Abstract
Tunicates are the only animals that perform cellulose biosynthesis. The tunicate gene for cellulose synthase, Ci-CesA, was likely acquired by horizontal transfer from bacteria and was a key innovation in the evolution of tunicates. Transposon-based mutagenesis in an ascidian, Ciona intestinalis, has generated a mutant, swimming juvenile (sj). Ci-CesA is the gene responsible for the sj mutant, in which a drastic reduction in cellulose was observed in the tunic. Furthermore, during metamorphosis, which in ascidians convert the vertebrate-like larva into a sessile filter feeder, sj showed abnormalities in the order of metamorphic events. In normal larvae, the metamorphic events in the trunk region are initiated after tail resorption. In contrast, sj mutant larvae initiated the metamorphic events in the trunk without tail resorption. Thus, sj larvae show a “swimming juvenile” phenotype, the juvenile-like trunk structure with a complete tail and the ability to swim. It is likely that ascidian cellulose synthase is required for the coordination of the metamorphic events in the trunk and tail in addition to cellulose biosynthesis.
Keywords: Minos, swimming juvenile, Ci-CesA, metamorphosis
Tunicates are the only animals able to perform cellulose biosynthesis (1–3). Cellulose synthase is an enzyme that has a central role in cellulose biosynthesis; the gene (Ci-CesA) for cellulose synthase is encoded in the Ciona intestinalis genome and expressed in the larval ectoderm (4). Recent molecular phylogenetic analyses of Ci-CesA and Cs-CesA of Ciona savignyi suggest that the gene has been acquired by horizontal transfer from bacteria (5, 6). Ascidian cellulose synthase may be involved in the formation of the tunic, the cellulose-containing structure that surrounds the surface of the body to protect against predators. However, functional analysis showing that ascidian cellulose synthase contributes to the tunic formation in vivo has not been reported.
Mutant analysis is a powerful way to understand gene function. In an ascidian, C. savignyi, mutagenesis with a chemical mutagen N-ethyl-N-nitrosourea, successfully revealed the function of prickle (7) and tyrosinase (8). Recently, germ-line transgenesis has been achieved in two ascidians, C. intestinalis and C. savignyi, with a Tc1/mariner superfamily transposon Minos (9–14). The insertion of Minos into the Ciona genome indicates that Minos can be used as a mutagen, because Minos disrupts the gene function if it is inserted into a genomic region that encodes a gene.
Here, we have isolated a mutant, swimming juvenile (sj), which was caused by an insertion of Minos into the C. intestinalis genome. sj shows defects in the tunic structure of swimming larvae. In addition to the tunic phenotype, they showed an abnormal order of metamorphic events, suggesting that the regulation of the timing of these events is affected in sj mutants. Ci-CesA is the gene responsible for the sj mutant, and the drastic loss of cellulose was observed in the tunic of sj mutants. These results indicate that ascidian cellulose synthase is necessary for normal metamorphic events in addition to cellulose biosynthesis.
Materials and Methods
Constructs. The PCR-amplified transcription termination sequence of hsp 70 of Drosophila melanogaster (derived from pGaTB) was ligated as a PstI fragment in the PstI site of pMiFr3dTPO-gfp (12) to generate pMiTFr3dTPO-gfp.
Insertional Mutant Screening. The generation of founder animals was performed as described (11), except that the concentration of plasmid DNA in the injection solution was 10 ng/μl. When F1 animals grew both sperm and eggs, each animal was put separately into a small bottle to spawn eggs and sperm. Fertilized eggs were cultured in Petri dishes. The phenotypes of these F2 families were examined to isolate mutants.
TUNEL Staining and Incorporation of BrdUrd. Larvae [16, 26, and 96 hours postfertilization (hpf) old] were fixed with seawater containing 3.7% formaldehyde for 20 min at room temperature, and TUNEL staining was performed as described (15, 16). In 26- and 96-hpf-old larvae, TUNEL-positive cells were observed. Three-day-postfertilization (dpf) larvae and juveniles were incubated in seawater containing 100 μM BrdUrd (Nacalai Tesque, Kyoto) for 30 min, and they were fixed with 3.7% formaldehyde overnight at 4°C. The subsequent procedures are described elsewhere (17).
Southern Blot and PCR Analyses. To isolate genomic DNA (18), sperm of wild-type or sj heterozygous animals were incubated in HMW buffer (10 mM Tris·Cl, pH 8.0/150 mM NaCl/10 mM EDTA/0.1% SDS) containing 400 μg/ml Proteinase K (Merck) overnight at 50°C, followed by extractions with phenol, phenol-chloroform, and chloroform. Isolated DNA was cut completely with EcoRI or HindIII, electrophoresed, and blotted on Hybond N+ nylon membranes (Amersham Biosciences). Digoxigenin-labeled probes were synthesized by DIG High Prime (Roche Diagnostics), and membranes were hybridized with the probes and washed under high-stringency conditions. Signals were detected by CDP-Star (Amersham Biosciences).
Approximately 3 μg of the mixture of genomic DNA and nucleotides isolated from sperm of sj heterozygous mutants was digested completely with HindIII and self-ligated with T4 DNA ligase (Takara Shuzo, Kyoto). DNA was precipitated with ethanol and dissolved in 50 μl of 0.1 × TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). Thermal asymmetric interlaced (TAIL)-PCR was performed as described (11, 19) using 1 μl of DNA solution as a template for each PCR.
Genomic DNA of mutant or normal larvae and juveniles was isolated with HMW buffer containing Proteinase K followed by phenol, phenol-chloroform, and chloroform extraction. After ethanol precipitation, DNA was dissolved in 40 μl of D3W. One microliter of DNA solution was used as a template for PCR with AccuTaq DNA polymerase (Sigma). The volume of PCR solution was 20 μl, which was prepared according to the instruction manual. PCR conditions were 30 s at 98°C, 40 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min at 68°C, followed by 10 min at 68°C. If bands did not appear, 1 μl of the first PCR product was dissolved in 50 μl of D3W, and 1 μl of the solution was subjected to a second PCR. The cycles of the second PCR were 30 s at 98°C, 25 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min at 68°C, followed by 10 min at 68°C. Primers were 5′-tgcagtcattcaagcatgccac-3′ and 5′-ctacggccttcagagaatccgc-3′ for the first PCR, 5′-cgtcataatctgtgtagattgc-3′ and 5′-aggcagctggtgatatttacgg-3′ for the second PCR, and 5′-cgggatccggtgagcaagggcgaggagctg-3′ and 5′-gcattgaattcttacttgtac-3′ for detection of gfp. Primers used for RT-PCR were 5′-agcagtgggaagaacgggagag-3′ and 5′-tcctggatatacgtcgacgctg-3′ for Ci-CesA, 5′-tcaggtccaaatggtgatgtgc-3′ and 5′-aaccgaatccaatgcggatacc-3′ for Ci-Epi1, acacgtatcagtatgaagacac-3′ and cgcattctgtgacatgttcttc-3′for ciad083d01, respectively. The second round of PCR was done with primers 5′-actgtaacattagttggtcaaa-3′ and 5′-ccgcactgacaagcttcagaac-3′ for Ci-CesA, 5′-gagcaacgacagtctcgtcatc-3′ and 5′-ccgccacagcagtgccatggtg-3′ for Ci-Epi1, and 5′-tgtaacgaccgacttatagcac-3′ and 5′-agtattcgaatgtgccacgttg-3′ for ciad083d01, respectively.
Whole-Mount in Situ Hybridization. Whole-mount in situ hybridization was performed as described (20).
Morpholino Oligonucleotide (MO) Injection. The sequences of MO specific to Ci-CesA and control MO are 5′-cttgatttcgatctggtctgctcat-3′ and 5′-cctcttacctcagttacaatttata-3′, respectively. The concentration of MO in the microinjection solution was 0.5–1.0 mM (21). MO was injected into fertilized eggs with chorion. The volume of injected solution was ∼5 pl.
Cellulose Staining. Larvae were fixed with 3.7% formaldehyde and dehydrated with ethanol before use. Rehydrated samples were pretreated with proteinase K (Merck), chitinase (Sigma), chondroitinase (Seikagaku Kyoto, Tokyo), and hyaluronidase (Sigma) to increase the permeability of the following reagents and to reduce the background signals. Samples were postfixed with 4% formaldehyde. For cellulose-binding domain (CBD) labeling, samples were incubated in PBST containing 10% goat serum (Funakoshi, Tokyo) (GS/PBST) for 10 h at room temperature (RT) and incubated in GS/PBST containing 0.1 mg/ml CBD from Clostridium cellulovorans (Sigma) for 1 h at RT.After washing with PBST containing 1% goat serum and 0.4 M NaCl (GS/PBST-W), samples were incubated in GS/PBST containing 1/10,000 mouse anti-CBD monoclonal antibody (Sigma) for 45 min at RT. After washing with GS/PBST-W, the primary antibody was detected with the Mouse ExtrAvidin alkaline phosphatase staining kit (Sigma). Alkaline phosphatase activity was visualized with nitro blue tetrazolium/bromo-chloro-indoryl phosphate.
Results
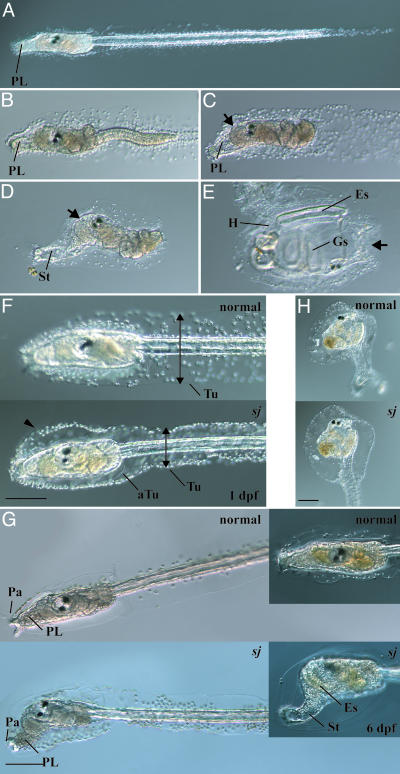
Phenotypes of swimming juvenile Mutant. After early embryogenesis, ascidians become typical tadpole-like larvae resembling those of vertebrates (Fig. 1A). Tadpole larvae dramatically change their shape to be sessile adults through metamorphosis (22). The larvae do not have the ability to initiate metamorphosis soon after hatching. This stage is called “precompetent” (23). Several hours after hatching, they acquire the competence for metamorphosis (24). They secrete a mucous substance from the papillae (Fig. 1 A) and adhere to the substrate. After adhesion, sequential metamorphic events take place, which are divided into 10 events, including the retraction of papillae, tail resorption, body axis rotation, and development of adult organs (Fig. 1 B–E) (22). These events are performed with strict timing in the above order.
Fig. 1.
Metamorphosis of C. intestinalis and phenotypes of the sj mutant. (A) A swimming larva. PL, preoral lobe. (B) A larva during tail resorption. (C) After the completion of tail resorption, the preoral lobe becomes long and transparent and is called the stalk (St in D). (D) During the formation of the stalk, body axis rotation is initiated, and the trunk rotates by 90°. The oral siphon is shown by an arrow. (E) A juvenile that has completed body axis rotation. Adult organs such as gill slits (Gs), endostyle (Es), and heart (H) are formed. The position of the oral siphon (arrow) is completely different from the initial position (an arrow in D). (F) At 1 dpf, the larval tunic (Tu) of a sj mutant larva (sj) was narrower at the tail (arrows), whereas it swelled in the trunk region (arrowhead), compared with a normal larva (normal). The adult tunic (aTu) of a sj larva was normally formed inside the larval tunic. (Bar, 100 μm.) (G) At 2 dpf, a sj mutant started body axis rotation without tail resorption. (Bar, 100 μm.) (Insets) At 6 dpf, the preoral lobe of a sj mutant larva became long and transparent to form the stalk (St). The endostyle (Es) became longer in the sj larva than a normal larva. (H) A metamorphosed sj mutant (Lower) showed normal body organization compared with normal juveniles (Upper). (Bar, 100 μm.)
The sj mutant underwent normal embryogenesis to produce normal swimming larvae. The first detectable abnormality in sj animals was the structure of the tunic (Fig. 1F). In normal larvae, a thin tunic layer expands in the dorsal and ventral directions to form a fin at the tail (22, 25). In sj mutant larvae, the tail tunic was narrower than that of wild-type larvae, and the normal fin did not develop (Fig. 1F, arrows). In the trunk region, the larval tunic of sj larvae swelled (Fig. 1F, arrowhead). After the acquisition of competence, the sj larvae initiated the first event of metamorphosis, the retraction of the adhesive papillae, even though the larvae continued swimming. At 2 dpf, the papillae of sj larvae were almost degenerated, and the body axis rotation was initiated (Fig. 1G). Therefore, the trunk region of sj mutants (Fig. 1G) looked like normal young juveniles (Fig. 1D) even though the sj larvae continued swimming with a tail (we therefore named this mutant swimming juvenile). Over several days after hatching, the preoral lobe of sj mutants became longer and transparent, and it looked like the stalk of young juveniles (Fig. 1 D and G Insets). These changes in morphology suggest that sj mutants skipped the event of tail resorption but continued metamorphic events in the trunk region.
The frequency of sj mutants with the defect of the tunic at 1 dpf met well with Mendel's law of a single heterozygous locus (Table 1), suggesting that one gene is responsible for this phenotype. On the other hand, the frequency of larvae showing abnormal metamorphic events at 2 dpf was far from the score expected from Mendel's law (Table 2). At 2 dpf, a certain number of individuals completed metamorphosis, and we could not judge whether these metamorphosed juveniles had defects in the order of metamorphic events. To examine whether the two defects, one in the tunic and the other in the metamorphic events, are caused by the same mutation, the mutant larvae showing the defect in the tunic at 1 dpf were separated from normal larvae. These mutant larvae were cultured 1 more day and observed to see whether they showed abnormal metamorphic events. All larvae that continued swimming with the abnormal tunic showed the papillae retraction and body axis rotation without tail resorption, whereas no normal larvae showed such abnormal metamorphosis (Table 3). Therefore, we concluded that the two defects, one in the tunic and the other in the metamorphosis, are strongly correlated, and they probably derive from the same single causal gene.
Table 1. Ratio of sj mutants with swelled tunic at 1 dpf.
| Family identification* | No. of larvae examined | Normal larvae, % | Mutant larvae, % | Unknown,† % |
|---|---|---|---|---|
| 1 | 24 | 75 | 25 | 0 |
| 2 | 145 | 73 | 26 | 0 |
| 3 | 182 | 74 | 24 | 1 |
| 4 | 45 | 75 | 24 | 0 |
All families were generated by the crossing of two sj heterozygous animals.
Animals whose phenotypes could not be judged because of abnormal development.
Table 2. Ratio of sj mutants showing abnormal metamorphic events at 2 dpf.
| Family identification* | No. of larvae examined | Normal larvae, % | Mutant larvae, % |
|---|---|---|---|
| 5 | 60 | 58 | 41 |
| 6 | 77 | 61 | 38 |
| 7 | 142 | 66 | 33 |
| 8 | 290 | 34 | 65 |
Only the number of swimming larvae was counted, and metamorphosed animals were excluded.
All families were generated by crossing two sj heterozygous animals.
Table 3. Correlation of the phenotypes in the tunic with those in the metamorphic events.
| Family identification | One-dpf phenotypes | No. of larvae examined | No. of normal larvae at 2 dpf | No. of larvae showing defects in metamorphosis at 2 dpf | No. of metamorphosed larvae at 2 dpf | No. of larvae lost by 2 dpf |
|---|---|---|---|---|---|---|
| 3 | Normal | 135 | 61 | 0 | 56 | 18 |
| 3 | Abnormal tunic | 44 | 0 | 38 | 3 | 3 |
| 4 | Normal | 34 | 11 | 0 | 23 | 0 |
| 4 | Abnormal tunic | 11 | 0 | 10 | 1 | 0 |
If sj and normal larvae have the same frequency to settle and initiate tail resorption, their proportion in 2-dpf-old larvae should obey Mendel's law. But the frequency of sj mutant larvae with defects in metamorphosis at 2 dpf was always higher than that expected from Mendel's law (Table 2). One possible explanation of this phenomenon is that sj mutant larvae undergo the settlement and tail resorption less frequently than normal larvae, and consequently the population of sj larvae became higher than expected. To examine whether sj larvae have a defect in the ability to perform the tail resorption, we separated sj larvae at 1–2 dpf from normal larvae to observe their metamorphosis. Although the frequency was <10% in the two experiments shown in Table 3 and other experiments whose data were not shown, some of the sj larvae started and completed the tail resorption, suggesting that sj larvae have the ability of tail resorption (Fig. 1H). Because apoptosis is necessary to initiate tail resorption (15), apoptosis in sj larvae was examined by TUNEL. Clear TUNEL-positive cells were observed in the tail of the sj mutants (Fig. 4A, which is published as supporting information on the PNAS web site), suggesting that apoptosis takes place normally in sj mutants. Therefore, it is highly likely that sj larvae do not have a deficiency in the mechanism responsible for tail resorption. Rather, the pathway that initiates the metamorphic events in the trunk may be triggered without tail resorption. The loss of papillae before settlement may affect the efficiency of the settlement, which triggers tail resorption. To address this question, normal and sj larvae were sorted into different Petri dishes, and the number of attached animals was counted. The number of attached sj mutants was far less than that of normal larvae (Table 4, which is published as supporting information on the PNAS web site), suggesting less-efficient attachment of sj larvae.
In the sj larvae that continued swimming, body axis rotation was initiated without tail resorption, but body axis rotation was never completed, and thus they ceased further metamorphic events in the trunk, including adult organ formation (Fig. 1G Insets). In contrast, sj juveniles that performed tail resorption completed body axis rotation and formed adult organs, so that they became juveniles with normal morphology (Fig. 1H). This suggests that an additional pathway associated with tail resorption is necessary for the completion of the metamorphic events in the trunk. Recently, we found that the initiation of body axis rotation is independent of cell proliferation, whereas the completion of body axis rotation and adult organ formation depends on cell proliferation (17). It is possible that cell proliferation is arrested in swimming sj larvae, and this causes the arrest of metamorphic events. To examine this possibility, cell proliferation in sj larvae was observed by monitoring the incorporation of BrdUrd. Only a few cells in the trunk of 3-dpf-old sj swimming larvae showed BrdUrd incorporation, compared with normal larvae and juveniles at the same age (Fig. 4B). Therefore, the tail resorption-associated pathway may reset the initiation of cell proliferation to complete metamorphic events.
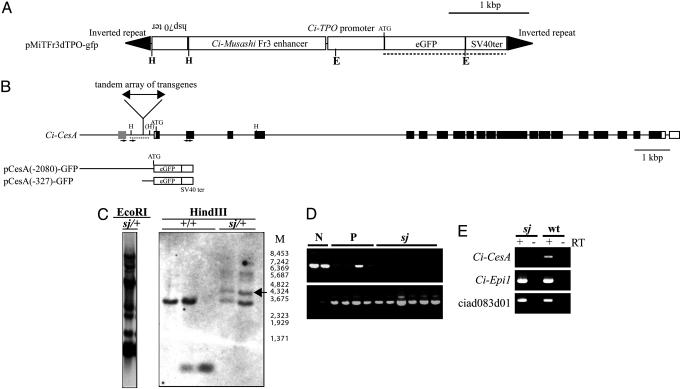
Ci-CesA Is the Gene Responsible for the sj Mutant. The sj mutant was obtained by Minos-based insertional mutagenesis (10–14). Our Minos construct contains gfp as a reporter gene (Fig. 2A), and sj heterozygous animals have a single position of Minos insertion according to the inheritance frequency of gfp in the next generations (data not shown). All sj mutants showed GFP expression; namely, 100% of sj larvae were GFP-positive (n = 70), whereas 73.9% of normal larvae and metamorphosed juveniles were GFP-positive (n = 165). This strong correlation between the sj phenotype and GFP expression suggests that sj is an insertional mutant, or that the sj locus is located very close to the Minos insertion. Southern blotting indicated that a large tandem repeat array of transgenes was inserted into a single locus (Fig. 2C Left). To identify the gene responsible for the sj mutation, the insertion site of Minos was identified by thermal asymmetric interlaced-PCR (19), which amplified flanking sequences of the transgene. Results showed that the inverted repeats of Minos were at both ends of the tandem array of transgenes, and the array was inserted into a TA dinucleotide, which was located 327–328 bp upstream of the transcription start site of Ci-CesA (Fig. 2B). The insertion manner suggests that the transgene was inserted as a large transposon. The insertion position was confirmed by genomic Southern blot analysis (Fig. 2C Right). PCR analysis with primers designated to distinguish homozygous or heterozygous/nontransgenic larvae revealed that sj animals were homozygous with respect to the insertion in the Ci-CesA promoter (Fig. 2D), supporting that this insertion causes the sj mutant. There are no predicted gene models around the Minos insertion in the public genome browser of C. intestinalis (4). Only one EST (rciad083d01) is present around the region (Fig. 2B, gray box). The expression of ciad083d01 was not affected in sj mutants (Fig. 2E), indicating that this gene is not responsible for sj.
Fig. 2.
Mapping of the insertion site of Minos transposon in the sj mutant genome. (A) The design of a Minos transposon used for the insertional mutagenesis. E and H indicate the restriction sites of EcoRI and HindIII, respectively. (B) Location of the Minos insertion site in the sj mutant genome. Open and filled boxes represent exons of Ci-CesA corresponding to the UTR and ORF, respectively. A gray box indicates the position of an EST (rciad083d01) in the public genome browser of C. intestinalis (http://genome.jgi-psf.org/ciona4/ciona4.home.html). H, the restriction sites of HindIII. One restriction site showed polymorphism in the Japanese population of C. intestinalis and is shown as (H). pCesA(-2080)-GFP and pCesA(-327)-GFP indicate constructs used for the promoter analyses in Fig. 3C. (C) Genomic Southern blot analyses of sj mutants. (Left) Genomic DNA from a sj heterozygous animal (sj/+) digested with EcoRI was detected with the probe shown in A (dotted line). Multiple bands indicate the formation of a tandem repeat array. (Right) Genomic DNA from wild-type animals (+/+) and sj heterozygous animals (sj/+) digested with HindIII were detected by the probe shown in B (dotted line). M indicates the size marker. sj heterozygous animals showed specific bands (arrow) that were expected from the fusion of a part of pMiTFr3dTPO-gfp and genomic DNA. (D) An example of PCR analysis showing the homozygosity of the Minos insertion in sj mutants. DNA from larvae with normal appearance without GFP signal (N), normal appearance with GFP expression (P), and sj mutants (sj) were subjected to PCR by primers shown in B (small arrows). The absence of a band indicates the homozygosity of insertion. (Lower) The results of PCR that detects gfp (positive control). (E) A dramatic reduction of Ci-CesA expression in sj mutants (sj) revealed by RT-PCR. Ci-Epi1, which is expressed in epidermis (36), was used as a positive control. The expression of ciad083d01 was not affected in sj mutants. Negative controls without reverse transcription are shown by RT – lanes.
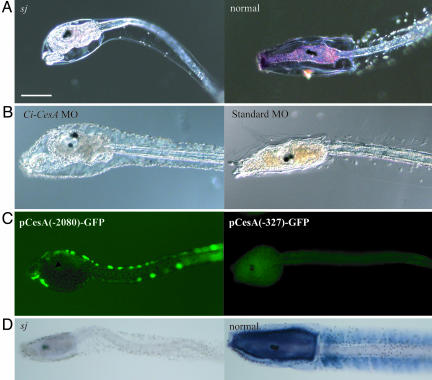
To examine the expression of Ci-CesA in the sj mutant, whole-mount in situ hybridization was performed. Ci-CesA mRNA was not detected in sj mutant larvae (Fig. 3A). RT-PCR also showed a dramatic reduction of Ci-CesA transcripts in sj mutants (Fig. 2E). To confirm that Ci-CesA is the gene responsible for the sj mutant, knockdown of Ci-CesA was performed by microinjection of MO into wild-type fertilized eggs. The MO-injected animals showed both phenotypes, the abnormality in the structure of the tunic and abnormal metamorphic events in the trunk without tail resorption (Fig. 3B). These results provide additional evidence that Ci-CesA is the gene responsible for the sj phenotypes. Namely, Minos insertion in the gene promoter destroys its function. The disruption of Ci-CesA expression by Minos insertion was further confirmed by promoter analyses. The Ci-CesA promoter with the 2,080-bp upstream region (Fig. 2B) drove reporter gene expression in the epidermis of swimming larvae (Fig. 3C). When the promoter was deleted at the position of Minos insertion (Fig. 2B), the expression of the reporter gene was lost (Fig. 3C), supporting the notion that Minos insertion at the promoter region of Ci-CesA impaired its expression.
Fig. 3.
Ci-CesA is the gene responsible for the sj mutation. (A) Expression of Ci-CesA was not detected in a sj mutant (sj), whereas the expression is evident at the trunk of a normal larva at the same age (normal). (Bar, 100 μm.) (B) Microinjection of MO specific to Ci-CesA caused the same phenotype as the sj mutant. A larva (Left) that developed from an egg into which Ci-CesA MO was injected showed the abnormal tunic, retraction of papillae, rotation of body axis, and stalk formation. A larva injected with control MO did not show such phenotypes (Right). (C) GFP expression from pCesA(-2080)-GFP and pCesA(-327)-GFP. (D) The reduction in cellulose in the sj mutant larva (sj), compared with the presence of cellulose in a normal larva at the same age (normal), as shown by dark blue signals.
Cellulose Biosynthesis in sj Mutants. It was unexpected that both larval and adult tunics were formed in sj mutants. Ci-CesA is the only gene encoding cellulose synthase in the C. intestinalis genome (4, 6), and cellulose synthase plays a critical role in cellulose synthesis in plants and bacteria (26, 27). Hence, it is expected that the loss of cellulose from the tunic of sj mutants would cause malformation of the tunic. To examine the presence or absence of cellulose in sj mutants, we visualized cellulose microfibrils by using the cellulose-binding domain (CBD) peptide and its specific antibody (28, 29). A clear difference in the signal appeared between sj mutants and normal larvae at the same age (Fig. 3D), suggesting that cellulose microfibrils were lost in the tunic of sj mutants. The presence of tunics in both larvae and adults of sj mutants therefore suggests that cellulose is not the only critical component for tunic formation. We successfully cultured a few metamorphosed sj mutants until sperm maturation. The present study could not examine the proportion of sj homozygous animals that grew to mature adults, because culture of them was difficult due to the deficiency in settlement even after metamorphosis. However, the sexual maturation of sj homozygous mutants indicates that the cellulose-containing tunic may not always be necessary for animal viability itself. The tunic of these mutants was very soft and jelly-like compared with that of the wild-type (data not shown), suggesting that cellulose may give the tunic the strength to withstand physical force.
Discussion
In the present study, we analyzed a recessive mutant of animal cellulose synthase, Ci-CesA, in C. intestinalis. We showed that Ci-CesA has three functions: cellulose biosynthesis, proper formation of the tunic, and requirement for the normal procedure of metamorphic events. The lack of cellulose in the tunic of sj mutants clearly shows that animal cellulose synthase functions in cellulose biosynthesis as it does in the endogenous system. Among the functions of Ci-CesA, its involvement in metamorphic events is surprising. The disruption of Ci-CesA leads to uncoordinated metamorphic events in the trunk in which the retraction of papillae, the conversion of the preoral lobe into the stalk, and body axis rotation take place independently of tail resorption. Therefore, Ci-CesA is necessary to coordinate the metamorphic events in the tail and trunk. sj mutant larvae had lower efficiency to settle and to initiate tail resorption. This may be because their adhesive papillae are lost before settlement, which may lessen the adhesive materials and result in lower activity to adhere to the substrate. The lower activity of sj mutants in the settlement suggests that the metamorphic events in the trunk commence after settlement to settle efficiently, and ascidian metamorphosis was evolved with cellulose synthase. It is necessary to reveal which of the components, cellulose or Ci-CesA protein, functions in the metamorphic events. In plants, the reduction in cellulose synthesis caused by mutations of cellulose synthase genes or treatment with chemicals that block cellulose biosynthesis results in the constitutive activation of two signaling pathways, jasmonate and ethylene signaling pathways, which act for defense responses (30–32). Similarly, in yeast, involvement of the cell wall has been suggested in some signaling in response to environmental stimuli (33–35). Therefore, it is possible that the reduction in the amount of cellulose from ascidian tunic might affect the timing of signaling event(s), which might cause the abnormal order of metamorphic events in the larval trunk. It is notable that ascidian metamorphosis is triggered in response to stimuli from the environment (physical contact with the substrate). Further isolation of Minos-mediated insertional mutants with phenotypes similar to the sj mutant will be useful to uncover the relationship between Ci-CesA and metamorphosis. Nevertheless, the present study shows that a gene acquired by horizontal transfer has been incorporated into the system of developmental events. It would be interesting to ask when cellulose synthase was recruited in the system of metamorphosis of tunicates during evolution, and how its recruitment affected the system of metamorphosis. Studies of cellulose biosynthesis in other tunicates will give us insights into these questions. Cellulose is observed in all three classes of tunicates, namely ascidians, thaliaceans, and larvaceans, but only ascidians and thaliaceans perform tail resorption. Future studies of cellulose synthase in C. intestinalis and in other tunicates will reveal how this gene functions in tunicate metamorphosis and how the acquisition of this gene affected the evolution of tunicates.
Supplementary Material
Acknowledgments
We thank Prof. Charalambos Savakis (Crete University, Crete, Greece) and Minos BioSystems, Ltd., for providing the Minos plasmids. We are grateful to the Maizuru Fishery Research Station of Kyoto University; the International Coast Research Center of the Ocean Research Institute of the University of Tokyo; the Marine Biological Laboratory, Graduate School of Science, University of Hiroshima; the Education and Research Center of Marine Bio-resources, Tohoku University; and Dr. Shigeki Fujiwara for collection of Ciona adults. We thank Dr. Tatsuya Oshika and members of Kobe Municipal Suma Aqualife Park for providing the seawater. Dr. Yutaka Satou, Ms. Kazuko Hirayama, and Mr. Yoshikazu Okada are acknowledged for supporting us with experiments and Ciona culturing. Y.S., K.N., and S.A. were Predoctoral and Postdoctoral Fellows of Japan Society for the Promotion of Science with Research Grants 0200967 (to Y.S.), 14001123 (to K.N.), and 1600873 (to S.A.). A.N. was a researcher of the Biodiversity Research of the 21st Century Centers of Excellence (A14). This research was also supported in part by Grants in Aid from the Ministry of Education, Culture, Sports, Science, and Technology Japan and Core Research for Evolutional Sciences and Technology Project (Japan Science and Technology Corporation, to N.S.).
Author contributions: Y.S., S.A., K.N., and N.S. designed research; Y.S., K.N., S.A., T.M., and A.N. performed research; Y.S., K.N., S.A., T.M., and J.-i.A. contributed new reagents/analytic tools; Y.S., K.N., S.A., A.N., J.-i.A., and N.S. analyzed data; Y.S. and N.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: dpf, days postfertilization; MO, morpholino oligonucleotide; sj, swimming juvenile.
References
- 1.Ranby, B. G. (1952) Ark. Kemi. 4, 241–248. [Google Scholar]
- 2.Hirose, E., Kimura, S., Itoh, T. & Nishikawa, J. (1999) Biol. Bull. 196, 113–120. [DOI] [PubMed] [Google Scholar]
- 3.Kimura, S., Ohshima, C., Hirose, E., Nishikawa, J. & Itoh, T. (2001) Protoplasma 216, 71–74. [DOI] [PubMed] [Google Scholar]
- 4.Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M., et al. (2002) Science 298, 2157–2167. [DOI] [PubMed] [Google Scholar]
- 5.Matthysse, A. G., Deschet, K., Williams, M., Marry, M., White, A. R. & Smith, W. C. (2003) Proc. Natl. Acad. Sci. USA 101, 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakashima, K., Yamada, L., Satou, Y., Azuma, J. & Satoh, N. (2004) Dev. Genes Evol. 214, 81–88. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, D, Munro, E. M. & Smith W. C. (2005) Curr. Biol. 15, 79–85. [DOI] [PubMed] [Google Scholar]
- 8.Jiang, D., Tresser, J. W., Horie, T., Tsuda, M. & Smith, W. C. (2005) J. Exp. Biol. 208, 433–438. [DOI] [PubMed] [Google Scholar]
- 9.Franz, G. & Savakis, C. (1991) Nucleic Acids Res. 19, 6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasakura, Y., Awazu, S., Chiba, S., Kano, S. & Satoh, N. (2003) Gene 308, 11–20. [DOI] [PubMed] [Google Scholar]
- 11.Sasakura, Y., Awazu, S., Chiba, S. & Satoh, N. (2003) Proc. Natl. Acad. Sci. USA 100, 7726–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Awazu, S., Sasaki, A., Matsuoka, T., Satoh, N. & Sasakura, Y. (2004) Dev. Biol. 275, 459–472. [DOI] [PubMed] [Google Scholar]
- 13.Matsuoka, T., Awazu, S., Satoh, N. & Sasakura, Y. (2004) Dev. Growth Differ. 46, 249–255. [DOI] [PubMed] [Google Scholar]
- 14.Matsuoka, T., Awazu, S., Shoguchi, E., Satoh, N. & Sasakura, Y. (2005) Genesis 41, 67–72. [DOI] [PubMed] [Google Scholar]
- 15.Chambon, J., Soule, J., Pomies, P., Fort, P., Sahuquet, A., Alexandre, D., Mangeat, P. & Baghdiguian, S. (2002) Development (Cambridge, U.K.) 129, 3105–3114. [DOI] [PubMed] [Google Scholar]
- 16.Jeffery, W. R. (2002) Mech. Dev. 118, 111–124. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama, A., Satoh, N. & Sasakura, Y. (2005) Zool. Sci. 22, 301–309. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning, A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 19.Liu, Y., Mitsukawa, N., Oosumi, T. & Whittier, R. F. (1995) Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama, A., Satou, Y. & Satoh, N. (2002) Differentiation 70, 429–437. [DOI] [PubMed] [Google Scholar]
- 21.Satou, Y., Imai-Satou, K. & Satoh, N. (2001) Genesis 30, 103–106. [DOI] [PubMed] [Google Scholar]
- 22.Cloney, R. A. (1982) Am. Zool. 22, 817–826. [Google Scholar]
- 23.Jackson, G. A. & Strathmann, R. R. (1981) Am. Nat. 118, 16–25. [Google Scholar]
- 24.Ishikawa, M. & Numakunai, T. (1972) Dev. Growth Differ. 14, 75–83. [DOI] [PubMed] [Google Scholar]
- 25.Sato, Y., Terakado, K. & Morisawa, M. (1997) Dev. Growth Differ. 39, 117–126. [DOI] [PubMed] [Google Scholar]
- 26.Delmer, D. P. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 245–276. [DOI] [PubMed] [Google Scholar]
- 27.Doblin, M. S., Kurek, I., Jacob-Wilk, D. & Delmer, D. P. (2002) Plant Cell Physiol. 43, 1407–1420. [DOI] [PubMed] [Google Scholar]
- 28.Goldstein, M. A., Takagi, M., Hashida, S., Shoseyov, O., Doi, R. H. & Segel, I. H. (1993) J. Bacteriol. 175, 5762–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCartney, L., Gilbert, H. J., Bolam, D. N., Boraston, A. B. & Knox, J. P. (2004) Anal. Biochem. 326, 49–54. [DOI] [PubMed] [Google Scholar]
- 30.Ellis, C., Karafyllidis, I., Wasternack, C. & Turner, J. G. (2002) Plant Cell 14, 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cano-Delgado, A., Penfield, S., Smith, C., Catley, M. & Bevan, M. (2003) Plant J. 34, 351–362. [DOI] [PubMed] [Google Scholar]
- 32.Scheible, W.-R. & Pauly, M. (2004) Curr. Opin. Plant Biol. 7, 285–295. [DOI] [PubMed] [Google Scholar]
- 33.Davenport, K. R., Sohaskey, M., Kamada, Y., Levin, D. E. & Gustin, D. P. (1995) J. Biol. Chem. 270, 30157–30161. [DOI] [PubMed] [Google Scholar]
- 34.Kamada, Y., Jung, U. S., Piotrowski, J. & Levin, D. E. (1995) Genes Dev. 9, 1559–1571. [DOI] [PubMed] [Google Scholar]
- 35.Buehrer, B. M. & Errede, B. (1997) Mol. Cell. Biol. 17, 6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamada, L., Shoguchi, E., Wada, S., Kobayashi, K., Mochizuki, Y., Satou, Y. & Satoh, N. (2003) Development (Cambridge, U.K.) 130, 6485–6495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.