, 1991; reviewed in Wilson and Barsky, 2001), followed by a coordinated reaction in which DNA polymerase β (Pol β) adds one nucleotide to the 3′ end of the incised AP site, simultaneously removing a 5′-sugar phosphate (5′-deoxyribosephosphate; dRP) residue by β-elimination (Matsumoto and Kim, 1995; Prasad et al., 1998a; Lindahl and Wood, 1999). Recently, human (Arai et al., 1997; Radicella et al., 1997; Roldan-Arjona et al., 1997) and mouse (Rosenquist et al., 1997) 8-oxoguanine-DNA glycosylases and human NTH1 (Aspinwall et al., 1997; Ikeda et al., 1998) have been cloned and characterized. Each demonstrated an AP lyase activity in addition to the glycosylase activity (Shinmura et al., 1997; Ikeda et al., 1998; Zharkov et al., 2000), suggesting a different pathway for repair of AP sites generated after removal of oxidative base lesions. In this scenario, the AP lyase activity of the glycosylase cleaves the phosphodiester bond 3′ to the AP site by catalysing a β-elimination reaction (Klungland et al., 1999). The subsequent action of APE1 generates a single nucleotide gap that may be filled by any polymerase because theoretically there is no need for the dRP lyase activity of Pol β at this reaction stage (Figure 1, pathway B). Although it is generally accepted that Pol β is involved in repair of 8-oxoguanine and oxidized pyrimidines in DNA (Dianov et al., 1998, 2000; Fortini et al., 1999; Klungland et al., 1999; Boiteux and Radicella, 2000; Nilsen and Krokan, 2001; Scharer and Jiricny, 2001), there is no evidence for the identity of the AP lyase activity involved. To identify the AP lyase activity involved in processing of oxidative DNA lesions, we characterized the repair in mammalian cell extracts of closed-circular DNA constructs containing a single 8-oxoguanine or dihydrouracil at a defined site. We find that the dRP lyase activity of Pol β is essential for its role in the repair of oxidative base lesions.
Fig. 1. Model for repair pathways involved in processing of 8-oxoguanine. Repair of 8-oxoguanine is initiated by 8-oxoguanine-DNA glycosylase; however, there are two possible scenarios for further steps. Processing through pathway A would involve cleavage of the AP site by AP endonuclease, removal of the 5′-dRP residue by dRP lyase activity of the Pol β, incorporation of one nucleotide by the same polymerase and ligation of the DNA ends broken during repair. Processing through pathway B would involve generation of a single nucleotide gap by the combined action of the AP lyase activity of DNA glycosylase and AP endonuclease. DNA polymerase then would fill the gap and finally DNA ligase would seal the DNA ends.
Results
Characterization of the substrates
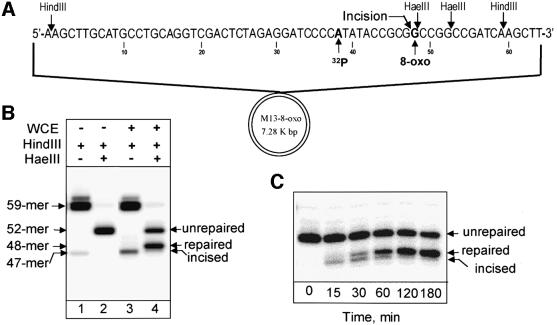
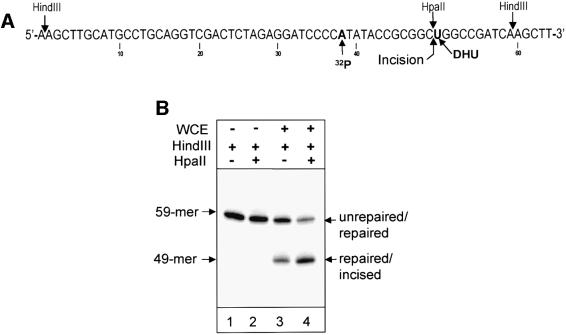
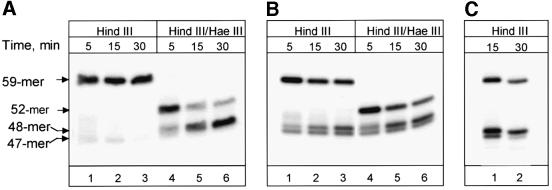
In this study, we used closed-circular double-stranded DNA substrates bearing a single 8-oxoguanine/cytosine or dihydrouracil/guanine base pair at a defined position (Figures 2A and 3A). The damage-containing strand of each substrate was 32P-labelled upstream of the damage site. HindIII cleavage of the DNA substrates released a 59mer labelled fragment containing the damage (Figures 2B, lane 1 and 3B, lane 1). In the case of the 8-oxoguanine-containing substrate, this fragment has two HaeIII restriction sites, but one of them is blocked by 8-oxoguanine and thus simultaneous cleavage with HaeIII and HindIII restriction endonucleases generated only a 52mer labelled product, containing 8-oxoguanine (Figure 2B, lane 2). Following repair of the 8-oxoguanine-containing substrate, the second HaeIII site will be restored and should give rise to a 48mer (repaired fragment). Similarly, in the dihydrouracil-containing substrate, the HpaII restriction site is blocked by dihydrouracil and thus simultaneous cleavage with HpaII and HindIII restriction endonucleases generated only a 59mer labelled product, containing dihydrouracil (Figure 3B, lane 2). However, once repair of the dihydrouracil-containing substrate has taken place, HindIII–HpaII cleavage will generate a 49mer fragment.
Fig. 2. Repair of 8-oxoguanine-containing substrate. (A) Schematic presentation of 8-oxoguanine-containing substrate. The sites of cleavage by the restriction enzymes HindIII and HaeIII and the position of the 32P label are shown. The site of incision by AP endonuclease subsequent to glycosylase processing of 8-oxoguanine (8-oxo) is indicated with an arrow. (B) Restriction digestion of untreated substrate DNA (lanes 1 and 2) and substrate incubated with 100 µg of human WCE at 37°C for 2 h (lanes 3 and 4). (C) Time-dependent repair of 8-oxoguanine by human WCE. Reactions contained 50 ng of plasmid DNA and 100 µg of WCE and were incubated at 37°C for the indicated time prior to isolation of the substrate DNA followed by digestion with HindIII and HaeIII. Reaction products were analysed by electrophoresis in a 10% denaturing polyacrylamide gel.
Fig. 3. Repair of dihydrouracil-containing substrate. (A) Schematic presentation of dihydrouracil-containing substrate. The sites of cleavage by the restriction enzymes HindIII and HpaII and the position of the 32P label are shown. The site of incision by AP endonuclease subsequent to glycosylase processing of dihydrouracil is indicated with an arrow. (B) Restriction digestion of untreated substrate DNA (lanes 1 and 2) and substrate incubated with 100 µg of human WCE at 37°C for 2 h (lanes 3 and 4).
When the 8-oxoguanine-containing substrate DNA was incubated with whole-cell extract (WCE) for 2 h and subsequently cleaved with HindIII after isolation from the reaction mixture, we observed majority release of a 59mer labelled product (containing both repaired and unrepaired DNA). Accumulation of 47 or 48mer fragments after HindIII cleavage of repaired DNA would indicate incomplete repair blocked after incision (47mer) or after addition of the first nucleotide during repair synthesis (48mer). Only a very small amount of 47mer repair incision intermediate was detected (Figure 2B, lane 3). However, simultaneous cleavage with HindIII and HaeIII of the extract-treated substrate generated approximately equal amounts of 52mer (unrepaired) and 48mer (repaired) products (Figure 2B, lane 4).
The kinetics of processing of 8-oxoguanine are shown in Figure 2C. Around 50% of the 8-oxoguanine lesions were repaired by human WCE within 2 h and repair proceeded to completion within 4–5 h. Repair of the dihydrouracil-containing substrate was noticeably faster, with ∼80% of dihydrouracil being removed within 2 h (Figure 3B, lane 4). Interestingly, only a minor amount of repair incision intermediates accumulate during repair of 8-oxoguanine, indicating that the sequential repair steps are highly coordinated (Figure 2C). In contrast, more repair intermediates accumulate during repair of dihydrouracil (Figure 3B, lane 3).
Repair of 8-oxoguanine by normal cell extracts
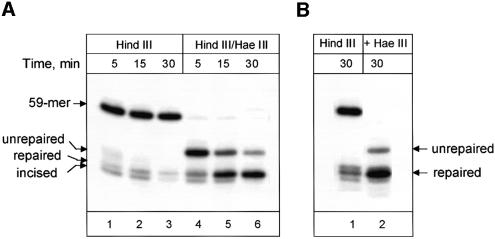
A combination of either APE1–Pol β or OGG1–APE1 may be involved in processing of the AP site, thus determining the repair pathway (Figure 1). To discriminate between these pathways, we used an 8-oxoguanine-containing substrate to develop reaction conditions under which only Pol β-dependent single nucleotide replacement repair was allowed, and then addressed the role of dRP lyase activity of Pol β under these conditions. Mouse WCEs, at concentrations similar to those used for human cell extracts, rapidly removed 8-oxoguanine from DNA. Repair was almost complete after only 30 min of incubation, as revealed by HindIII–HaeIII hydrolysis (Figure 4A, lanes 4–6). The Pol β dependence of the repair process was controlled by using the DNA synthesis inhibitor aphidicolin. Three major mammalian polymerases involved in DNA replication, α, δ and ε, are very sensitive to aphidicolin. In contrast, Pol β is only slightly sensitive to aphidicolin (Wang, 1996); thus, in the presence of aphidicolin, mainly Pol β-dependent repair will be monitored. Repair synthesis in mouse cell extracts was not affected by aphidicolin and, as revealed after HindIII cleavage of the DNA, very few repair intermediates were accumulated (Figure 4A, lanes 1–3). We next changed repair reaction conditions so that only Pol β-dependent short-patch repair was allowed [aphidicolin, dGTP and dideoxyribonucleotide triphosphates (ddNTPs)]. Under these conditions, any extension of the repair gap beyond one nucleotide will lead to incorporation of ddCMP (see Figure 1) and termination of both DNA synthesis and ligation, detectable by accumulation of incised, but unligated, products after HindIII hydrolysis. As in human cell extracts (Dianov et al., 1998; Fortini et al., 1999), repair in normal mouse cell extracts was accomplished mostly through insertion of a single nucleotide, as it was not blocked under these ‘short-patch repair’ conditions (Figure 4B).
Fig. 4. Repair of 8-oxoguanine by wild-type mouse cell extract. (A) Time-dependent repair of 8-oxoguanine in the presence of dNTPs. Reaction mixtures (50 µl) contained 50 ng of substrate DNA, 100 µg of mouse WCE, 100 µg/ml aphidicolin and 20 µM dNTPs and were incubated at 37°C for the indicated time periods. The substrate DNA was subsequently purified and then treated with either HindIII (lanes 1–3) or HindIII and HaeIII (lanes 4–6). (B) Repair is able to proceed to completion via single nucleotide insertion. The composition of reactions was altered such that dNTPs were replaced with 50 µM dGTP, ddCTP, ddATP and ddTTP. The reactions were incubated at 37°C for the indicated time periods. The substrate DNA subsequently was purified and then treated with either HindIII (lane 1) or HindIII and HaeIII (lane 2). Reaction products were analysed in a 10% denaturing polyacrylamide gel.
Repair of 8-oxoguanine by Pol β-deficient mouse cell extracts
If Pol β is an essential enzyme for single nucleotide BER of oxidative DNA lesions, then disruption of this gene should lead to the disruption of this pathway and engagement of other means for DNA repair. In the absence of aphidicolin, Pol β-deficient extracts were able to carry out repair (Figure 5A), most probably through a long-patch repair mechanism supported by an aphidicolin-sensitive DNA polymerase (Fortini et al., 2000). Indeed, in the presence of aphidicolin and normal dNTPs, WCE derived from Pol β-deficient mouse cells showed the time-dependent accumulation of repair intermediates (Figure 5B, lanes 1–3). Simultaneous digestion with HindIII and HaeIII confirmed that under these conditions, very little of the substrate had undergone full repair since approximately the same amount of the 48mer fragment accumulated after HindIII cleavage alone (Figure 5B, 48mer at lanes 3 and 6). When reactions were carried out under conditions that completely block the long-patch pathway (aphidicolin, dGTP and the rest ddNTPs), repair was stalled within 15 min after incision and insertion of the first nucleotide (Figure 5C, lane 1) and, after 30 min of incubation in cell extract, most of the substrate had undergone further degradation (Figure 5C, lane 2). These data demonstrate that, although there is some residual aphidicolin-resistant repair synthesis in the absence of Pol β, under these conditions Pol β-deficient WCEs are unable to support efficient BER via the characteristic single nucleotide insertion pathway.
Fig. 5. Pol β-null mouse cell extracts are unable to complete single nucleotide patch repair. (A) Time-dependent repair of 8-oxoguanine in the presence of dNTPs. Reaction mixtures (50 µl) contained 50 ng of substrate DNA, 100 µg of Pol β-deficient mouse WCE and 20 µM dNTPs and were incubated at 37°C for the indicated time periods. The substrate DNAs subsequently were purified and then treated with either HindIII (lanes 1–3) or HindIII and HaeIII (lanes 4–6). (B) Repair of 8-oxoguanine by Pol β-deficient cell extracts is blocked by aphidicolin. Reaction mixtures (50 µl) contained 20 ng of substrate DNA, 100 µg of mouse pol β–/– WCE, 100 µg/ml aphidicolin and 20 µM dNTPs and were incubated at 37°C for the indicated time periods. The substrate DNA subsequently was purified and then treated with either HindIII (lanes 1–3) or HindIII and HaeIII (lanes 4–6). (C) Single nucleotide repair synthesis. Reactions were carried out as in (B) with the exception of the dNTPs being replaced with dGTP/ddCTP/ddATP/ddTTP. The reactions were incubated at 37°C for the indicated time periods. The substrate DNA subsequently was purified and then treated with either HindIII (lane 1) or HindIII and HaeIII (lane 2). Reaction products were analysed in a 10% denaturing polyacrylamide gel.
Complementation of Pol β-deficient mouse cell extracts: essential role of Pol β dRP lyase activity
As we mentioned above, a combination of either APE1–Pol β or glycosylase–APE1 may be involved in processing of the AP site generated after removal of an oxidative base lesion (Figure 1). We hypothesized that if the removal of the AP site is initiated by the AP lyase activity of the glycosylase, then the inability of Pol β-deficient cell extracts to carry out short-patch repair could be complemented by addition of a mutant of Pol β, proficient in DNA synthesis but deficient in dRP lyase activity. Such a polymerase should be able efficiently to fill the gap generated by sequential action of AP lyase and APE1 (Figure 1). To test this hypothesis, we carried out repair reactions under conditions where only Pol β-dependent short-patch repair was allowed (dGTP and the rest ddNTPs, in the presence of aphidicolin) and complemented Pol β-deficient extracts with purified, recombinant Pol β or Pol β K72A, a dRP lyase-deficient mutant (Prasad et al., 1998b). We analysed the efficiency of repair by monitoring the accumulation of repair intermediates after HindIII cleavage of substrate DNA repaired in a Pol β-deficient mouse cell extract. Under these conditions, as described in detail above (Figure 5C), we detected accumulation of a 48mer intermediate in Pol β-deficient cell extracts but the ligation step was blocked (Figure 6A, lane 1). When the extract was complemented with Pol β K72A, we observed addition of a second nucleotide to the 3′ end of the incised AP site (49mer). However, because the second nucleotide was ddCMP, further repair was blocked at this stage (Figure 6A, lane 3). These data indicated that Pol β K72A was shifting repair to the long-patch pathway and was not able to stimulate repair via a short-patch mechanism. However, the addition of Pol β completely restored repair capability to Pol β-deficient extracts (Figure 6A, lane 2). Removal of 8-oxoguanine from substrate DNA in the reaction complemented with Pol β was confirmed by simultaneous HindIII–HaeIII cleavage (Figure 6A, lane 4).
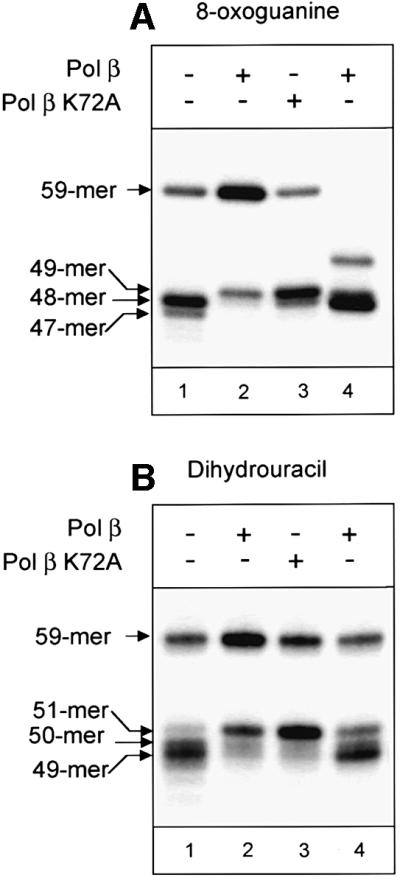
Fig. 6. Complementation of Pol β-deficient cell extracts with Pol β and Pol β K72A mutant. Pol β-deficient mouse WCEs (100 µg) were incubated with 50 ng of DNA substrate and 50 µM dGTP/ddCTP/ddATP/ddTTP in the case of 8-oxoguanine-containing substrate (A) and 50 µM dCTP/ddGTP/ddATP/ddTTP in the case of dihydrouracil-containing substrate (B) in the presence of aphidicolin (100 µg/ml). Pol β and Pol β K72A (2 ng) were included in the reactions shown in lanes 2 and 3, respectively. The substrate DNA was purified and then treated with HindIII. To demonstrate complete restoration of repair after addition of Pol β, a sample identical to that in lane 2 was cleaved simultaneously with HindIII and HaeIII (A, lane 4) or with HindIII and HpaII (B, lane 4). Reaction products were analysed in a 10% denaturing polyacrylamide gel.
In a similar experiment with dihydrouracil-containing substrate, we also observed failure to carry out complete short-patch repair by Pol β-deficient cell extract. Again, repair activity could be restored by complementation with Pol β but not by dRP lyase-deficient Pol β K72A (Figure 6B). The requirement for the dRP lyase function of Pol β in the complementation reactions indicates that dRP is an intermediate product during repair of both 8-oxoguanine and dihydrouracil, substrates for the bifunctional glycosylases OGG1 and NTH1, respectively. We therefore conclude that processing of the AP sites generated after removal of these lesions is initiated by APE1, directing the repair process through pathway A (Figure 1).
Repair of 8-oxoguanine-containing substrate in a reconstituted system
The presence of a 5′-dRP incision intermediate and the essential role of the dRP lyase activity of Pol β were confirmed further by experiments with purified enzymes. The reconstitution of repair of the 8-oxoguanine-containing substrate was performed with mOGG1, APE1, Pol β and DNA ligase I. Substrate DNA was incubated with the indicated enzymes and then treated with the restriction endonuclease HindIII to release a 59mer labelled fragment from the substrate DNA. Incubation of the substrate DNA with mOGG1 and APE1 generated a 47mer incision product (Figure 7, lane 2). Pol β added one nucleotide to the incised DNA (Figure 7, lane 3) and finally DNA ligase sealed the ends to restore the 59mer repaired product (Figure 7, lane 4). Removal of 8-oxoguanine in the complete reaction was confirmed by simultaneous HindIII–HaeIII cleavage (Figure 7, lane 5). However, although Pol β K72A (dRP lyase mutant) in a similar reaction was able to add one nucleotide to the incised substrate DNA (Figure 7, lane 6), DNA ligase was not able to seal the ends (Figure 6, lane 7). These data demonstrate the essential role of the dRP lyase activity of Pol β in removal of the dRP residue prior to ligation, indicating the presence of the dRP residue at the 5′ end of the incised substrate and thus confirming bypass of the AP lyase activity of OGG1. We therefore conclude that, in agreement with our experiments performed with cell extracts, the reconstituted repair of 8-oxoguanine proceeds through incision of the AP site by APE1 (Figure 1, pathway A).
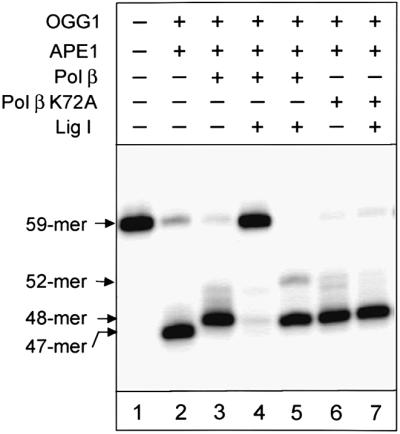
Fig. 7. Repair of 8-oxoguanine reconstituted with purified enzymes. mOGG1 (1 ng, 25 fmol), APE 1 (2 ng, 58 fmol), Pol β or Pol β K72A (0.5 ng, 12.5 fmol) and DNA ligase I (10 ng, 80 fmol) were mixed and incubated for 5 min on ice. The buffer containing magnesium, dNTPs and 50 ng of substrate DNA was then added and incubation was carried out for 20 min at 37°C. The substrate DNA was purified as described in Materials and methods, treated with HindIII and analysed by electrophoresis in a 10% denaturing polyacrylamide gel. The sample in lane 5 was digested with HaeIII in addition to HindIII.
Discussion
Pol β is a major repair DNA polymerase involved in both short- and long-patch repair of AP sites generated by monofunctional glycosylases (Wiebauer and Jiricny, 1990; Dianov et al., 1992, 1999; Singhal et al., 1995; Sobol et al., 1996; Podlutsky et al., 2001). The role of this enzyme in repair of G/T mismatches and single strand breaks has also been well documented (Wiebauer and Jiricny, 1990; Whitehouse et al., 2001). However, there is some controversy about the role of Pol β and its dRP lyase activity in the repair of oxidative lesions. In eukaryotes, the 8-oxoguanine-DNA glycosylase OGG1 and NTH1 proteins are central players in the repair of oxidative base lesions (Lindahl and Wood, 1999). In addition to their glycosylase activity, each has also been shown to catalyse a β-elimination (lyase) reaction, incising the phosphodiester backbone 3′ to the abasic site (McCullough et al., 1999). The resultant 3′-unsaturated aldehyde terminus can be removed by APE1 (Klungland, 1999), leaving a one-nucleotide gap with a 3′-hydroxyl and a 5′-phosphate, although the 3′-phosphodiesterase activity of APE1 is significantly (∼100-fold) weaker than its AP endonuclease activity (Wilson and Barsky, 2001). Theoretically, repair could then proceed simply through insertion of a single nucleotide followed by ligation. This would then imply that there were two different pathways for single nucleotide patch repair, one dRP lyase-dependent pathway initiated by monofunctional glycosylases (Figure 1, pathway A) and another dRP lyase-independent pathway initiated by bifunctional glycosylases (Figure 1, pathway B). However, there has been a recent accumulation of evidence demonstrating the stimulation of hOGG1 (Hill et al., 2001; Saitoh et al., 2001; Vidal et al., 2001) activity by AP endonuclease and it has been reported that this stimulation of glycosylase activity may occur at the expense of AP lyase activity (Vidal et al., 2001). We therefore set out to address the question of whether Pol β/dRP lyase is involved in processing of the AP site arising after removal of either 8-oxoguanine or dihydrouracil, or whether the OGG1/NTH1 AP lyase activity entails a different mechanism for bifunctional glycosylases in which the dRP lyase activity of Pol β is redundant.
We constructed 32P-labelled closed-circular plasmid substrates, containing either a single site-specific 8-oxoguanine or dihydrouracil residue, which subsequently were treated with mammalian WCEs in the presence of the relevant cofactors. Restriction analysis of the substrates allowed us to follow the progress of the repair reactions. Addition of aphidicolin and limitation of the pool of dNTPs available for strand resynthesis to the first nucleotide alone, with the remaining nucleotides present as dideoxyribonucleoside triphosphates, prevented completion of repair through any pathway other than Pol β-dependent single nucleotide patch repair. Our experiments showed that wild-type mouse WCE is capable of carrying out full BER of 8-oxoguanine and dihydrouracil under such conditions, with only a trace amount of product corresponding to insertion of more than one nucleotide detectable. We therefore concluded that under these conditions, the majority of oxidative base lesion repair in normal WCEs proceeds through the single nucleotide insertion mechanism and is supported by Pol β. This result is in good accordance with previously reported experiments where restriction analysis of repair incorporation was performed (Dianov et al., 1998; Fortini et al., 1999). In contrast, repair in extracts derived from Pol β knockout mouse cells was inhibited substantially in the presence of aphidicolin, even under conditions where a complete pool of dNTPs was provided (Figure 5). However, in Pol β-deficient cell extracts, we always observed some residual DNA polymerase activity even in the presence of aphidicolin, suggesting that in the absence of Pol β other DNA polymerases may become involved in repair synthesis. The repair defect in Pol β-deficient cells is evident: they are highly sensitive to hydrogen peroxide (Fortini et al., 2000) and alkylating agents (Sobol et al., 1996) and have increased rates of chromosomal abnormalities (Ochs et al., 1999). Although our data demonstrate that the short-patch BER pathway does not operate in Pol β-null cells, the viability of these cells indicates that some alternative DNA repair pathways (most probably long-patch BER; Fortini et al., 2000; G.L.Dianov, unpublished data) must operate. Nevertheless, since the knockout mice are not viable (Gu et al., 1994; Sugo et al., 2000), it is likely that the efficiency or quality of this backup repair is insufficient to maintain genome integrity during development of a multicellular organism. However, as we demonstrate, in cell extracts this repair defect can be complemented by wild-type Pol β and these cells may therefore be used for complementation studies.
In order to establish which of either the AP lyase activity of OGG1/NTH1 or the dRP lyase activity of Pol β was responsible for supporting single nucleotide patch repair, we complemented Pol β-deficient extract with either Pol β or Pol β K72A mutant deficient in dRP lyase activity. Whereas Pol β was able to restore repair capability to the extract, the mutant enzyme, which is unable to catalyse β-elimination of a 5′-dRP residue (Prasad et al., 1998b), did not. It therefore seems reasonable to conclude that dRP remains as a 5′-block, with the implication that, under our experimental conditions, BER of 8-oxoguanine and dihydrouracil proceeds via the same fundamental mechanism as repair initiated by monofunctional glycosylases (Figure 1, pathway A). This conclusion is also strongly supported by our data obtained in a reconstituted repair reaction.
What is the role of a glycosylase’s AP lyase activity? It has been proposed that BER proceeds via a highly orchestrated mechanism in which the enzyme involved at one step directly communicates with the enzyme of the next (Lindahl and Wood, 1999; Mol et al., 2000; Wilson and Kunkel, 2000). The possibility exists that the covalent interaction between bifunctional DNA glycosylase and the DNA substrate, which necessarily precedes lyase activity, is required for some other function such as ensuring that abasic sites are not left unprotected during BER.
It has also been hypothesized that the AP lyase pathway for repair of oxidative lesions may play a role in prevention of double-strand break formation during repair of clustered lesions induced by ionizing radiation (Klungland et al., 1999). Since the mechanism for repair of clustered oxidative lesions may be distinct from that of repair of single base lesions (Dianov et al., 2001), the AP lyase activity of OGG1 may play a role in repair of such lesions.
In conclusion, our data suggest that BER involved in repair of simple base lesions has one major pathway strictly depending on the dRP lyase activity of Pol β. As has been shown before, elimination of the dRP lyase activity of Pol β, or Pol β itself, leads to increased sensitivity to DNA-damaging agents (Sobol et al., 2000), genetic instability (Ochs et al., 1999) and premature termination of embryonic development (Gu et al., 1994; Sugo et al., 2000).
Materials and methods
Materials
Synthetic oligodeoxyribonucleotides purified by high-performance liquid chromatography were obtained from Midland (8-oxoguanine) or Synthegen (5,6-dihydrouracil). [γ-32P]ATP (3000 Ci/mmol) was purchased from NEN Life Science Products.
Proteins
Recombinant human wild-type Pol β and K72A mutant deficient in dRP lyase activity were kindly provided by Drs S.Wilson and R.Prasad. Mouse 8-oxoguanine-DNA glycosylase (mOGG1) was a gift from Dr D.Zharkov, and human DNA ligase I was a gift from Dr A.Tomkinson. Histidine-tagged human APE1 was purified on Ni2+-charged His-Bind resin (Novagen, Cambridge, MA) as recommended by the manufacturer.
DNA substrates
The oligonucleotides 5′-ATATACCGCG[8-oxo]GCCGATCAAGCTTATT-3′ (30 pmol) and 5′-ATATACCGCGGCUGATCAAGCTTATT-3′ (where U stands for dihydrouracil, 30 pmol) were 5′-end labelled with 100 µCi (33 pmol) of [γ-32P]ATP and used for construction of substrates containing single 8-oxoguanine or dihydrouracil in closed-circular double-stranded DNA as previously described (Dianov et al., 1998).
BER reactions
The BER reactions were carried out in a reaction mixture (50 µl) containing 50 mM HEPES–KOH pH 7.8, 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 1.5 mM dithiothreitol (DTT), 2 mM ATP, 0.4 mg/ml bovine serum albumin (BSA), 25 mM phosphocreatine (di-Tris salt, Sigma), 2.5 µg creatine phosphokinase (type I, Sigma), 8.5% glycerol (Fluka), 20 µM each of the indicated dNTPs or ddNTPs, 50 ng (10 fmol) of 32P-labelled single 8-oxoguanine-containing DNA substrate and 400 ng of carrier plasmid DNA (pUC18). When aphidicolin (Sigma) was added to a final concentration of 100 µg/ml, the buffer also contained 1% dimethylsulfoxide (DMSO). Reactions were initiated by the addition of WCEs (100 µg) and incubated for the indicated time at 37°C. The reactions were stopped by addition of 2 µl of 0.5 M EDTA. Substrate DNA was purified from the reaction mixture by phenol–chloroform extraction and ethanol precipitation, and treated with 10–40 U of the indicated restriction endonuclease(s) for 2 h at 37°C in buffers supplied by the manufacturer. An equal volume of gel loading buffer was then added (95% formamide, 20 mM EDTA, 0.02% bromophenol blue and 0.02% xylene cyanol). Following incubation at 90°C for 2–5 min, the reaction products were separated by electrophoresis in a 10% polyacrylamide gel containing 7 M urea in 89 mM Tris–HCl, 89 mM boric acid and 2 mM EDTA pH 8.0.
Reconstituted BER
The BER was reconstituted with purified enzymes in a reaction mixture (20 µl) that contained 45 mM HEPES pH 7.8, 70 mM KCl, 7.5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 2 mM ATP, 20 µM each of dATP, dGTP, dCTP and dTTP, 0.5 mg/ml BSA and a 32P-labelled single 8-oxoguanine-containing substrate DNA (∼50 ng, 10 fmol). The reaction was initiated by adding BER proteins at the amount indicated in the figure legends. After incubation for the indicated time at 37°C, the reaction was stopped by addition of 20 µl of phenol–chloroform. Following extraction, the aqueous phase was collected and passed through a G-25 spin column. The samples were evaporated and the pellets were dissolved in 10 µl of a HindIII buffer supplied by the manufacturer. DNA was treated with 40 U of the indicated restriction endonuclease(s) for 1 h at 37°C. Reactions were stopped by addition of 10 µl of gel loading buffer (95% formamide, 20 mM EDTA, 0.02% bromophenol blue and 0.02% xylene cyanol). Following incubation at 80°C for 2 min, the reaction products were separated by electrophoresis in a 10% denaturing polyacrylamide gel.
Cells and extracts
Normal human lymphoid cells AG09387 were obtained from the Human Genetic Mutant Cell Repository (Coriell Institute, Camden, NJ). Cells were grown in medium recommended by the Repository. The normal mouse fibroblast and DNA Pol β knockout mouse fibroblast cell lines were obtained from Dr R.Sobol and were grown as described (Sobol et al., 1996). WCEs were prepared by the method of Manley et al. (1980) and dialysed overnight against buffer containing 25 mM HEPES–KOH pH 7.9, 2 mM DTT, 12 mM MgCl2, 0.1 mM EDTA, 17% glycerol and 0.1 M KCl. Extracts were aliquoted and stored at –80°C.
All experiments were repeated at least 3–5 times and representative gels are shown.
Acknowledgments
Acknowledgements
We thank Drs Samuel Wilson, Rajendra Prasad, Robert Sobol, Dmitry Zharkov and Alan Tomkinson for providing reagent proteins, expression constructs and cell lines. Dr.Yoshihiro Matsumoto and Helen Budworth are thanked for critical reading of the manuscript.
References
- Arai K., Morishita,K., Shinmura,K., Kohno,T., Kim,S.-R., Nohmi,T., Taniwaki,S., Ohwada,S. and Yokota,J. (1997) Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene, 14, 2857–2861. [DOI] [PubMed] [Google Scholar]
- Aspinwall R. et al. (1997) Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl Acad. Sci. USA, 94, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S. and Radicella,J.P. (2000) The human OGG1 gene: structure, functions and its implication in the process of carcinogenesis. Arch. Biochem. Biophys., 377, 1–8. [DOI] [PubMed] [Google Scholar]
- Clark J.M. and Beardsley,G.P. (1987) Functional effects of cis-thymine glycol lesions on DNA synthesis in vitro. Biochemistry, 26, 5398–5403. [DOI] [PubMed] [Google Scholar]
- Demple B. and Harrison,L. (1994) Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem., 63, 915–948. [DOI] [PubMed] [Google Scholar]
- Demple B., Herman,T. and Chem,D.S. (1991) Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl Acad. Sci. USA, 88, 11450–11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov G., Price,A. and Lindahl,T. (1992) Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol. Cell. Biol., 12, 1605–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov G., Bischoff,C., Piotrowski,J. and Bohr,V.A. (1998) Repair pathways for processing of 8-oxoguanine in DNA by mammalian cell extracts. J. Biol. Chem., 273, 33811–33816. [DOI] [PubMed] [Google Scholar]
- Dianov G.L., Prasad,R., Wilson,S.H. and Bohr,V.A. (1999) Role of DNA polymerase β in the excision step of long patch mammalian base excision repair. J. Biol. Chem., 274, 13741–13743. [DOI] [PubMed] [Google Scholar]
- Dianov G.L., Thybo,T., Dianova,I.I, Lipinski,L.J. and Bohr,V.A. (2000) Single nucleotide patch base excision repair is the major pathway for removal of thymine glycol from DNA in human cell extracts. J. Biol. Chem., 275, 11809–11813. [DOI] [PubMed] [Google Scholar]
- Dianov G.L., O’Neill,P. and Goodhead,D.T. (2001) Securing genome stability by orchestrating DNA repair: removal of radiation-induced clustered lesions in DNA. BioEssays, 23, 745–749. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M. (1992) Oxidative damage to DNA in mammalian chromatin. Mutat. Res., 275, 331–342. [DOI] [PubMed] [Google Scholar]
- Fortini P., Parlanti,E., Sidorkina,O.M., Laval,J. and Dogliotti,E. (1999) The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J. Biol. Chem., 274, 15230–15236. [DOI] [PubMed] [Google Scholar]
- Fortini P., Pascucci,B., Belisario,F. and Dogliotti,E. (2000) DNA polymerase β is required for efficient DNA strand break repair induced by methyl methanesulfonate but not by hydrogen peroxide. Nucleic Acids Res., 28, 3040–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Marth,J.D., Orban,P.C., Mossman,H. and Rajewsky,K. (1994) Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science, 265, 103–106. [DOI] [PubMed] [Google Scholar]
- Hill J.W., Hazra,T.K., Izumi,T. and Mitra,S. (2001) Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res., 29, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide H., Kow,Y.W. and Wallace,S.S. (1985) Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res., 13, 8035–8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Biswas,T., Roy,R., Izumi,T., Boldogh,I., Kurosky,A., Sarker,A.H., Seki,S. and Mitra,S. (1998) Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III—direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem., 273, 21585–21593. [DOI] [PubMed] [Google Scholar]
- Klungland A., Hoss,M., Gunz,D., Constantinou,A., Clarkson,S.G., Doetsch,P.W., Bolton,P.H., Wood,R.D. and Lindahl,T. (1999) Base excision repair of oxidative DNA damage activated by XPG protein. Mol. Cell, 3, 33–42. [DOI] [PubMed] [Google Scholar]
- Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- Manley J.L., Fire,A., Cano,A., Sharp,P.A. and Gefter,M.L. (1980) DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl Acad. Sci. USA, 77, 3855–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y. and Kim,K. (1995) Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science, 269, 699–702. [DOI] [PubMed] [Google Scholar]
- McCullough A.K., Dodson,M.L. and Lloyd,R.S. (1999) Initiation of base excision repair: glycosylase mechanism and structure. Annu. Rev. Biochem., 68, 255–285. [DOI] [PubMed] [Google Scholar]
- Mol C.D., Izumi,T., Mitra,S. and Tainer,J.A. (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature, 403, 451–456. [DOI] [PubMed] [Google Scholar]
- Nilsen H. and Krokan,H.E. (2001) Base excision repair in a network of defence and tolerance. Carcinogenesis, 22, 987–998. [DOI] [PubMed] [Google Scholar]
- Ochs K., Sobol,R.W., Wilson,S.H. and Kaina,B. (1999) Cells deficient in DNA polymerase β are hypersensitive to alkylating agent-induced apoptosis and chromosomal breakage. Cancer Res., 59, 1544–1551. [PubMed] [Google Scholar]
- Podlutsky A.J., Dianova,I., Podust,V.N., Bohr,V.A. and Dianov,G. (2001) Human DNA polymerase β initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. EMBO J., 20, 1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., Beard,W., Strauss,P. and Wilson,S. (1998a) Human DNA polymerase β deoxyribose phosphate lyase. J. Biol. Chem., 273, 15263–15270. [DOI] [PubMed] [Google Scholar]
- Prasad R., Beard,W.A., Chyan,J.Y., Maciejewski,M.W., Mullen,G.P. and Wilson,S.H. (1998b) Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase β as revealed by site-directed mutagenesis—DNA binding and 5′-deoxyribose phosphate lyase activities. J. Biol. Chem., 273, 11121–11126. [DOI] [PubMed] [Google Scholar]
- Radicella J.P., Dherin,C., Desmaze,C., Fox,M.S. and Boiteux,S. (1997) Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 8010–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson C.N. and Hickson,I.D. (1991) Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in Escherichia coli xth (exonuclease III) mutants. Nucleic Acids Res., 19, 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan-Arjona T., Wei,Y.F., Carter,K.C., Klungland,A., Anselmino,C., Wang,R.P., Augustus,M. and Lindahl,T. (1997) Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl Acad. Sci. USA, 94, 8016–8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist T.A., Zharkov,D.O. and Grollman,A.P. (1997) Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl Acad. Sci. USA, 94, 7429–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Shinmura,K., Yamaguchi,S., Tani,M., Seki,S., Murakami,H., Nojima,Y. and Yokota,J. (2001) Enhancement of OGG1 protein AP lyase activity by increase of APEX protein. Mutat. Res., 486, 31–40. [DOI] [PubMed] [Google Scholar]
- Scharer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. BioEssays, 23, 270–281. [DOI] [PubMed] [Google Scholar]
- Shibutani S., Takeshita,M. and Grollman,A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434. [DOI] [PubMed] [Google Scholar]
- Shinmura K., Kasai,H., Sasaki,A., Sugimura,H. and Yokota,J. (1997) 8-Hydroxyguanine (7,8-dihydro-8-oxoguanine) DNA glycosylase and AP lyase activities of hOGG1 protein and their substrate specificity. Mutat. Res., 385, 75–82. [DOI] [PubMed] [Google Scholar]
- Singhal R.K., Prasad,R. and Wilson,S.H. (1995) DNA polymerase β conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J. Biol. Chem., 270, 949–957. [DOI] [PubMed] [Google Scholar]
- Sobol R.W., Horton,J.K., Kuhn,R., Gu,H., Singhal,R.K., Prasad,R., Rajewsky,K. and Wilson,S.H. (1996) Requirement of mammalian DNA polymerase-β in base-excision repair. Nature, 379, 183–186. [DOI] [PubMed] [Google Scholar]
- Sobol R.W., Prasad,R., Evenski,A., Baker,A., Yang,X.P., Horton,J.K. and Wilson,S.H. (2000) The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature, 405, 807–810. [DOI] [PubMed] [Google Scholar]
- Sugo N., Aratani,Y., Nagashima,Y., Kubota,Y. and Koyama,H. (2000) Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase β. EMBO J., 19, 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal A.E., Hickson,I.D., Boiteux,S. and Radicella,J.P. (2001) Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res., 29, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace S.S. (1997) Oxidative damage to DNA and its repair. In Scandalios,J.G. (ed.), Oxidative Stress and the Molecular Biology of Antioxidant Defences. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 49–90.
- Wang T.S.-F. (1996) Cellular DNA polymerases. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 461–463.
- Whitehouse C.J., Taylor,R.M., Thistlethwaite,A., Zhang,H., Karimi-Busheri,F., Lasko,D.D., Weinfeld,M. and Caldecott,K.W. (2001) XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell, 104, 107–117. [DOI] [PubMed] [Google Scholar]
- Wiebauer K. and Jiricny,J. (1990) Mismatch-specific thymine DNA glycosylase and DNA polymerase β mediate the correction of G–T mispairs in nuclear extracts from human cells. Proc. Natl Acad. Sci. USA, 87, 5842–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.M. III, and Barsky,D. (2001) The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat. Res., 485, 283–307. [DOI] [PubMed] [Google Scholar]
- Wilson S.H. and Kunkel,T.A. (2000) Passing the baton in base excision repair. Nature Struct. Biol., 7, 176–178. [DOI] [PubMed] [Google Scholar]
- Wood R.D., Mitchell,M., Sgouros,J. and Lindahl,T. (2001) Human DNA repair genes. Science, 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- Zharkov D.O., Rosenquist,T.A., Gerchman,S.E. and Grollman,A.P. (2000) Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem., 275, 28607–28617. [DOI] [PubMed] [Google Scholar]