Abstract
The nonsense-mediated decay (NMD) RNA surveillance pathway detects and degrades mRNAs containing premature termination codons (PTCs). T-cell receptor (TCR) and immunoglobulin transcripts, which commonly harbor PTCs as a result of programmed DNA rearrangement during normal development, are down-regulated much more than other known mammalian gene transcripts in response to nonsense codons. Here, we demonstrate that this is not because of promoter or cell type but instead is directed by regulatory sequences within the rearranging VDJ exon and immediately flanking intron sequences of a Vβ8.1 TCR-β gene. Insertion of these sequences into a heterologous gene elicited strong down-regulation (>30-fold) in response to PTCs, indicating that this region is sufficient to trigger robust down-regulation. The rearranging Vβ5.1 exon and the flanking intron sequences from another member of the TCR-β family also triggered strong down-regulation, suggesting that down-regulatory-promoting elements are a conserved feature of TCR genes. Importantly, we found that the Vβ8.1 down-regulatory-promoting element was position dependent, such that it failed to function when positioned downstream of a PTC. To our knowledge, this is the first class of down-regulatory elements identified that act upstream of nonsense codons.
Keywords: down-regulatory elements/nonsense-mediated decay/premature termination codon/T-cell receptor/triose phosphate isomerase
Introduction
Nonsense-mediated decay (NMD) is an RNA surveillance pathway that safeguards gene expression by eliminating abnormal mRNAs (Maquat, 1995; Ruiz-Echevarria et al., 1996; Li and Wilkinson, 1998; Culbertson, 1999; Hentze and Kulozik, 1999). The substrates for this pathway are aberrant transcripts harboring premature termination codons (PTCs) generated by frameshifts, nonsense mutations, splicing errors and transcriptional errors. This pathway is important because it protects eukaryotic cells from the deleterious dominant-negative or gain-of-function effects of the truncated proteins that can be generated from aberrant mRNAs harboring PTCs (Pulak and Anderson, 1993; Cali and Anderson, 1998; Mitrovich and Anderson, 2000).
Although NMD is clearly an important quality control mechanism that ensures high fidelity of gene expression, the basic mechanisms by which this pathway operates are poorly understood. To understand the underlying molecular mechanism of NMD, we used the T cell-receptor (TCR)-β gene as a model system. TCR and immunoglobulin (Ig) genes are unique among vertebrates in that they undergo programmed DNA rearrangement during normal lymphocyte development (Li and Wilkinson, 1998). The variable (V), diversity (D) and joining (J) segments of TCR and Ig genes rearrange in different combinations during lymphocyte development, generating a diverse set of receptors that recognize different antigens. During these recombination events, the enzyme terminal transferase introduces non-template-coded nucleotides (so-called ‘N’ nucleotides) at the junction of the rearranging V, D and J elements. Additional receptor diversity is generated by transfer of ‘P’ nucleotides from the complementary strand to the coding strand as a result of endonucleolytic cleavage of the hairpins generated at an intermediate step of recombination. Small deletions also commonly occur at the junctions of the V, D and J elements as a consequence of the imprecise junctions of these elements, thereby further increasing receptor diversity (Li and Wilkinson, 1998).
Although the addition and deletion of nucleotides at V, D and J junctions increases the receptor repertoire, on two out of three occasions, these genes undergo a non-productive rearrangement, resulting in the generation of a downstream PTC. These aberrant transcripts must be degraded to avoid the translation of truncated non-functional receptors that may engender deleterious effects on lymphocytes. In agreement with this, we and others have discovered that the steady-state levels of Ig and TCR transcripts from non-productively rearranged Ig and TCR genes are dramatically reduced as a result of NMD (Baumann et al., 1985; Jäck et al., 1989; Connor et al., 1994; Lozano et al., 1994; Carter et al., 1995, 1996; Li et al., 1997; Buzina and Shulman, 1999). Several lines of evidence suggest that this down-regulation is mediated by a post-transcriptional mechanism. First, nuclear run-on analysis performed on Ig-µ heavy chain genes indicated that PTCs do not influence their rate of transcription and therefore the down-regulation is not transcriptional (Jäck et al., 1989; Mühlemann et al., 2001). Secondly, nuclear run-off analysis of a PTC-bearing TCR-β gene in the SL12.4 T-cell clone showed that this gene is highly transcriptionally active even in the absence of the protein synthesis inhibitor cycloheximide, which reverses NMD (Wilkinson and MacLeod, 1988). Thirdly, sequence analysis of RT–PCR products from fetal and adult thymus revealed that while PTC-bearing (PTC+) TCR-β mature transcripts rarely accumulate as a result of NMD, PTC+ TCR-β pre-mRNAs are present at levels that reflect the high frequency of PTC+ genes present in the thymic cells (Carter et al., 1995).
TCR and Ig transcripts undergo robust down-regulation in response to PTCs whereas triose phosphate isomerase (TPI), β-globin, v-src, dihydrofolate reductase (DHFR) and adenine phosphoribosyl transferase (APRT) transcripts are down-regulated more modestly (Table I). We previously hypothesized that the dramatic down-regulation of TCR and Ig transcripts is a result of the strong selection pressure that results from their frequent acquisition of PTCs during normal lymphoid development (Li and Wilkinson, 1998). In the study reported here, we examined the mechanism underlying the dramatic down-regulation of TCR-β transcripts. In particular, we addressed whether the robust down-regulatory response of TCR-β transcripts is cell type-, promoter- or transcript-specific. Our investigations demonstrated that the regulation is mediated specifically by sequences within TCR-β transcripts. We localized the regulatory sequences to a region composed of the rearranged V(D)J exon and the immediately flanking intron sequences from two different TCR-β genes. We showed that this region is both necessary and sufficient to trigger robust down-regulation in response to nonsense codons. Importantly, PTCs located within or downstream, but not upstream, of the VDJ exon triggered strong down-regulation. We provide two models to explain the novel properties of these TCR cis elements that greatly amplify the down-regulatory response elicited by nonsense codons.
Table I. NMD in mammalian cells.
| Transcript | Fold down-regulation | Reference |
|---|---|---|
| Triose phosphate isomerase (TPI) | 3–5 | Cheng et al. (1990) |
| Avian sarcoma virus v-src | 2–4 | Simpson and Stoltzfus (1994) |
| β-globin | 2–5 | Baserga and Benz (1988) |
| Dihydrofolate reductase (DHFR) | 5–12a | Urlaub et al. (1989) |
| Adenine phosphoribosyl transferase (APRT) | 5–10a | Kessler and Chasin (1996) |
| T-cell receptor β (TCR-β) | ≥30 | this report |
| Immunoglobulin (Ig) | 10–1000a | Connor et al. (1994); Buzina and Shulman (1999) |
aResults obtained with mutant endogenous gene, not with transfected genes.
Results
Cell types and promoters are not responsible for the differential down-regulation of TCR-β and TPI transcripts in response to PTCs
Differences in promoters and cell types have been reported by others to play a role in NMD. For example, the nature of the promoter driving the expression of the β-globin gene dictates whether it is susceptible to NMD. β-globin transcripts are down-regulated by PTCs when they are expressed from either the β-globin or cytomegalovirus (CMV) immediate early promoters but not when they are expressed from the thymidine kinase promoter (Enssle et al., 1993; Kugler et al., 1995). B-cell nuclear extracts, but not nuclear extracts from several other cell types, have been reported to selectively decrease the levels of mature transcripts transcribed and spliced from Ig-κ genes harboring PTCs (Aoufouchi et al., 1996).
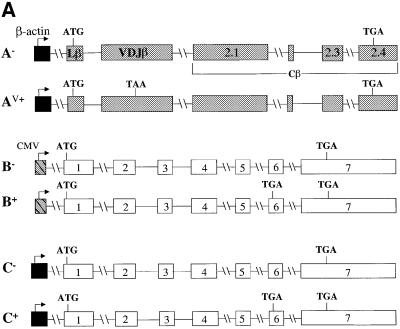
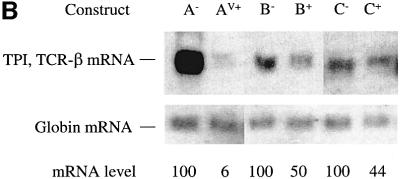
Here, we investigated whether cell type or promoter type are responsible for the dramatic difference in TCR-β and TPI mRNA down-regulation in response to PTCs. This point had not been addressed before, as past studies examining TCR-β and TPI expression used constructs containing different promoters expressed in different cell types. In these earlier studies, TPI transcripts driven by a metallothionin (MT) and CMV immediate early promoter were down-regulated ∼4-fold in response to PTCs in mouse Ltk– and NIH 3T3 fibroblast cells, respectively (Belgrader et al., 1994; Zhang et al., 1998a). In contrast, TCR-β transcripts driven by TCR-β and human β-actin promoters were down-regulated by PTCs by ≥10-fold in T-cell lines and HeLa epithelial cells, respectively (Carter et al., 1995, 1996). To examine whether a difference in cell type is responsible for this difference in the degree of down-regulation, we compared the down-regulatory responses of TCR-β and TPI in the same cell type, NIH 3T3 cells. Northern analysis of total cellular RNA from NIH 3T3 cells transiently transfected with PTC– and PTC+ versions of TCR-β and TPI genomic constructs (Figure 1A) revealed that the transcripts from these two genes exhibited dramatically different down-regulatory responses. Figure 1B shows that TCR-β transcripts were down-regulated by ∼17-fold (compare A– with AV+) whereas TPI transcripts were down-regulated only 2-fold (compare B– with B+) in NIH 3T3 cells. In both cases, the degree of down-regulation was similar to that reported previously for other cell types. These results indicate that the more robust down-regulatory response of TCR-β transcripts is not the result of differences in cell type.
Fig. 1. Cell type and promoter are not responsible for the dramatic down-regulation of TCR-β transcripts in response to nonsense codons. (A) TCR-β and TPI genomic constructs used for transfection. A– and AV+ are PTC– and PTC+ versions, respectively, of the full-length Vβ8.1–Dβ2–Jβ2.3–Cβ2 TCR-β gene, driven by the β-actin promoter. B– and B+ are PTC– and PTC+ versions, respectively, of the TPI gene, driven by the CMV promoter. C– and C+ are PTC– and PTC+ versions, respectively, of the TPI gene, driven by the β-actin promoter. (B) Northern blot analysis of total RNA (10 µg) isolated from NIH 3T3 cells transiently transfected with the constructs shown. The numbers below the blot are relative PTC+ mRNA levels compared with PTC– mRNA levels (PTC– levels are 100) normalized against globin mRNA levels. Comparable results were obtained in two independent transfection experiments.
We next tested whether the nature of promoter was responsible for the more dramatic down-regulatory response of TCR-β transcripts as compared with TPI transcripts. To test this, TPI was placed under the control of the human β-actin promoter and transiently transfected into NIH 3T3 cells. Figure 1B shows that TPI transcripts expressed in this way were down-regulated only 2-fold (compare C– with C+) whereas TCR-β transcripts driven from the same promoter were strongly down-regulated (compare A– with AV+). These results indicate that the robust down-regulatory response of TCR-β transcripts is not dictated by the nature of the promoter. These observations lead us to hypothesize that TCR genes may have evolved cis elements to promote robust down-regulation at the post-transcriptional level. To test this hypothesis, we undertook a search for such elements in TCR-β transcripts, as described in the following sections.
Localization of a down-regulatory-promoting element to the 5′ end of TCR-β
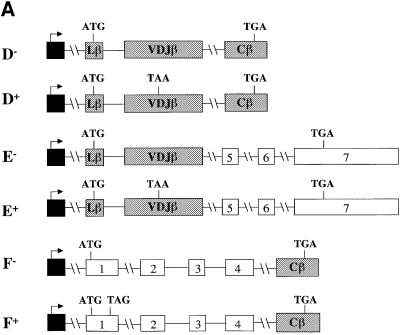
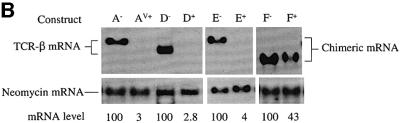
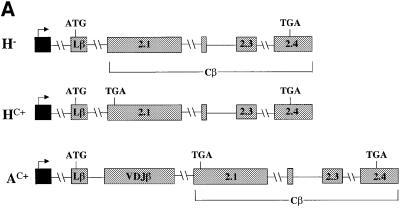
Figure 1 shows that the rearranged full-length Vβ8.1– Dβ2–Jβ2.3–Cβ2 TCR gene used in this study is comprised of six exons: a leader exon (Lβ), a rearranged variable region exon (VDJβ) and four constant region exons (Cβ). To localize elements that augment down-regulation, we first tested whether a mini-gene lacking a 1443 nucleotide (nt) region, encompassing the sequences from the 3′ half of the Cβ2.1 exon to the 5′ half of the Cβ2.4 exon (most of the constant region) of TCR-β, retained the ability to confer robust down-regulation. PTC– and PTC+ versions of this mini-gene (constructs D– and D+, respectively) and the full-length gene (constructs A– and AV+, respectively) were transiently transfected into HeLa cells (Figure 2A). Northern blot analysis of the transfectants revealed that mRNA from the TCR-β mini-gene was as strongly down-regulated by a PTC (compare D– with D+) as mRNA from the full-length TCR-β gene harboring a PTC at the same position (Figure 2B, compare A– with AV+). These results demonstrated that there are one or more elements responsible for enhanced down-regulation within the TCR-β mini-gene and that most of the constant region is dispensable for robust down-regulation.
Fig. 2. The 5′ portion of TCR-β is responsible for robust down-regulation in response to nonsense codons. (A) Constructs D– and D+ are PTC– and PTC+ versions of the TCR-β mini-gene, respectively. E– and E+ are PTC– and PTC+ versions, respectively, of a TCR-β and TPI chimeric gene comprised of the 5′ half of TCR-β fused to the 3′ half of TPI. F– and F+ are PTC– and PTC+ versions, respectively, of a TCR-β and TPI chimeric gene comprised of the 5′ half of TPI fused to the 3′ half of TCR-β. (B) Northern blot analysis of total RNA (10 µg) isolated from HeLa cells transiently transfected with the constructs shown. The numbers below the blots are relative PTC+ mRNA levels compared with PTC– mRNA levels (PTC– levels are 100). mRNA levels were normalized for differences in transfection efficiency and RNA loading by measurement of the level of neomycin mRNA, which is expressed as a separate transcription unit from all the plasmids in (A). Comparable results were obtained in at least two independent transfection experiments.
To localize sequences that confer robust down-regulation within the TCR-β mini-gene, we generated PTC– and PTC+ versions of two chimeric constructs: the 5′ half of the TCR-β mini-gene fused to the 3′ half of TPI (Figure 2A, constructs E– and E+) and the 5′ half of TPI fused to the 3′ half of the TCR-β mini-gene (constructs F– and F+). Transient transfection of these constructs revealed that transcripts from the chimeric constructs harboring the 5′ half of the TCR-β mini-gene were at least as strongly down-regulated (∼25-fold; Figure 2B, compare E– with E+) in response to PTCs as that of the parent TCR-β mini-gene (compare D– with D+). In contrast, transcripts from the chimeric constructs harboring the 3′ portion of the TCR-β mini-gene were only modestly down-regulated (∼3-fold; compare F– with F+). These results indicate that the 5′ portion of TCR-β mini-gene harbors one or more elements required for robust down-regulation.
The TCR-β leader exon and initiation codon are not essential for robust down-regulation in response to nonsense codons
To localize down-regulatory-promoting elements within the 5′ portion of TCR-β mini-gene, we first swapped exon 1 from the TCR-β mini-gene (the leader exon) for exon 1 from the TPI gene (Figure 3A). We found that transcripts from this chimeric gene were as strongly down-regulated in response to a PTC (>30-fold; Figure 3B, compare GV+ with G–) as transcripts from the parent TCR-β mini-gene harboring the same PTC (compare D– with D+). This indicates that the leader exon from the TCR-β gene, which harbors the initiation codon and its immediate flanking sequences, is not essential for robust down-regulation. Because the sequences surrounding initiation codons dictate translation efficiency, these results also suggest (but do not prove) that the translation initiation efficiency of TCR-β transcripts is not responsible for robust down-regulation. Surprisingly, a PTC located upstream of the VDJ exon did not elicit strong down-regulation (Figure 3B, compare G1+ with G–), suggesting that the magnitude of down-regulation depends upon the location of a PTC with respect to the down-regulatory-promoting element. This issue will be discussed in the last section.
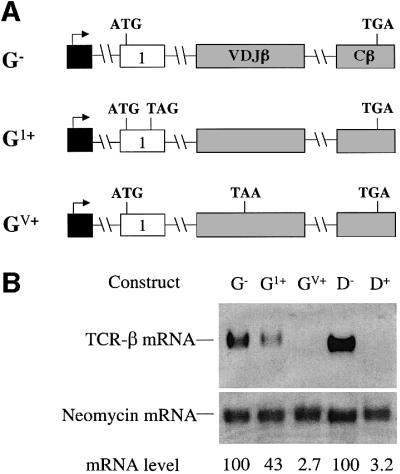
Fig. 3. The TCR-β leader exon and its initiation codon are not essential for robust down-regulation in response to nonsense codons. (A) All of the constructs have the Lβ exon from the TCR-β mini-gene replaced with TPI exon 1. G– lacks a PTC, whereas G1+ and GV+ each contain a single nucleotide mutation that generates a PTC in the exons shown. (B) Transfection, northern blot analysis and quantitation were performed as in Figure 2. Comparable results were obtained in two independent transfection experiments.
The data from Figures 2 and 3 demonstrate that TCR-β exons 1, 3, 4, 5 and 6 are not essential for robust down-regulation and therefore do not contain elements that are essential to increase the magnitude of down-regulation. This suggested the possibility that the region comprising the rearranging VDJ exon (exon 2) and flanking intron sequences is responsible for robust down-regulation.
The region comprising the rearranging VDJ exon and flanking sequences is essential for strong down-regulation in response to nonsense codons
To test whether the VDJ exon and the flanking sequences harbor elements required for robust down-regulation in response to nonsense codons, we deleted these sequences from the full-length TCR-β gene (Figure 4A). Transcripts expressed from this deleted gene were down-regulated weakly in response to a nonsense codon (∼3-fold; Figure 4B, compare H– with HC+) whereas those expressed from a construct harboring the same PTC but without the deletion were strongly down-regulated (∼50-fold; compare A– with AC+). These results provided direct evidence that the region encompassing the VDJ exon and flanking sequences is necessary for the strong down-regulation of PTC-bearing TCR-β transcripts.
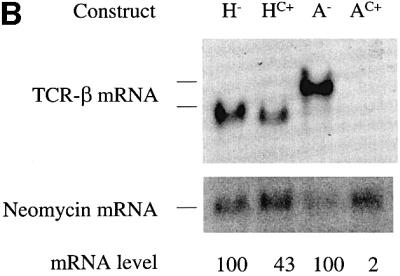
Fig. 4. The region encompassing a rearranged Vβ8.1DJ exon and flanking intron sequences is essential for strong down-regulation in response to nonsense codons. (A) H– and HC+ are TCR-β constructs that lack the VDJ exon and adjacent intron sequences. H– lacks a PTC, whereas HC+ contains a single nucleotide mutation that creates a PTC in the exon shown. AC+ is a full-length TCR-β construct identical to A– in Figure 1 except that it contains a single nucleotide mutation that creates a PTC in the exon shown. (B) Transfection, northern blot analysis and quantification were performed as in Figure 2. Comparable results were obtained in two independent transfection experiments.
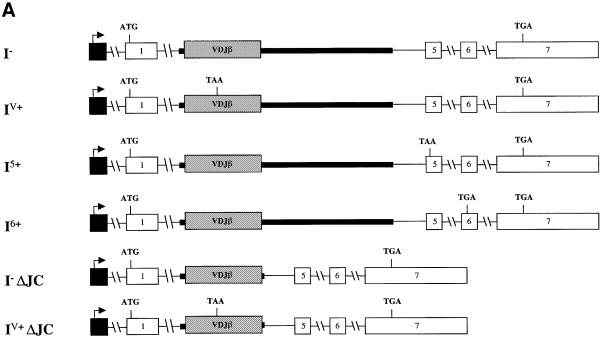
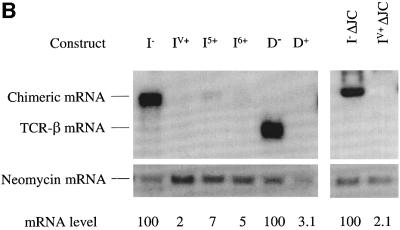
To test whether the VDJ exon and flanking sequences are sufficient to induce strong down-regulation, we introduced these sequences into the TPI gene. This was accomplished by replacing TPI exons 2–4 with the TCR-β VDJ exon and immediately flanking intron sequences (Figure 5A). Transcripts expressed from this chimeric construct were down-regulated by 50-fold in response to PTCs (compare I– with IV+ in Figure 5B) suggesting that either the VDJ exon, flanking sequences or both are necessary and sufficient to trigger robust down-regulation in response to nonsense codons.
Fig. 5. The region containing a rearranged Vβ8.1DJ exon and flanking intron sequences is sufficient to trigger robust down-regulation in response to nonsense codons. (A) All of the constructs have TPI exons 2–4 and flanking intron sequences replaced by the VDJ exon and flanking intron sequences from a rearranged Vβ8.1–Dβ2–Jβ2.3–Cβ2 TCR gene. I– lacks a PTC, whereas IV+ and I6+ each contain a single nucleotide mutation that creates a PTC in the exons shown. I5+ harbors a frameshift mutation in the VDJ exon that creates a PTC in the exon shown. I–ΔJC and IV+ΔJC are the same as I– and IV+, respectively, except that TCR-β IVS-JC intron sequences are deleted. (B) Transfection, northern blot analysis and quantification were performed as in Figure 2. Comparable results were obtained in at least two independent transfection experiments.
We next asked whether the robust down-regulation caused by the VDJ exon and flanking intron sequences is dictated by the sequence context of the PTC. To explore this, we introduced PTCs at different positions in the chimeric construct. The original construct contained a PTC in the VDJ exon; we also tested PTCs in the two downstream TPI exons (Figure 5A, I5+ and I6+). Transcripts expressed from these constructs were down-regulated by >15-fold (Figure 5B, compare I5+ and I6+ with I–), indicating that the robust down-regulatory response induced by TCR-β sequences is independent of the sequence context of the PTC. Importantly, this result also indicated that it is not necessary for a PTC to be in a TCR-β exon for the TCR-β down-regulatory-promoting element to decrease mRNA levels in response to nonsense codons.
To determine whether sequences in the TCR-β intron downstream of the VDJ exon were necessary to trigger robust down-regulation, we removed them from the chimeric construct. All but the 5′ splice-site sequences (668 of 674 nt) were deleted from IVS-JC (Figure 5A, constructs I– ΔJC and IV+ ΔJC). Transcripts expressed from this TCR-β intron-deleted genomic construct were down-regulated as strongly (∼50-fold; Figure 5B, compare lane I– ΔJC with IV+ ΔJC) as those from construct IV+ in response to a PTC. This indicated that the IVS-JC intron is not essential for the robust down-regulatory response and that cis-acting sequences sufficient to elicit strong mRNA down-regulation in response to nonsense codons are likely to be located within the 383 nt sequence comprising the VDJ exon and immediately flanking 3′ and 5′ splice sites.
Down-regulatory-promoting elements are a conserved feature of TCR-β genes
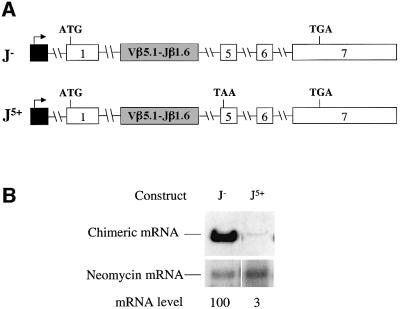
TCR-β genes display tremendous variability in the V(D)J exon. During programmed rearrangements in the thymus, >30 different Vβ segments, 12 different Jβ segments and two different Dβ segments contribute to generating this variable region exon. To determine whether the particular rearranged gene that we used in the present study (Vβ8.1–Dβ2–Jβ2.3–Cβ2) is unique in containing cis-acting elements that promote strong down-regulation or instead these elements are a conserved feature of TCR-β genes, we examined the functional properties of another rearranged TCR-β gene. We elected to test a Vβ5.1–Jβ1.6–Cβ1 gene that is highly transcribed from its normal endogenous site in the SL12.4 T-lymphoma cell clone but gives rise to transcripts that are dramaticaly down-regulated as a result of a naturally occurring frameshift in the VJ exon that generates a downstream PTC (Wilkinson and MacLeod, 1988; Carter et al., 1995). To examine whether the VJ exon region of this Vβ5.1–Jβ1.6–Cβ1 gene contained down-regulatory-promoting activity, we first isolated a genomic clone containing these sequences from SL12.4 cells. We then generated an in-frame version of the VJ exon by site-specific mutagenesis. Lastly, we introduced both the in-frame and out-of-frame versions of this Vβ5.1–Jβ1.6 exon and flanking intron sequences into the TPI gene (Figure 6A). We found that transcripts expressed from this chimeric gene were down-regulated by ∼30-fold in response to the PTC (Figure 6B, compare J– with J5+), indicating that Vβ5.1–Jβ1.6 and adjacent sequences, like the Vβ8.1–Dβ2–Jβ2.3 exon and flanking sequences, can dramatically decrease mRNA levels in response to nonsense codons and therefore probably harbor one or more down-regulatory-promoting elements.
Fig. 6. A rearranged Vβ5.1–Jβ1.6 exon and flanking intron sequences promotes strong down-regulation in response to nonsense codons. (A) Both constructs have TPI exons 2–4 and flanking intron sequences replaced with the VJ exon and flanking intron sequences from a rearranged Vβ5.1–Jβ1.6–Cβ1 gene. J– lacks a PTC and J5+ has a frameshift mutation in the VJ exon that creates a PTC in the exon shown. (B) Transfection, northern blot analysis and quantification were performed as in Figure 2. Comparable results were obtained in two independent transfection experiments.
Only nonsense codons within or downstream of the VDJ exon trigger dramatic down-regulation in response to nonsense codons
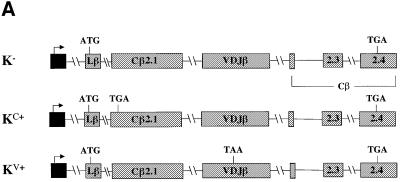
We next examined whether the location of a PTC with respect to a down-regulatory-promoting element determines the magnitude of down-regulation. We therefore placed PTCs upstream and downstream of the VDJ exon in the full-length Vβ8.1–Dβ2–Jβ2.3–Cβ2 gene and determined the down-regulatory response. To achieve this, we moved the VDJ exon and the flanking sequences downstream of the constant region exon Cβ2.1. The resulting constructs consisted of the Lβ exon followed by the Cβ2.1, VDJ, Cβ2.2, Cβ2.3 and Cβ2.4 exons (Figure 7A). Transient transfection analysis of these constructs indicated that a PTC upstream of the VDJ exon caused only a basal level of down-regulation (∼2-fold; Figure 7B, compare K– with KC+). In contrast, the same PTC in the same position of the Cβ2.1 exon but with this exon situated downstream of the VDJ exon (Figure 4A, construct AC+) caused dramatic down-regulation (∼50-fold; Figure 7B, compare A– with AC+). To test whether the reduction in the down-regulatory response in KC+ was a result of moving the VDJ exon to a different location in the TCR-β gene, we generated a construct that was identical to KC+ except that it contained a PTC in the VDJ exon rather than the upstream Cβ2.1 exon (KV+; Figure 7A). Transcripts from KV+ were down-regulated strongly (>17-fold; Figure 7B, compare K– with KV+), indicating that moving the VDJ exon was not responsible for the differences in the magnitude of the down-regulatory response. Instead, these results suggest that the down-regulatory-promoting element causes robust down-regulation if the PTC is located within or downstream of the VDJ exon. Our data from construct G1+ (Figure 3A), in which the PTC is in a TPI exon upstream of the VDJ exon, also support this conclusion (Figure 3B, compare G– with G1+).

Fig. 7. Position-dependent effect of nonsense codons on the down-regulatory-promoting element. (A) All of the constructs have the Vβ8.1–Dβ2–Jβ2.3 exon and flanking intron sequences moved downstream of the Cβ2.1 exon. K– lacks a PTC, whereas KC+ and KV+ each contain a single nucleotide mutation that creates a PTC in the same positions as in AC+ and AV+, respectively. (B) Transfection, northern blot analysis and quantitation were performed as in Figure 2. Comparable results were obtained in three independent transfection experiments.
Discussion
By examining mRNA expression from a series of chimeric and deletion constructs, we have identified a region of a TCR-β gene that dramatically down-regulates transcript levels in response to nonsense codons (Figures 2–4). We found that this region was sufficient to confer robust down-regulation on a heterologous gene as long as the nonsense codons were within or downstream of it (Figure 5). Such down-regulatory elements may be a conserved feature of TCR-β genes, as we found strong down-regulatory activity in sequences from two different Vβ family members (Figure 6). Although such TCR-β down-regulatory elements would normally only function in T cells, we found that they also cause strong down-regulation in response to nonsense codons when expressed from non-T-cell-specific promoters in non-lymphoid cells. Thus, this type of element appears to respond to NMD-promoting factors found in many different cell types. To our knowledge, the type of cis element we have identified in this study is the first to be shown to act upstream of a nonsense codon.
To engage the NMD pathway in response to nonsense codons, the element that we identified here works together with a downstream cis element. Previously, we showed that this downstream element in TCR-β transcripts is an intron (Carter et al., 1996). Many other studies have also demonstrated that an intron is necessary to trigger NMD, including for TPI, β-globin, mouse urinary protein and gpx-1 (Li and Wilkinson, 1998; Nagy and Maquat, 1998; Hilleren and Parker, 1999; Sun et al., 2000). How an intron acts as a second signal to trigger the NMD pathway is not known. Introns could act in the nucleus before mRNAs are completely spliced (the nuclear scanning model) or they could generate a ‘mark’ left after RNA splicing that is recognized in either the nucleus or the cytoplasm by the translation machinery (the marker model; Carter et al., 1996; Li and Wilkinson, 1998; Nagy and Maquat, 1998). Recently, an exon junction complex (EJC) left behind at exon–exon junctions as a signature of splicing was identified that could act as the mark to trigger NMD (Le Hir et al., 2000). This EJC contains the UPF2 and UPF3 NMD proteins, as well as several proteins that may function in RNA export or RNA splicing, including Aly(REF), RNPS1, Y14, SRm160 and DEK (Shyu and Wilkinson, 2000; Le Hir et al., 2000, 2001; Lykke-Andersen, 2001; Lykke-Andersen et al., 2001; Kim et al., 2001a,b; Wilkinson and Shyu, 2001). To test which proteins of the EJC are important for NMD, individual members of this complex have been tethered downstream of a stop codon in β-globin transcripts using the MS2 system. This tethering experiment has suggested that all of the UPF proteins, RNPS1 and possibly Y14 serve as a mark for NMD if encountered downstream of a stop codon by the post-termination surveillance complex, which has been hypothesized to form after a ribosome meets a stop codon (Lykke-Andersen et al., 2001).
Exonic downstream sequence elements (DSEs) that act as the second signal for NMD have also been identified. These are prevalent in Saccharomyces cerevisae genes, which rarely have introns. Such yeast DSEs have the following characteristics: (i) they are located within the coding region upstream of the normal stop codon, (ii) they trigger NMD when a nonsense codon is introduced upstream of them, (iii) they have a loosely defined consensus motif and (iv) they interact with the RNA-binding protein Hrp1–Nab4, which promotes NMD (Peltz et al., 1993, 1994; Ruiz-Echevarría et al., 1998; Hellerin and Parker, 1999; González et al., 2000). Some vertebrate genes also appear to have downstream exonic elements. Although not well characterized, their existence is implied by the finding that an intron downstream of a nonsense codon is, in some cases, not required to trigger NMD. For example, Rous sarcoma virus v-src (Simpson and Stoltzfus, 1994), Xenopus laevis XLPOU-60 (Whitfield et al., 1994) and human HEXA (Rajavel and Neufeld, 2001) transcript levels have all been shown to be down-regulated in response to nonsense codons without a downstream intron. In the case of β-globin, a so-called fail-safe downstream exonic element has been identified that permits NMD, even when the intron downstream of a PTC is deleted (Zhang et al., 1998b).
Two general models have been proposed to explain how the sequence information downstream of the termination codon elicits NMD. The scanning surveillance model posits that after premature translation termination, the surveillance complex assembles, scans and interacts with the DSE or intron mark, which elicits RNA decay (Peltz et al., 1993, 1994; Ruiz-Echevarría et al., 1998). The termination–mRNP context model posits that the mRNA transcript is stabilized by the proper remodeling of the mRNP complex that occurs when the 3′-terminal region of an mRNA interacts with a terminating ribosome. Premature termination does not allow the establishment of this interaction and consequently mRNP remodeling does not occur and the mRNA is targeted for degradation (Hellerin and Parker, 1999).
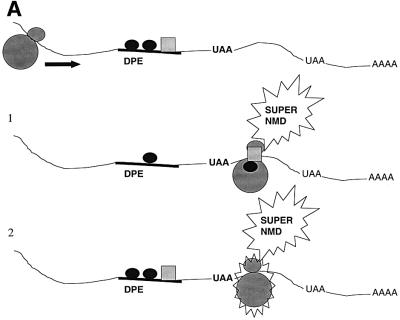
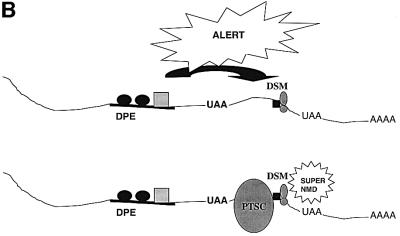
How does the upstream regulatory element that we identified in this report act to greatly stimulate the magnitude of down-regulation in response to nonsense codons? We suggest two models. In the ribosome sensitization model, the act of scanning past the upstream regulatory element alters the characteristics of the translation apparatus such that it becomes capable of triggering robust down-regulation when it later encounters a nonsense codon (Figure 8A). Two possible alterations that could occur are picking up a positive regulatory protein bound to the down-regulatory-promoting element (such as a UPF protein or an SR protein; Figure 8A, scheme 1) or post-translational alterations (such as phosphorylation) that activate proteins already bound to the translation apparatus (Figure 8A, scheme 2). According to this model, the ribosome ‘sensitized’ by such modifications can recognize the stop codons more efficiently or attract release factors to a greater degree. More release factors may in turn generate more active post-termination surveillance complexes, which in turn would make more productive contacts with the EJC. In the cross-talk model, the upstream regulatory element communicates with the downstream mark (the second signal; Figure 8B). According to this model, the factors binding to the upstream regulatory element could influence the composition of the downstream mark, making it highly sensitive to interactions with the post-termination surveillance complex. Alternatively, the down-regulatory-promoting element could increase the affinity by which the downstream mark binds to the exon–exon junction, thereby increasing the magnitude of the second signal for NMD.
Fig. 8. Models to explain how down-regulatory-promoting elements could stimulate the decay of transcripts containing a downstream nonsense codon. (A) Ribosome sensitization models. Scheme 1, the ribosome picks up positive regulatory proteins from the down-regulatory-promoting element that primes the ribosome for strong NMD when it later encounters a PTC. Scheme 2, the ribosome undergoes a post-translational alteration in response to passage over the down-regulatory-promoting element that primes it for strong NMD when it later encounters a PTC. (B) The cross-talk model, in which the down-regulatory-promoting element communicates with a downstream marker protein to augment its ability to trigger NMD. Abbreviations: DPE, down-regulatory-promoting element; PTSC, post-termination surveillance complex; DSM, downstream mark.
It remains for future studies to determine whether the down-regulatory-promoting element we have defined here is a modulatory element that enhances the rate of NMD or whether it directs a down-regulatory mechanism different from the one that acts on most other transcripts. The former possibility is supported by the fact that TCR-β transcripts are degraded in response to nonsense codons by a mechanism displaying some characteristics similar to that which acts on other transcripts, such as the requirement for a downstream intron. The latter possibility is supported by the fact that TCR and Ig genes are unique in that they very frequently acquire nonsense or frameshift mutations as a result of programmed rearrangements in T cells. Because the only TCR and Ig exon that acquires frameshifts and point mutations is the V(D)J exon, perhaps this particular exon has evolved to recognize special factors that permit efficient nonsense codon surveillance within and downstream of it. If indeed TCR and Ig transcripts are scanned by a down-regulatory mechanism with unique properties, both this pathway and the classical NMD pathway may operate simultaneously; TCR and Ig transcripts that escape one pathway are degraded by the other, thereby causing the robust down-regulatory response that we have characterized in this report.
In this report, we demonstrated that PTCs located downstream but not upstream of the VDJ exon elicit a robust down-regulatory response. This indicates that the down-regulatory-promoting element functions when upstream of a nonsense codon. However, our data does not rule out the possibility that the down-regulatory element can function downstream of a nonsense codon when they are in the same exon. An important step towards elucidating the mechanism for robust NMD will be to identify and locate more precisely the specific sequences responsible. This may be facilitated by sequence comparison of different TCR-β genes. However, there is only limited sequence identity between the corresponding region of the two TCR-β genes that we have so far demonstrated to have down-regulatory-promoting activity (the Vβ8.1–Dβ2–Jβ2.3 and Vβ5.1–Jβ1.6 exons have only 42% sequence identity and there is no discernable sequence identity in the flanking intron sequences besides the splice junctions). Our future studies will be devoted to determining the precise location of the TCR-β cis elements that stimulate the down-regulatory response, identifying the trans-acting factors that bind to these elements even in non-lymphoid cell types and elucidating the mechanism by which these factors act upstream of a nonsense codon to stimulate down-regulation in response to nonsense codons.
Materials and methods
Plasmid construction
Plasmid constructs A– (β-290) and AV+ (β-367) are pIF and pFS1, respectively (Carter et al., 1995). B– (G-256–) and B+ (G-256+) are PTC– and PTC+ (codon 189) versions of human TPI, respectively, cloned in a puc13-based vector (Daar and Maquat, 1988). C– (G-266) and C+ (G-268) were prepared by subcloning the TPI gene from B– and B+, respectively, downstream of the β-actin promoter into the pHβ Apr-1-neo vector (Gunning et al., 1987). D– (β-556) and D+ (β-337) are pAc–IFΔ (Carter et al., 1995) and construct C (Li et al., 1997), respectively. E– (β-567) and E+ (β-568) were prepared by amplifying the sequences from the 3′ half of TPI intron 4 (nt 2568) to the 3′ end of TPI exon 7 (nt 3870) by PCR and then subcloning them into the ClaI and BamHI sites of constructs D– and D+, respectively. F– (β-571) and F+ (β-572) were generated by amplifying the 5′ half of TPI (nt 601–2618) from construct C– and a PTC+ (codon 23) TPI gene (G-267; Daar and Maquat, 1988), respectively, by PCR and subcloning the products into the SalI and ClaI sites of D–. G– (β-642) and GV+ (β-634) were made by amplifying the sequences between the 3′ end of the intron IVS-LV (nt 1311) and the 3′ end of exon 3 (nt 2905) of D– and D+, respectively, and subcloning the PCR products into the BglII and BamHI sites of C–. G1+ (β-633) was generated by subcloning the above-described PCR product from D– into the BglII and BamHI sites of PTC+ (codon 23) TPI. H– (β-780) and HC+ (β-781) were generated by deleting TCR-β nt 1301–2364 by inverse PCR from A– and AC+ (β-189), respectively. I– (β-652) and IV+ (β-653) were generated by subcloning the SalI–ClaI fragment from G– and GV+, respectively, into the SalI and ClaI sites of E–. I5+ (β-725) was generated by replacing the VDJβ exon and flanking sequences (BglII–ClaI fragment) in I– with that derived from construct pRV harboring a frameshift mutation in the VDJ exon (Carter et al., 1995). To generate I6+ (β-726), the sequences from the 3′ half of TPI intron 4 (bp 2568) to the 3′ end of TPI exon 7 (nt 3870; C+) were amplified by PCR and subcloned into the ClaI and BamHI sites of I–. I–ΔJC (β-839) and IV+ΔJC (β-840) were generated by deleting IVS-JC intron sequences (nt 1694–2364) from I– and IV+, respectively, by inverse PCR. To prepare J– (β-829) and J5+ (β-830), the VJ exon and the immediately flanking intron sequences (51 nt of the 3′ end of IVS-LV and 28 nt of the 5′ end of IVS-JC) from in-frame (β-809) and out-of-frame (β-808) versions, respectively, of a Vβ5.1–Jβ1.6–Cβ1 gene obtained from SL12.4 T-lymphoma cells (Carter et al., 1995) were amplified by PCR and subcloned into the BglII and ClaI sites of I–. β-809 was prepared by deleting a single nucleotide (nt 311; Carter et al., 1995) from the Vβ5.1–Jβ1.6 exon in β-808 by inverse PCR. To generate K– (β-782), KC+ (β-783) and KV+ (β-784), the VDJ exon and the flanking intron sequences (nt 1307–2368) from A– (to generate K– and KC+) and AV+ (to generate KV+) were amplified by PCR and inserted at the MunI site of H– or HC+.
PCR
PCR was performed in a 50 µl reaction mixture with Pfu polymerase (Stratagene, La Jolla, CA). Amplification was performed as follows: 95°C for 3 min followed by 30 cycles of 94°C for 1 min, 58°C for 90 sec and 72°C for 3 min (increasing by 20 sec for each cycle) and a final elongation step at 72°C for 10 min. Inverse PCR was performed by using 5′ end-phosphorylated primers in a 50 µl reaction as follows: 95°C for 3 min, followed by 30 cycles of 95°C for 30 sec, 55°C for 1 min, 68°C for 10–14 min and a final elongation step at 72°C for 10 min.
Cell culture and transfection
NIH 3T3 and HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. HeLa cells cultured to 50–70% confluency in 10 cm plates were transiently transfected with plasmid DNA (1–3 µg) using lipofectamine reagent (Gibco BRL Life Technologies, NY) according to the manufacturer’s instructions. For NIH 3T3 cell transfections, the calcium phosphate method (Chen and Okayama, 1988) was used to transfect 50–70% confluent cells; a β-globin plasmid was co-transfected to normalize for transfection efficiency. Cells were harvested 40–45 h after the start of transfection.
RNA isolation and northern blot analysis
Total cellular RNA was isolated by guanidinium isothiocyanate lysis and centrifugation over a 5.7 M CsCl cushion (Wilkinson, 2000). To perform northern blot analysis, 10 µg of RNA were electrophoresed, blotted and hybridized as previously described (Qian et al., 1993). TCR-β mRNA levels were assessed by hybridization with a 0.3 kb DNA fragment corresponding to either the Vβ8.1 or Vβ5.1 exons. To assess the levels of chimeric mRNAs, we used either Vβ exon probes or a constant region probe (0.7 kb). A neomycin gene probe was used to assess neomycin mRNA levels to normalize for transfection efficiency. Relative mRNA levels were determined with an Instant Imager (Packard Instrument Company, IL).
Acknowledgments
Acknowledgements
We are grateful to Dr Lynne Maquat for providing plasmids pmCMV-TPI Norm, pmCMV-TPI 189ter and pMT-TPI 23ter and Dr John Hamilton for providing the Vβ5.1 plasmids. We also thank Ann-Bin Shyu, Dr Gilbert Cote, Dr Maureen Goode and members of the Wilkinson laboratory for their useful suggestions on this manuscript. This work was supported by National Institutes of Health grant GM58595 and National Science Foundation grant MCB-9808936.
References
- Aoufouchi S., Yélamos,J. and Milstein,C. (1996) Nonsense mutations inhibit RNA splicing in a cell-free system: recognition of mutant codon is independent of protein synthesis. Cell, 85, 415–422. [DOI] [PubMed] [Google Scholar]
- Baserga S.J. and Benz,E.J.,Jr (1988) Nonsense mutations in the human β-globin gene affect mRNA metabolism. Proc. Natl Acad. Sci. USA, 85, 2056–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B., Potash,M.J. and Köhler,G. (1985) Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J., 4, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Cheng,J., Zhou,X., Stephenson,L.S. and Maquat,L.E. (1994) Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol. Cell. Biol., 14, 8219–8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzina A. and Shulman,M.J. (1999) Infrequent translation of a nonsense codon is sufficient to decrease mRNA level. Mol. Biol. Cell, 10, 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali B.M. and Anderson,P. (1998) mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol. Gen. Genet., 260, 176–184. [DOI] [PubMed] [Google Scholar]
- Carter M.S., Doskow,J., Morris,P., Li,S., Nhim,R.P., Sandstedt,S. and Wilkinson,M.F. (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem., 270, 28995–29003. [DOI] [PubMed] [Google Scholar]
- Carter M.S., Li,S. and Wilkinson,M.F. (1996) A splicing-dependent regulatory mechanism that detects translation signals. EMBO J., 15, 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Chen C.A. and Okayama,H. (1988) Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stable transforming cells with plasmid DNA. Biotechniques, 6, 632–638. [PubMed] [Google Scholar]
- Cheng J., Fogel-Petrovic,M. and Maquat,L.E. (1990) Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol. Cell. Biol., 10, 5215–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor A., Wiersma,E. and Shulman,M.J. (1994) On the linkage between RNA processing and RNA translatability. J. Biol. Chem., 269, 25178–25184. [PubMed] [Google Scholar]
- Culbertson M.R. (1999) RNA surveillance: unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet., 15, 74–80. [DOI] [PubMed] [Google Scholar]
- Daar I.O. and Maquat,L.E. (1988) Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol. Cell. Biol., 8, 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enssle J., Kugler,W., Hentze,M.W. and Kulozik,A.E. (1993) Determination of mRNA fate by different RNA polymerase II promoters. Proc. Natl Acad. Sci. USA, 90, 10091–10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González C.I., Ruiz-Echevarria,M.J., Vasudevan,S., Henry,M.F. and Peltz,S.W. (2000) The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell, 5, 489–499. [DOI] [PubMed] [Google Scholar]
- Gunning P., Leavitt,J., Muscat,G., Ng,S.-Y. and Kedes,L. (1987) A human β-actin expression vector system directs high-level accumulation of antisense transcripts. Proc. Natl Acad. Sci. USA, 84, 4831–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (1999) Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet., 33, 229–260. [DOI] [PubMed] [Google Scholar]
- Jäck H.-M., Berg,J. and Wabl,M. (1989) Translation affects immunoglobulin mRNA stability. Eur. J. Immunol., 19, 843–847. [DOI] [PubMed] [Google Scholar]
- Kessler O. and Chasin,L. (1996) Effects of nonsense mutations on nuclear and cytoplasmic adenine phosphoribosyltransferase RNA. Mol. Cell. Biol., 16, 4426–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Yong,J., Kataoka,N., Abel,L., Diem,M.D. and Dreyfuss,G. (2001a) The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J., 20, 2062–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Kataoka,N. and Dreyfuss,G. (2001b) Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science, 293, 1832–1836. [DOI] [PubMed] [Google Scholar]
- Kugler W., Enssle,J., Hentze,M.W. and Kulozik,A.E. (1995) Nuclear degradation of nonsense mutated β-globin mRNA: a post-transcriptional mechanism to protect heterozygotes from severe clinical manifestations of β-thalassemia? Nucleic Acids Res., 23, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000) The spliceosome deposits multiple protein 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield,D., Izaurralde,E. and Moore,M.J. (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J., 20, 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. and Wilkinson,M.F. (1998) Nonsense surveillance in lymphocytes? Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- Li S., Leonard,D. and Wilkinson,M.F. (1997) T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J. Exp. Med., 185, 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano F., Maertzdorf,B., Pannell,R. and Milstein,C. (1994) Low cytoplasmic mRNA levels of immunoglobulin κ light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J., 13, 4617–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. (2001) mRNA quality control: marking the message for life or death. Curr. Biol., 11, R88–R91. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M-D. and Steitz,J.A. (2001) Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science, 293, 1836–1839. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA, 1, 453–465. [PMC free article] [PubMed] [Google Scholar]
- Mitrovich Q.M. and Anderson,P. (2000) Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C.elegans. Genes Dev., 14, 2173–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlemann O., Mock-Casagrande,C.S., Wang,J., Li,S., Custódio,N., Carmo-Fonseca,M., Wilkinson,M.F. and Moore,M.J. (2001) Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell, 8, 33–44. [DOI] [PubMed] [Google Scholar]
- Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Peltz S.W., Brown,A.H. and Jacobson,A. (1993) mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev., 7, 1737–1754. [DOI] [PubMed] [Google Scholar]
- Peltz S.W., He,F., Welch,E. and Jacobson,A. (1994) Nonsense-mediated mRNA decay in yeast. Prog. Nucleic Acid Res. Mol. Biol., 47, 271–298. [DOI] [PubMed] [Google Scholar]
- Pulak R. and Anderson,P. (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev., 7, 1885–1897. [DOI] [PubMed] [Google Scholar]
- Qian L., Theodor,L., Carter,M., Vu,M.N., Sasaki,A.W. and Wilkinson,M.F. (1993) T cell receptor-β mRNA splicing: regulation of unusual splicing intermediates. Mol. Cell. Biol., 13, 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajavel K.S. and Neufeld,E.F. (2001) Nonsense-mediated decay of human HEXA mRNA. Mol. Cell. Biol., 21, 5512–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J., Czaplinski,K. and Peltz,S.W. (1996) Making sense of nonsense in yeast. Trends Biochem. Sci., 21, 433–438. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J., González,C.I. and Peltz,S.W. (1998) Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J., 17, 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A-B. and Wilkinson,M.F. (2000) The double lives of shuttling mRNA binding proteins. Cell, 102, 135–138. [DOI] [PubMed] [Google Scholar]
- Simpson S.B. and Stoltzfus,C.M. (1994) Frameshift mutations in the v-src gene of avian sarcoma virus act in cis to specifically reduce v-src mRNA levels. Mol. Cell. Biol., 14, 1835–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Moriarty,P. and Maquat,L.E. (2000) Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position. EMBO J., 19, 4734–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Mitchell,P.J., Ciudad,C.J. and Chasin,L.A. (1989) Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol. Cell. Biol., 9, 2868–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield T.T., Sharpe,C.R. and Wylie,C.C. (1994) Nonsense-mediated mRNA decay in Xenopus oocytes and embryos. Dev. Biol., 165, 731–734. [DOI] [PubMed] [Google Scholar]
- Wilkinson M.F. (2000) Purification of RNA. In Brown,T.A. (ed.), Essential Molecular Biology. Oxford University Press, Oxford, UK, pp. 69–88.
- Wilkinson M.F. and MacLeod,C.L. (1988) Induction of T-cell receptor-α and -β mRNA in SL12 cells can occur by transcriptional and post-transcriptional mechanisms. EMBO J., 7, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.F. and Shyu,A-B. (2001) Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. BioEssays, 23, 775–787. [DOI] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y., LaDuca,J.P. and Maquat,L.E. (1998a) At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol., 18, 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y. and Maquat,L.E. (1998b) Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA splicing in the cytoplasm. RNA, 4, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]