Abstract
An external stem, essential for the release of small nucleolar RNAs (snoRNAs) from their pre-mRNAs, flanks the majority of yeast intron-encoded snoRNAs. Even if this stem is not a canonical Rnt1p substrate, several experiments have indicated that the Rnt1p endonuclease is required for snoRNA processing. To identify the factors necessary for processing of intron-encoded snoRNAs, we have raised in vitro extracts able to reproduce such activity. We found that snoRNP factors are associated with the snoRNA- coding region throughout all the processing steps, and that mutants unable to assemble snoRNPs have a processing-deficient phenotype. Specific depletion of Nop1p completely prevents U18 snoRNA synthesis, but does not affect processing of a dicistronic snoRNA-coding unit that has a canonical Rnt1p site. Correct cleavage of intron-encoded U18 and snR38 snoRNAs can be reproduced in vitro by incubating together purified Nop1p and Rnt1p. Pull-down experiments showed that the two proteins interact physically. These data indicate that cleavage of U18, snR38 and possibly other intron-encoded snoRNAs is a regulated process, since the stem is cleaved by the Rnt1p endonuclease only when snoRNP assembly has occurred.
Keywords: intron-encoded snoRNA/RNase III/snoRNP/U18/yeast
Introduction
Small nucleolar RNAs (snoRNAs) belong to an abundant class of RNAs localized in the nucleolus of eukaryotic cells, where they direct processing and modification of several substrate molecules. Few snoRNAs participate in pre-rRNA cleavage events, while the vast majority are involved in guiding 2′-O-methylation (box C/D family) and pseudouridylation (box H/ACA family) of rRNAs (Maxwell and Fournier, 1995; Kiss, 2001), snRNAs (Tycowski et al., 1998; Jady and Kiss, 2001) and possibly mRNAs (Cavaillé et al., 2000). The conserved sequence elements that characterize each family function as protein binding sites and are essential for snoRNA stability, biogenesis, localization and function (reviewed in Maxwell and Fournier, 1995; Smith and Steitz, 1997; Tollervey and Kiss, 1997; Filipowicz et al., 1999; Weinstein and Steitz, 1999); all snoRNAs exist, in fact, as RNP particles and share sets of class-specific proteins. So far, snoRNP protein composition has been much better characterized in yeast than in vertebrate systems (Kressler et al., 1999). Cbf5p, Gar1p, Nhp2p and Nop10p associate to box H/ACA snoRNPs (Lubben et al., 1995; Henras et al., 1998; Lafontaine and Tollervey, 1998; Watkins et al., 1998), while Nop1p, Nop58p, Nop56p and Snu13p are the common components to all box C/D snoRNPs (Schimmang et al., 1989; Gautier et al., 1997; Wu et al., 1998; Lafontaine and Tollervey, 1999, 2000; Watkins et al., 2000). Of the box C/D snoRNP proteins, Nop1p, Snu13p and Nop58p are required for snoRNA stability and accumulation, suggesting that they form the basic core complex with the snoRNA. Among these, Snu13p and Nop1p are good candidates as RNA binding proteins, since their vertebrate homologs, p15.5 and fibrillarin, bind directly to the conserved box C/D motif (Fatica et al., 2000a; Watkins et al., 2000). In contrast, Nop56p has no effect on snoRNAs stability or accumulation and depends on Nop1p for its association with the snoRNP (Lafontaine and Tollervey, 2000). However, Nop56p is suggested as being required for U3 and/or U14 snoRNAs function, since its depletion inhibits rRNA processing at sites A0, A1 and A2.
snoRNAs are produced through a variety of different pathways, which reflect their heterogeneous genomic organization. Most yeast and a few vertebrate snoRNAs derive from independently transcribed units, either monocistronic or polycistronic (Maxwell and Fournier, 1995; Chanfreau et al., 1998a,b). On the other hand, the majority of metazoan and seven yeast snoRNAs are encoded in introns of protein-coding genes (reviewed in Weinstein and Steitz, 1999).
Despite this diversity, in both cases snoRNA synthesis relies on the generation of entry sites for 5′–3′ and 3′–5′ exonucleases. Such activities convert the snoRNA into its mature form, the mature ends being defined by the formation of snoRNP-specific complexes that protect snoRNA termini from further exonucleolytic digestion. Independently transcribed units often display target sites for the Rnt1p endoribonuclease (Chanfreau et al., 1998a,b; Qu et al., 1999); in the 5′ portion, Rnt1p cleavage eliminates the cap structure, allowing Rat1p 5′–3′ digestion (Ooi et al., 1998; Petfalski et al., 1998), while in the 3′ portion the endonuclease creates entry sites for 3′–5′ trimming by the exosome (Allmang et al., 1999; Van Hoof et al., 2000). Also, intron-encoded snoRNAs are produced via two different pathways. The major pathway is dependent upon splicing and relies on the linearization of the spliced lariat and subsequent exonucleolytic digestion of the snoRNA flanking sequences. In the alternative pathway, entry sites for exonucleases are generated by endonucleolytic cleavages of the unspliced pre-mRNA, producing processing intermediates that are promptly trimmed to the mature ends of the snoRNA. This endonucleolytic processing, initially characterized as the only biosynthetic pathway for Xenopus laevis U16 snoRNA, has recently been shown to be the secondary pathway for most yeast intron-encoded snoRNAs (Ooi et al., 1998; Villa et al., 2000). In these cases, snoRNA processing is alternative to splicing, raising the issue of how the choice between the two pathways is made. By comparative analyses, five out of six yeast intron-encoded box C/D snoRNAs were shown to lack the 5′–3′ terminal stem described to be part of the conserved core motif of box C/D snoRNAs (Bachellerie et al., 1995). Instead, complementary regions within the flanking intron sequences have been found in these five cases. Formation of this stem, ‘external’ to the snoRNA coding region, was shown to be essential for U18 and snR38 release and was supposed to compensate for the lack of the 5′–3′ ‘terminal’ stem (Villa et al., 2000). This peculiar structure of yeast intron-encoded snoRNAs is likely to represent the key element for understanding how the alternative expression of the snoRNA and of the co-transcribed mRNA is regulated.
In this study, we have reproduced in vitro U18 snoRNA biosynthesis from its host intron and we have characterized the trans-acting factors that control such a process. We show that specific snoRNP proteins associate to U18 through all the processing steps and that this interaction is a prerequisite for the cleavage reaction to occur. We also show that Rnt1p and Nop1p interact physically and that cleavage of the U18 external stem is a regulated process: when snoRNP assembly has occurred on the pre-mRNA, the Nop1 protein recruits the Rnt1p endonuclease addressing its activity on the external stem, which is a non-canonical Rnt1p substrate.
Results
Yeast extracts are able to reproduce the processing of the intron-encoded U18 snoRNA
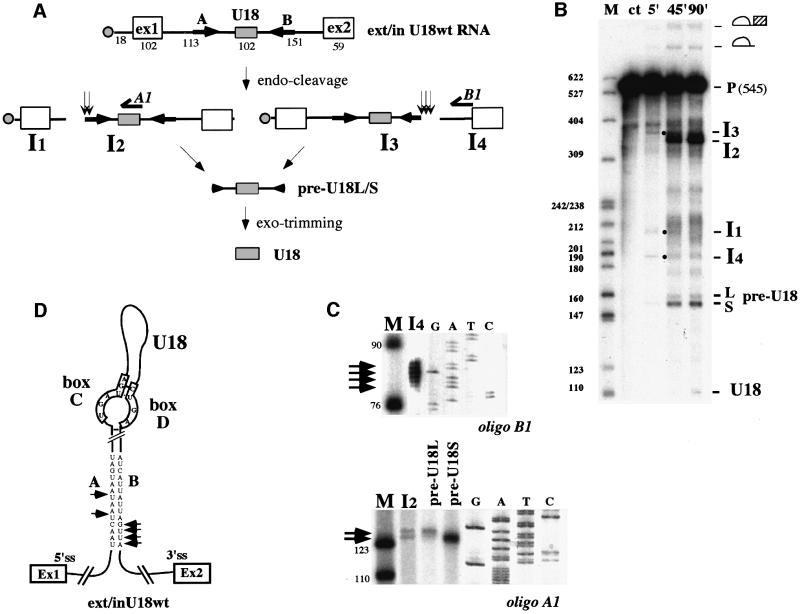
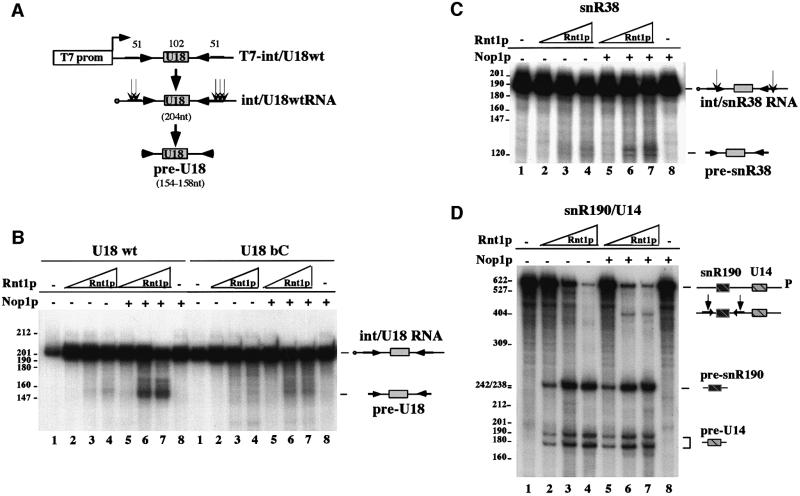
A model radiolabeled precursor RNA, containing the U18 snoRNA plus the flanking intron and exon sequences (Figure 1A), was generated by in vitro transcription of the T7-ex/intU18 construct (Materials and methods) and incubated over time in a yeast whole-cell extract from strain CH1462. This extract is particularly suited for the analysis of the U18 splicing-independent biosynthetic pathway, since it is has poor splicing activity and lacks debranching activity. Figure 1B shows that this extract is in fact able to reproduce the pattern of U18 endonucleolytic processing from its host intron; in particular, the precursor RNA undergoes cleavages upstream and downstream of U18 producing the different intermediate products already identified in vivo (Villa et al., 1998). Cleavage upstream of U18 produces the I1 and I2 products, while downstream cleavage produces the I3 and I4 molecules. When double cleavage occurs on the same molecule the pre-U18 is generated (see schematic representation in Figure 1A). Since extracts have poor trimming activity, mature U18 is produced at very low levels; therefore, in the present study, the pre-U18 RNA will be considered the species diagnostic of cleavage. The occurrence of endo-cleavage is proved by the identification of 5′ and 3′ cut-off products (bands I1 and I4). The I2, I3 and pre-U18 intermediates extend inside the external stem (see below), while in vivo they are trimmed to the 5′ and 3′ end of the snoRNA, respectively, and are very probably stabilized by the interaction with snoRNP-specific factors (Terns et al., 1995; Caffarelli et al., 1996; Xia et al., 1997; Villa et al., 1998). Mapping of the cleavage sites, made by primer extension on gel-purified bands, is shown in Figure 1C. Oligo B1 (see Figure 1A) was utilized for the I4 molecules (lane I4) and oligo A1 for the I2 and pre-U18 RNAs (lanes I2 and pre-U18). The two bands corresponding to the pre-U18 species were eluted and analyzed separately. They appear to differ in their 5′ end by three nucleotides, similar to the I2 molecules. The I4 RNA gives a population of four to five extended products differing by one nucleotide. In Figure 1D, the location of the endo-cuts on the external stem of the U18-containing intron is shown: two cuts, three nucleotides apart, are made in the A sequence, while in the B element a region of four to five nucleotides is target for cleavage. The cut-off products I1 and I3, which extend from the 5′ end of the transcript, have a size in agreement with that expected for cleavage occurring in the identified regions.
Fig. 1. U18 snoRNA is faithfully processed in vitro from its host pre-mRNA. (A) Schematic representation of the ex/intU18 RNA containing the U18 host intron and part of the flanking EFB1 exons. In vitro transcription from this construct produces a 546 nucleotide transcript. Open boxes represent the first and second exons, while the U18 snoRNA coding region is represented by a gray box. The cap structure is represented by a filled circle. Arrows indicate the 14-nucleotide external stem [A and B sequences as indicated in (D)]. The different processing intermediates are indicated together with the formation of the mature U18 molecule. Vertical arrows indicate the position of the cleavage sites. Numbers refer to nucleotide lengths. (B) 32P-labeled ex/intU18 RNA was incubated, for the times indicated above each lane, in a yeast extract raised from strain CH1462. Input RNA was loaded on lane ct. Processing products were resolved on a 6% polyacrylamide–urea gel and visualized by autoradiography. Splicing and endonucleolytic processing intermediates are indicated on the right-hand side of the panel. Some of the intermediates (indicated by dots) are better visualized in the 5′ lane. Lane M: molecular weight marker (pBR322 plasmid DNA, MspI digested). (C) Primer extension analysis performed, with oligos A1 and B1 [see (A)], on gel-purified I2 and I4 molecules as well as on the short (S) and long (L) forms of the pre-U18 species. Extended products (indicated by horizontal arrows) are run in parallel with sequencing reactions and with molecular weight markers [as in (B)]. (D) Secondary structure for ex/intU18 RNA; the terminal conserved core structure is according to Watkins et al. (2000). The cleavage sites are indicated by arrows on the A/B external stem.
Box C/D-specific factors are associated in vitro to U18-containing precursors
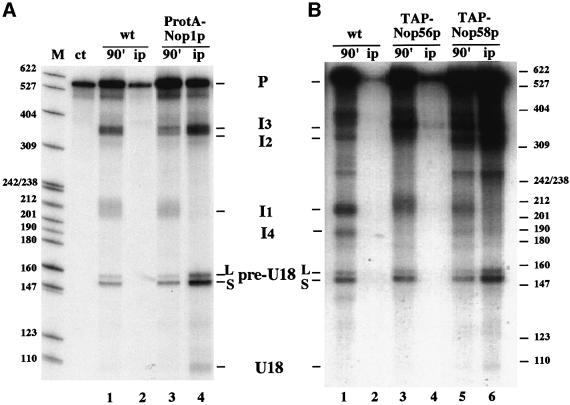
We previously showed that mutations in the conserved C and D box elements prevented cleavage both in Saccharomyces cerevisiae (Villa et al., 2000) and in X.laevis (Caffarelli et al., 1996). These sequences were shown to be essential for the assembly of snoRNP-specific factors with snoRNAs (Caffarelli et al., 1998; Watkins et al., 1998; Fatica et al., 2000a; Filippini et al., 2000; Watkins et al., 2000) and to snoRNA-containing precursors (Caffarelli et al., 1998), suggesting that such early interactions may be important in committing the precursor molecule to the processing pathway. To test this scenario, we investigated both the binding of snoRNP factors and the behavior of different mutant substrates. We took advantage of extracts from a strain carrying a protein A-tagged version of the Nop1p protein (strain ProtA-NOP1). This extract exhibited processing of the ex/intU18 RNA similar to the control (Figure 2A, compare lanes 1 and 3). Immunoprecipitation of the processing reactions with IgG–Sepharose shows that Nop1p is associated with both the precursor RNA and the different intermediates containing the U18 sequence (lane 4). The specificity of the reaction is shown by the absence of the I1 species in the pellet. Since three times more material was used in the immunoprecipitation sample, the mature U18 snoRNA was better visualized. The control untagged strain (Figure 2A, lanes wt) shows that no RNA species are detected in the immunoprecipitates, besides some background pre-mRNA (lane 2). The same approach was utilized for testing the association of other box C/D-specific factors: Nop56p and Nop58p (Figure 2B). Strains were produced in which each protein was tagged with a TAP epitope (Rigaut et al., 1999); extracts were prepared and tested for U18 processing. In comparison with the control (Figure 2B, lane 1), both extracts are active in processing as indicated by the accumulation of the pre-U18 species (lanes 3 and 5). Surprisingly, while Nop58p appears to be associated with all U18-containing molecules (lane 6), the Nop56p protein has a very weak, almost undetectable, association with the processing products (lane 4). Since we verified that in TAP-Nop56p extracts the protein is associated with box C/D snoRNAs (data not shown), the absence of interaction with the U18-processing intermediates in the in vitro reaction indicates either that Nop56p is a late assembly factor or that its binding to the U18 RNA is not retained under these experimental conditions. Snu13p was identified as the fourth box C/D snoRNP-specific factor and described to be a core component of box C/D snoRNPs (Watkins et al., 2000; S.Galardi, personal communication). Due to the fact that we did not manage to obtain a tagged version of this protein, and antibodies were not available to us, we could not analyze its association to the different U18 processing intermediates.
Fig. 2. snoRNP factors associate during processing. 32P-labeled ex/intU18 RNA was incubated for 90 min with extracts from strains CH1462 (lanes wt), ProtA-NOP1 (lanes ProtA-Nop1p, A), YAF2 (lanes TAP-Nop56p, B) and YAF3 (lanes TAP-Nop58p, B). One-third of each processing reaction was directly extracted (lanes 90′: 1, 3 and 5) and the remaining samples were immunoprecipitated with IgG–Sepharose (lanes ip: 2, 4 and 6). RNA was extracted and run on a 6% polyacrylamide–urea gel and visualized by autoradiography. Schematic representation of the different processing intermediates are shown at the sides of the panels.
Since the ProtA-Nop1p extract was found to be active in U18 processing (Figure 2A), it was used to test both the behavior and the association of different cis-acting mutants derived from the U18-containing precursor with Nop1p (Figure 3A).
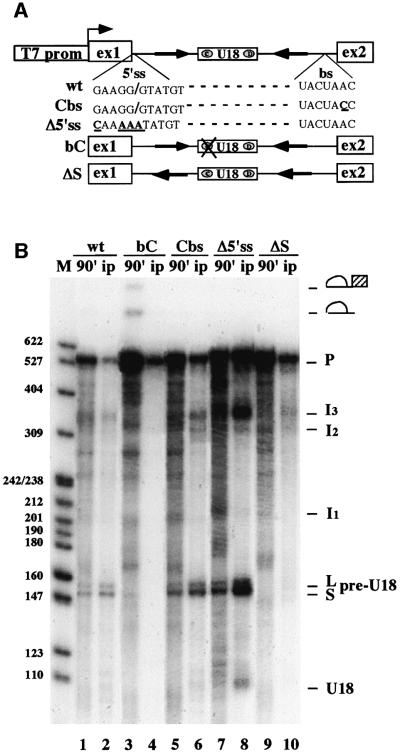
Fig. 3. Effect of cis-acting mutations on U18 processing. (A) Schematic representation of the T7-ex/intU18 construct and of the mutant derivatives used in this study. Δ5′ss and Cbs contain site-specific mutations known to affect splicing, bC has nucleotide substitutions in the conserved box C (UGAUGA-CAUUGA) that affect snoRNP assembly and ΔS disrupts A–B pairing by inverting sequence A. (B) 32P-labeled ex/intU18 RNA, or its mutant derivatives, were incubated in a ProtA–Nop1p yeast extract and incubation was allowed to proceed for 90 min. One-third of each processing reaction was extracted directly (lanes 1, 3, 5, 7 and 9) and the remaining samples were immunoprecipitated with IgG–Sepharose (lanes 2, 4, 6, 8 and 10). RNA from total and pellet samples was resolved on a 6% polyacrylamide–urea gel and visualized by autoradiography. Splicing and processing intermediates are represented schematically at the side of the panel.
All these constructs behaved in the extract exactly as they did in vivo (Villa et al., 2000): a mutation in the conserved box C (construct bC) completely abolished processing, while increasing splicing, as indicated by lariat forms being visible only in this sample (Figure 3B, lane 3). As expected, no Nop1p-specific immunoprecipitation is obtained with this RNA, besides some background of the input RNA (lane 4). This is a further demonstration that the absence of snoRNP-specific factors correlates with the inability to process the precursor RNA. When splicing was inhibited by mutations in either the branch site (construct Cbs) or the 5′ splice site (construct Δ5′ss), processing increased (lanes 5 and 7). In these two constructs, the association of Nop1p with the different intermediate molecules can be very well appreciated (lanes 6 and 8). The immunoprecipitation lanes are useful to distinguish the processing intermediates from degradation molecules which comigrate with the processing intermediates. In particular, the I2 and I3 intermediates, as well as pre-U18 and U18, are clearly visualized only in the pellet fractions. Construct ΔS, which lacks the external stem, confirmed that the absence of this element is also detrimental to U18 processing in vitro (lane 9).
Rnt1p together with Nop1p are required for U18 in vitro processing
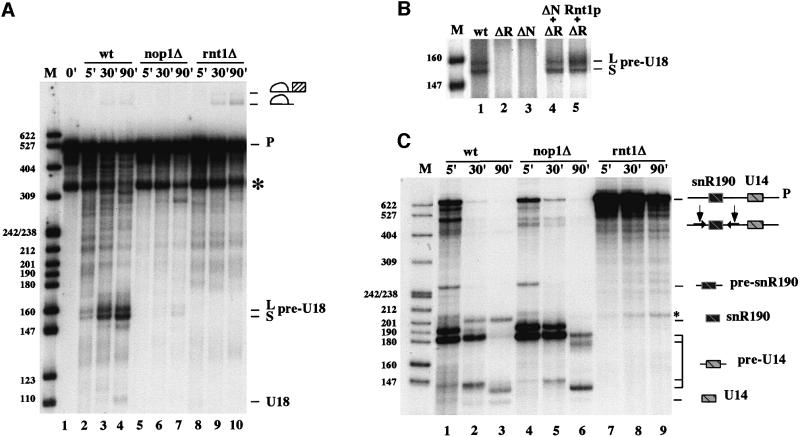
Recent experiments (Fatica et al., 2000b) showed that the external stem of U18 is susceptible to Rnt1p activity even if it does not appear to be a canonical Rnt1p substrate (Chanfreau et al., 2000; Nagel and Ares, 2000). In order to investigate the role played by the Rnt1p endonuclease and by the box C/D-specific factors in U18 processing, we took advantage of strains in which the genes coding for these factors are either deleted (rnt1Δ) or can be functionally inactivated (D255). The first strain has a deletion of the RNT1 gene. The second contains the NOP1-coding region under the GAL10 promoter; when these cells are grown in galactose and then shifted to glucose for 30 h, the synthesis of Nop1p is blocked. It was previously reported that, under these conditions, both Nop56p and Nop58p proteins are still accumulated in the cell (Lafontaine and Tollervey, 2000). Figure 4A, lanes 2–4, shows the control processing reaction with a wild-type extract; in this gel the I2 and I3 intermediates co-migrate with a breakdown product already present in the input RNA (lane 1), while the pre-U18 species is very well visible. Extracts from the two mutant strains do not show any cleavage activity, as demonstrated by the absence of the pre-U18 species, indicating that Nop1p and Rnt1p are both required for U18 processing. If the two extracts are mixed together, rescue of U18 processing is obtained as witnessed by the accumulation of the pre-U18 species (Figure 4B, lane 4). In order to prove that in the rnt1Δ strain the functional missing component is indeed the Rnt1p factor, we supplemented the extract with recombinant Rnt1p protein according to Chanfreau et al. (1998b); Figure 4B, lane 5 shows again, rescue of U18 processing. Figure 4C shows a control processing reaction with the dicistronic snR190/U14 snoRNAs precursor that harbors a canonical Rnt1p site (Chanfreau et al., 1998b). Differently from the U18-containing precursor, processing of this RNA is only affected in extracts depleted of the Rnt1p endonuclease (Figure 4C, lanes rnt1Δ, see the absence of the primary cleavage product pre-snR190 and of the other intermediate products). The absence of the Nop1p protein has no effect on cleavage of this precursor RNA (lanes nop1Δ). As expected, only small amounts of mature snR190 and U14 snoRNAs are accumulated (lanes 3 and 6) due to the poor trimming activity of the extracts, even if all the input RNA is converted to cleaved products.
Fig. 4. Nop1p and Rnt1p proteins are required for U18 snoRNA processing. (A) 32P-labeled ex/intU18 RNA was incubated with extracts from strains CH1462 (lanes wt), D255 (lanes nop1Δ) and rnt1Δ (lanes rnt1Δ) for the time indicated above the lanes. The band indicated by an asterisk corresponds to a breakdown product already present in the input RNA (lane 1) and overlaps with molecules I2 and I3. (B) 32P-labeled ex/intU18 RNA was incubated with extracts from strains CH1462 (lane wt), rnt1Δ (lane ΔR), D255 + rnt1Δ (ΔN + ΔR) and rnt1Δ + recombinant GST–Rnt1p (5 ng/20 µl, lane ΔR + Rnt1). After 90 min of incubation, RNA was extracted and run a 6% polyacrylamide–urea gel and visualized by autoradiography. The panel shows only the region of the gel containing the pre-U18 molecule, which is diagnostic of cleavage. (C) The experiment is the same as in (A), with the difference that the input RNA is the dicistronic precursor containing the snR190 and U14 snoRNAs. The asterisk indicates an unspecific cleavage product. The products of the reactions are schematically represented at the sides of the panels.
These data suggest that even if Rnt1p is required for cleavage of the U18 precursor, it is not sufficient alone, in contrast to what happens for the dicistronic snR190/U14 transcript, where the endonuclease is able to process the precursor independently from Nop1p.
Since the experiments with the Nop1p-depleted extracts indicated that this protein is required for cleavage, we investigated the ability to reproduce the cleavage reaction in vitro with recombinant purified factors. Both Rnt1p and Nop1p were expressed as glutathione S-transferase (GST) fusion proteins. A substrate RNA, containing U18, the external stem and an additional 20 nucleotides on both sides (see schematic representation of Figure 5A), was incubated with increasing amounts of purified GST–Rnt1p in the presence or in the absence of purified GST–Nop1p. Figure 5B, lanes U18wt, shows that the endonuclease alone cleaves the precursor int/U18wt RNA very inefficiently: only low levels of conversion are observed (lanes 2–4). Addition of Nop1p strongly enhances the production of a molecule of the size expected if double cleavage in the opposite strands of the stem had occurred (lanes 5–7). The incubation with Nop1p alone does not produce any cleavage (lane 8). As control for this type of experiment we used a mutant substrate in which one of the two boxes required for snoRNP-assembly was mutated (bC). This mutation had already been shown to inhibit processing and to prevent snoRNP assembly and in particular Nop1p-binding in vivo and in vitro (Figure 2; Villa et al., 1998, 2000; Fatica et al., 2000a). Figure 5B, lanes U18bC, shows that on this substrate, the co-incubation of Rnt1p and Nop1p produces only minimal levels of cleavage (lanes 5–7), close to those observed on the wild-type template with Rnt1p alone. Altogether, these data indicate that the U18 external stem cannot be cleaved by the Rnt1p enzyme alone unless the Nop1p factor is bound to the snoRNA substrate. At the enzyme concentrations used in this experiment, cleavage of the snR190/U14 dicistronic precursor occurs very efficiently and no enhancement due to Nop1p addition is observed (Figure 5D, compare lanes 2–4 with lanes 5–7). In order to analyze whether co-operation between Nop1p and Rnt1p is acting on other substrates, we selected the snR38 snoRNA, another intron-encoded snoRNA flanked by an 11 nucleotide external stem. This stem is also not a canonical substrate for Rnt1p; in fact, when tested in a rnt1Δ strain, the snR38 precursor displayed the same processing-deficient phenotype as U18 (not shown). A precursor RNA (int/38 RNA), containing the snR38 coding region plus the external stem and flanking intron sequences, was transcribed in vitro and incubated as in Figure 5B). The results are similar to those obtained with U18 (Figure 6C), as conversion of the precursor molecule by Rnt1p depends on the presence of the Nop1 protein.
Fig. 5. Purified Nop1p and Rnt1p are able to process pre-U18 molecules. (A) Schematic representation of the int/U18wt construct utilized: the U18 coding region plus the flanking intronic regions containing the external stem were cloned under the T7 promoter. The transcribed RNA is 204 nucleotides long. (B) In vitro cleavage of the int/U18wt RNA (lanes U18wt) and of its mutant derivative in the box C (lanes U18bC) (see construct bC in Figure 3). 32P-labeled RNAs were incubated in a 10 µl reaction with increasing concentrations of GST–Rnt1p protein (0.05 ng, lanes 2 and 5; 0.5 ng, lanes 3 and 6; 1 ng, lanes 4 and 7). In lanes 5–7, 10 ng of GST–Nop1p protein were added. Lanes 1: input RNA; lanes 8: RNAs were incubated only with GST–Nop1p (10 ng in 10 µl reaction). Incubations were allowed to proceed for 60 min. The RNA was extracted and run on a 6% polyacrylamide–urea gel and visualized by autoradiography. The products of the reaction are represented schematically at the side of the panel. (C) In vitro cleavage of a model snR38 precursor RNA. The 32P-labeled int/38 RNA containing the snR38 snoRNA coding sequences plus the external stem and flanking intron sequences was treated as the RNA in (B). The int/snR38 RNA is 190 nucleotides long and the pre-snR38 is ∼120 nucleotides long. (D) In vitro cleavage of 32P-labeled snR190/U14dicistronic precursor RNA. The RNA transcript is the same as that used in Figure 4C. The reactions were performed as in (B). Reactions and lanes in (C) and (D) are numbered as in (B).
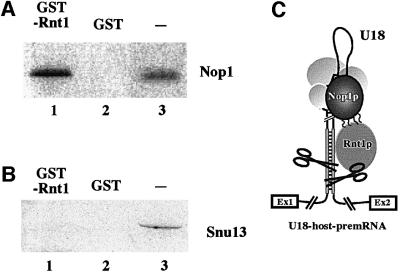
Fig. 6. Nop1p and Rnt1p proteins interact physically. In vitro-translated 35S-labeled Nop1p (A) and Snu13p (B) were loaded on a GST–Rnt1p column (lanes 1) and, as control, on a GST only column (lanes 2). Proteins recovered from each column were analyzed on a SDS–polyacrylamide gel in parallel with the in vitro translated input proteins (lanes 3). (C) Proposed model of how cleavage of the non-canonical Rnt1p-substrate is stimulated by the snoRNP-specific factor Nop1p.
These observations indicate that Nop1p regulates Rnt1p activity in U18 snoRNA endonucleolytic processing and that this processing mechanism may be generalized to all yeast intron encoded snoRNAs that are flanked by a non-canonical Rnt1 stem.
Nop1p and Rnt1p interact physically
In order to understand the molecular basis of the co-operativity between Nop1p and Rnt1p, we tested whether these factors interact physically. A GST–Rnt1 fusion protein was immobilized on a glutathione–Sepharose column and was tested for its ability to bind recombinant Nop1p and, as control, Snu13p. In vitro-translated [35S]methionine-labeled Nop1p and Snu13p proteins were loaded on GST–Rnt1p columns and, as control, on GST only columns. Figure 6 shows that Rnt1p specifically retains Nop1p (Figure 6A, lane 1), whereas no retention was observed for the Snu13 protein (Figure 6B, lane 1). Snu13p was chosen out of all snoRNP factors as the control, since, like Nop1p, is thought to be an early RNA-binding protein. We conclude that Nop1p, when bound to snoRNA-containing precursors, enhances Rnt1p activity through direct interaction with this factor (Figure 6C).
Discussion
The double-strand endoribonuclease Rnt1p was shown to be responsible for processing of monocistronic and polycistronic snoRNA transcripts (Chanfreau et al., 1998a,b). In these units canonical Rnt1p substrates are present, which consist of stem structures with terminal tetraloops showing the consensus sequence AGNN. This loop is located at a distance of 13–16 bp from the Rnt1p cleavage sites (Chanfreau et al., 1997). It was shown that when this sequence is altered, the rate of Rnt1p cleavage is reduced due to reduced binding of the protein to the mutant substrate (Nagel and Ares, 2000). In yeast, five of the six intron-encoded snoRNAs possess an ‘external’ stem, which was shown to be required for the snoRNA release from the pre-mRNA through the endonucleolytic pathway (Ooi et al., 1998; Villa et al., 2000). Even though this stem did not appear to be a canonical target for Rnt1p (Chanfreau et al., 2000; Nagel and Ares, 2000), recent experiments with ad hoc constructs indicated that this stem element is indeed a substrate for the Rnt1p activity (Fatica et al., 2000b).
Two connected questions are raised at this point: (i) why are canonical Rnt1p substrates present in independently transcribed snoRNA coding units and not in intron containing snoRNAs; and (ii) in the second case, is the presence of non-canonical Rnt1p sites correlated to the alternative choice between pre-mRNA cleavage and splicing?
In this paper we have addressed these questions, and the results obtained have allowed us to draw a possible model able to explain these differential choices.
We have set up in vitro extracts able to reproduce the processing of U18 snoRNA from its host intron: an RNA precursor, containing U18 and the flanking intron and exon sequences, is converted into intermediate and final products similar to what was shown previously in vivo (Villa et al., 1998, 2000). The use of mutant substrates has allowed us to show that in vitro also, splicing of the intron and cleavage of the pre-mRNA are alternative pathways and that mutations which prevent snoRNP assembly block the endonucleolytic processing.
Immunoprecipitation of tagged snoRNP factors showed that at least two of the box C/D core proteins, Nop1p and Nop58p, are present on the precursor RNA and remain associated with the U18 coding region throughout the different processing steps. On the other hand, in our experimental conditions, Nop56p was not found associated with the processing products. Previous work by Lafontaine and Tollervey (2000) showed that while Nop1p and Nop58p associate independently on the snoRNA and are indispensable for its stability, Nop56p requires the presence of Nop1p. Furthermore, it was shown that Nop56p depletion does not affect snoRNA accumulation and stability (Gautier et al., 1997; Lafontaine and Tollervey, 2000). In line with these data, our results can be interpreted as confirmation that Nop56p is a late assembly factor, not likely to be required for commitment to processing. Nevertheless, we cannot rule out the possibility that in our experimental conditions Nop56p binding to Nop1p- and Nop58p-containing particles is not stable. A fourth factor, Snu13p, was recently described to be part of box C/D snoRNPs and to be required for snoRNA accumulation (Watkins et al., 2000; S.Galardi, personal communication). The presence of this factor during U18 processing was not assessed due to the lack of antibodies and of a Snu13p-tagged strain. Since this protein was described to bind RNA (Watkins et al., 2000), it is likely to be an early assembly factor and to be associated to pre-snoRNA molecules. Further work is necessary to clarify this point.
In order to test whether the formation of a snoRNP is a fundamental prerequisite for snoRNA processing, we prepared extracts depleted of the Nop1 protein. In parallel, we tested U18 production in an extract lacking the Rnt1p endonuclease. The results indicated that both factors are required for U18 snoRNA processing, while for the maturation of the snR190/U14 dicistronic transcript containing a canonical Rnt1 site, only the endonuclease was found to be essential. Experiments performed with purified Nop1p and Rnt1p proteins indicated that while each factor alone was not able to reconstitute U18 processing, when they were mixed together efficient cleavage was obtained. This effect was shown to depend on the ability of Nop1p to bind the substrate snoRNA. Interestingly, the same co-operativity was shown to be required for the in vitro processing of the snR38 snoRNA precursor, another snoRNA flanked by a non-canonical Rnt1p substrate (Chanfreau et al., 2000; Nagel and Ares, 2000). In addition, pull-down experiments with GST–Rnt1p have proven that Nop1p interacts physically with Rnt1p.
These results suggest that, in those cases where canonical Rnt1p cleavage sites are absent, Nop1p binding to the snoRNA stimulates Rnt1p substrate recognition and cleavage (Figure 6C). We propose a model in which snoRNP assembly regulates the cleavage of the snoRNA from its host pre-mRNA. This has very important regulatory implications since non-canonical Rnt1p substrates are present in those transcription units where the synthesis of a snoRNA is often alternative to that of an mRNA. Our findings explain how the commitment to the processing pathway is controlled: only when snoRNP factors assemble on the snoRNA does cleavage by Rnt1p occur, alternatively, the pre-mRNA is available for splicing.
Materials and methods
Strains
Growth and handling of S.cerevisiae were carried out using standard techniques. Strains used in this study were the following: CH1462: MATα, ade2, ade3, leu2, ura3, his3, can1 (Kranz and Holm, 1990); rnt1Δ: MATa, his3, lys2, leu2-3,112, trp1, ura3-52, pep4, prb1, prc1, rnt1::HIS3 (Chanfreau et al., 1997); YAF2: MATα, trp-Δ his3-Δ ura3-52, lys2-801, ade2-101, URA3::U24, NOP56::TAP::TRP1 (Fatica et al., 2000b); YAF3: MATα, trp-Δ, his3-Δ, ura3-52, lys2-801, ade2-101, URA3::U24, NOP58::TAP::TRP1; ProtA-NOP1: MATα, ade, leu, trp, lys, ura3, nop1::HIS3, pUN100–ProtA-NOP1; D255: MATa, ura3-52, leu2-3, 112, ade1-100, his4-519, URA3-pGAL10::NOP1 (Tollervey et al., 1991). Epitope TAP tagging of Nop1p (strain YAF1) and Nop58p (strain YAF3) was performed as described in Rigaut et al. (1999). Nop1p-depleted extract was prepared from strain D255; cells growing exponentially in permissive SRG complete medium at 30°C were harvested by centrifugation, washed, resuspended in YPD medium and constantly maintained in exponential phase. After 30 h cells were collected by centrifugation and the whole-cell extract prepared.
Plasmids and templates for RNA transcription
Oligonucleotides YU18up and YU18down were used to amplify a PCR fragment from plasmid pGALU18wt (Villa et al., 2000). The PCR product was digested with SacI and HindIII and cloned under the T7 promoter of the Bluescript KS vector to obtain plasmid pBSyU18wt. This plasmid was then digested with EcoRI to generate construct T7-ex/intU18wt for the in vitro T7 transcription.
Constructs T7-ex/intU18Cbs, T7-ex/intU18ΔS, T7-ex/intU18 bC and T7-ext/inU18Δ5′ss were obtained by PCR on plasmids pGALU18Cbs, pGALU18ΔS, pGALU18bC (Villa et al., 2000) and pGALU18Δ5′ss, respectively, with the oligonucleotides EIF1B and yU18 down. Plasmid pGALU18wt was used as starting material to generate pGALU18Δ5′ss by inverse PCR with the oligonucleotides U185′ssUP and U185′ssDOWN. Constructs T7-intU18wt and T7-intU18bC were obtained by PCR on plasmids pGALU18wt and pGALU18bc with the oligonucleotides T7-U18in and 3′stemU18. Construct T7-int38wt was obtained by PCR on plasmids pGAL38wt (Villa et al., 2000) with oligonucleotides T7-38in and 3′stem38.
Plasmid PSP64-snR190-U14 (Chanfreau et al., 1998b) digested with PstI was used as a template for in vitro transcription of snR190-U14 dicistronic pre-RNA.
In vitro processing and cleavage assays
Whole-cell extracts were prepared as described in Ansari and Schwer (1995). All RNA transcripts used in this study were obtained by in vitro transcription of the templates constructs described above. In vitro transcription was performed as described previously (Caffarelli et al., 1998b).
In vitro processing reactions were performed in a 10 µl reaction containing: 10 fmol gel-purified 32P-labeled RNA, 6/8 µg/µl of appropriate yeast extract, 60 mM KCl, 3% PEG, 2 mM ATP, 2.5 mM MgCl2, 2 mM spermidine, 1 U/µl RNase inhibitor, 1 mM DTT. Incubation was allowed to proceed at 24°C for the time indicated. RNAs were extracted with phenol-chloroform, precipitated with ethanol and loaded on a 6% polyacrylamide–7 M urea gel. For immunoprecipitations, processing reactions were performed in extracts carrying the appropriate ProtA- or TAP-tagged protein. After 90 min at 24°C, one-third of the reaction was directly subjected to proteinase K digestion and phenol-chloroform RNA extraction, while the residual two-thirds of the reaction were incubated for 1 h at 4°C with IgG–Sepharose beads (Pharmacia) in 400 µl of NET150 buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40). Beads were then washed four times with NET150 and subjected to proteinase K digestion and phenol-chloroform extraction. The recovered RNAs were analyzed on a 6% polyacrylamide–7 M urea gel. Processing in the rnt1Δ extract was rescued either by adding 5 ng of purified GST–Rnt1p to the 20 µl processing reaction or performing the reaction with an equimolar mixture of the nop1Δ and rnt1Δ extracts.
In vitro cleavage reactions were performed incubating 10 fmol gel-purified 32P-labeled RNA transcript with 0.05, 0.5 or 1 ng of purified GST–Rnt1p and/or with 10 ng of purified GST–Nop1 protein in a 10 µl reaction containing 5 mM KCl, 30 mM Tris–HCl pH 7.4, 0.1 mM EDTA, 10 mM MgCl2, 2 mM spermidine, 0.1 µg/µl tRNA, 1 U/µl RNase inhibitor, 0.1 mM DTT. Incubation was allowed to proceed for 60 min at 26°C. RNAs were extracted with phenol-chloroform, precipitated with ethanol and loaded on a 6% polyacrylamide–7 M urea gel.
Primer extension analysis
A standard processing reaction was performed in extract from strain CH1462 by incubating a mixture of 5 fmol of 32P-labeled ex/intU18wt RNA with 1 µg of cold ex/intU18wt RNA. This cold RNA was obtained by standard T7-transcription reaction in the presence of 500 µM unlabeled UTP. Resulting processing products were extracted and run on 6% polyacrylamide–7 M urea gel. Bands, corresponding to those indicated as I2, I4 and pre-U18S/L in Figure 1B, were cut from the gel and the eluted RNA was subjected to primer extension analysis as described in Villa et al. (2000). The A1 oligonucleotide was utilized as primers for the reverse transcription of gel-purified I2 and pre-U18S/L RNAs. The B1 oligonucleotides were used for reverse transcription of the I4 molecule.
GST fusion proteins and in vitro interaction assays
A PCR fragment corresponding to the NOP1 open reading frame (ORF) was cloned into pBluescript KS vector (Stratagene) to obtain Nop1-BS plasmid and into pGEX-3X vector (Smith and Johnson, 1988) to generate GST–NOP1 fusion vector. Snu13 was cloned by PCR from genomic DNA as a BamHI–HindIII fragment and inserted into plasmid pBluescript KS.
[35S]methionine-labeled Nop1p and Snu13p were made by in vitro T7-transcription/translation (TNT, Promega) using as template Nop1-BS plasmid and Snu13-BS plasmid, respectively. The GST–RNT1 fusion vector (Abou Elela et al., 1996) was kindly provided by M.J.Ares. Expression of GST–NOP1 and GST–RNT1 fusion proteins was performed in a RNase III– strain of Escherichia coli BL2114 to avoid contaminations with RNase III. This E.coli strain was kindly provided by M.J.Ares. Both proteins were purified using glutathione agarose as described in Abou Elela et al. (1996).
For the Rnt1p/Nop1p in vitro interaction, GST or GST–Rntp was incubated with 25 µl bed volume of glutathione–Sepharose beads in 300 µl of NET buffer (20 mM Tris pH 8, 100 mM NaCl, 0.2 mM PMSF) for 30 min at 4°C. After extensive washing in 1 M NaCl and then in 100 mM NaCl, [35S]methionine-labeled Nop1p and Snu13p were incubated with GST and GST–RNT pre-bound beads. Incubation was allowed to proceed for 30 min at 4°C. Beads were treated with RNase A and washed four times with NET100. Bound proteins were then recovered by heating for 4 min at 95°C in loading buffer and analyzed by SDS–PAGE.
Oligonucleotides
The sequences of the oligonucleotides used in this study are (5′–3′):
yU18 up, GCGAGCTCGTCCAACCGAATATA;
yU18 down, GCAAGCTTGTTGAACCATCTGAA;
A1, CGTCAGATACTGTGATAG;
B1, ATGAGAACTTTTTTCTT
T7-EIFB/ex1, TAATACGACTCACTATAGGGCATCCACCGATTTCTCCAAG;
U185′ssUP, AATATGTTCCGATTTAGTTTACTTTATAGATCG;
U185′ssDOWN, TTTGAATGTATGACTTGTCTCGG;
T7-U18in, TAATACGACTCACTATAGGGTTACATGTAAAGGGAA;
3′stemU18, CACTCCATTTCCCTTCAGATAACTAATAATGATACTC;
T7-38in, TAATACGACTCACTATAGGCCCGTTCGTTTATTTGGC; and
3′stem38, GGACACGCTTTGTACTTCC.
Acknowledgments
Acknowledgements
We are particularly grateful to Manny Ares for helpful suggestions and for providing purified GST–Rnt1p, GST–RNT1 plasmid and the rnt1Δ strains. We thank Tommaso Villa for helpful advice and all members of the laboratory for collaboration. We wish to thank M.Arceci and G.Ricci for skillful technical help. R.N. was supported by NIH grant GM55557 to M.Ares. This work was partially supported by grants from ‘MURST-CNR Biotechnology Program L.95/95’, from MURST ‘Biotecnologie’ and PRIN 40%, and from CNR ‘Target Project on Biotechnology’ and ‘Tecnologie di base della post-genomica’. This paper is dedicated to the memory of our friend and colleague Professor Franco Tato.
References
- AbouElela S.A., Igel,H. and Ares,M.,Jr (1996) RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell, 85, 115–124. [DOI] [PubMed] [Google Scholar]
- Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A. and Schwer,B. (1995) SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J., 14, 4001–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J.-P., Michot,B., Nicoloso,M., Balakin,A., Ni,J. and Fournier,M.J. (1995) Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci., 20, 261–264. [DOI] [PubMed] [Google Scholar]
- Caffarelli E., Fatica,A., Prislei,S., De Gregorio,E., Fragapane,P. and Bozzoni,I. (1996) Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J., 15, 1121–1131. [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E., Losito,M., Giorgi,C., Fatica,A. and Bozzoni,I. (1998) In vivo identification of nuclear factors interacting with the conserved elements of box C/D small nucleolar RNAs. Mol. Cell. Biol., 18, 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillé J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Abou Elela,S., Ares,M.,Jr and Guthrie,C. (1997) Alternative 3′-end processing of U5 snRNA by RNaseIII. Genes Dev., 11, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Legrain,P. and Jacquier,A. (1998a) Yeast RnaseIII as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol., 284, 975–988. [DOI] [PubMed] [Google Scholar]
- Chanfreau G., Rotondo,G., Legrain,P. and Jacquier,A. (1998b) Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J., 17, 3726–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Buckle,M. and Jacquier,A. (2000) Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl Acad. Sci. USA, 97, 3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Galardi,S., Altieri,F. and Bozzoni,I. (2000a) Fibrillarin binds directly and specifically to U16 box C/D snoRNA. RNA, 6, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Morlando,M. and Bozzoni,I. (2000b) Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J., 19, 6218–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Pelczar,P., Pogacic,V. and Dragon,F. (1999) Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochim. Pol., 46, 377–389. [PubMed] [Google Scholar]
- Filippini D., Bozzoni,I. and Caffarelli,E. (2000) p62, a novel Xenopus laevis component of box C/D snoRNPs. RNA, 6, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier T., Berges,T., Tollervey,D. and Hurt,E. (1997) Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol., 17, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A., Henry,Y., Bousquet-Antonelli,C., Noaillac-Depeyre,J., Gelugne,J.P. and Caizergues-Ferrer,M. (1998) Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J., 17, 7078–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady B.E. and Kiss,T. (2001) A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz J.E. and Holm,C. (1990) Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc. Natl Acad. Sci. USA, 87, 6629–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Linder,P. and de La Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.L. and Tollervey,D. (1998) Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem. Sci., 23, 383–388. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L.J. and Tollervey,D. (1999) Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA, 5, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.L. and Tollervey,D. (2000) Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubben B., Fabrizio,P., Kastner,B. and Luhrmann,R. (1995) Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J. Biol. Chem., 270, 11549–11554. [DOI] [PubMed] [Google Scholar]
- Maxwell E.S. and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- Nagel R. and Ares,M.,Jr (2000) Substrate recognition by a eukaryotic RNase III: the double-stranded RNA-binding domain of Rnt1p selectively binds RNA containing a 5′-AGNN-3′ tetraloop. RNA, 6, 1142–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S.L., Samarsky,D., Fournier,M.J. and Boeke,J.D. (1998) Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA, 4, 1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petfalski E., Dandekar,T., Henry,Y. and Tollervey,D. (1998) Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol., 18, 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L.-H. et al. (1999) Seven novel methylation-guide snoRNAs are processed from a common polycistronic transcript by Rat1 and Rnase III in yeast. Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko,A., Rutz,B., Wilm,M., Mann,M. and Séraphin,B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol., 10, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Schimmang T., Tollervey,D., Kern,H., Frank,R. and Hurt,E.C. (1989) A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J., 8, 4015–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- Smith D.B. and Johnson,K.S. (1988) Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene, 67, 31–40. [DOI] [PubMed] [Google Scholar]
- Terns M.P., Grimm,C., Lund,E. and Dahlberg,J.E. (1995) A common maturation pathway for small nucleolar RNAs. EMBO J., 14, 4860–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen,H., Carmo-Fonseca,M. and Hurt,E.C. (1991) The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J., 10, 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., You,Z.H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- VanHoof A., Lennertz,P. and Parker,R. (2000) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol., 20, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T., Ceradini,F., Presutti,C. and Bozzoni,I. (1998) Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol. Cell. Biol., 18, 3376–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T., Ceradini,F. and Bozzoni,I. (2000) Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J., Newman,D.R., Kuhn,J.F. and Maxwell,E.S. (1998) In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65-kDa box C/D-binding protein. RNA, 4, 582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J. et al. (2000) A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell, 103, 457–466. [DOI] [PubMed] [Google Scholar]
- Weinstein L.B. and Steitz,J.A. (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- Wu P., Brockenbrough,S., Metcalfe,A., Chen,S. and Aris,J.P. (1998) Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J. Biol. Chem., 273, 16453–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L., Watkins,N.J. and Maxwell,E.S. (1997) Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA, 3, 17–26. [PMC free article] [PubMed] [Google Scholar]