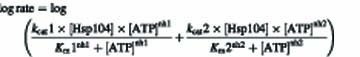Abstract
AAA proteins share a conserved active site for ATP hydrolysis and regulate many cellular processes. AAA proteins are oligomeric and often have multiple ATPase domains per monomer, which is suggestive of complex allosteric kinetics of ATP hydrolysis. Here, using wild-type Hsp104 in the hexameric state, we demonstrate that its two AAA modules (NBD1 and NBD2) have very different catalytic activities, but each displays cooperative kinetics of hydrolysis. Using mutations in the AAA sensor-1 motif of NBD1 and NBD2 that reduce the rate of ATP hydrolysis without affecting nucleotide binding, we also examine the consequences of keeping each site in the ATP-bound state. In vitro, reducing kcat at NBD2 significantly alters the steady-state kinetic behavior of NBD1. Thus, Hsp104 exhibits allosteric communication between the two sites in addition to homotypic cooperativity at both NBD1 and NBD2. In vivo, each sensor-1 mutation causes a loss-of-function phenotype in two assays of Hsp104 function (thermotolerance and yeast prion propagation), demonstrating the importance of ATP hydrolysis as distinct from ATP binding at each site for Hsp104 function.
Keywords: AAA superfamily/ATP hydrolysis/Hsp104/[PSI]/sensor-1
Introduction
The AAA superfamily of proteins couple ATP binding and hydrolysis to an extraordinary variety of cellular activities, including membrane fusion, organelle biogenesis, DNA replication, proteolysis and protein folding. The AAA ATPase module consists of two physical domains, with five conserved sequence motifs, Walker A, Walker B, sensor-1, Box VII (also known as the second region of homology) and sensor-2, which form the active site for ATP hydrolysis (Neuwald et al., 1999). Current models for function of the AAA proteins suggest that ATP binding and hydrolysis at the AAA module function to aid in unfolding proteins or disrupting protein–protein interactions (for reviews see Vale, 2000; Ogura and Wilkinson, 2001). The function of individual AAA sequence motifs in the ATP hydrolysis cycle has been described for some proteins (Karata et al., 1999, 2001; Steel et al., 2000; Putnam et al., 2001; Su’Etsugu et al., 2001), yet the mechanisms that link ATP binding and hydrolysis to protein remodeling remain largely undefined. This is a particularly difficult problem for proteins with multiple AAA modules, as it requires obtaining site-specific information regarding the catalytic properties of each module and how these properties are regulated by the activity of the other site.
Hsp104 of Saccharomyces cerevisiae is a member of the AAA superfamily that was first described as a heat-inducible protein required for thermotolerance, an adaptive response by which a mild stress conditions organisms to survive an otherwise lethal severe stress (Sanchez and Lindquist, 1990; Sanchez et al., 1992). Aggregation of cellular proteins is one consequence of severe stress, and Hsp104 acts directly on protein aggregates to return them to the soluble state (Parsell et al., 1994b; Glover and Lindquist, 1998). Complete recovery of active proteins from the aggregated state by Hsp104 and its Escherichia coli homolog, ClpB, requires the assistance of the Hsp70–Hsp40 chaperone system (Glover and Lindquist, 1998; Goloubinoff et al., 1999; Mogk et al., 1999; Zolkiewski, 1999). At normal growth temperatures, Hsp104 is required for the propagation of the yeast prions [PSI+], [URE3] and [RNQ+] (Chernoff et al., 1995; Moriyama et al., 2000; Sondheimer and Lindquist, 2000). Prions cause heritable phenotypic changes via the propagation of aggregated, inactive protein conformations, and Hsp104 acts in prion maintenance by mediating conversions between the aggregated and soluble states (Patino et al., 1996; Paushkin et al., 1996; Sondheimer and Lindquist, 2000; Ferreira et al., 2001; Wegrzyn et al., 2001). Here too, chaperone proteins of the Hsp70 and Hsp40 families are involved (Chernoff et al., 1999; Newnam et al., 1999; Jung et al., 2000; Sondheimer et al., 2001).
Although the biological functions of Hsp104 are well characterized, due to difficulties of working with aggregated protein substrates, the molecular details of its function remain obscure. It is reasonable to expect that it might function in a manner similar to the related E.coli ClpA and ClpX proteins, which unfold soluble protein substrates for proteolysis (Weber-Ban et al., 1999; Hoskins et al., 2000; Y.I.Kim et al., 2000; Singh et al., 2000). Like Hsp104, these proteins assemble into ring-shaped hexamers with an axial pore (Parsell et al., 1994a; Kessel et al., 1995; Grimaud et al., 1998; Sousa et al., 2000). The substrates that ClpA and ClpX unfold are passed to the ClpP protease via translocation through the axial pore (Ortega et al., 2000; Ishikawa et al., 2001). For ClpA, translocation is a multi-step process with intermediates observable in the axial pore by electron microscopy (Ishikawa et al., 2001), suggesting that the unfolding reaction requires a series of interactions of the protein substrate with distinct binding sites on the chaperone. Alternatively, Hsp104/ClpB might function in a manner distinct from ClpA and ClpX. For example, rather than unfolding substrates by a threading mechanism, it was proposed by Goloubinoff et al. (Diamant et al., 2000; Ben-Zvi and Goloubinoff, 2001) that ClpB uses ATP hydrolysis to disrupt the surface of a protein aggregate, exposing binding sites for the E.coli Hsp70 homolog (DnaK), which extracts proteins via multiple rounds of binding and release.
The Clp/Hsp100 subfamily of AAA proteins is divided into two classes: class I members, such as ClpA, ClpB and Hsp104, have two AAA modules (termed NBD1 and NBD2), while class II members, such as ClpX and HslU, have only one. The presence of 12 ATP binding sites per hexamer of ClpA and Hsp104/ClpB is suggestive of complex allosteric behavior, whereby there may be cooperativity around the ring of NBD1 and NBD2, in addition to regulatory interactions between the two types of binding sites. Indeed, measurements of ATP hydrolysis, fitted to a single-site model in S.cerevisiae Hsp104 (Schirmer et al., 2001) and Thermus thermophilus ClpB (Schlee et al., 2001), identified positive cooperativity. In the case of Hsp104, this was interpreted as cooperative interactions through the NBD1 ring, while for T.thermophilus ClpB, an allosteric interaction between NBD1 and NBD2 was proposed. Site-specific descriptions of the catalytic properties of each site in the hexamer are needed to distinguish between these different interpretations.
A general mechanism for protein recognition by ATPases (and other NTPases) is that ATP and ADP promote different conformational states of the protein with different affinities for a protein substrate or cofactor. Under steady-state conditions, the lifetime of each conformational state is controlled by the dissociation constants for ATP and ADP binding, and by the rate of ATP hydrolysis. Understanding the molecular mechanisms of the Clp/Hsp100 proteins with two ATPase domains requires experimental manipulation of their ATP hydrolysis cycles in a site-specific manner. However, the presence of more than one AAA module precludes one common approach: the use of non-hydrolysable ATP analogs. A second approach is the use of site-directed mutations to reduce the rate of hydrolysis without affecting binding, effectively trapping a given site in the ATP-bound state.
The design of such mutants in the class I Clp/Hsp100 proteins requires a quantitative understanding of the catalytic properties of both ATPase domains. Experiments to define these properties are complicated by three factors. First, the catalytic rates of the two active sites may be so similar that it is difficult to distinguish between them, or extremely different, such that a site with a fast rate masks hydrolysis by a second site whose rate is much slower. In no case have the basal activities of each site in a wild-type class I Clp/Hsp100 hexamer been defined. In Hsp104, NBD1 seems to be responsible for the vast majority of measured ATP hydrolysis (Schirmer et al., 1998) and no catalytic activity has been assigned to NBD2. Secondly, mutations in the Walker A sequence motif often impair hexamer formation. Because the ATP hydrolysis activity of AAA proteins can depend on hexamerization (Akiyama and Ito, 2001; Schirmer et al., 2001) such mutations can reduce ATPase activity from both sites, complicating attempts to assign an observed activity to a given site. Thirdly, the effects of mutations studied to date are not consistent for different class I Clp/Hsp100 proteins. For example, in the ClpA protein, a mutation in NBD1 impairs oligomerization (Singh and Maurizi, 1994), while for Hsp104, it is a mutation in NBD2 that causes assembly defects (Parsell et al., 1994a).
To understand further the linkage between ATP binding and hydrolysis and unfolding of substrate proteins by Hsp104, we developed an assay for ATP hydrolysis that measures the steady-state kinetic parameters (Km, kcat and Hill coefficient) for NBD1 and NBD2 of Hsp104 in the hexamer. Mutations in the AAA sensor-1 motif were introduced to slow ATP hydrolysis at NBD1 and NBD2. These sensor-1 mutations do not impair nucleotide binding or hexamerization, allowing us to assign each observed catalytic activity to NBD1 or NBD2, determine the importance of ATP hydrolysis per se in Hsp104 function in vivo, and identify allosteric communication between NBD1 and NBD2.
Results
Dependence of Hsp104 hexamer formation on ionic strength
ATP hydrolysis by Hsp104 is greatly stimulated upon hexamerization (Schirmer et al., 2001), and hexamerization is affected by the presence of ATP (Parsell et al., 1994a). This complicates kinetic measurements, as there may be variation in the proportion of Hsp104 that is in the hexameric state at the different ATP concentrations that are used for kinetic measurements. Therefore, we first sought conditions where Hsp104 is assembled even in the absence of ATP to focus analysis of nucleotide hydrolysis on protein that is already in the hexameric state.
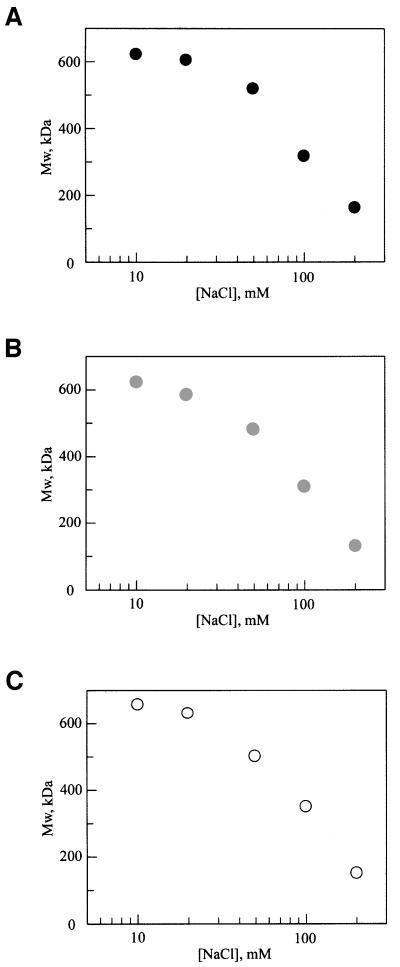
Since the assembly of the Hsp104 homolog ClpB is known to depend on salt concentration (K.I.Kim et al., 2000; Schlee et al., 2001), we directly measured the molecular weight of Hsp104 in increasing NaCl concentrations by static light scattering. Scattering was measured in an unfractionated solution to provide a determination of the average molecular weight of the Hsp104 oligomer at equilibrium. As shown in Figure 1A, wild-type Hsp104 was hexameric at ≤20 mM NaCl, while at higher salt concentrations, the hexamer dissociated. Thus, in a low-salt buffer, the kinetics of ATP hydrolysis by Hsp104 should not be affected by nucleotide-induced assembly.
Fig. 1. The oligomeric state of Hsp104 and sensor-1 mutants depends on ionic strength. The molecular weight determined by static light scattering of an unfractionated protein solution (200 nM monomer concentration) is plotted as a function of NaCl concentration for (A) wild-type Hsp104, (B) Hsp104T317A and (C) Hsp104N728A.
ATP hydrolysis by NBD1 and NBD2 in the Hsp104 hexamer
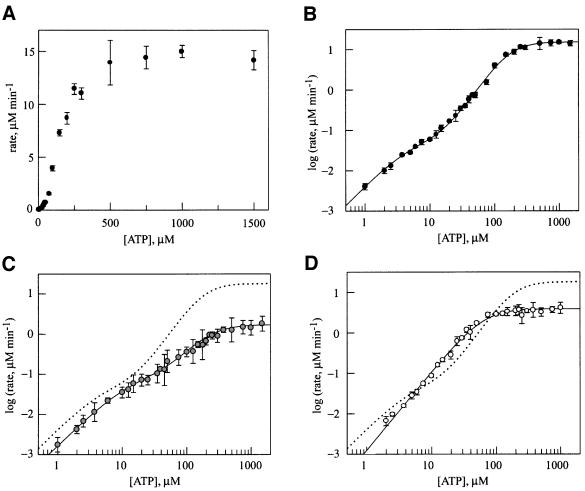
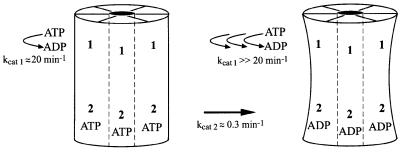
In contrast to previous studies (Schirmer et al., 1998; D.A.Parsell and S.L.Lindquist, unpublished data), our use of conditions to stabilize the hexamer in the absence of nucleotide and a more sensitive radioactive assay for hydrolysis allowed us to distinguish the contribution of both NBD1 and NBD2 to ATP hydrolysis by Hsp104. Because the properties of the two sites are very different, one site is obscured by the other (Figure 2A) unless the data are plotted as the log of the observed initial rate of ATP hydrolysis as a function of ATP concentration (Figure 2B). To provide an initial, general sense of the properties of the two sites, the data in Figure 2B were fitted to an equation describing hydrolysis at two independent allosteric sites. One is a low-affinity site with a relatively high turnover (Km1 = 170 µM, kcat1 = 76 min–1; Table I, top), similar to the previously observed hydrolysis activity suggested to be due to Hsp104 NBD1 (Schirmer et al., 1998) (the Km for NBD1 differs from that determined in the previous study due to the lower NaCl concentration used here). The second site, whose hydrolysis activity was not observed previously, has much higher affinity and a 300-fold slower turnover (Km2 = 4.7 µM, kcat2 = 0.27 min–1; Table I, bottom). Both sites show positive cooperativity, with Hill coefficients of 2.3 and 1.6 for the low- and high-affinity sites, respectively (Table I).
Fig. 2. Steady-state kinetics of ATP hydrolysis by (A) wild-type Hsp104 (linear plot), (B) wild-type Hsp104 (log–log plot), (C) Hsp104T317A (log–log plot) and (D) Hsp104N728A (log–log plot). In each case, the protein concentration was 200 nM (monomer). Data in (B), (C) and (D) were fitted to equations describing either two independent allosteric sites (wild type and Hsp104T317A) or a single allosteric site (Hsp104N728A). In (C) and (D), the dashed line represents ATP hydrolysis by wild-type Hsp104.
Table I. Kinetics of ATP hydrolysis by wild-type Hsp104 and the NBD1 and NBD2 sensor-1 mutants.
| Wild type | T317A | N728A | |
|---|---|---|---|
| kcat1 (min–1) | 75.5 ± 4.5 | 8.2 ± 1.2 | 19.5 ± 1.3 |
| Km1 (µM) | 170 ± 12 | 218 ± 27 | 64 ± 4 |
|
nh1 |
2.3 ± 0.1 |
1.9 ± 0.3 |
1.9 ± 0.1 |
| kcat2 (min–1) | 0.27 ± 0.05 | 0.43 ± 0.20 | n.d. |
| Km2 (µM) | 4.7 ± 1.1 | 13 ± 5.4 | n.d. |
| nh2 | 1.6 ± 0.2 | 1.5 ± 0.1 | n.d. |
Effects of sensor-1 mutations on ATP hydrolysis
The sensor-1 motif that is common to the nucleotide-binding sites of AAA proteins is characterized by a highly conserved hydrogen-bonding amino acid (Neuwald et al., 1999). This residue in the NSF D2 domain (Ser655) is part of a hydrogen-bonding network that positions a water 4 Å from the γ-phosphate of ATP or AMP–PNP (Lenzen et al., 1998; Yu et al., 1998), and this water has been suggested to be the nucleophile for ATP hydrolysis in AAA proteins (Lenzen et al., 1998). This predicts that conservative mutations in this residue should have a strong effect on ATP hydrolysis without deleterious effects on nucleotide binding or protein stability. Thus, for determining the importance of ATP hydrolysis for function of AAA proteins, sensor-1 mutations may provide a more specific tool than mutations in the Walker A motif, which often show undesirable effects on nucleotide binding (Schlee et al., 2001) and quaternary structure (Parsell et al., 1994a).
In Hsp104, the hydrogen-bonding residues of sensor-1 of NBD1 and NBD2 are predicted to be Thr317 and Asn728, respectively (Figure 3). We constructed and purified two mutant proteins, Hsp104T317A and Hsp104N728A, for determining the role of these amino acids in hydrolysis at each NBD. Significantly, neither mutation affected hexamer stability, as the salt dependence of assembly in the absence of nucleotide was identical to that of wild-type Hsp104 (Figure 1B and C).

Fig. 3. Multiple sequence alignment of the sensor-1 motif of E.coli ClpA, ClpB and HslU, S.cerevisiae Hsp104 and Cricetulus griseus NSF. The conserved hydrogen-bonding residue predicted to position the nucleophilic water for ATP hydrolysis is colored red and the two conserved hydrophobic residues are blue. The Hsp104 and ClpB sequences, which were not included in the original AAA superfamily alignment (Neuwald et al., 1999), were aligned with ClpA NBD1 and NBD2 using PSI-BLAST (Altschul et al., 1997).
In comparison to wild-type Hsp104, the most significant effect of the T317A mutation was a nearly 10-fold decrease in the rate of ATP hydrolysis (kcat1; Table I, top) at the low-affinity, high-turnover site (Figure 2C). No significant effects on either the Km or the Hill coefficient for this site were observed. The residual catalytic activity in this mutant may reflect the role of other residues in positioning the water nucleophile, so that it is still present but with lower occupancy. This mutation caused slight increases in the kcat and Km of the second site (kcat2, Km2; Table I, bottom), suggesting that the activity of NBD2 is affected by the hydrolysis cycle at NBD1.
ATP hydrolysis by Hsp104 was more dramatically affected by the N728A mutation (Figure 2D). The primary defect appeared to be a near total loss of ATP hydrolysis at the high-affinity site (NBD2), such that the data could only be fitted to a single kinetic transition. The loss of hydrolysis at NBD2 also affected the steady-state parameters of NBD1; kcat1 was reduced by a factor of four and Km1 changed from 170 to 60 µM (Table I, top). This result clearly reveals a level of allostery between the two active sites of Hsp104, whereby the activity of NBD1 is regulated by ATP hydrolysis at NBD2.
Sensor-1 mutations do not affect nucleotide binding affinity
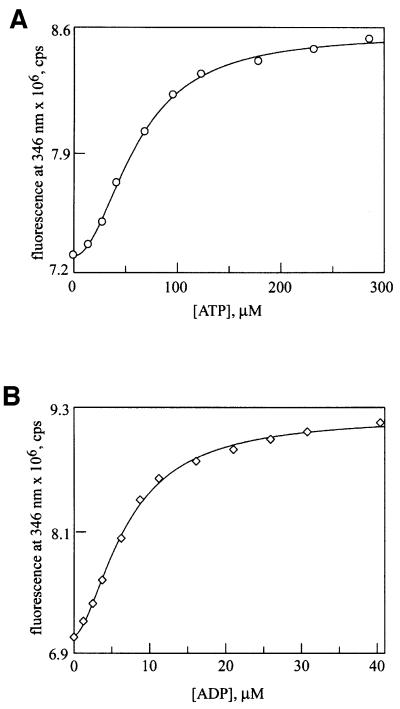
To characterize further the role of the AAA sensor-1 motif, we measured the effects of the N728A mutation on ATP and ADP binding. We took advantage of another mutant, Y819W, which places a single trypto phan residue in the C-terminal domain of Hsp104 and has no effect on its function in thermotolerance (A.Cashikar and S.L.Lindquist, in preparation), and only minor effects on ATP hydrolysis activity (D.A.Hattendorf and S.L.Lindquist, in preparation). The fluorescence of Hsp104Y819W changes in response to nucleotide binding at NBD2 (D.A.Hattendorf and S.L.Lindquist, in preparation). A double mutant, Hsp104N728A,Y819W, was constructed and purified from E.coli. When either ATP (Figure 4A) or ADP (Figure 4B) was titrated into a solution of purified Hsp104N728A,Y819W, a saturable increase in fluorescence was observed. For each nucleotide, the Kd and Hill coefficient determined from a fit of the fluorescence data were nearly identical to those determined for the wild-type protein (D.A.Hattendorf and S.L.Lindquist, in preparation; Table II). Thus, the N728A mutation has no major effect on ATP or ADP binding. Since no ATP hydrolysis is observed at NBD2 in the Hsp104N728A protein, this mutation keeps NBD2 in the ATP-bound state under steady-state conditions.
Fig. 4. Nucleotide binding is unaffected by the N728A mutation. Binding of (A) ATP and (B) ADP to Hsp104N728A,Y819W (400 nM monomer concentration) was monitored by changes in intrinsic tryptophan fluorescence. Data were fitted to a single-site Hill equation.
Table II. Cooperative ATP and ADP binding by wild-type Hsp104 and the NBD2 sensor-1 mutant.
| Y819W | N728A,Y819W | ||
|---|---|---|---|
| ATP | Fmax × 106 | 1.39 ± 0.01 | 1.27 ± 0.03 |
| Kd (µM) | 69 ± 1 | 61 ± 2 | |
| |
nh |
1.8 ± 0.1 |
2.0 ± 0.1 |
| ADP | Fmax × 106 | 2.05 ± 0.06 | 2.14 ± 0.06 |
| Kd (µM) | 9.1 ± 0.3 | 6.9 ± 0.3 | |
| nh | 1.7 ± 0.1 | 1.7 ± 0.1 |
A site-specific probe of nucleotide binding to NBD1 has not been developed, but the residual ATPase activity of NBD1 in the Hsp104T317A mutant provided an indirect measure of the role of sensor-1 in nucleotide binding to this site. Under steady-state conditions, Km is a complex parameter describing the on and off rates for the substrate and all products in an enzymatic reaction. The Km of NBD1 was essentially unaffected by the T317A mutation (Km1, Table I, top), suggesting that the sensor-1 of NBD1 also does not contribute to ATP or ADP binding.
Effects of sensor-1 mutations on Hsp104 activity in vivo
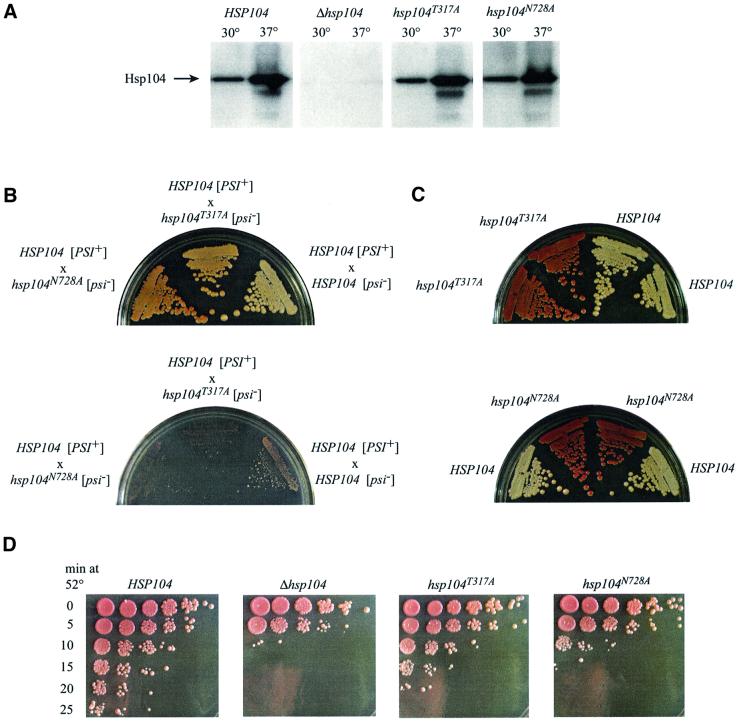
The sensor-1 mutants provide a means to determine the importance of ATP hydrolysis at NBD1 and at NBD2 for Hsp104 function in vivo. The effects of the T317A and N728A mutations were measured in two in vivo assays: propagation of the [PSI+] prion and thermotolerance. We created S.cerevisiae strains containing the T317A or N728A mutation at the Hsp104 locus by the pop-in/pop-out method of gene replacement, yielding strains that contained the mutant as the only copy of the HSP104 gene, under the regulation of its endogenous promoter. Neither sensor-1 mutation caused a significant change in either the basal or the heat-induced expression of the Hsp104 protein (Figure 5A). This is particularly important for analysis of the effects of these mutations on [PSI+] propagation, as methods that alter the normal expression level of Hsp104 by just a few fold can have profound effects on prion maintenance. [PSI+] can be cured not only by deletion of the Hsp104 gene, but also by transient overexpression of wild-type Hsp104 from a single-copy plasmid under the control of its own promoter (Chernoff et al., 1995). As a result, it is difficult to distinguish curing by a strong loss-of-function Hsp104 allele from that due to an allele with no deleterious effect (or only a mild effect) whose expression is slightly higher than endogenous Hsp104.
Fig. 5. In vivo phenotypes of Hsp104 sensor-1 mutants. (A) Hsp104 expression in wild-type and mutant strains, determined by western blot analysis of cell lysates. (B) Growth of diploids with one copy of the T317A or N728A mutant allele and one copy of wild-type HSP104 on YPD (top) or SD ade– medium (bottom) at 30°C. (C) Growth of progeny from sporulation of the HSP104–hsp104T317A diploid (top) or HSP104–hsp104N728A diploid (bottom) on YPD at 30°C. (D) Survival of wild-type and mutant cells after exposure to 52°C for various times. Five-fold serial dilutions of treated cells were spotted on agar and incubated at 30°C to determine colony-forming ability. Pigmentation on these plates is less intense than in (B) and (C) because they were grown for a shorter period of time.
The effects of the Hsp104 sensor-1 mutants on propagation of the [PSI+] prion were determined by mating a haploid [psi–] strain (lacking the prion) carrying either the T317A or N728A mutation to a wild-type haploid [PSI+] strain. The diploids were sporulated and tetrads were analyzed for their [PSI] status. Because the [PSI+] element causes low-level read-through of nonsense codons, its presence can be detected by the ability of a strain with a nonsense codon in the ade1 gene to grow on synthetic medium lacking adenine (SD ade–) or by growth as a white colony on YPD. Because of defects in adenine biosynthesis, [psi–] strains grow as red colonies on YPD due to build-up of a colored intermediate on the biosynthesis pathway, and they cannot grow on SD ade–. In control experiments, where both the [psi–] and [PSI+] haploids were wild type at the HSP104 locus, the diploid and all four spores were [PSI+] (Figure 5B and data not shown), reflecting the dominant, non-Mendelian inheritance that is characteristic of yeast prion-like genetic elements (Cox et al., 1988).
Streaks of the diploids resulting from a cross between either the hsp104T317A [psi–] strain or the hsp104N728A [psi–] strain with a wild-type HSP104 [PSI+] strain are shown in Figure 5B. Partial dominant-negative behavior of both mutants on Hsp104 function was evident, reflected as a slight pink color on YPD and a growth defect on SD ade– medium, but the magnitudes of their effects were different. The N728A mutation, which eliminates ATP hydrolysis at NBD2, inhibited wild-type Hsp104 function most strongly, seen in the darker pink color on YPD. The effect of the T317A mutation was weaker, presumably reflecting the residual hydrolysis at NBD1 in the Hsp104T317A protein. These effects were not simply a result of decreasing the level of active Hsp104 protein, since a HSP104/Δhsp104 diploid maintains the [PSI+] element and grows white, not pink, on YPD (L.Li and S.L.Lindquist, unpublished observations). Because neither mutation affects Hsp104 hexamer assembly, the most likely explanation for their dominance is formation of mixed hexamers with wild-type Hsp104.
Upon sporulation of these heterozygous diploids, 2:2 segregation of the [PSI+] element was observed instead of the 4:0 expected for a wild-type diploid. A small percentage of tetrads from the hsp104N728A/HSP104 diploid gave rise to four red colonies on YPD (data not shown), reflecting the stronger inhibition of wild-type Hsp104 by the N728A mutant relative to the T317A mutant. When 2:2 segregation occurred, the [PSI+] phenotype segregated with the wild-type HSP104 locus and the [psi–] phenotype segregated with the sensor-1 mutant allele (Figure 5C). Because the two progeny with wild-type HSP104 exhibited the strong [PSI+] phenotype of the parent, the intermediate color phenotypes observed in the heterozygous mutant diploids reflect a transient increase in the amount of soluble Sup35 in these strains (Patino et al., 1996; Paushkin et al., 1996) and not a heritable change in the type of [PSI+] element (e.g. weak versus strong; Derkatch et al., 1996).
The haploid hsp104T317A and hsp104N728A progeny of the heterozygous diploid were genuinely cured of [PSI+] and were not maintaining it in a cryptic state, as can occur in some genetic backgrounds and crosses (Cox et al., 1988). Cryptic [PSI+] refers to a condition where enough Sup35 protein remains in the prion state to seed the conversion of more protein to this state, but a sufficient level of Sup35 is in the soluble state to fulfill its normal role in translation termination, covering the [PSI+] phenotype (Patino et al., 1996). When the [psi–] cells with the hsp104T317A or the hsp104N728A allele were crossed with a wild-type [psi–] strain, the meiotic progeny produced upon sporulation were all [psi–] (data not shown). If the [PSI+] element had been preserved in a cryptic state, progeny with wild-type HSP104 would have inherited the prion and displayed the [PSI+] phenotype. Thus, neither Hsp104 sensor-1 mutant can support propagation of the [PSI+] prion.
For thermotolerance measurements, [psi–] yeast cultures were given a 39°C pre-treatment to induce the heat-shock response, then incubated at 52°C for increasing amounts of time. The extent of survival at each time point was determined from 5-fold serial dilutions spotted to YPD plates (Figure 5D). Relative to the wild-type strain, the Δhsp104 strain showed ∼625-fold less survival, as seen in the 10 min time point. Each of the sensor-1 mutations decreased survival at high temperature, but again the severity of their phenotypes differed. The hsp104N728A strain exhibited a thermotolerance defect nearly as severe as that of the Δhsp104 strain, while the hsp104T317A strain showed a much milder loss-of-function phenotype. Here too, both sensor-1 alleles were semi-dominant, as both heterozygous sensor-1 diploids showed decreased thermotolerance relative to either a wild-type diploid or a HSP104/Δhsp104 diploid (data not shown). As observed for [PSI+] propagation, the hsp104N728A allele showed stronger dominance than the hsp104T317A allele, which had a very weak effect.
Discussion
For nucleotide-binding proteins involved in protein– protein interactions, the nucleotide hydrolysis cycle regulates switching between distinct conformations that have different affinities for substrates and cofactor proteins. The inactivation of nucleotide hydrolysis in these proteins can be a powerful method to understand how conformational switching and the concomitant change in protein binding specificity are achieved. Here, we individually inactivated both NBD1 and NBD2 of Hsp104 by introducing mutations in the AAA sensor-1 motif of each domain. Notably, each mutation (T317A and N728A) causes a strong reduction in kcat with no significant effect on nucleotide binding. This indicates that in Hsp104, sensor-1 acts in catalysis and perhaps not, as has been suggested for other AAA ATPases (Jeruzalmi et al., 2001; Putnam et al., 2001), to distinguish between ATP and ADP in the binding pocket. The hydrogen-bonding residue in sensor-1 thus would appear to be functionally analogous to Gln61 in the GTPase Ras (Pai et al., 1990), acting to position the nucleophilic water molecule. This suggests that the AAA module hydrolyses ATP by a mechanism similar to other Walker-type NTPases (for reviews see Maegley et al., 1996; Rees and Howard, 1999). We suggest that using mutations with these specific properties may be a general approach to understanding the mechanisms of AAA proteins. It is particularly valuable for proteins with more than one ATPase domain, as it is the only way to manipulate their ATP hydrolysis cycles in a site-specific manner.
Since Hsp104 is a hexamer with two binding sites per monomer, the timing of any conformational changes linked to the ATP hydrolysis reaction is likely to be regulated at both the homotypic (between identical sites on different subunits) and heterotypic (between NBD1 and NBD2) levels. Our analysis of the kinetics of ATP hydrolysis of NBD1 and NBD2 shows that the two sites are very different with respect to kcat and Km, but they each exhibit homotypic cooperativity. In addition, there is communication between the two sites, most strikingly seen in the effect of a mutation in the sensor-1 motif of NBD2 on the activity of NBD1. This provides a basic kinetic framework for further experiments to describe the linkage of ATP binding and hydrolysis at NBD1 and NBD2 to the interaction of Hsp104 with its substrates and cofactor proteins.
The effect of the N728A mutation in NBD2 on the catalytic activity of NBD1 indicates that ATP hydrolysis by wild-type Hsp104 can be roughly described by two independent allosteric transitions, but the activity of NBD1 is in fact regulated by the nucleotide-bound state of NBD2. Because N728A blocks ATP hydrolysis without affecting nucleotide binding, in this mutant, NBD2 should exist primarily in the ATP-bound state during our kinetic measurement. Thus, when NBD2 is bound to ATP, the conformation of NBD1 may be shifted to a state with a distinct catalytic activity (kcat = 19.5 min–1, Km = 64 µM, nh = 1.9; Figure 6). In the wild-type catalytic cycle, switching of NBD2 to the ADP-bound state should cause a corresponding switch in NBD1 to a different state with an increased kcat and Km (this activity cannot be measured directly as it requires trapping NBD2 in the ADP-bound state). A more accurate description of the wild-type Hsp104 ATP hydrolysis cycle may therefore be a model incorporating these different catalytic activities of NBD1, weighted by the fraction of time that NBD2 is in the ATP- and ADP-bound states. However, such fits cannot be applied to the present data (D.A.Hattendorf and S.L.Lindquist, unpublished data). In the wild-type protein, the average of the two catalytic activities of NBD1 can apparently be approximated as a single site. Future development of such a model will require measurement of the rate constants associated with ATP binding, ATP hydrolysis and ADP release at NBD2 to determine the correct weighting factors.
Fig. 6. Model for regulation of ATP hydrolysis at Hsp104 NBD1 by the hydrolysis cycle at NBD2. When ATP is bound to NBD2, hydrolysis at NBD1 is relatively slow; this state predominates in the Hsp104N728A mutant. Upon ATP hydrolysis at NBD2, a presumed conformational change occurs, resulting in a state in which the rate at NBD1 is elevated.
Our results provide the first direct measurement of the catalytic activity of Hsp104 NBD2; moreover, the sensor-1 mutation in NBD2 (N728A) demonstrates that this activity is necessary for Hsp104 function in vivo. Previous descriptions of ATP hydrolysis by Hsp104 and its bacterial homolog ClpB failed to show evidence for two distinct catalytic sites, probably due to the very low turnover number of NBD2 relative to NBD1 and to difficulties associated with nucleotide-induced hexamer assembly. To avoid such problems, we used a more sensitive assay for hydrolysis and a low-salt buffer to reduce variation in the fraction of protein that was in the hexameric state at different ATP concentrations. The earlier apparent lack of ATP hydrolysis by NBD2, taken together with the observation that a mutation in the Walker A motif of NBD2 (K620T) caused defects in nucleotide-induced hexamer assembly (Parsell et al., 1994a), was suggestive of a function for NBD2 reminiscent of the NSF D2 AAA module. In NSF, the D1 ATPase domain has high enzymatic activity, while D2 has no detectable catalytic activity and appears to serve a purely structural role in hexamer stability, as mutations predicted to eliminate hydrolysis in D2 have little effect on its biological activity (May et al., 2001). Such observations led to a general assumption that nucleotide binding to NBD2 of Hsp104 might serve simply to promote hexamerization. However, our recent analysis of hexamer assembly of Hsp104K620T in the absence of nucleotides revealed defects in both protein concentration- and salt concentration-dependent assembly (D.A.Hattendorf and S.L.Lindquist, unpublished data), indicative of an assembly defect that is not related to any nucleotide binding defect. This observation, together with our analysis of the N728A mutation, which is a total loss-of-function allele in vivo and apparently eliminates ATP hydrolysis at NBD2 without affecting nucleotide binding affinity or hexamer stability, clearly indicates that a revised model for the role of NBD2 is required.
In particular, the requirement for catalytic activity at NBD2, together with the extreme differences in catalytic rates of NBD1 and NBD2 of Hsp104, puts constraints on models for Hsp104-mediated aggregate resolubilization. We suggest that hydrolysis at NBD2 regulates switching between two distinct conformations of a protein binding site. NBD1 may regulate a second protein interaction site and, because of its much higher turnover rate, its hydrolysis activity is likely to provide the energy for disruption of aggregates. In one possible manifestation, based on the proposed mechanism of ClpA (Ishikawa et al., 2000), both sites interact with a protein substrate, and communication between them would regulate the processive translocation of protein substrates through the axial pore of the Hsp104 hexamer. Alternatively, NBD2 may act to direct Hsp104 to a protein substrate (aggregate), so that the site controlled by NBD1 can exert a ‘crow bar’-like action to disrupt the aggregate via a ‘bind → conformational change → release’ mechanism that does not involve threading substrates through the pore. NBD2 might also serve a purely regulatory role, such that cofactors that modulate its ATP hydrolysis cycle control the activity of NBD1 and thus the protein remodeling activity of Hsp104.
From a technical standpoint, our data indicate that results of site-directed mutagenesis on AAA proteins with multiple active sites must be interpreted with caution if the kinetics of each site are not defined. We observed heterotypic allosteric effects using mutations in the sensor-1 motif; significantly, both T317A and N728A reduced the rate of hydrolysis at NBD1 to an extent that would have made interpretation very difficult without site-specific information. These heterotypic effects were observed with mutations that specifically reduced kcat without affecting binding affinity. Other types of mutations, such as those in the Walker A motif, which can affect binding affinity and hydrolysis (Schlee et al., 2001), might show different types of heterotypic effects. For ClpB from E.coli, analogous mutations in each Walker A motif caused an increase in bulk ATPase activity (Kim et al., 1998), while for ClpB from T.thermophilus, they caused a near total loss of activity (Schlee et al., 2001). Notably, these effects are distinct from another technical problem: that of mutations in one active site, such as the K620T mutation in the Walker A motif of Hsp104 NBD2 (Schirmer et al., 2001), which affect catalysis at a second site by destabilizing the active, hexameric form of the protein. Because of such problems, we suggest that for determining the mechanism of the Clp/Hsp100 proteins and other AAA proteins it is essential not only to define the kinetic parameters of the wild-type protein in the fully assembled state, but also to employ mutants with very specific catalytic defects.
It is notable that the best studied protein-folding factor, GroEL, is also a ring-shaped oligomer with multiple ATP binding sites. GroEL and Hsp104 share some similarities (e.g. positive cooperativity around the ring and communication between the two ‘rings’), but otherwise the details of their mechanisms are quite distinct. The two rings of GroEL are composed of identical subunits, but in Hsp104, the two ATPase domains actually share very limited sequence homology and have very different kinetic properties. In GroEL, negative cooperativity between the two rings clearly serves to set up the ‘two-stroke’ mechanism of this protein (for review see Horovitz et al., 2001). Whether these two proteins will prove to have fundamentally different mechanisms or to be variations of a similar theme will have significant impact on our concepts of the evolution of protein remodeling mechanisms.
Materials and methods
Mutagenesis
For integrating plasmids, the Hsp104R gene (Schirmer et al., 1998) was subcloned into pRS306 (Sikorski and Hieter, 1989) as a BamHI–SacI fragment to create pRS306Hsp104. For T317A, a mutant cassette was created by two-step PCR mutagenesis and subcloned into pRS306Hsp104 as a EagI–SacII fragment. For N728A, the mutant cassette was inserted as a SpeI–SacI fragment. The T317A mutation created a BstUI restriction site and the N728A mutation created a BsrBI restriction site.
For bacterial expression of the sensor-1 mutant proteins, the relevant mutant cassettes from pRS306Hsp104T317A and pRS306Hsp104N728A were subcloned into pNOTAGHsp104 (D.A.Hattendorf and S.L.Lindquist, in preparation), yielding pNOTAGHsp104T317A and pNOTAGHsp104N728A. For recombinant expression of the N728A Y819W double mutant, a mutant cassette was created by two-step PCR using pNOTAG Hsp104Y819W (D.A.Hattendorf and S.L.Lindquist, in preparation) as a template, then inserted into pNOTAG Hsp104 as a SpeI–SacI fragment. The fidelity of all mutagenesis was verified by DNA sequencing.
Protein purification
Hsp104, Hsp104T317A, Hsp104N728A and Hsp104N728A,Y819W were purified as untagged proteins from E.coli, as described for Hsp104Y819W (D.A.Hattendorf and S.L.Lindquist, in preparation).
Static light scattering
Molecular weight determinations were performed in an unfractionated solution with a DAWN DSP Laser Photometer (Wyatt Technology) at room temperature. Buffer solution (5 ml; 20 mM HEPES pH 7.5, 10 mM MgCl2, 10–200 mM NaCl) was placed into a glass vial and static light scattering at 90° was measured. A protein solution (1 ml) was then added to give a final protein concentration of 200 nM (monomer) and scattering was again measured. The weight-average molecular weight (Mw) was calculated from the buffer-corrected scattering using a Debye plot (Astra v. 4.5; Wyatt Technology). Each data point represents the mean of two independent measurements.
To remove any large particles, all protein and buffer solutions were filtered with 0.22 µm filters and the glass vial was washed with 0.22 µm filtered ethanol. A Sephacryl S400 (Pharmacia) gel filtration column was used as a final step in protein purification to further ensure that no protein aggregates were present.
ATP hydrolysis
All measurements of ATP hydrolysis were performed in 20 mM HEPES pH 7.5, 20 mM NaCl and 10 mM MgCl2 at 30°C. Hydrolysis reactions were initiated by mixing 12.5 µl of an Hsp104 solution (final concentration 200 nM) and 12.5 µl of an ATP solution containing 10 µCi of [γ-32P]ATP (3000 Ci/mmol; Amersham), both pre-equilibrated to 30°C. At varying times (5 min for [ATP] <5 µM; 2 min for 5 µM <[ATP] <15 µM and 1 min for 15 µM <[ATP]), reactions were stopped by addition of 175 µl of 1 M HClO4 and 1 mM NaPO4. Ammonium molybdate was added (400 µl of 20 mM) and the molybdate–inorganic phosphate complex was extracted by addition of 400 µl of isopropyl acetate followed by vortexing. The amounts of radioactivity in the organic phase and in the aqueous phase were determined by scintillation counting and used to determine the percentage of ATP hydrolysis. After subtraction of free phosphate in a blank reaction, the initial reaction rate was calculated. Each data point represents the average of four independent measurements. For fitting, each data point was weighted by the standard deviation of this average.
Plots of the log of initial reaction rates versus ATP concentration were fitted using Grafit v. 4.0 (Erithacus software) to a sum of two independent cooperative kinetic transitions:
In the case of Hsp104N728A, the data were fitted to a single cooperative kinetic transition. Values of kcat are defined per individual subunit in the Hsp104 hexamer.
Nucleotide binding
Nucleotide binding measured by intrinsic tryptophan fluorescence was performed as described (D.A.Hattendorf and S.L.Lindquist, manuscript in preparation). Essentially, ATP or ADP was titrated into a solution of 400 nM Hsp104Y819W,N728A in 20 mM HEPES pH 7.5, 10 mM MgCl2 and 20 mM NaCl at 4°C. After each addition, the solution was equilibrated for 3 min with stirring, then fluorescence at 346 nm (excitation wavelength 295 nm) was measured. Fluorescence was corrected for dilution, then plotted versus nucleotide concentration. Data were fitted using Grafit v. 4.0 (Erithacus software) to the Hill equation:
Yeast strain construction
For gene replacement by the pop-in/pop-out method (Rothstein, 1991), the relevant integrating plasmid was linearized with Eco81I (yielding a single cut 3′ of both mutations), then transformed into S.cerevisiae strain 74D-694a [PSI+] (Mata, ade1-14, trp1-289, his3Δ-200, ura3-52, leu2-3, lys2). Transformants were selected on SD Ura– plates, then grown to saturation in SD Ura– liquid. PCR was performed on the transformants using genomic DNA as a template, with two sets of primers. For T317A, primer sets A (5′-CAAGTAACGCTTGGCTAA and 5′-GGCAAAGGGGCGCAAACTTATG) and B (5′-CAAGTAACGCTTGGCTAA and 5′-GACTGAGCAGGCTCGTCAAG) were used. For N728A, primer sets C (5′-TTTTTTGGATCCGGGATGACAGCTGCAAGATTG and 5′-GGTAACGCCAGGGTTTTCC) and D (5′-TTTTTTGGATCCGGGATGACAGCTGCAAGATTG and 5′-CTAAACGTTAAAATGTGAGCTC) were used. For T317A, transformants (pop-ins) were judged as positive if PCR product B contained a BstUI restriction site and PCR product A did not. For N728A, pop-ins were positive if product D was cut with BsrBI and product C was uncut. Thus, in both pop-ins, only the wild-type Hsp104 gene was under control of a promoter. These pop-ins were grown to saturation in YPD liquid, then plated to 5-FOA to select for recombination events resulting in only one copy of Hsp104 in the genome. Colonies that grew on 5-FOA (pop-outs) were screened by PCR using primer set B or D, followed by restriction digestion. Colonies were also screened by their color (red or white) on YPD, which is a sensitive indicator of the presence of the [PSI+] prion in this strain. For both T317A and N728A, eight pop-outs were screened. Without exception, those colonies with the mutation were red on YPD, while those that were wild type were white on YPD. Thus, the red and white screening is a convenient method for identification of loss-of-function Hsp104 alleles generated by this method in this strain.
For creation of diploids, the hsp104T317A [psi–] and hsp104N728A [psi–] strains were each mated to 74D-694α [PSI+]. Matings and sporulation were performed by standard techniques (Adams et al., 1997). Sporulation plates contained 100 mg/ml adenine. The genotypes of spores were identified by PCR on genomic DNA, using primer set B for T317A and primer set D for N728A. PCR products were analyzed by restriction digestion for the presence of the BstUI or BsrBI site.
Thermotolerance
Strains were grown in YPD liquid at 30°C to log phase (5 × 106 cells/ml), then given a 39°C pre-treatment for 1 h. Following the pre-treatment, aliquots of the cultures were placed in pre-warmed glass tubes at 52°C for increasing times. Heat shock was stopped by removing the tubes from 52°C and placing them on ice. Aliquots (5 µl) of 5-fold serial dilutions of the cultures were plated to YPD and allowed to recover for 3 days at 30°C to assess viability. For western blot analysis of Hsp104 expression, cells were lysed either before or after the pre-treatment and lysates from 1 × 106 cells were separated by SDS–PAGE, then transferred to PVDF membrane. Hsp104 was detected by western blotting with the monoclonal antibody 17B (A.Cashikar and S.L.Lindquist, unpublished data).
Acknowledgments
Acknowledgements
We would like to thank Anneli Asikainen for excellent technical assistance with protein purification and enzymatic assays, Anil Cashikar for helpful discussions and members of the Lindquist laboratory for critical reading of the manuscript. This work was supported by the Howard Hughes Medical Institute and NIH grants to S.L.L. D.A.H. was supported by NIH training grant 5 T32 GM07183.
References
- Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Akiyama Y. and Ito,K. (2001) Roles of homooligomerization and membrane association in ATPase and proteolytic activities of FtsH in vitro. Biochemistry, 40, 7687–7693. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi A.P. and Goloubinoff,P. (2001) Review: mechanisms of disaggregation and refolding of stable protein aggregates by molecular chaperones. J. Struct. Biol., 135, 84–93. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist,S.L., Ono,B., Inge-Vechtomov,S.G. and Liebman,S.W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science, 268, 880–884. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Newnam,G.P., Kumar,J., Allen,K. and Zink,A.D. (1999) Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability and toxicity of the [PSI] prion. Mol. Cell. Biol., 19, 8103–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B.S., Tuite,M.F. and McLaughlin,C.S. (1988) The psi factor of yeast: a problem in inheritance. Yeast, 4, 159–178. [DOI] [PubMed] [Google Scholar]
- Derkatch I.L., Chernoff,Y.O., Kushnirov,V.V., Inge-Vechtomov,S.G. and Liebman,S.W. (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics, 144, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant S., Ben-Zvi,A.P., Bukau,B. and Goloubinoff,P. (2000) Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J. Biol. Chem., 275, 21107–21113. [DOI] [PubMed] [Google Scholar]
- Ferreira P.C., Ness,F., Edwards,S.R., Cox,B.S. and Tuite,M.F. (2001) The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol., 40, 1357–1369. [DOI] [PubMed] [Google Scholar]
- Glover J.R. and Lindquist,S. (1998) Hsp104, Hsp70 and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell, 94, 73–82. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P., Mogk,A., Zvi,A.P., Tomoyasu,T. and Bukau,B. (1999) Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl Acad. Sci. USA, 96, 13732–13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud R., Kessel,M., Beuron,F., Steven,A.C. and Maurizi,M.R. (1998) Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem., 273, 12476–12481. [DOI] [PubMed] [Google Scholar]
- Horovitz A., Fridmann,Y., Kafri,G. and Yifrach,O. (2001) Review: allostery in chaperonins. J. Struct. Biol., 135, 104–114. [DOI] [PubMed] [Google Scholar]
- Hoskins J.R., Singh,S.K., Maurizi,M.R. and Wickner,S. (2000) Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc. Natl Acad. Sci. USA, 97, 8892–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Beuron,F., Kessel,M., Wickner,S., Maurizi,M.R. and Steven,A.C. (2001) Translocation pathway of protein substrates in ClpAP protease. Proc. Natl Acad. Sci. USA, 98, 4328–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H. et al. (2000) Mutational analysis of the functional motifs of RuvB, an AAA+ class helicase and motor protein for Holliday junction branch migration. Mol. Microbiol., 36, 528–538. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D., O’Donnell,M. and Kuriyan,M. (2001) Crystal structure of the processivity clamp loader gamma (γ) complex of E.coli DNA polymerase III. Cell, 106, 429–441. [DOI] [PubMed] [Google Scholar]
- Jung G., Jones,G., Wegrzyn,R.D. and Masison,D.C. (2000) A role for cytosolic hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics, 156, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karata K., Inagawa,T., Wilkinson,A.J., Tatsuta,T. and Ogura,T. (1999) Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J. Biol. Chem., 274, 26225–26232. [DOI] [PubMed] [Google Scholar]
- Karata K. et al. (2001) Probing the mechanism of ATP hydrolysis and substrate translocation in the AAA protease FtsH by modelling and mutagenesis. Mol. Microbiol., 39, 890–903. [DOI] [PubMed] [Google Scholar]
- Kessel M., Maurizi,M.R., Kim,B., Kocsis,E., Trus,B.L., Singh,S.K. and Steven,A.C. (1995) Homology in structural organization between E.coli ClpAP protease and the eukaryotic 26S proteasome. J. Mol. Biol., 250, 587–594. [DOI] [PubMed] [Google Scholar]
- Kim K.I., Woo,K.M., Seong,I.S., Lee,Z.W., Baek,S.H. and Chung,C.H. (1998) Mutational analysis of the two ATP-binding sites in ClpB, a heat shock protein with protein-activated ATPase activity in Escherichia coli. Biochem. J., 333, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.I., Cheong,G.W., Park,S.C., Ha,J.S., Woo,K.M., Choi,S.J. and Chung,C.H. (2000) Heptameric ring structure of the heat-shock protein ClpB, a protein-activated ATPase in Escherichia coli. J. Mol. Biol., 303, 655–666. [DOI] [PubMed] [Google Scholar]
- Kim Y.I., Burton,R.E., Burton,B.M., Sauer,R.T. and Baker,T.A. (2000) Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell, 5, 639–648. [DOI] [PubMed] [Google Scholar]
- Lenzen C.U., Steinmann,D., Whiteheart,S.W. and Weis,W.I. (1998) Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell, 94, 525–536. [DOI] [PubMed] [Google Scholar]
- Maegley K.A., Admiraal,S.J. and Herschlag,D. (1996) Ras-catalyzed hydrolysis of GTP: a new perspective from model studies. Proc. Natl Acad. Sci. USA, 93, 8160–8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A.P., Whiteheart,S.W. and Weis,W.I. (2001) Unraveling the mechanism of the vesicle transport ATPase NSF, the N-ethylmaleimide-sensitive factor. J. Biol. Chem., 276, 21991–21994. [DOI] [PubMed] [Google Scholar]
- Mogk A., Tomoyasu,T., Goloubinoff,P., Rudiger,S., Roder,D., Langen,H. and Bukau,B. (1999) Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J., 18, 6934–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H., Edskes,H.K. and Wickner,R.B. (2000) [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol., 20, 8916–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind,L., Spouge,J.L. and Koonin,E.V. (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation and disassembly of protein complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- Newnam G.P., Wegrzyn,R.D., Lindquist,S.L. and Chernoff,Y.O. (1999) Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol., 19, 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T. and Wilkinson,A.J. (2001) AAA+ superfamily ATPases: common structure–diverse function. Genes Cells, 6, 575–597. [DOI] [PubMed] [Google Scholar]
- Ortega J., Singh,S.K., Ishikawa,T., Maurizi,M.R. and Steven,A.C. (2000) Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol. Cell, 6, 1515–1521. [DOI] [PubMed] [Google Scholar]
- Pai E.F., Krengel,U., Petsko,G.A., Goody,R.S., Kabsch,W. and Wittinghofer,A. (1990) Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J., 9, 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell D.A., Kowal,A.S. and Lindquist,S. (1994a) Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J. Biol. Chem., 269, 4480–4487. [PubMed] [Google Scholar]
- Parsell D.A., Kowal,A.S., Singer,M.A. and Lindquist,S. (1994b) Protein disaggregation mediated by heat-shock protein Hsp104. Nature, 372, 475–478. [DOI] [PubMed] [Google Scholar]
- Patino M.M., Liu,J.J., Glover,J.R. and Lindquist,S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science, 273, 622–626. [DOI] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Putnam C.D., Clancy,S.B., Tsuruta,H., Gonzalez,S., Wetmur,J.G. and Tainer,J.A. (2001) Structure and mechanism of the ruvb Holliday junction branch migration motor. J. Mol. Biol., 311, 297–310. [DOI] [PubMed] [Google Scholar]
- Rees D.C. and Howard,J.B. (1999) Structural bioenergetics and energy transduction mechanisms. J. Mol. Biol., 293, 343–350. [DOI] [PubMed] [Google Scholar]
- Rothstein R. (1991) Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol., 194, 281–301. [DOI] [PubMed] [Google Scholar]
- Sanchez Y. and Lindquist,S.L. (1990) HSP104 required for induced thermotolerance. Science, 248, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Taulien,J., Borkovich,K.A. and Lindquist,S. (1992) Hsp104 is required for tolerance to many forms of stress. EMBO J., 11, 2357–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer E.C., Queitsch,C., Kowal,A.S., Parsell,D.A. and Lindquist,S. (1998) The ATPase activity of Hsp104, effects of environmental conditions and mutations. J. Biol. Chem., 273, 15546–15552. [DOI] [PubMed] [Google Scholar]
- Schirmer E.C., Ware,D.M., Queitsch,C., Kowal,A.S. and Lindquist,S.L. (2001) Subunit interactions influence the biochemical and biological properties of Hsp104. Proc. Natl Acad. Sci. USA, 98, 914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee S., Groemping,Y., Herde,P., Seidel,R. and Reinstein,J. (2001) The chaperone function of ClpB from Thermus thermophilus depends on allosteric interactions of its two ATP-binding sites. J. Mol. Biol., 306, 889–899. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K. and Maurizi,M.R. (1994) Mutational analysis demonstrates different functional roles for the two ATP-binding sites in ClpAP protease from Escherichia coli. J. Biol. Chem., 269, 29537–29545. [PubMed] [Google Scholar]
- Singh S.K., Grimaud,R., Hoskins,J.R., Wickner,S. and Maurizi,M.R. (2000) Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc. Natl Acad. Sci. USA, 97, 8898–8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N. and Lindquist,S. (2000) Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell, 5, 163–172. [DOI] [PubMed] [Google Scholar]
- Sondheimer N., Lopez,N., Craig,E.A. and Lindquist,S. (2001) The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J., 20, 2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M.C., Trame,C.B., Tsuruta,H., Wilbanks,S.M., Reddy,V.S. and McKay,D.B. (2000) Crystal and solution structures of an HslUV protease-chaperone complex. Cell, 103, 633–643. [DOI] [PubMed] [Google Scholar]
- Steel G.J., Harley,C., Boyd,A. and Morgan,A. (2000) A screen for dominant negative mutants of SEC18 reveals a role for the AAA protein consensus sequence in ATP hydrolysis. Mol. Biol. Cell, 11, 1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su’Etsugu M., Kawakami,H., Kurokawa,K., Kubota,T., Takata,M. and Katayama,T. (2001) DNA replication-coupled inactivation of DnaA protein in vitro: a role for DnaA arginine-334 of the AAA+ Box VIII motif in ATP hydrolysis. Mol. Microbiol., 40, 376–386. [DOI] [PubMed] [Google Scholar]
- Vale R.D. (2000) AAA proteins. Lords of the ring. J. Cell Biol., 150, F13–F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Ban E.U., Reid,B.G., Miranker,A.D. and Horwich,A.L. (1999) Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature, 401, 90–93. [DOI] [PubMed] [Google Scholar]
- Wegrzyn R.D., Bapat,K., Newnam,G.P., Zink,A.D. and Chernoff,Y.O. (2001) Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol., 21, 4656–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R.C., Hanson,P.I., Jahn,R. and Brunger,A.T. (1998) Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nature Struct. Biol., 5, 803–811. [DOI] [PubMed] [Google Scholar]
- Zolkiewski M. (1999) ClpB cooperates with DnaK, DnaJ and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J. Biol. Chem., 274, 28083–28086. [DOI] [PubMed] [Google Scholar]