Abstract
A primary thermodynamic goal in protein biochemistry is to attain predictive understanding of the detailed energetic changes that are responsible for folding/unfolding. Through use of recently determined free energies of side-chain and backbone transfer from water to osmolytes and Tanford's transfer model, we demonstrate that the long-sought goal of predicting solvent-dependent cooperative protein folding/unfolding free-energy changes (m values) can be achieved. Moreover, the approach permits dissection of the folding/unfolding free-energy changes into individual contributions from the peptide backbone and residue side chains.
Keywords: group transfer free energies, m value, transfer model
A primary thermodynamic goal in the field of protein folding is to understand, at the amino acid level, the energetic changes accompanying protein folding/unfolding. As early as the 1930s, Edwin Cohn (1) developed a way to evaluate interactions between protein groups and solute molecules. His work framed the protein-folding question in terms of transfer free energies, the free-energy changes that accompany the transfer of amino acid side chains and peptide backbone units from water to various solvents (2). Historically, such measurements have contributed substantially to the concepts and measurement of hydrophobicity, hydrogen bonding, and aqueous solvation of proteins: key forces that are fundamental to our understanding of protein structure and stability (1, 3–5).
These earlier ideas were ultimately consolidated into Tanford's transfer model, an approach based on the proposition that the transfer of a native (N) or denatured (D) state of a protein from water to denaturants could be quantified simply by summing the group transfer free energies (GTFEs) of the constituent solvent-exposed parts of N and D (2, 5). By all accounts then, GTFEs should have become a mainstay in the analysis of protein structure and folding. Yet, despite this long history, the potential for GTFEs to quantitatively rationalize native-to-denatured transitions in proteins has yet to be fulfilled. Tanford's approach was conceptually attractive, but it failed to achieve quantitative success. Although Tanford found side-chain transfer to be additive (6), failure in quantification came about in large part because his GTFEs for the peptide backbone unit did not exhibit additivity, and they depended critically on both the concentration scale used in obtaining the quantity and the chemical model of the backbone unit (2, 7, 8).
Recently, we have overcome these obstacles by developing the means to obtain GTFEs for the peptide backbone unit that are additive and independent of both concentration scale and backbone model (9). Our findings provide a powerful and productive means to evaluate the energetics of solvent-induced transitions between native and unfolded proteins, thereby realizing the long-sought goal of applying GTFEs to predict such energetic effects for solvents of interest.
Since Tanford's pioneering work on denaturants, a seminal discovery identified a host of small intracellular organic molecules, called protecting osmolytes, which protect cells against water stress conditions encountered by the organism (10, 11). Water stress environments include extremes of temperature, desiccation, high external osmotic pressure, and even the occurrence of urea inside cells (e.g., mammalian kidney) (10, 12). Organisms that use organic osmolytes for protection against such stresses are widespread in nature, with examples occurring in essentially all taxa (10, 11).
Protecting osmolytes and the classical chemical denaturant urea, a denaturing osmolyte, have opposite effects on proteins. A unifying theme in our work (shown below) is that regardless of whether the solvent is a protecting osmolyte or a denaturing osmolyte, it is the peptide backbone that makes the dominant contribution to the free-energy change between the native and denatured states.
In cases of environmental stresses that are denaturing, nature's incorporation of protecting osmolytes within the cellular milieu stabilizes intracellular proteins while protecting the cell (organism) against water stress (13). This vital strategy, discovered largely by Somero, Yancey, and colleagues (10, 11), establishes a direct link between cellular function and folding energetics, underscoring yet again the central role of solvent–protein interactions in living systems.
The array of naturally occurring osmolytes affect protein stability to differing extents. These compounds include both the protecting osmolytes [e.g., proline, glycinebetaine, sorbitol, sucrose, sarcosine, and trimethylamine N-oxide (TMAO)] and the denaturing osmolyte urea. Osmolytes chosen for the present study range from TMAO, which is strongly structure-inducing, to urea, which is strongly denaturing.
Tanford's transfer model (2), shown in Scheme 1, is a thermodynamic strategy for evaluating the energetics of the overall N→D transition in terms of the residue-specific participants. The N→D conversion at the top of the scheme represents the transition in the absence of osmolyte, and its free-energy change is given by  ; the transition at the bottom of the scheme is the corresponding reaction in the presence of 1 M osmolyte. Free-energy differences (
; the transition at the bottom of the scheme is the corresponding reaction in the presence of 1 M osmolyte. Free-energy differences (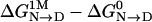 ) for osmolytes of interest are readily obtained from experiment by analyzing data from either denaturation by urea or forced folding of intrinsically unstructured proteins by protecting osmolytes (14, 15) using the linear extrapolation method (LEM). The above free-energy difference obtained from the LEM is equal to the slope (m value) of the linear extrapolation (14). The m value is a measure of the osmolyte-dependent cooperativity of N→D or D→N transitions; it has units of kcal/mol·M–1 and measures the efficacy of the osmolyte in either forcing folding (protecting osmolyte) or unfolding (denaturing osmolyte).
) for osmolytes of interest are readily obtained from experiment by analyzing data from either denaturation by urea or forced folding of intrinsically unstructured proteins by protecting osmolytes (14, 15) using the linear extrapolation method (LEM). The above free-energy difference obtained from the LEM is equal to the slope (m value) of the linear extrapolation (14). The m value is a measure of the osmolyte-dependent cooperativity of N→D or D→N transitions; it has units of kcal/mol·M–1 and measures the efficacy of the osmolyte in either forcing folding (protecting osmolyte) or unfolding (denaturing osmolyte).
Scheme 1.
Transfer model.
The transfer model is a thermodynamic cycle, and the relationship 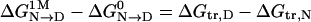 is a resulting consequence. Therefore, the m value must also equal ΔGtr,D – ΔGtr,N, and this equality provides a way to test the transfer model in terms of the additivity of GTFEs (1, 2, 7, 8).
is a resulting consequence. Therefore, the m value must also equal ΔGtr,D – ΔGtr,N, and this equality provides a way to test the transfer model in terms of the additivity of GTFEs (1, 2, 7, 8).
Methods
Specifically, we calculate ΔGtr,D and ΔGtr,N by using GTFEs. These calculated values predict protein m values for N→D or D→N conversion in the presence of various naturally occurring osmolytes. An additional advantage of this procedure is that it explicitly reveals which newly exposed groups contribute energetically and by how much. The successful implementation of this approach has been enabled by solution of the lingering methodological problem of obtaining transfer free energies that are independent of the chosen concentration scale and backbone model (9).
The detailed procedure, provided in the supporting information, which is published on the PNAS web site, is straight-forward. The method of determination of free energy for transfer of the native state from water to 1 M osmolyte, ΔGtr,N, has been previously illustrated (15–17) from knowledge of the three-dimensional structure. The Lee and Richards algorithm (18) is used to calculate solvent-exposed surface from atomic coordinates (18) with a solvent probe of 1.4 Å. Application of the algorithm gives the number of solvent-exposed peptide backbone units along with the numbers of each type of solvent-exposed amino acid side chain. From model compound studies, we have determined GTFE values for side chains and peptide backbone units from water to 1 M concentrations of a substantial number of the naturally occurring organic osmolytes (9, 16, 17, 19). Assuming group additivity, the numbers of backbone units and each type of side chain exposed in the native state are multiplied by their respective GTFEs and summed, providing an estimate of ΔGtr,N for native-state transfer from water to 1 M osmolyte. For example, native RNase T1 transfer to 1 M TMAO gives 2.02 kcal/mol·M–1. The same type of summation is carried out with the denatured state to give ΔGtr,D, except that, in this case, surface areas are based on denatured-state models. Two extreme models are used, one representing a random coil-like denatured state in a good solvent and the other representing a compact denatured state in a poor solvent (20, 21). These models are upper and lower limits that serve to bracket the expected solvent accessibility for a denatured ensemble. By way of example, for the protein RCAM-T1 (reduced and carboxyamidated RNase T1) in 1 M TMAO, to be discussed below, the upper and lower limits are 5.19 and 2.42 kcal/mol·M–1, respectively. The range is not a fault of the transfer model but of the inability in the protein-folding field to thermodynamically define, with accuracy, the denatured state of a protein. Faced with this, Schellman (22) used the arithmetic mean of the limits to represent the denatured ensemble, and for the present work, we do the same. The average exposure of protein groups is available from published tables (21), and Schellman's denatured-state model is derived from these numbers by interpolation.
Results and Discussion
Fig. 1 shows our calculated m values versus corresponding m values determined experimentally by using the LEM. The experimental data are from the osmolyte-induced folding of three proteins that are intrinsically unstructured in aqueous buffer at 25°C: RCAM-T1, the protein component of RNase P (P protein), and the T62P mutant of staphylococcal nuclease (SN) (15, 23, 24). In the absence of salt and/or divalent anions, P protein is by nature intrinsically unstructured. Sulfate ion, high salt, or the RNA component of RNase P causes P protein to fold (24). By contrast, RCAM-T1 and SN T62P are derived from thermodynamically stable proteins and have been made to be intrinsically unstructured by reducing and blocking the two disulfide bonds in RNase T1 and by replacing threonine with proline in the α-helix of SN. These intrinsically unstructured proteins are the only ones we've studied thus far that have known native wild-type crystal structures, thus enabling application of the transfer model. The denatured ensemble of RCAM-T1 is more expanded than the denatured ensemble of its parent, RNase T1, and the denatured ensemble of SN T62P is considered to be similar to the somewhat compact denatured state of SN (25).
Fig. 1.
Calculated m values versus experimentally determined m values. Calculated m values for folding/unfolding of RCAM-T1 (○) as induced by the osmolytes indicated are shown versus m values experimentally determined by means of the LEM. Also included are calculated m values versus experimental TMAO-induced m values determined by Henkels et al. (24) for P protein (⋄) and by Baskakov and Bolen (15) for T62P SNase (□). The slope of the line shown is 1.15 ± 0.07.
As the temperature decreases below 25°C, RCAM-T1 becomes increasingly stable, and urea-induced unfolding m values can be determined (26). Given that the m values are temperature-independent, a result observed with other proteins as well (24, 27, 28), the m value for urea represents the quantity determined from urea-induced unfolding at low temperature (5–15°C) and extended to 25°C.
Fig. 1 shows a strong correlation between calculated and observed m values over a range of ≈5 kcal/mol·M–1. The ability to predict the sign and magnitude of the m value in the presence of seven different osmolytes for three proteins indicates that the assumption of group additivity is valid. Accordingly, model compound transfer free energies are shown to quantitatively predict this key molecular quantity that measures the cooperativity of the N→D transition.
The wealth of detail inherent in the transfer model is readily seen upon dissection into the interactions that account for the m values (Fig. 2). For simplicity, the transition is taken in the direction of denaturation (i.e., N→D); the free-energy contributions of newly exposed groups for RCAM-T1 are shown in 1 M urea (Fig. 2a), 1 M proline (Fig. 2b), and 1 M TMAO (Fig. 2c). These three osmolytes represent the full range of osmolyte efficacies: urea forces proteins to unfold, proline is a weakly stabilizing osmolyte, and TMAO, the most effective protecting organic osmolyte, forces proteins to fold.
Fig. 2.
Side-chain and backbone m value contributions (per Å2) as a function of total surface area (6,520 Å2) newly exposed on denaturation of RCAM-T1. The m value contributions per Å2 were determined as described in the text. Colored bars are for side chains (nonpolar, orange; polar, green; acidic, red; basic, blue), and black bars are for peptide backbone units. The algebraic sum of areas shown in the colored bars equals the calculated m value for the protein. Transfer from water to 1 M urea (a), 1 M proline (b), and 1 M TMAO (c) is shown.
In greater detail, the overall osmolyte-dependent free-energy difference, 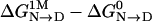 , can be dissected into GTFEs that represent individual free-energy contributions to the m value by constituent groups newly exposed on unfolding, using native-state atomic coordinates and Schellman's model for the denatured state. We define these individual group contributions as ΔΔgtransfer; the groupwise free-energy contribution per unit change in newly exposed accessible surface area is then given by ΔΔgtransfer/ΔASA. Fig. 2 is a plot of (ΔΔgtransfer/ΔASA) against the total newly exposed surface area (6,520 Å2) for RCAM-T1. The area under each bar represents the groupwise free-energy contribution to the m value, and the algebraic sum over all areas in a panel is the calculated m value (see Fig. 1) for that osmolyte.
, can be dissected into GTFEs that represent individual free-energy contributions to the m value by constituent groups newly exposed on unfolding, using native-state atomic coordinates and Schellman's model for the denatured state. We define these individual group contributions as ΔΔgtransfer; the groupwise free-energy contribution per unit change in newly exposed accessible surface area is then given by ΔΔgtransfer/ΔASA. Fig. 2 is a plot of (ΔΔgtransfer/ΔASA) against the total newly exposed surface area (6,520 Å2) for RCAM-T1. The area under each bar represents the groupwise free-energy contribution to the m value, and the algebraic sum over all areas in a panel is the calculated m value (see Fig. 1) for that osmolyte.
The individual panels in Fig. 2 are instructive. In 1 M urea (Fig. 2a), the signs of the groupwise free-energy contributions are all negative, meaning that both newly exposed side chains and backbone interact favorably with solvent, which is why urea is an effective denaturant. In 1 M proline (Fig. 2b), solvent interactions are favorable for newly exposed side chains but are unfavorable for the peptide unit (the osmophobic effect) (23). As given by the experimental m value, the latter contribution slightly outweighs the former, tipping proline toward acting as a protecting osmolyte. In 1 M TMAO (Fig. 2c), solvent interactions with most side chains are favorable, but unfolding is opposed by the large and dominant unfavorable interaction with the peptide backbone. Consequently, TMAO is a highly effective protecting osmolyte.
It is important to recognize the determinative role that the peptide backbone plays in all three osmolytes. Although backbone units represent only a quarter (23%) of the total area newly exposed on unfolding (corresponding to the width of backbone bars in Fig. 2), they make a disproportionately large contribution to the overall free energy (corresponding to the area of the bars) in all cases.
Fig. 2c underscores this point vividly. The TMAO-induced folding of RCAM-T1 is driven by burial of the peptide backbone and, in fact, side-chain burial opposes folding. This result is opposite to the current view regarding protein folding in osmolyte-free buffer, where burial of the nonpolar side chains is the driving force, and burial of the peptide backbone opposes folding (3, 4).
In essence, the driving forces for folding in buffer appear to provide no basis for anticipating the corresponding driving forces in the presence of protecting osmolytes. But when 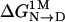 is considered relative to
is considered relative to  , as it is in the transfer model, one gains the perspective that osmolytes act by selective adjustment of the folding forces of unfavorable backbone burial and favorable apolar group burial that are said to dominate in aqueous buffer. A note of caution is in order: Success of the transfer model depends entirely on group additivity, which may not hold in some instances, such as effects arising from clustering of electrostatic surface groups (29), complex configurations (30), perturbed pKs, the presence of disulfide bonds, and variation of solution components such as pH and salt that affect m values.
, as it is in the transfer model, one gains the perspective that osmolytes act by selective adjustment of the folding forces of unfavorable backbone burial and favorable apolar group burial that are said to dominate in aqueous buffer. A note of caution is in order: Success of the transfer model depends entirely on group additivity, which may not hold in some instances, such as effects arising from clustering of electrostatic surface groups (29), complex configurations (30), perturbed pKs, the presence of disulfide bonds, and variation of solution components such as pH and salt that affect m values.
To our knowledge, the preceding examples represent the first demonstration that the transfer model can quantitatively predict the osmolyte-dependent energetics of proteins. The virtue of the transfer model over the denaturant-binding model (5), the LEM (14), Kirkwood–Buff treatment (31), and the two-domain model (29, 32) is that it provides a residue-specific free-energy profile, which, in turn, illuminates the roles and actions of naturally occurring protein denaturants and stabilizers. These results pave the way for more comprehensive investigations of solvent effects.
Supplementary Material
Acknowledgments
Special thanks are extended to Dr. George Rose for editing the manuscript. We also thank Drs. V. Hilser, J. Rösgen, G. Rose, and R. L. Baldwin for their comments on the work. This work was supported by National Institutes of Health Grant GM49760 and Welch Foundation Grant H-1444.
Author contributions: M.A. and D.W.B. designed research, performed research, analyzed data, and wrote the paper.
Abbreviations: LEM, linear extrapolation method; TMAO, trimethylamine N-oxide; GTFE, group transfer free energy; RCAM-T1, reduced and carboxyamidated RNase T1; SN, staphylococcal nuclease.
References
- 1.Cohn, E. J. & Edsall, J. T. (1943) in Proteins, Amino Acids, and Peptides as Ions and Dipolar Ions, eds. Cohn, E. J. & Edsall, J. T. (Reinhold, New York), pp. 196–216.
- 2.Tanford, C. (1964) J. Am. Chem. Soc. 86, 2050–2059. [Google Scholar]
- 3.Dill, K. A. (1990) Biochemistry 29, 7133–7155. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin, R. L. (2005) in Protein Folding Handbook, eds. Buchner, J. & Kiefhaber, T. (Wiley–VCH, Weinheim, Germany), Vol. 1, pp. 127–162. [Google Scholar]
- 5.Tanford, C. (1970) Adv. Protein Chem. 24, 1–95. [PubMed] [Google Scholar]
- 6.Tanford, C. (1962) J. Am. Chem. Soc. 84, 4240–4247. [Google Scholar]
- 7.Nozaki, Y. & Tanford, C. (1963) J. Biol. Chem. 238, 4074–4081. [PubMed] [Google Scholar]
- 8.Nozaki, Y. & Tanford, C. (1965) J. Biol. Chem. 240, 3568–3575. [PubMed] [Google Scholar]
- 9.Auton, M. & Bolen, D. W. (2004) Biochemistry 43, 1329–1342. [DOI] [PubMed] [Google Scholar]
- 10.Yancey, P. H., Clark, M. E., Hand, S. C., Bowlus, R. D. & Somero, G. N. (1982) Science 217, 1214–1222. [DOI] [PubMed] [Google Scholar]
- 11.Hochachka, P. W. & Somero, G. N. (2002) Biochemical Adaptation: Mechanism and Process in Physiological Evolution (Oxford Univ. Press, Oxford), pp. 217–289.
- 12.Yancey, P. H. & Burg, M. B. (1989) Am. J. Physiol. 257, F602–F607. [DOI] [PubMed] [Google Scholar]
- 13.Yancey, P. H. (2005) J. Exp. Biol. 208, 2819–2830. [DOI] [PubMed] [Google Scholar]
- 14.Greene, R. F. J. & Pace, C. N. (1974) J. Biol. Chem. 249, 5388–5393. [PubMed] [Google Scholar]
- 15.Baskakov, I. V. & Bolen, D. W. (1998) J. Biol. Chem. 273, 4831–4834. [DOI] [PubMed] [Google Scholar]
- 16.Liu, Y. & Bolen, D. W. (1995) Biochemistry 34, 12884–12891. [DOI] [PubMed] [Google Scholar]
- 17.Wang, A. & Bolen, D. W. (1997) Biochemistry 36, 9101–9108. [DOI] [PubMed] [Google Scholar]
- 18.Lee, B. & Richards, F. M. (1971) J. Mol. Biol. 55, 379–400. [DOI] [PubMed] [Google Scholar]
- 19.Qu, Y., Bolen, C. L. & Bolen, D. W. (1998) Proc. Natl. Acad. Sci. USA 95, 9268–9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Creamer, T. P., Srinivasan, R. & Rose, G. D. (1995) Biochemistry 34, 16245–16250. [DOI] [PubMed] [Google Scholar]
- 21.Creamer, T. P., Srinivasan, R. & Rose, G. D. (1997) Biochemistry 36, 2832–2835. [DOI] [PubMed] [Google Scholar]
- 22.Schellman, J. A. (2003) Biophys. J. 85, 108–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolen, D. W. & Baskakov, I. V. (2001) J. Mol. Biol. 310, 955–963. [DOI] [PubMed] [Google Scholar]
- 24.Henkels, C. H., Kurz, J. C., Fierke, C. A. & Oas, T. G. (2001) Biochemistry 40, 2777–2789. [DOI] [PubMed] [Google Scholar]
- 25.Baskakov, I. V. & Bolen, D. W. (1998) Biochemistry 37, 18010–18017. [DOI] [PubMed] [Google Scholar]
- 26.Baskakov, I. V. & Bolen, D. W. (1999) Protein Sci. 8, 1314–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giletto, A. & Pace, C. N. (1999) Biochemistry 38, 13379–13384. [DOI] [PubMed] [Google Scholar]
- 28.Pace, C. N. & Laurents, D. V. (1989) Biochemistry 28, 2520–2525. [DOI] [PubMed] [Google Scholar]
- 29.Felitsky, D. J., Cannon, J. G., Capp, M. W., Hong, J., Van Wynsberghe, A. W., Anderson, C. F. & Record, M. T. J. (2004) Biochemistry 43, 14732–14743. [DOI] [PubMed] [Google Scholar]
- 30.Shimizu, S. & Chan, H. S. (2002) Proteins Struct. Funct. Genet. 48, 15–30. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu, S. (2004) Proc. Natl. Acad. Sci. USA 101, 1195–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timasheff, S. N. (2002) Biochemistry 41, 13473–13482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





