Abstract
Signal transducers and activators of transcription (STATs) play a central role in cytokine signaling. Activating and repressing gene transcription is a dynamic process involving chromatin remodeling by histone acetylases and deacetylases, yet the role of this process in STAT-dependent transcription remains largely unknown. In a search for STAT5-interacting proteins by yeast two-hybrid screening, we identified the nuclear receptor co-repressor SMRT (silencing mediator for retinoic acid receptor and thyroid hormone receptor) as a potential STAT5-binding partner. SMRT binds to both STAT5A and 5B, and strongly repressed STAT5-dependent transcription in vitro. SMRT binds to the N-terminal coiled-coil domain of STAT5 and a mutation within this region previously found to render STAT5 hyperactive in response to cytokines abolished the interaction with SMRT. Overexpression of SMRT suppressed the induction of STAT5 target genes by interleukin-3, whereas the histone deacetylase inhibitor trichostatin A effectively enhanced and prolonged their expression. Together, these findings illuminate the potential role of SMRT in down-regulating STAT5 activity, with a consequent reduction of STAT5 target gene expression.
Keywords: co-repressor/cytokine/SMRT/STAT/transcription
Introduction
Cytokines regulate a variety of biological responses through their interaction with their cognate receptors, which belong to a cytokine receptor superfamily. The ligand binding to the receptor activates receptor-associated Janus kinase (Jak) and initiates an intracellular signaling cascade (Ihle et al., 1994). Activated Jaks phosphorylate tyrosine residues on the cytoplasmic domain of the receptors, which then serve as docking sites for the Src-homology 2 (SH2) domains of a variety of signaling molecules. Common substrates of Jak tyrosine phosphorylation include one or more members of the signal transducers and activators of transcription (STATs; Darnell et al., 1994; Darnell, 1997). Among the seven mammalian family members, the two highly related STAT5 gene products STAT5A and STAT5B have been of particular interest because of the broad spectrum of cytokines that induce their activation. These include interleukin (IL)-2, IL-3, IL-5, IL-7, erythropoietin (Epo), thrombopoietin (Tpo), granulocyte–macrophage colony-stimulating factor (GM-CSF), prolactin and growth hormone (Ihle, 1996; Darnell, 1997; Leonard and O’Shea, 1998).
STAT5 is activated by the phosphorylation of a critical tyrosine residue near its C-terminus by Jaks. This allows STAT5 to form homo- or hetero-dimers by intermolecular interaction involving the phosphotyrosine and SH2 domain. Dimerized STATs translocate to the nucleus by an unknown mechanism and activate transcription of target genes by binding to a consensus DNA sequence (TTCNNNGAA) on their promoters (Ihle, 1996).
Remodeling chromatin structure by histone acetylation is one of the critical processes in transcriptional regulation. A number of transcriptional co-activators including CREB-binding protein (CBP)/p300, p/CAF and p/CIP, which possess histone acetyltransferase (HAT) activity, have been shown to associate with various transcription factors and act as general integrators of the transcription machinery (Torchia et al., 1997; Korzus et al., 1998). On the other hand, transcription is negatively regulated by co-repressor complexes comprising SMRT (silencing mediator for retinoid and thyroid hormone receptors)/N-CoR (nuclear receptor co-repressor), mSin3A/B, c-Ski and histone deacetylases (Heinzel et al., 1997; Nagy et al., 1997; Nomura et al., 1999). SMRT and N-CoR were first cloned as a co-repressor protein bound to unliganded retinoic acid receptor or thyroid hormone receptor (Chen and Evans, 1995; Horlein et al., 1995). Recent studies have revealed that SMRT/N-CoR interacts not only with nuclear receptors, but also with other transcription factors or nuclear proteins such as Bcl-6 (Dhordain et al., 1997; Huynh and Bardwell, 1998), the homeodomain proteins Rpx2 and Pit-1 (Xu et al., 1998), and leukemic fusion protein partners PLZF and ETO (Lin et al., 1998; Wong and Privalsky, 1998). STATs have been shown to bind to CBP/p300 co-activator through its C-terminal trans- activation domain (Gingras et al., 1999; Paulson et al., 1999) and the role of p/CIP has also been suggested for STAT1 function (Torchia et al., 1997). However, the role of co-repressors in the function of STATs remains unclear.
Here we report on the functional interaction between STAT5 and SMRT. SMRT interacts with the N-terminal coiled-coil region of STAT5, which is distinct from the C-terminal activation domain and represses STAT5-dependent transcription. This suggests that SMRT modulates STAT5 function independently of co-activators that interact with the activation domain, and proposes a novel concept that transcription of STAT5 could be regulated through a balance of co-activators and co-repressors.
Result
Identification of SMRT as a potential STAT5-interacting protein
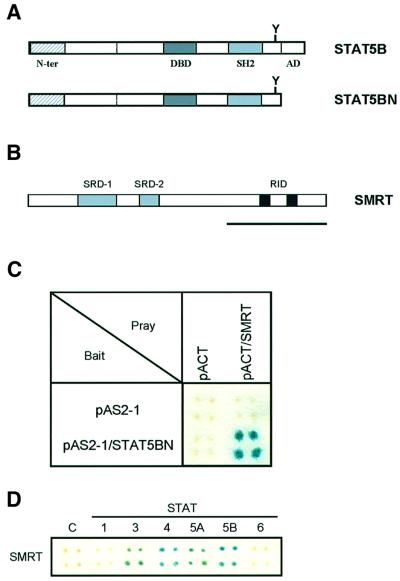
In order to clarify further the role of STAT5 in cytokine signaling, we set out to identify proteins that interact with STAT5 by using a yeast two-hybrid system. Since the C-terminus of STAT5 contains a strong trans-activation domain and shows high background activity in yeast, we used carboxyl-truncated STAT5B as bait (Figure 1A; STAT5BN). We screened 1 × 107 clones from a mouse T-cell lymphoma library. One positive clone encoded a 900 bp fragment that showed very high homology to the C-terminal receptor-interacting domain of human SMRT (Figure 1B, bold line, and C). We investigated further the interaction of SMRT with other STATs using the two-hybrid assay (Figure 1D). Interestingly, SMRT also associated with STAT3, 4 and 5A, but not with STAT1 and 6, demonstrating that SMRT binds specifically to a subset of STAT family members.
Fig. 1. (A) Schematic presentation of carboxyl-truncated STAT5B (STAT5BN) used in the yeast two-hybrid screening. The carboxyl trans-activation domain of STAT5B was deleted at amino acid 713 and subcloned into pAS2-1 vector. N-ter, N-terminus; DBD, DNA-binding domain; SH2, Src-homology 2 domain; AD, activation domain. (B) Schematic structure of SMRT protein. The bold line indicates a 900 bp cDNA fragment that was pulled out by two-hybrid screening. SRD, suppressor domain; RID, receptor-interacting domain. (C) Interaction of STAT5BN and SMRT in yeast two-hybrid assay. pAS2-1, bait vector; pACT, prey vector. The blue color indicates a positive interaction. (D) Interaction of various STATs and SMRT. Carboxyl truncated forms of each STAT were subcloned into pAS2-1 and examined for interaction with SMRT. C indicates a negative control, which is a bait vector (pAS2-1).
The STAT5–SMRT interaction was confirmed by protein binding assays in vitro. As shown in Figure 2A, Sf9-expressed STAT3 and STAT5A proteins immobilized on protein A–Sepharose beads efficiently and specifically pulled down in vitro translated SMRT. Conversely, SMRT-Flag protein specifically pulled down STAT5A by immunoprecipitation with a Flag-specific antibody (Figure 2B). Similar results were obtained for STAT5B (data not shown). Consistent with the two-hybrid data, SMRT also interacted with STAT3 and STAT4 in vitro (Figure 2C).
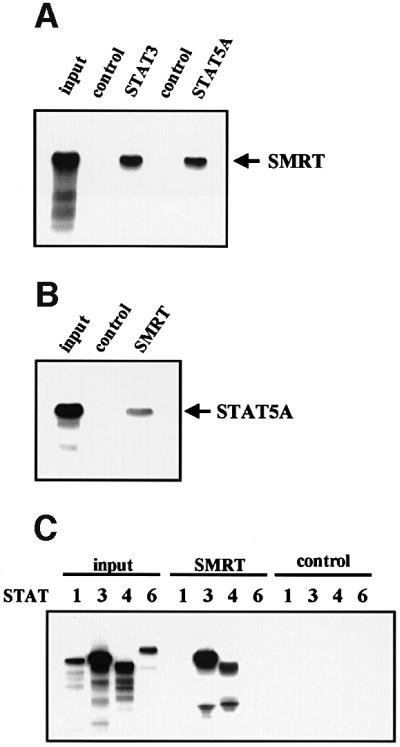
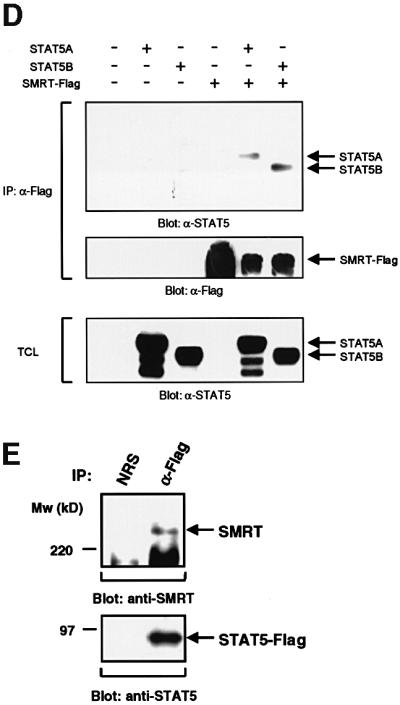
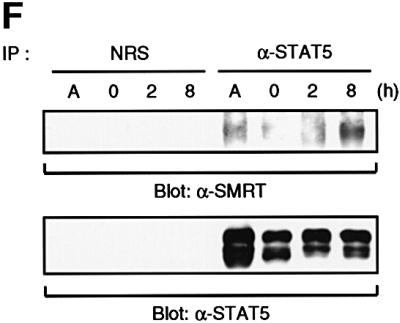
Fig. 2. Interaction of STAT5 and SMRT in vitro and in vivo. (A) In vitro translated [35S]methionine-labeled SMRT was incubated with STAT3 and STAT5A proteins immobilized on Sepharose beads in vitro. STAT3 and STAT5A proteins were generated in Sf9 cells as described previously (Quelle et al., 1996). Bound proteins were eluted and analyzed by SDS–PAGE. Input indicates 20% of 35S-labeled protein used in the reaction. Sepharose beads alone were used as a control. (B and C) In vitro translated [35S]methionine-labeled STAT1, 3, 4, 5A and 6 were incubated in vitro with SMRT-Flag protein immobilized on Sepharose beads. SMRT-Flag protein was expressed in Cos7 cells and immobilized on Sepharose beads by anti-Flag antibody. Bound proteins were eluted and analyzed on 4–20% SDS–PAGE gel. Input indicates 20% of 35S-labeled protein used in the reaction. Sepharose beads alone were used as a control. (D) In vivo interaction of STAT5 and SMRT. Extracts of 293 cells expressing SMRT-Flag and STAT5A or 5B were immunoprecipitated with anti-Flag antibody. Precipitated proteins were resolved on 4–20% SDS–PAGE gel, transferred to nitrocellulose membrane and immunoblotted with anti-STAT5 or anti-Flag monoclonal antibodies. Total cell lysates (TCL) were also run on 4–20% SDS–PAGE gel and immunoblotted with anti-STAT5 monoclonal antibody. (E) 32D cells expressing STAT5A-Flag were lysed for immunoprecipitation with non-immunized rabbit serum (NRS) or anti-Flag antibody. Precipitated proteins were eluted and analyzed by SDS–PAGE. Co-precipitated SMRT protein was detected by anti-SMRT polyclonal antibody. (F) The IL-3-dependent cell line DA3 was starved overnight and stimulated with 10 ng/ml IL-3 for the time period indicated. Cells were lysed, immunoprecipitated with NRS or anti-STAT5 antibody and analyzed by SDS–PAGE. A, asynchronously growing cells.
The specific interaction of STAT5 and SMRT was also demonstrated in vivo by transient transfection of 293 cells. Flag-tagged SMRT was co-expressed either with an empty vector, STAT5A or STAT5B and the resulting cell lysates were immunoprecipitated with anti-Flag antibody. STAT5A and STAT5B proteins were found specifically in the immunoprecipitates from cells co-transfected with SMRT (Figure 2D). Moreover, a STAT5–SMRT complex was demonstrated by co-immunoprecipitation from 32D cells expressing Flag-tagged STAT5A or native DA3 cells (Figure 2E and F). In addition, we assessed the association kinetics of endogenous STAT5 and SMRT upon cytokine stimulation (Figure 2F). A significant amount of SMRT protein associated with STAT5 in asynchronously growing cells; however, most of them dissociated from STAT5 when cells were deprived of IL-3. Upon nuclear translocation of STAT5 by IL-3 stimulation, SMRT became associated with STAT5 and maximal association was observed after 8 h of stimulation.
These data indicate that SMRT can physically interact with STAT5A and 5B in vitro and in vivo, and their association occurs upon nuclear translocation of STAT5 by cytokine stimulation.
SMRT represses STAT5-dependent transcription
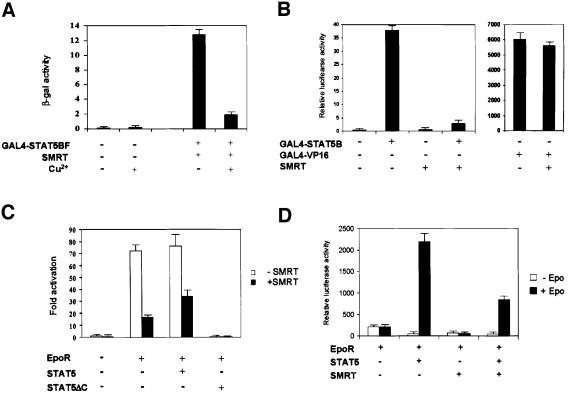
The interaction of SMRT with STAT5 suggests that SMRT could act as a transcriptional repressor for STAT5. We first tested this possibility by using a GAL4-driven LacZ reporter in yeast. Full-length STAT5B fused to the GAL4 DNA-binding domain and was co-transfected with a Cu2+-inducible SMRT expression vector into yeast strain Y190, which carries an integrated GAL4-driven LacZ reporter gene. Transfection of GAL4–STAT5B resulted in high β-galactosidase (β-gal) activity due to the C-terminal transcriptional activation domain of STAT5B. However, when SMRT expression was induced by CuSO4, GAL4– STAT5B activity was repressed ∼6-fold (Figure 3A), whereas basal activity of the reporter gene was unaffected.
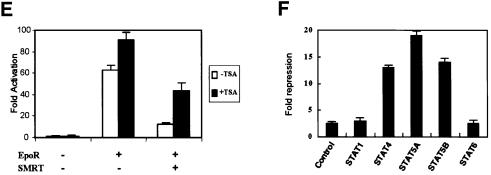
Fig. 3. Effects of SMRT on transcriptional activity of STAT5. (A) Full-length STAT5B fused to the GAL4 DNA-binding domain (GAL4–STAT5BF) and SMRT were co-transfected into Y190, which bears the LacZ reporter driven by GAL4. SMRT expression was induced by addition of CuSO4 and the β-gal activities induced by GAL4–STAT5BF were compared. β-gal activity was measured according to the protocol described in Materials and methods. (B) GAL4–STAT5B or GAL4-VP-16 and SMRT were co-transfected into 293 cells with luciferase reporter plasmid bearing five copies of the GAL4-binding site (GAL4-Luc). Cells were harvested and lysed after 48 h of transfection. Luciferase activity of the extract was examined by luciferase assay systems (Promega). Data were normalized according to the β-gal activity by co-transfected RSV-β-gal plasmid. (C) 293 cells were transfected with a reporter plasmid bearing four copies of optimal STAT5 binding site (ST5BS-Luc), Epo receptor (EpoR), SMRT and wild-type STAT5 or STAT5ΔC. Cells were treated with or without 25 U/ml recombinant human Epo after 24 h of transfection. After 24 h of Epo stimulation, cells were harvested and lysed for luciferase assay. Data are shown as fold activation. (D) Luciferase assay using β-casein promoter. EpoR, STAT5, SMRT and β-casein reporter construct were transfected into 293 cells. Epo stimulation and cell lysis were performed similarly as in (C). Data are shown as relative luciferase activity. (E) Effects of TSA on the repressive effect of SMRT. EpoR, SMRT and ST5BS-Luc construct were transfected into 293 cells. Cells were treated with 100 ng/ml TSA together with 25 U/ml Epo for 24 h before cell harvest. Data are shown as fold activation. (F) Effects of SMRT on transcription by various STATs. Full-length STAT1, 4, 5A, 5B and 6 were fused to the GAL4 DNA-binding domain and transfected into 293 cells with SMRT and GAL4-Luc reporter. Repressive effects of SMRT are shown as fold repression.
Similar observations were made in mammalian cells. When fused to the GAL4 DNA-binding domain, full-length STAT5B again showed high transcriptional activity on a heterologous promoter, which bears five copies of the GAL4-binding site in 293 cells (Figure 3B). Co-expression of SMRT with GAL4–STAT5B dramatically repressed its activity by ∼10- to 20-fold. This repressive effect of SMRT was specific to STAT5A and STAT5B, since it did not affect the activities of STAT1 and STAT6 (Figure 3F), consistent with the association data (Figure 2). In addition, SMRT did not affect the activities of other transcription factors such as VP-16 and Ets-1 (Figure 3B; data not shown).
We also examined promoters containing natural STAT5-binding sites. As shown in Figure 3C and D, SMRT efficiently repressed STAT5-dependent transcription from a promoter containing four copies of an optimal STAT5-binding site (Figure 3C) or the β-casein promoter (Figure 3D). This effect was partly relieved by overexpression of wild-type STAT5 (Figure 3C, third column). SMRT-dependent repression was stronger on the GAL4 promoter than on the promoters with natural STAT5-binding sites (10-fold repression versus 3- to 4-fold repression). This suggests that either SMRT associates less efficiently with dimerized STAT5 on the native promoter compared with the GAL4–STAT5 on the heterologous promoter or, conversely, co-activator complexes associate more strongly with dimerized STAT5 than the monomer.
To confirm that SMRT-dependent repression of STAT5 depends on histone deacetylase activity, we examined the effect of the histone deacetylase inhibitor trichostatin A (TSA). TSA efficiently reversed the repressive effect of SMRT on transcription from a natural STAT5-binding site promoter (Figure 3E). TSA also slightly increased STAT5 activity in the absence of SMRT overexpression, suggesting that endogenous SMRT and STAT5 interact in cells. The TSA effect appears to be specific since control promoter activity was unaffected.
Lastly, we tested the effect of SMRT on various STATs fused to the GAL4 DNA-binding domain (Figure 3F). SMRT effectively repressed the activities of STAT4, 5A and 5B, but not those of STAT1 and STAT6. We were unable to assess the effect on STAT3 since it did not show significant transcriptional activity in this system. This result confirms the differential association of SMRT and various STATs revealed by the in vitro association study.
SMRT associates with the N-terminal coiled-coil region of STAT5
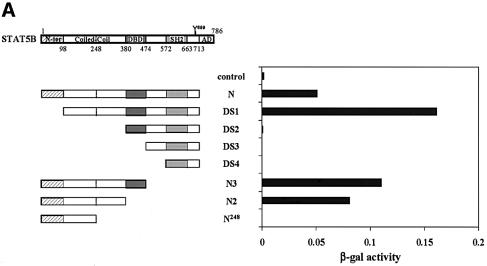
To delineate the region in STAT5 required for association with SMRT, we made a series of STAT5B mutants and assayed their interaction with SMRT by using the yeast two-hybrid system. As shown in Figure 4A, the N, DS1, N3 and N2 mutants, all of which retained the C-terminal half of a coiled-coil region, showed interaction with SMRT, whereas the mutants that lacked this domain did not. This result clearly indicates that the C-terminal half of the coiled-coil region of STAT5 is critical for the interaction with SMRT.
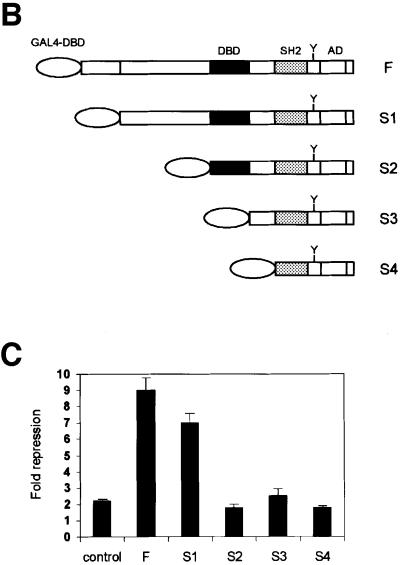
Fig. 4. SMRT interacts with the coiled-coil region of STAT5. (A) Analysis of the SMRT-interacting domain in STAT5B. Various deletion mutants of STAT5B were assessed for their ability to interact with SMRT by two-hybrid assay. Numbers denote the position of amino acids in STAT5B. The interaction is shown as β-gal activity. N-ter, N-terminus; Coiled-Coil, coiled-coil domain; DBD, DNA-binding domain; SH2, Src-homology 2 domain; AD, activation domain. (B and C) Repressive effect of SMRT depends on the coiled-coil region of STAT5. Various STAT5B deletion mutants were fused to the GAL4 DNA-binding domain at their N-terminus and transfected into 293 cells with SMRT and the GAL4-Luc reporter for luciferase assay. Effects of SMRT on their transcriptional activity are presented as fold repression. Control shows the activity of an empty vector that harbors the GAL4 DNA-binding domain alone.
We also made a series of N-terminal truncation mutants of STAT5 fused to the GAL4 DNA-binding domain and tested the repressive effect of SMRT (Figure 4B). SMRT equally repressed transcriptional activity of the F and S1 mutants; however, it did not repress S2, S3 and S4, which lacked the coiled-coil domain (Figure 4C).
Together, these data show that the C-terminal half of the coiled-coil domain of STAT5 (amino acids 248–380) is necessary for the association with SMRT.
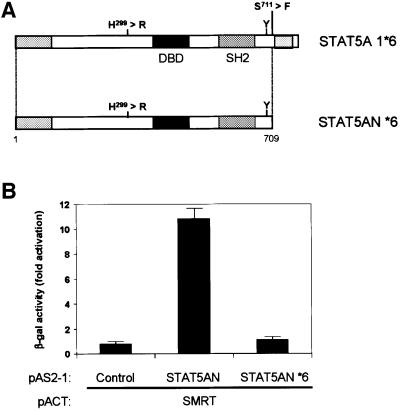
A point mutation in the coiled-coil domain found in a constitutively active form of STAT5 disrupts the association with SMRT
If STAT5–SMRT association plays a regulatory role in STAT5-dependent transcription in vivo, a mutation that disrupts this association should result in increased transcriptional activity of STAT5. Onishi et al. (1998) previously reported a transcriptionally active mutant of STAT5 that promotes cellular proliferation. They identified two point mutations: one located right before the C-terminal activation domain (*1 mutation, S711F) and the other located in the coiled-coil domain (*6 mutation, H299R; Figure 5A). They showed that the mutation in the C-terminus (*1 mutation) inhibits protein turnover and stabilizes the activated form of the protein. However, they did not know the underlying molecular mechanism of how the mutation in the coiled-coil domain (*6 mutation) contributes to the transcriptional enhancement, although this mutation itself potentiates transcription ∼2- to 4-fold (Onishi et al., 1998). Since this mutation falls right into the region that is required for the interaction with SMRT, we speculated that it might disrupt the association with SMRT, resulting in higher transcriptional activity because of its inability to associate with a repressor. We tested this hypothesis in a two-hybrid assay. As shown in Figure 5, the *6 mutation in the context of carboxyl-truncated STAT5A (STAT5AN *6) clearly abrogated the interaction with SMRT (Figure 5). These data indicate that the interaction between STAT5 and SMRT is relevant in a physiological context and SMRT is playing a negative regulatory role in STAT5-dependent transcription in vivo.
Fig. 5. Active STAT5 mutation in the coiled-coil region disrupts interaction with SMRT. (A) Schematic presentation of STAT5A 1*6 and C-terminal truncated mutant of STAT5A 1*6 (STAT5AN *6) used in the interaction study with SMRT. Sites and amino acid changes of each mutation are shown. Numbers denote the position of amino acids. DBD, DNA-binding domain; SH2, Src-homology 2 domain. (B) Interaction of SMRT and STAT5AN or STAT5AN *6. STAT5AN is carboxyl-truncated STAT5A without mutation. Interaction was assessed in the yeast two-hybrid assay and data are shown as β-gal activity. Empty vector (pAS2-1) was used as control.
Overexpression of SMRT or treatment with TSA alters the induction profile of STAT5 target genes
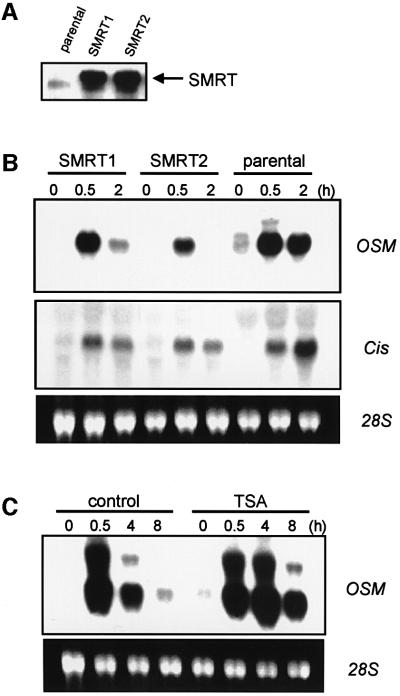
To investigate further the role of STAT5–SMRT association in vivo, we overexpressed SMRT in IL-3-dependent 32D cells and examined the effects on the induction of STAT5 target genes by IL-3. As shown in Figure 6A, SMRT was expressed in these cells at a significantly higher level than endogenous protein. When stimulated with IL-3, one STAT5 target gene, Cis (Yoshimura et al., 1995), was induced in 30 min and expression levels increased further at 2 h in parental cells (Figure 6B). In contrast, in cells overexpressing SMRT, Cis reached maximal expression to the same degree as parental cells at 30 min after stimulation and then decreased significantly at 2 h (Figure 6B). Another STAT5 target gene, oncostatin M (OSM; Yoshimura et al., 1996), reached maximal induction in 30 min and stayed at the same level after 2 h in parental cells. In SMRT-expressing cells, OSM was induced similarly or slightly less at 30 min and mRNA was almost absent after 2 h (Figure 6B). These data indicate that overexpression of SMRT suppresses the expression of endogenous STAT5 target genes, mainly by facilitating their down-regulation after the initial induction phase.
Fig. 6. SMRT represses STAT5 target genes in vivo. (A) Expression of SMRT in 32D cells. Total cell lysates were prepared as described in Materials and methods and resolved on 4–20% SDS–PAGE gel. Expression of SMRT was examined by western blotting using anti-SMRT antibody. SMRT1 and SMRT2 represent two independent clones of 32D cells expressing SMRT. (B) Induction of two STAT5 target genes Cis and osm were assessed by northern blot analysis in parental 32D cells and 32D cells expressing SMRT (SMRT1 and SMRT2). Cells were starved for 5 h and then stimulated with 10 ng/ml IL-3 for the times indicated. RNA extraction and northern blot analysis were performed as described in Materials and methods. (C) 32D cells were starved for 5 h, treated with or without 100 ng/ml TSA for 30 min and then stimulated with 10 ng/ml IL-3 for the times indicated. Induction of osm was analyzed by northern blot analysis as in (B).
In order to investigate the possibility that histone deacetylase activity is involved in the down-regulation of STAT5-dependent transcription, the effect of TSA on the transcription of STAT5 target genes was examined. OSM was chosen for this experiment since its down-regulation during the first 8 h was more evident than Cis. As shown in Figure 6C, induction of OSM by IL-3 was markedly sustained in cells treated with TSA, whereas it was almost non-existent at 8 h in non-treated cells. This indicates that histone deacetylase activity is involved in the negative regulation of STAT5 transcription in vivo, supporting a functional role of SMRT in this process.
Discussion
In this study, we presented evidence for the functional interaction of STAT5 with SMRT. To date, abundant evidence has been generated to show the molecular basis of how STAT5 activates target genes; however, little is known about negative regulation of STAT5. Chung et al. (1997) and Liu et al. (1998) previously reported a novel protein family whose members PIAS1 and PIAS3 negatively regulate STAT1- or STAT3-dependent transcription through interactions with the N-terminus of STATs. These proteins later turned out to be homologs of a protease that binds to an RNA helicase and are now speculated to terminate STAT-dependent transcription through degradation of STATs themselves. Association of SMRT with STAT5 not only provides evidence of novel negative regulatory mechanisms acting directly on transcriptional levels, but also reveals an intriguing possibility that negative regulation of Jak–STAT and nuclear receptor signaling pathways are directed through a common set of chromatin remodeling complexes containing histone deacetylase.
It has been shown that transcriptional co-activators like CBP and p300 are important in STAT-dependent transcription (Gingras et al., 1999; Paulson et al., 1999). These co-activators bind to the C-terminal activation domain of STATs. In this regard, it is interesting that SMRT interacts with the N-terminal coiled-coil region of STAT5. Since CBP/p300 and SMRT bind to different regions of STAT5, the question is how are these factors assembled when STAT5 binds to DNA. One possibility is that both CBP/p300 and SMRT can bind to STAT5 at the same time and regulate transcription through the functional balance between them. This hypothesis is partly supported by the observation that the interaction of SMRT is disrupted by a STAT5 mutation known to result in higher transcriptional activity (Onishi et al., 1998). In addition, the concept that the balance of a co-activator and a co-repressor regulates the transcriptional activity of certain transcription factors was previously proposed for the POU homeodomain factor Pit-1 by Xu et al. (1998). It was shown that Pit-1 can associate with both CBP and N-CoR, and disruption of the N-CoR interaction actually resulted in the enhancement of Pit-1 transcription. They also identified a point mutation in Pit-1 that enhanced the N-CoR interaction and showed that this mutation caused decreased transcriptional activity of Pit-1. These observations possess a striking similarity to the results presented in this paper for STAT5. STAT5 can associate with co-activators and co-repressors through different domains and may regulate transcription by a similar mechanism to Pit-1. In the case of STAT5, we were unable to show the association of endogenous N-CoR and STAT5 by co-immunoprecipitation (H.Nakajima, unpublished data), suggesting that SMRT and N-CoR selectively interact with different transcription factors.
Another possibility is that one of the co-activators or co-repressors associates with STAT5, depending on the promoter it binds to. This predicts that STAT5 either activates or represses promoter activity by associating with CBP/p300 or SMRT and this selective association is determined by the promoter context by itself. Although none of the STAT5 target genes we tested was shown to be repressed by STAT5, this possibility is also likely considering that STAT5 is exerting its biological functions through complex gene regulations.
The last and most intriguing possibility is that SMRT predominantly associates with C-terminal truncated STAT5 (STAT5ΔC) and is involved in the negative regulation of STAT5-dependent transcription. STAT5ΔC is naturally generated in vivo, either by alternative splicing or protein processing (Wang et al., 1996; Azam et al., 1997), and acts as a dominant-negative protein in vitro (Mui et al., 1996; Wang et al., 1996). However, little is known about the physiological role of this isoform and the mechanism of how it functions in vivo. The data presented here predict that STAT5ΔC would act as an active repressor of transcription since it lacks an activation domain while still retaining the ability to associate with SMRT. Therefore, it is reasonably speculated that the dominant-negative function of STAT5ΔC is achieved not only by replacing wild-type STAT5 with the transcriptionally inactive variant, but also by tethering the repressor complex to the target gene promoter through SMRT.
In addition, the experiments presented here show that SMRT is mainly involved in the attenuation of STAT5 target gene expression rather than the initial up-regulation. Like many signal-dependent pathways, cytokine-dependent gene expression displays burst-attenuation kinetics, yet the attenuation phase of this process is not well understood. This study suggests that co-repressors like SMRT may play an important part in turning off STAT5-dependent target genes after the initial burst of signal-dependent transcription. Kinetic analysis shows that the formation of the STAT5–SMRT complex occurs during the first few hours of cytokine stimulation, parallel to the down-regulation of target genes. In addition, STAT5ΔC is generated mainly by nuclear translocation of full-length STAT5 upon ligand stimulation (Azam et al., 1997), which coincides with the attenuation of target gene expression. Taken together, this evidence proposes that the negative regulation of STAT5-dependent transcription could be directed through a protein complex containing STAT5ΔC and SMRT. Further studies will be required to test this intriguing possibility.
STAT is an evolutionarily conserved protein throughout eukaryotes, including Caenorhabditis elegans, Droso phila, Dictyostelium and mammals (Darnell, 1997). Previously, a Dictyostelium STAT protein (Dd-STAT) was cloned and, surprisingly, did not possess a C-terminal activation domain, although other functional domains (DNA-binding domain, SH2 domain) were conserved (Kawata et al., 1997). Dd-STAT acts as both an activator and repressor on two different genes, ecmA and ecmB. Thus, it is tempting to speculate that, in lower eukaryotes, STAT originally regulated transcription by associating with various co-factors (i.e. co-activator or co-repressor) through its N-terminus, and acquired the trans-activation domain on the C-terminus during evolution. In this context, it is of interest that STAT1 has been reported to associate with CBP not only through its C-terminus, but also through its N-terminus (Zhang et al., 1996). Furthermore, a protein named Nmi was reported to interact with the coiled-coil region of STATs and augment transcription by tethering the CBP/p300 protein complex (Zhu et al., 1999). Irrespectively, these studies underscore the importance of the N-terminal coiled-coil region of STAT in transcriptional regulation, possibly through various protein–protein interactions. The molecular basis for the differential interaction of SMRT with various STAT proteins is not clear at present. We could not find any sequence similarity or difference in the critical coiled-coil region that may explain this differential association between STATs. We suspect this might be based on the difference in their ternary structures and this is an interesting area for future study.
In summary, this study revealed a functional interaction of STAT5 and SMRT, and provided evidence that SMRT could be involved in the negative regulation of STAT5-dependent transcription in vivo. The concept that transcription is regulated by the balance of co-activators and co-repressors seems to be fundamental not only in the nuclear receptor or POU homeodomain factor systems, but also in STATs.
Materials and methods
Yeast two-hybrid screening
C-terminal truncated STAT5B (amino acids 1–713) was created by PCR using Pfu polymerase (Stratagene) and subcloned into pAS2-1 vector (designated pAS2-1–STAT5BN). The integrity of the amplified sequence was confirmed by DNA sequencing. The plasmid was transfected into the yeast strain Y190 by the lithium acetate method (yeast transformation system; Clontech). Resultant Y190 cells expressing pAS2-1-STAT5BN were further transfected with a mouse T-cell lymphoma library (Clontech). A total of 1 × 107 transformants were screened and positive clones were identified according to the manufacturer’s protocol (Matchmaker 2 yeast two-hybrid system; Clontech).
Quantitative interaction analysis by the yeast two-hybrid system was performed according to the manufacturer’s protocol (Matchmaker 2 yeast two-hybrid system; Clontech). Briefly, plasmids of interest were co-transfected into Y190 and then several colonies were inoculated and cultured in appropriate SD medium overnight. The culture was further continued for 3 h after adding YPD media. Cells were harvested and broken up by three freeze–thaw cycles in liquid nitrogen. β-gal activity was measured by using O-nitrophenyl β-d-galactopyranoside (OPNG) as the substrate.
In vitro protein binding assay
STAT3 and STAT5A proteins were generated in Sf9 cells using the baculovirus expression system as described previously (Quelle et al., 1996). Flag-SMRT was expressed in Cos7 cells by transient transfection with LipofectAmine Plus (Life Technologies). Proteins generated were immunoprecipitated by anti-STAT3, STAT5 rabbit polyclonal antibody (Santa Cruz) or anti-Flag M2 monoclonal antibody (Kodak), respectively, and immobilized on protein A– or protein G–Sepharose beads (Amersham Pharmacia). [35S]methionine labeling was performed by TNT-reticulocyte coupled system (Promega). Labeled proteins were incubated with STAT3, STAT5A or SMRT protein beads in a binding buffer [50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 0.1% Tween-20, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF)] at room temperature for 1 h. Beads were washed extensively in binding buffer and pulled-down proteins were resolved on 4–20% gradient SDS–PAGE.
Plasmids
Various STAT5 mutants were generated by PCR using Pfu polymerase (Stratagene) and subcloned into appropriate vectors. The Flag epitope was introduced to the 5′-end of SMRT cDNA by PCR and resulting Flag-SMRT cDNA was subcloned into pcDNA3. The integrity of the amplified sequence was confirmed by DNA sequencing.
Cell lines
32Dcl3 cells and DA3 cells were cultured in RPMI1640 (Life Technologies) supplemented with 10% fetal bovine serum (FBS; Hyclone), 100 U/ml penicillin G, 100 µg/ml streptomycin, 2 mM l-glutamine and 2.5 U/ml recombinant murine IL-3. 293 cells were cultured in Dulbecco’s modified Eagle’s medium (Life Technologies) supplemented with 10% FBS, 100 U/ml penicillin G and 100 µg/ml streptomycin. All cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
Establishment of 32D transfectants
To obtain 32D cells stably expressing SMRT, 1 × 107 32Dcl3 cells were electroporated with 20 µg of pcDNA3-Flag-SMRT and selected in the presence of G418 (750 µg/ml) for 14 days. The clones were obtained by limiting dilution and the expression of SMRT was examined by western blot analysis as described below. Three independent clones that showed a high level of expression were further analyzed. All clones showed essentially the same phenotype and the representative data are shown in each experiment. 32D cells expressing STAT5A-Flag have been described elsewhere (Wang et al., 1996).
Northern blot analysis
Total RNA was extracted from ∼1 × 107 cells by RNAzol-B according to the manufacturer’s protocol (Tel-test Inc.). The RNA samples (20 µg/lane) were separated on 1.0% formaldehyde-denaturing agarose gel and transferred onto Hybond N+ membrane (Amersham). Probes were labeled with [α-32P]dCTP using a Rediprime kit (Amersham). Hybridizations with 32P-labeled probes were carried out in ExpressHyb buffer (Clontech) according to the manufacturer’s protocol. The membranes were washed in 2× standard saline citrate (SSC) and 0.1% SDS wash buffer for 30 min at room temperature with several buffer changes, followed by two washes in 0.1× SSC, 0.1% SDS for 15 min at 42°C. The membranes were exposed on XAR films (Kodak) at –80°C for 1–5 days.
Immunoprecipitation and western blotting
Cells (1 × 107) were lysed in extraction buffer (10 mM Tris–HCl, 50 mM NaCl, 5 mM EDTA, 50 mM NaF, 30 mM sodium pyrophosphate, 100 µM sodium orthovanadate, 1% Triton X-100, 1 mM PMSF). For co-immunoprecipitation studies, 0.5% NP-40 buffer (0.5% NP-40, 20 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM PMSF) was used for cell lysis. Lysates were centrifuged at 12 000 g for 15 min at 4°C to remove debris and protein concentrations were measured by the BCA method (Pierce Chemicals). Immunoprecipitation was carried out at 4°C for 2 h by adding 3 µg of primary antibody and protein A–Sepharose beads. Bound proteins were extracted and run on 4–20% SDS–PAGE gel. For direct western blotting, 50 µg of each cell extract were resolved on 4–20% SDS–PAGE gel and transferred to Hybond ECL (Amersham). The membrane was blocked in 5% non-fat milk in TBS-T (20 mM Tris–HCl, 137 mM NaCl, 0.1% Tween-20) and hybridized sequentially with primary antibodies and horseradish peroxidase-conjugated secondary antibody (Amersham). Primary antibodies used in this study were anti-STAT5 monoclonal antibody (Transduction Laboratory), anti-Flag monoclonal antibody (M2, Kodak) and anti-SMRT polyclonal antibody (a generous gift from Dr R.Evans, Salk Institute). Bound antibodies were detected by enhanced chemiluminescence (ECL) western blotting kit (Amersham).
Transient transfection and luciferase assay
293 cells were transfected by LipofectAmine plus reagent (Life Technologies) according to the manufacturer’s protocol. The total amount of vectors for each transfection was adjusted to 5 µg by adding empty vectors. If needed, cells were stimulated by adding 25 U/ml recombinant human Epo (Amgen) 24 h after transfection. Cells were harvested and lysed 48 h after transfection, and luciferase assays were performed by luciferase assay systems (Promega) according to the manufacturer’s protocol. A β-gal assay was performed using the β-galactosidase enzyme assay system (Promega) according to the manufacturer’s protocol. Transfections were performed in duplicate and data were averaged from three independent experiments.
Acknowledgments
Acknowledgements
We are grateful for the excellent technical assistance of Lynda Snyder. We thank Dr Ronald Evans for SMRT cDNA and antibody. This work is supported by the grant from the Ministry of Education, Science and Culture of Japan to H.N., the Cancer Center CORE grant CA21765, the grant RO1 DK42932 to J.N.I., the grant PO1 HL53749 and the American Lebanese Syrian Associated Charities (ALSAC).
References
- Azam M., Lee,C., Strehlow,I. and Schindler,C. (1997) Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity, 6, 691–701. [DOI] [PubMed] [Google Scholar]
- Chen J.D. and Evans,R.M. (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature, 377, 454–457. [DOI] [PubMed] [Google Scholar]
- Chung C.D., Liao,J., Liu,B., Rao,X., Jay,P., Berta,P. and Shuai,K. (1997) Specific inhibition of Stat3 signal transduction by PIAS3. Science, 278, 1803–1805. [DOI] [PubMed] [Google Scholar]
- Darnell J.E. Jr, (1997) STATs and gene regulation. Science, 277, 1630–1635. [DOI] [PubMed] [Google Scholar]
- Darnell J.E. Jr, Kerr,I.M. and Stark,G.R. (1994) Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science, 264, 1415–1421. [DOI] [PubMed] [Google Scholar]
- Dhordain P., Albagli,O., Lin,R.J., Ansieau,S., Quief,S., Leutz,A., Kerckaert,J.P., Evans,R.M. and Leprince,D. (1997) Co-repressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl Acad. Sci. USA, 94, 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras S., Simard,J., Groner,B. and Pfitzner,E. (1999) p300/CBP is required for transcriptional induction by interleukin-4 and interacts with Stat6. Nucleic Acids Res., 27, 2722–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T. et al. (1997) A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature, 387, 43–48. [DOI] [PubMed] [Google Scholar]
- Horlein A.J. et al. (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature, 377, 397–404. [DOI] [PubMed] [Google Scholar]
- Huynh K.D. and Bardwell,V.J. (1998) The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene, 17, 2473–2484. [DOI] [PubMed] [Google Scholar]
- Ihle J.N. (1996) STATs: signal transducers and activators of transcription. Cell, 84, 331–334. [DOI] [PubMed] [Google Scholar]
- Ihle J.N., Witthuhn,B., Tang,B., Yi,T. and Quelle,F.W. (1994) Cytokine receptors and signal transduction. Baillieres Clin. Haematol., 7, 17–48. [DOI] [PubMed] [Google Scholar]
- Kawata T., Shevchenko,A., Fukuzawa,M., Jermyn,K.A., Totty,N.F., Zhukovskaya,N.V., Sterling,A.E., Mann,M. and Williams,J.G. (1997) SH2 signaling in a lower eukaryote: a STAT protein that regulates stalk cell differentiation in Dictyostelium. Cell, 89, 909–916. [DOI] [PubMed] [Google Scholar]
- Korzus E., Torchia,J., Rose,D.W., Xu,L., Kurokawa,R., McInerney,E.M., Mullen,T.M., Glass,C.K. and Rosenfeld,M.G. (1998) Transcription factor-specific requirements for co-activators and their acetyltransferase functions. Science, 279, 703–707. [DOI] [PubMed] [Google Scholar]
- Leonard W.J. and O’Shea,J.J. (1998) Jaks and STATs: biological implications. Annu. Rev. Immunol., 16, 293–322. [DOI] [PubMed] [Google Scholar]
- Lin R.J., Nagy,L., Inoue,S., Shao,W., Miller,W.H.,Jr and Evans,R.M. (1998) Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature, 391, 811–814. [DOI] [PubMed] [Google Scholar]
- Liu B., Liao,J., Rao,X., Kushner,S.A., Chung,C.D., Chang,D.D. and Shuai,K. (1998) Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl Acad. Sci. USA, 95, 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mui A.L., Wakao,H., Kinoshita,T., Kitamura,T. and Miyajima,A. (1996) Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: role of Stat5 in proliferation. EMBO J., 15, 2425–2433. [PMC free article] [PubMed] [Google Scholar]
- Nagy L., Kao,H.Y., Chakravarti,D., Lin,R.J., Hassig,C.A., Ayer,D.E., Schreiber,S.L. and Evans,R.M. (1997) Nuclear receptor repression mediated by a complex containing SMRT, mSin3A and histone deacetylase. Cell, 89, 373–380. [DOI] [PubMed] [Google Scholar]
- Nomura T., Khan,M.M., Kaul,S.C., Dong,H.D., Wadhwa,R., Colmenares,C., Kohno,I. and Ishii,S. (1999) Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev., 13, 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M., Nosaka,T., Misawa,K., Mui,A.L., Gorman,D., McMahon,M., Miyajima,A. and Kitamura,T. (1998) Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol., 18, 3871–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson M., Pisharody,S., Pan,L., Guadagno,S., Mui,A.L. and Levy,D.E. (1999) Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem., 274, 25343–25349. [DOI] [PubMed] [Google Scholar]
- Quelle F.W., Wang,D., Nosaka,T., Thierfelder,W.E., Stravopodis,D., Weinstein,Y. and Ihle,J.N. (1996) Erythropoietin induces activation of Stat5 through association with specific tyrosines on the receptor that are not required for a mitogenic response. Mol. Cell. Biol., 16, 1622–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia J., Rose,D.W., Inostroza,J., Kamei,Y., Westin,S., Glass,C.K. and Rosenfeld,M.G. (1997) The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature, 387, 677–684. [DOI] [PubMed] [Google Scholar]
- Wang D., Stravopodis,D., Teglund,S., Kitazawa,J. and Ihle,J.N. (1996) Naturally occurring dominant negative variants of Stat5. Mol. Cell. Biol., 16, 6141–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.W. and Privalsky,M.L. (1998) Components of the SMRT co-repressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF-RARα and BCL-6. J. Biol. Chem., 273, 27695–27702. [DOI] [PubMed] [Google Scholar]
- Xu L. et al. (1998) Signal-specific co-activator domain requirements for Pit-1 activation. Nature, 395, 301–306. [DOI] [PubMed] [Google Scholar]
- Yoshimura A., Ohkubo,T., Kiguchi,T., Jenkins,N.A., Gilbert,D.J., Copeland,N.G., Hara,T. and Miyajima,A. (1995) A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. EMBO J., 14, 2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A., Ichihara,M., Kinjyo,I., Moriyama,M., Copeland,N.G., Gilbert,D.J., Jenkins,N.A., Hara,T. and Miyajima,A. (1996) Mouse oncostatin M: an immediate early gene induced by multiple cytokines through the JAK-STAT5 pathway. EMBO J., 15, 1055–1063. [PMC free article] [PubMed] [Google Scholar]
- Zhang J.J., Vinkemeier,U., Gu,W., Chakravarti,D., Horvath,C.M. and Darnell,J.E.,Jr (1996) Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc. Natl Acad. Sci. USA, 93, 15092–15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., John,S., Berg,M. and Leonard,W.J. (1999) Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNγ-mediated signaling. Cell, 96, 121–130. [DOI] [PubMed] [Google Scholar]