Abstract
We show here that Ask1p, Dad2p, Spc19p and Spc34p are subunits of the budding yeast Duo1p–Dam1p– Dad1p complex, which associate with kinetochores and localize along metaphase and anaphase spindles. Analysis of spc34-3 cells revealed three novel functions of the Duo1–Dam1p–Dad1p subunit Spc34p. First, SPC34 is required to establish biorientation of sister kinetochores. Secondly, SPC34 is essential to maintain biorientation. Thirdly, SPC34 is necessary to maintain an anaphase spindle independently of chromosome segregation. Moreover, we show that in spc34-3 cells, sister centromeres preferentially associate with the pre-existing, old spindle pole body (SPB). A similar preferential attachment of sister centromeres to the old SPB occurs in cells depleted of the cohesin Scc1p, a protein with a known role in facilitating biorientation. Thus, the two SPBs are not equally active in early S phase. We suggest that not only in spc34-3 and Δscc1 cells but also in wild-type cells, sister centromeres bind after replication preferentially to microtubules organized by the old SPB. Monopolar attached sister centromeres are resolved to bipolar attachment in wild-type cells but persist in spc34-3 cells.
Keywords: Ask1p/centromere/Dad2p/Dam1p/Duo1p
Introduction
It is during mitosis that sister chromatids, generated by chromosome duplication during S phase, are segregated to opposite poles by the mitotic spindle. Budding yeast Saccharomyces cerevisiae has been particularly useful for the dissection of mitotic events (Winey and O’Toole, 2001). In yeast, microtubule (MT) organizing functions are provided by the spindle pole body (SPB), a multilayered structure that is embedded in the nuclear envelope throughout the cell cycle. The new SPB forms adjacent to the pre-existing old one by a conservative mechanism. SPB duplication is probably completed during S phase (Byers and Goetsch, 1975; Adams and Kilmartin, 1999). At this point, both adjacently positioned SPBs organize nuclear MTs that are required to assemble a short bipolar spindle at the end of S phase. At anaphase onset, the old SPB migrates into the bud, while the new one is retained in the mother cell (Pereira et al., 2001).
The kinetochore ensures high-fidelity chromosome segregation in mitosis and meiosis by mediating the attachment and movement of chromosomes along spindle MTs (Pidoux and Allshire, 2000). In budding yeast, proteins of the kinetochore assemble around a specific DNA region of only ∼125 bp, the centromere DNA (Fitzgerald-Hayes et al., 1982). Kinetochores consist of a number of relatively stable subcomplexes. Ndc10p, Cep3p, Ctf13p and Skp1p form the CBF3 complex, which directly interacts with centromere DNA (Lechner and Carbon, 1991; Goh and Kilmartin, 1993). The Okp1p complex, composed of Ctf19p, Mcm21p and Okp1p, localizes to yeast centromeres in a CBF3-dependent manner (Ortíz et al., 1999). Ndc80p, Nuf2p, Spc24p and Spc25p assemble into the Ndc80p complex with functions in kinetochore–MT binding and mitotic checkpoint control (Janke et al., 2001; Wigge and Kilmartin, 2001). Duo1p and Dam1p are part of a complex that associates with kinetochores and along nuclear MTs with an ill-defined role in chromosome segregation and spindle stability (Hofmann et al., 1998; Jones et al., 1999, 2001; Cheeseman et al., 2001). Recently, Dad1p was described as a third component of the Duo1p–Dam1p complex (Enquist-Newman et al., 2001), raising the possibility that this complex contains yet more subunits.
In the G1 phase of the cell cycle, the budding yeast centromeres are clustered near SPBs through the binding of kinetochores to MTs (Jin et al., 2000). In early S phase, centromere DNA is replicated (McCarroll and Fangmann, 1988), but it is unknown when and how the reassembled kinetochores bind to MTs. Sister kinetochores may first interact with MTs organized by only one of the two SPBs (monopolar attachment), although no evidence for monopolar binding has been obtained in budding yeast (discussed by Winey and O’Toole, 2001). It is also unclear whether the old and new SPBs are equally active in capturing sister kinetochores in early S phase. Eventually, both sister kinetochores have to attach to MTs organized by opposite SPBs (bipolar attachment) to ensure accurate chromosome segregation. The steps and proteins involved in establishing and maintaining bipolarity are poorly understood. It is only known that the cohesin complex and the aurora-like kinase Ipl1p are required for this process (Biggins et al., 1999; Tanaka et al., 2000).
We demonstrate that the proteins Ask1p, Dad2p, Spc19p and Spc34p are part of the Duo1p–Dam1p– Dad1p complex (here named the DDD complex). The phenotype of spc34-3 cells and cells depleted of the cohesin subunit Scc1p indicates that in budding yeast sister kinetochores bind first to the old SPB. While the monopolar attachment is resolved in wild-type cells, spc34-3 cells fail to establish biorientation. We suggest that one function of the Spc34p subunit of the DDD complex is to establish and maintain biorientation of sister kinetochores.
Results
Ask1p, Dad2p, Spc19p and Spc34p are components of the DDD complex
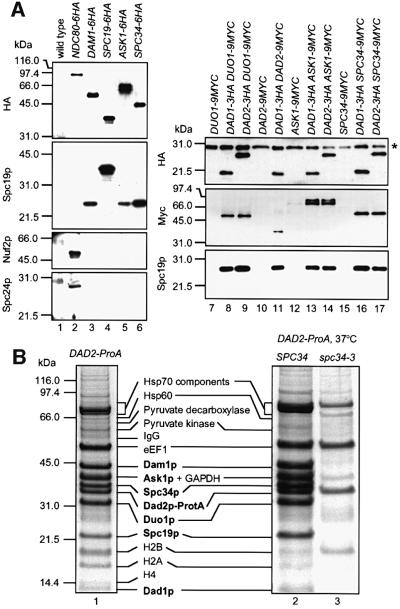
Spc19p and Spc34p are proteins of unknown function that co-purify with budding yeast spindles and locate along spindle MTs and at spindle poles (Wigge et al., 1998). Since two-hybrid interactions between Ask1p (YKL052c), Dad1p, Dad2p (Duo1p and Dam1p-interacting protein), Dam1p, Duo1p, Spc19p and Spc34p have been reported (Ito et al., 2000; Uetz et al., 2000), we analysed whether these proteins are present in common complexes. Immunoprecipitations were performed with cells expressing ASK1, DAM1, SPC19 and SPC34 fused to the haemagglutinin (HA) epitope. NDC80-6HA cells, coding for a component of the Ndc80p complex, were included as control. Analysis of the anti-HA precipitates revealed co-immunoprecipitation of Spc19p with Dam1p-6HA, Ask1p-6HA and Spc34p-6HA (Figure 1A, lanes 3–6) but not with Ndc80p-6HA (Figure 1A, lane 2). In contrast, the Ndc80p complex subunits Nuf2p and Spc24p were only detected in the Ndc80p-6HA (Figure 1A, lane 2) but not in the Dam1p-6HA, Spc19p-6HA, Ask1p-6HA or Spc34p-6HA precipitations (Figure 1A, lanes 3–6). Furthermore, Spc19p, Duo1p-9Myc, Dad2p-9Myc, Ask1p-9Myc and Spc34p-9Myc (Figure 1A, lanes 7–17) were found to co-immunoprecipitate with Dad1p-3HA and Dad2p-3HA, while Nuf2p and Spc24p were not detected in any of these immunoprecipitations (not shown). Together, Ask1p, Dad1p, Dad2p, Dam1p, Spc19p and Spc34p are part of common complexes, which do not include components of the Ndc80p complex.
Fig. 1. Ask1p, Dad2p, Spc19p and Spc34p are components of the DDD complex. (A) Immunoprecipitation of DDD complex components with anti-HA antibodies. Immunoblots are shown. The asterisk shows the IgG light chain that is detected by the secondary antibody. (B) Dad2p–ProA was purified from SPC34 DAD2-ProA or spc34-3 DAD2-ProA cells grown at 23°C (lane 1) or incubated for 4 h at 37°C (lanes 2 and 3). The purified proteins were analysed by SDS–PAGE and Coomassie Blue staining. Protein bands were identified by MALDI.
To address the subunit composition of the DDD complex, a functional, chromosomal gene fusion of DAD2 with protein A was constructed (DAD2-ProA). The Dad2p–ProA fusion protein was purified from yeast extracts using the property of protein A to bind with high affinity to IgG. Cells lacking a ProA tag were used as control. The bands of 44, 40, 37, 33, 32, 22 and 13 kDa were only present in the Dad2p–ProA purification (Figure 1B, lane 1, bold) but not in the control (not shown). These proteins were identified by matrix-assisted laser desorption/ionization (MALDI) analysis (Shevchenko et al., 1996) as Dam1p, Ask1p, Spc34p, Dad2p–ProA, Duo1p, Spc19p and Dad1p. The analysis of the protein bands also present in the control revealed no other kinetochore or spindle protein. Finally, we tested whether the DDD complex remained stable in spc34-3 DAD2- ProA cells incubated at 37°C. Affinity purification of Dad2p–ProA from spc34-3 cells yielded only Dad2p– ProA (lane 3), while the entire DDD complex was purified from SPC34 DAD2-ProA cells (lane 2). Co-purification of Ask1p, Dad1p, Dam1p, Duo1p, Spc19p and Spc34p with Dad2p–ProA in an Spc34p-dependent manner is consistent with one complex containing all seven proteins.
Ask1p, Dad1p, Dad2p, Spc19p and Spc34p associate with kinetochores and spindle MTs
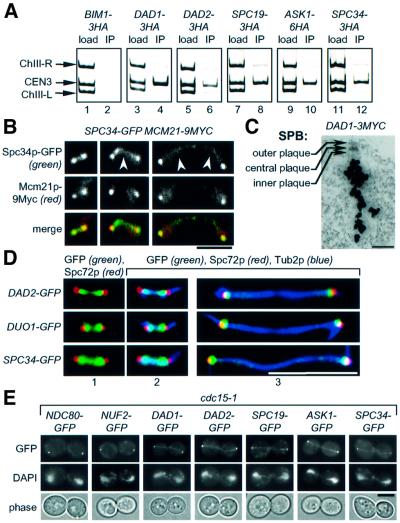
We tested by chromatin immunoprecipitation (ChIP) whether Ask1p, Dad1p, Dad2p, Spc19p and Spc34p are associated with centromere DNA. The spindle protein Bim1p was used as control (Schwartz et al., 1997). CEN3 DNA was enriched in the multiplex PCR analysis of the immunoprecipitates of Dad1p-3HA, Dad2p-3HA, Spc19p-3HA, Ask1p-6HA and Spc34p-3HA (Figure 2A, lanes 4, 6, 8, 10 and 12) but not of Bim1p-3HA (lane 2). These CEN3 ChIPs were specific because CEN3 DNA and control DNAs flanking CEN3 (ChIII-R and ChIII-L) were detected equally in the input material (Figure 2A, load), but only CEN3 was enriched in the immunoprecipitates (Figure 2A, IP of lanes 4, 6, 8, 10 and 12). Ask1p, Dad1p, Dad2p, Spc19p and Spc34p localize to centromere DNA in vivo.
Fig. 2. Ask1p, Dad1p, Dad2p, Spc19p and Spc34p are kinetochore components and localize along nuclear MTs. (A) ChIP analysis. CEN3 DNA was co-immunoprecipitated with Dad1p-3HA (lane 4), Dad2p-3HA (lane 6), Spc19p-3HA (lane 8), Ask1p-6HA (lane 10) and Spc34p-3HA (lane 12) but not with Bim1p-3HA (lane 2). CEN3 DNA and control DNAs flanking CEN3 (ChIII-R and ChIII-L) were detected equally in the input material (lanes 1, 3, 5, 7, 9 and 11, load). (B) Spc34p co-localizes with the kinetochore protein Mcm21p. Fixed SPC34-GFP MCM21-9Myc cells were subjected to indirect immunofluorescence with anti-Myc antibodies. The GFP signal was analysed by fluorescence microscopy. The arrows indicate localization of Spc34p–GFP at the nuclear MTs. (C) Localization of Dad1p-3Myc by immunoelectron microscopy. Bar: 0.25 µm. (D) Dad2p, Duo1p and Spc34p do not localize with SPBs. Cells of DAD2-GFP, DUO1-GFP and SPC34-GFP were analysed as in (B) except using anti-Spc72p and anti-Tub2p antibodies. Cells in panels 1 and 2 are identical but, in (1), the tubulin signal is not shown. (E) Cells of cdc15-1 with the indicated gene fusions were incubated for 3 h at 37°C. DNA was stained with DAPI. Cells were analysed by fluorescence microscopy. Bars in (B), (D) and (E): 5 µm.
Owing to the centromere clustering during most parts of the cell cycle, kinetochore components appear by fluorescence microscopy as one or two dots per cell, which are, at least in metaphase, clearly distinct from SPBs (Jin et al., 2000). We used this property to confirm kinetochore association of Spc34p. For this analysis, the chromosomal SPC34 was fused with the green fluorescence protein (GFP). Mcm21p-9Myc was employed as a marker for kinetochores (Ortíz et al., 1999). Mcm21p-9Myc was detected as one or two dots per cell, which co-localized in all inspected cells (n >100) with the strong dot-like Spc34p–GFP signals (Figure 2B). In addition, a weak Spc34p–GFP signal was also associated with the nuclear MTs (Figure 2B, arrows).
We tested whether Dad2p, Duo1p and Spc34p co-localize with the SPB marker Spc72p. The Spc72p signal failed to co-localize with the dot-like Dad2p–GFP, Duo1p–GFP and Spc34p–GFP in cells with a metaphase spindle (Figure 2D, panels 1 and 2). In anaphase cells, a partial overlap of the GFP and the Spc72p signals was observed (Figure 2D, panel 3, yellow colour), which is consistent with the close clustering of kinetochores with SPBs during this stage of the cell cycle (Jin et al., 2000). The relatively weak Dad2p–GFP, Duo1p–GFP and Spc34p–GFP signal along MTs could not be seen because of the strong tubulin signal. This experiment indicates that Dad2p, Duo1p and Spc34p are associated with kinetochores but not with SPBs.
To exclude that the staining between spindle poles reflects the DDD complex on chromosomes in the process of segregation, we studied localization of DDD proteins in cdc15-1 cells. Ndc80p and Nuf2p were included as controls. Cells of cdc15-1 arrested at the end of anaphase with fully segregated chromosomes but failed to disassemble the spindle and exit mitosis (Shou et al., 1999). Consistent with a complete clustering of kinetochores at spindle poles in cdc15-1 cells, the kinetochore proteins Ndc80p–GFP and Nuf2p–GFP appeared as two dots, which localized at the periphery of the DAPI-staining regions (Figure 2E). Similarly, two dot-like signals were observed in the cdc15-1 cells expressing ASK1, DAD1, DAD2, SPC19 and SPC34 GFP gene fusions. In addition, all fusion proteins localized along anaphase spindles (Figure 2E).
Spindle localization of Spc19p, Spc34p and Duo1p has been confirmed by immunoelectron microscopy (Hofmann et al., 1998; Wigge et al., 1998). Immunoelectron microscopic analysis of Dad1p-3Myc cells with anti-Myc antibodies revealed a band-like signal within the centre of yeast cells originating adjacent to the nuclear side of the SPB (Figure 2C). This pattern is typical for proteins that associate with nuclear MTs (Wigge et al., 1998).
Taken together, these results show that Ask1p, Dam1p, Spc19p and Spc34p are associated with kinetochores and nuclear MTs.
Kinetochore association of Dad2p and Spc34p requires the kinetochore proteins Ndc10p and Ndc80p and MTs
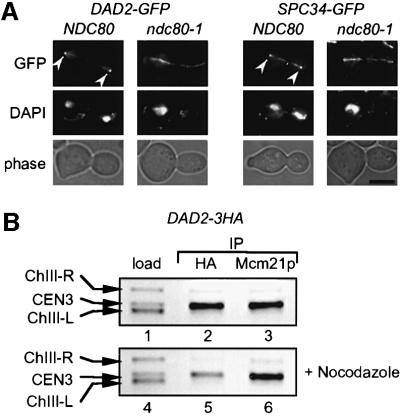
In ndc10-1 and ndc80-1 cells, kinetochores fail to interact with MTs (Goh and Kilmartin, 1993; Janke et al., 2001; Wigge and Kilmartin, 2001). We tested whether the association of Dad2p and Spc34p with kinetochores is affected in ndc10-1 and ndc80-1 cells. The strong Dad2p–GFP and Spc34p–GFP signals at the end of the spindle, representing clustered kinetochores, was lost upon shifting ndc80-1 but not NDC80 cells to 37°C (Figure 3A). However, Dad2p–GFP and Spc34p–GFP maintained spindle association. Similar results were obtained with ndc10-1 cells (not shown). Furthermore, the dissociation of Dad2p–GFP and Spc34p–GFP from centromere DNA in ndc10-1 and ndc80-1 cells incubated at 37°C was confirmed by ChIP (not shown). Thus, kinetochore but not MT association of Dad2p and Spc34p is dependent on Ndc10p and Ndc80p.
Fig. 3. Determinates of kinetochore association of Dad2p and Spc34p. (A) Association of Dad2p and Spc34p with kinetochores but not MTs is dependent on Ndc80p. Wild-type NDC80 and ndc80-1 cells with DAD2-GFP and SPC34-GFP were incubated at 37°C for 3 h. Cells were analysed by fluorescence microscopy. Arrows indicate kinetochores. Bar: 5 µm. (B) Centromere binding of Dad2p-3HA requires MTs. DAD2-3HA cells were incubated with (lanes 4–6) and without 10 µg/ml nocodazole (lanes 1–3) for 3 h at 30°C. ChIPs with anti-HA (lanes 2 and 5) and anti-Mcm21p antibodies (lanes 3 and 6) were performed as in Figure 2A. Lanes 1 and 4 represent the input material.
Kinetochore association of DDD complex components may depend on the binding of kinetochores to MTs. To test this possibility, DAD2-3HA cells were treated with the drug nocodazole, which depolymerizes most MTs. It is important to note that MT remnants at SPBs are probably responsible for the kinetochore clustering seen in some nocodazole-treated cells (Jin et al., 2000). Therefore, we expected only a reduction but no complete abolition of the interaction of Dad2-3HA with CEN3 DNA if MTs are required for the binding of the DDD complex to centromeres. We determined centromere association of Dad2p-3HA by ChIP in untreated and nocodazole-treated cells relative to the kinetochore protein Mcm21p. Dad2p-3HA and Mcm21p were precipitated with equal efficiency from extracts of cells not treated with nocodazole (Figure 3B, lanes 2 and 3). MT depolymerization did not affect centromere association of Mcm21p (lane 6) but reduced the precipitation of CEN3 DNA with Dad2p-3HA at least 3-fold (lane 5). Thus, efficient kinetochore association of Dad2p requires MTs.
Conditional lethal spc34-3 cells elongate spindles and distribute DNA masses in the presence of Pds1p
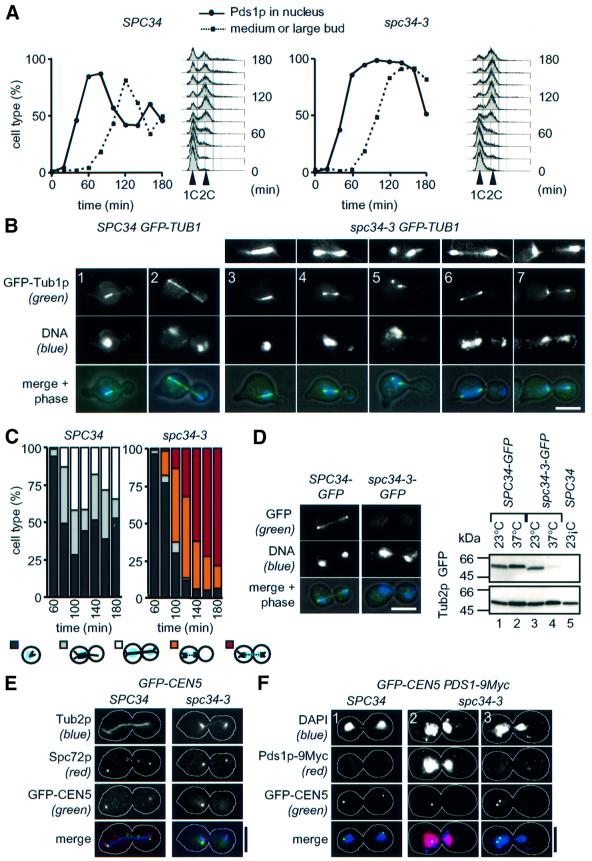
Temperature-sensitive mutants were analysed to investigate the as yet ill-defined function of the DDD complex in chromosome segregation and spindle formation. Since cells of dad1-9, dad2-9, spc19-4 and spc34-3 displayed very similar phenotypes (see Supplementary figure 9 available at The EMBO Journal Online), we only show the analysis of spc34-3 cells. Cell cycle progression of α-factor-synchronized wild-type SPC34 and spc34-3 cells carrying the α-tubulin gene TUB1 fused to GFP (GFP-TUB1) was followed at 37°C. Cells of spc34-3 replicated their DNA similarly to wild-type SPC34 cells but then arrested in the cell cycle with a large bud, 2C DNA content and non-degraded nuclear Pds1p. In contrast, SPC34 cells degraded Pds1p and started a new cell cycle (Figure 4A). The failure of spc34-3 cells to degrade Pds1p is typical for cells with an activated spindle checkpoint (Hwang et al., 1998).
Fig. 4. Conditional lethal cells of SPC34 are defective in anaphase spindle formation and biorientation of sister centromeres. (A–C) Cells of SPC34 and spc34-3 with PDS1-6HA or GFP-TUB1 were synchronized with α-factor (t = 0). After release from the cell cycle block by washing, cells were shifted to 37°C. (A) Samples were analysed every 20 min for DNA content, nuclear Pds1p by indirect immunofluorescence, and budding index (n >100). (B) Spindle morphology was determined 100 min after release from α-factor by fluorescence microscopy. The smaller panels in 3–7 are 2-fold magnifications of the spindles below. (C) Quantification of the spindle phenotypes of spc34-3 cells of (B); n >100. The blue area in the cartoons to the bottom symbolizes the DAPI-staining regions. The lines inside the cells indicate MTs and the dotted lines defective MTs. (D) Cells of SPC34-GFP and spc34-3-GFP were incubated for 3 h at 37°C. Cells were inspected by fluorescence microscopy. Protein extracts of the indicated cells were analysed by immunoblotting with anti-GFP and anti-Tub2p antibodies. (E) Duplicated CEN5 of spc34-3 cells fails to attach to the spindle in a bipolar manner. α-Factor-synchronized SPC34 GFP-CEN5 and spc34-3 GFP-CEN5 cells were fixed after 3 h at 37°C and analysed as in Figure 2D. (F) Chromosomes of spc34-3 cells are segregated in the presence of nuclear Pds1p. SPC34 GFP-CEN5 PDS1-9Myc and spc34-3 GFP-CEN5 PDS1-9Myc cells were shifted for 2 h (panels 1 and 2) or 3 h (panel 3) to 37°C. Cells were subjected to indirect immunofluorescence to stain Pds1p-9Myc. GFP–CEN5 was visualized by fluorescence microscopy. Bars: 5 µm.
The spindle structure of SPC34 and spc34-3 cells was determined by fluorescent microscopy. Cells of spc34-3 were able to form a short bipolar spindle (Figure 4B, panel 3, and C, light grey bars, 60–100 min), which was similar in appearance to SPC34 cells (Figure 4B, panel 1). At 80 min, spc34-3 cells appeared with a defective spindle positioned in the mother cell body (Figure 4B, panels 4–6, and C, orange bars). Another cell type, which was first observed after 80 min, displayed two separated DAPI staining regions, one in the mother cell and the other in the bud (Figure 4B, panel 7, and C, red bars). The spindle poles of these cells were only loosely connected by a thin bundle of nuclear MTs. Sometimes the two half spindles were even completely separated (not shown). The extended spindle structures of spc34-3 cells were quite different from the anaphase spindle of wild-type cells (Figure 4B, panel 2). Hence we asked the question whether mitotic checkpoint activation contributes to the broken spindle phenotype of spc34-3 cells. The analysis of Δmad2 spc34-3 cells revealed the same spindle defects as in spc34-3 cells (not shown). In summary, spc34-3 cells assemble a short spindle, which seems to break during the elongation process.
The defects of spc34-3 cells may be caused by the dissociation of the mutated Spc34p (spc34p-3) from spindle MTs and kinetochores. Indeed, the GFP-tagged spc34p-3 was no longer localized with the nuclear MTs and kinetochores (Figure 4D) when cells were incubated at 37°C. Analysis of SPC34-GFP and spc34-3-GFP cells by immunoblotting showed that most spc34p-3–GFP was degraded at the restrictive temperature (Figure 4D, lane 4). Considering that the DDD complex disassembles in spc34-3 cells (Figure 1B), it is likely that spc34p-3 becomes degraded after its release from the DDD complex.
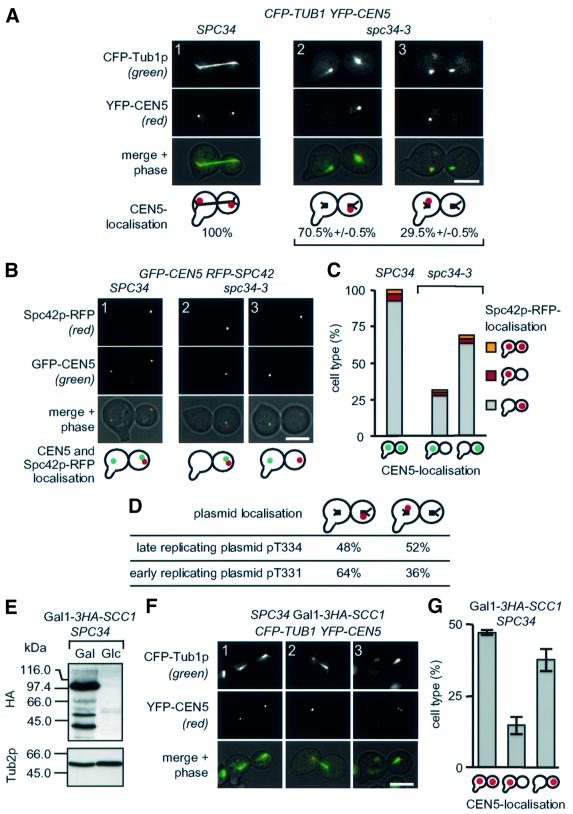
To study centromere localization in SPC34 and spc34-3 cells, we constructed strains with 112 copies of TetO-binding sites next to CEN5 DNA and which expressed the tetracycline repressor fused to GFP (TetR–GFP) (here referred to as GFP-CEN5). α-factor-synchronized SPC34 and spc34-3 cells with GFP-CEN5 were then shifted to 37°C. Fixed cells were subjected to indirect immunofluorescence using antibodies against yeast β-tubulin (anti-Tub2p) and the SPB component Spc72p. In SPC34 cells, the dot-like CEN5 DNA signal associated with the two spindle poles (Figure 4E). In contrast, in spc34-3 cells, the CEN5 DNA signal was always (100%, n >100) associated with only one spindle pole (Figure 4E, and F, panels 2 and 3). In addition, only one CEN5 signal was seen in spc34-3 cells inspected from 0.5 to 2 h after the temperature shift (Figure 4E, and F, panel 2) although the DNA replicated during this period (Figure 4A). This indicates that sister centromeres fail to establish biorientation in spc34-3 cells. Whether in these cells both or only one sister kinetochore binds to MTs organized by one SPB is not known.
The lack of two CEN5 DNA signals suggests that sister chromatid cohesion was maintained in spc34-3 cells, probably due to mitotic checkpoint activation preventing Pds1p degradation (Hwang et al., 1998). Consistently, in spc34-3 cells (2 h at 37°C) with two separated DAPI-staining regions, the nuclear Pds1p was not degraded (Figure 4F, panel 2), as is normally the case in wild-type cells in anaphase (Figure 4F, panel 1). However, Pds1p was degraded in half of the spc34-3 cells after ∼3 h at 37°C (Figure 4A). In these spc34-3 cells, the two separated CEN5 DNA signals located next to one SPB (Figure 4F, panel 3), confirming that paired sister chromatids were segregated to one of the two spindle poles. Thus, in spc34-3 cells, sister centromeres fail to establish biorientation.
Cells of spc34-3 fail to maintain biorientation of sister centromeres
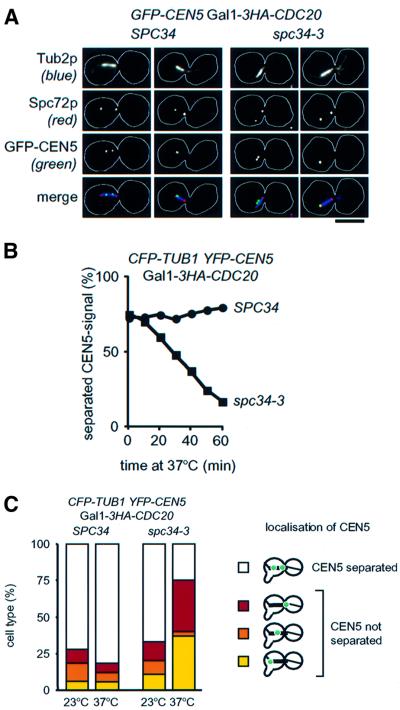
Cells of spc34-3 do not establish biorientation of sister centromeres (Figure 4F). This failure may be caused by spindle collapse or by a kinetochore defect. To discriminate both possibilities, we sought conditions to study biorientation without spindle collapse. Previously, cells of dam1-11 arrested in metaphase by Cdc20p depletion were shown to maintain a bipolar spindle when subsequently shifted to 37°C (Cheeseman et al., 2001). However, biorientation of sister centromeres has not been investigated in dam1-11 cells. Similarly, when Cdc20p was depleted at 23°C (ΔCdc20), SPC34 and spc34-3 cells arrest in metaphase with a short bipolar spindle. At least 72% of the sister CEN5 DNAs of SPC34 and spc34-3 cells were separated, indicative that most if not all sister centromeres were attached to spindle poles in a bipolar manner and that tension was applied to the kinetochores (Tanaka et al., 2000). When ΔCdc20 SPC34 and ΔCdc20 spc34-3 cells were shifted to 37°C (the restrictive temperature of spc34-3 cells) for up to 1 h, no major changes in the morphology of the spindle were seen by light microscopy (Figure 5A). CEN5 DNA separation in ΔCdc20 spc34-3 cells, despite maintaining a metaphase spindle, was gradually reduced from 72 to 17%, while it was not affected in ΔCdc20 SPC34 cells (Figure 5B). This phenotype suggests that spc34-3 cells fail to maintain biorientation at the restrictive temperature, as illustrated by the lack of sister centromere separation, even when a mitotic spindle is maintained.
Fig. 5. Spc34p is required to maintain bipolarity. (A) Cells of SPC34 and spc34-3 with GFP-CEN5 Gal1-CDC20 were grown in galactose medium at 23°C. Cells were then incubated for 2 h at 23°C in glucose medium to repress Gal1-CDC20 followed by a shift to 37°C for 1 h. Cells were analysed as in Figure 2D. Bar: 5 µm. (B) Sister CEN5 DNA biorientation gradually decreases in spc34-3 cells. Cells of SPC34 and spc34-3 with CFP-TUB1 YFP-CEN5 Gal1-CDC20 were incubated for 2 h at 23°C in glucose medium to arrest cells in metaphase. Cells were then shifted to 37°C. Separation of sister CEN5 of cells with a metaphase spindle was determined by fluorescence microscopy. (C) Cells of (B) were synchronized with α-factor at 23°C to mark the mother cell body with a mating projection. After washing the cells with glucose medium, they arrested in metaphase with a short spindle. The sample designated ‘23°C’ was taken and fixed. Cells were then incubated for 1 h at 37°C. Sister CEN5 localization was determined by fluorescent microscopy; n >100.
To investigate the localization of the CEN5 DNA, SPC34 and spc34-3 cells with Gal1-CDC20 were synchronized by α-factor, which marked the mother cell body with a mating projection. Cells were released from the α-factor block into glucose medium at 23°C to deplete Cdc20p. Sister CEN5 DNA of the metaphase-arrested cells was separated in >67% of SPC34 or spc34-3 cells (Figure 5C). Both cell types were then incubated for 1 h at 37°C, resulting in >75% of ΔCdc20 spc34-3 cells with unseparated CEN5 DNA. Of these, only 3% localized between both spindle poles. Most of the unseparated sister CEN5 DNAs were associated with either spindle pole, showing no preference between the two SPBs. Thus, when metaphase-arrested spc34-3 cells are incubated at the restrictive temperature, most of the sister centromeres lose bipolar attachment and locate unseparated with equal likelihood adjacent to either SPB.
Preferential association of the CEN5 DNA in spc34-3 cells with the old SPB is dependent on the timing of centromere DNA replication
In ΔCdc20 spc34-3 cells, in which biorientation was established before incubating the cells at the restrictive temperature (Figure 5C), the unseparated sister kinetochores associated with equal likelihood with one of the two SPBs. In contrast, when spc34-3 cells were shifted in G1 to the restrictive temperature, CEN5 DNA showed preferential association with the spindle pole in the cell body without a mating projection, the bud (Figure 6A, panel 2). This difference in behaviour provides additional evidence that spc34-3 cells shifted in G1 to 37°C never achieve a state of biorientation. Thus, spc34-3 cells may also be defective in a step leading to biorientation. To evaluate this notion, sister CEN5 distribution was investigated further in spc34-3 cells. Using the property of the SPB marker Spc42p–RFP to label the old SPB selectively, we recently showed that in unperturbed wild-type cells the old SPB always segregates into the bud (Pereira et al., 2001). To confirm that this is also the case in spc34-3 cells, the localization of CEN5 DNA was studied in SPC42-RFP spc34-3 cells. CEN5 DNA was detected in ∼70% of the mutant cells next to the red fluorescent old SPB in the bud (Figure 6B, panel 2, and C, last column). In only ∼30% of spc34-3 cells did the CEN5 DNA signal associate next to the newly formed SPB in the mother cell (Figure 6B, panel 3, and C, middle column). At anaphase, in all SPC34 cells, one CEN5 DNA signal was associated with the new SPB in the mother cell body and the other with the red fluorescent old SPB in the bud (Figure 6A, panel 1, B, panel 1, and C, first column). In summary, in spc34-3 cells, sister CEN5 DNAs preferentially associate with the ‘old’ SPB.
Fig. 6. Sister CEN5 DNAs of spc34-3 cells are associated preferentially with the old SPB. (A) SPC34 and spc34-3 cells with CFP-TUB1 YFP-CEN5 were arrested with α-factor to mark the mother cell. Washed cells were then shifted to 37°C for 3 h. CFP–Tub1p and YFP–CEN5 were analysed by fluorescence microscopy. The percentage of anaphase cells with YFP–CEN5 (red dot) at both or only one of the two spindle poles was determined in two independent experiments (n >100). The black lines in the drawn cells symbolize the MTs. The mating projection of the mother cell body is shown as an extension. (B) Cells of GFP-CEN5 RFP-SPC42 in which the old SPB is marked by the fluorescent RFP signal were grown as described in Materials and methods. The Spc42p–RFP and the GFP–CEN5 signals were detected by fluorescence microscopy. The red dot in the cartoons indicates the old SPB and the green dots the GFP–CEN5 signals. The mating projection marking the mother cell body is indicated as an extension. (C) Quantification of (B); n >100. Symbols are as in (B). (D) Preferential binding of sister chromatids to the old SPB is dependent on the time of centromere replication. Cells of spc34-3 with the late and early replicating plasmids pT334 and pT331 were analysed as in (A). Cartoon as in (A). (E) Gal1-3HA-SCC1 SPC34 cells carrying CFP-TUB1 YFP-CEN5 were grown and analysed as described in Figure 7A. (F) Cells of (E) were treated and analysed as in Figure 7B. (G) Quantification of (F). Shown is the average of two independent experiments; n >100. Cartoons are as in (A). Bars: 5 µm.
One explanation for the preference of sister CEN5s binding to the ‘old’ SPB is that centromere replication, which occurs early in S phase (McCarroll and Fangmann, 1988), is completed before the new SPB is fully active. As a consequence, kinetochores bind preferentially to MTs organized by the old SPB even in wild-type cells. Spc34p may then be required to establish biorientation. To address this possibility, we compared in α-factor-synchronized spc34-3 TetR–GFP cells the segregation of two mini-chromosomes labelled with 112 copies of TetO-binding sites that differ in the timing of their replication by ∼10 min (McCarroll and Fangmann, 1988). The late replicating plasmid pT334 was partitioned equally to both spindle poles, whereas the early replicating plasmid pT331 was associated preferentially with the spindle pole in the bud (Figure 6D). Thus, early centromere DNA replication favours monopolar attachment with the old SPB.
Cells depleted of the cohesin subunit Scc1p (ΔScc1) frequently are defective in bipolar segregation of sister centromeres. Time-lapse analysis revealed that in ΔScc1 cells, kinetochores remained stably associated with the spindle pole they first bind to (Tanaka et al., 2000). Thus, in ΔScc1 cells, the kinetochore distribution is a reflection of the initial binding of kinetochores to spindle poles. We tested whether in ΔScc1 cells kinetochores interact preferentially with the old SPB. The genomic copy of SCC1 was set under the control of the Gal1 promoter (Gal1-3HA-SCC1). Gal1-3HA-SCC1 SPC34 cells were first arrested with α-factor in glucose medium to allow 3HA-Scc1p degradation (Figure 6E). Following α-factor release in the presence of glucose, ∼45% of ΔScc1 SPC34 cells showed attachment of sister CEN5 DNAs with the two spindle poles (Figure 6F, panel 1, and G, first column). In ∼55% of cells, both separated CEN5 signals resided with only one spindle pole (Figure 6F, panels 2 and 3, and G, last two columns). As was the case with spc34-3 cells (Figure 6A), attachment to the SPB in the bud was clearly more frequent (Figure 6G, compare middle with last column). In ΔScc1p cells, sister CEN5 DNAs associate preferentially with the old SPB in the bud.
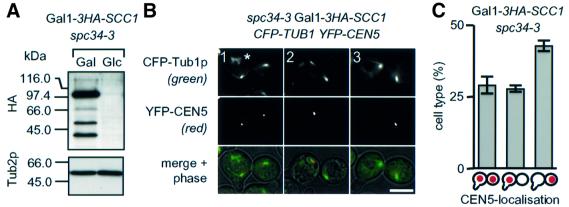
Sister kinetochores of spc34-3 cells lacking Scc1p are functionally equivalent
The failure of spc34-3 cells to maintain and establish bipolar attachment of sister centromeres may either be caused by the dysfunction of one of the two sister kinetochores or by the weakened kinetochore–MT interaction at both kinetochores. These possibilities were addressed by the depletion of the cohesin subunit Scc1p (ΔScc1) in spc34-3 cells, allowing the disjoined sister centromeres to bind independently to MTs (Tanaka et al., 2000). Gal1-3HA-SCC1 spc34-3 cells were arrested with α-factor in glucose medium at the permissive temperature to degrade Scc1p (Figure 7A). After removal of α-factor, cells were shifted to 37°C in glucose medium. Cells progressed through S phase without establishing sister chromatid cohesion (Tanaka et al., 2000). Similarly to spc34-3 cells, cells of ΔScc1 spc34-3 arrested in the cell cycle with a large bud and defective anaphase spindles (compare Figures 4 and 7B). However, unlike spc34-3 cells, in ∼30% of ΔScc1 spc34-3 cells the sister CEN5 DNA signals were separated to opposite poles and each associated adjacent to one spindle pole (Figure 7B, panel 1, and C, first column). This suggests that both kinetochores are attached to MTs. The residual ΔScc1 spc34-3 cells displayed attachment of the two CEN5 DNAs to the same spindle pole (Figure 7B, panels 2 and 3, and C, last two columns) with a clear preference for the SPB in the bud (Figure 7C, compare last two columns). Furthermore, both CEN5 DNAs (n >100) located adjacent to SPBs, suggesting that both kinetochores bind MTs. We conclude that both sister kinetochores of spc34-3 cells can be distributed to opposite poles in the absence of the tension provided by sister chromatid cohesion. Furthermore, this demonstrates that both sister kinetochores of spc34-3 cells are competent to interact with MTs.
Fig. 7. Sister kinetochores of spc34-3 cells are functional. (A) Gal1-3HA-SCC1 spc34-3 cells carrying CFP-TUB1 YFP-CEN5 were grown in galactose medium at 23°C (‘Gal’). Cells were resuspended in glucose medium with α-factor and incubated for 4 h at 23°C to suppress SCC1 expression (‘Glc’). Shown is an immunoblot with anti-HA antibodies. Tub2p was detected as loading control. (B) Cells of (A) with depleted Scc1p were washed with glucose medium to remove α-factor and simultaneously shifted to 37°C. After 2 h at 37°C, spindle formation (CFP–Tub1p) and CEN5 localization (CEN5–YFP) were analysed by fluorescence microscopy. The asterisk indicates CFP–Tub1p, which is not associated with the spindle. Bar: 5 µm. (C) Quantification of (B). Shown is the average of two independent experiments. The CEN5 DNA localization of at least 100 large budded cells was determined. Cartoons are as in Figure 6A.
Spc34p ensures spindle stability in anaphase
Whether components of the DDD complex are required to maintain an anaphase spindle has not been investigated. We addressed this possibility by arresting SPC34 and spc34-3 cells with GFP-TUB1 Gal1-lacI-TEM1 in late anaphase at 23°C by glucose-induced depletion of Tem1p (ΔTem1) (Figure 8A, lanes 2 and 6). ΔTem1 SPC34 or ΔTem1 spc34-3 cells displayed a large bud, an elongated anaphase spindle and separated DAPI-staining regions (Figure 8B and C). When these cells were incubated at 37°C, most ΔTem1 SPC34 cells remained in anaphase, as suggested by the maintenance of the anaphase spindle (Figure 8B and C), the failure to form an additional bud (not shown) and the constant Clb2p content (Figure 8A, compare lane 2 with lanes 3 and 4), the latter being degraded normally with mitotic exit (Shou et al., 1999). ΔTem1 spc34-3 cells also maintained Clb2p (compare lane 6 with lanes 7 and 8) and failed to from a bud (not shown), consistent with an arrest in anaphase. However, in most ΔTem1 spc34-3 cells, the anaphase spindle collapsed into two relatively stable half spindles (Figure 8B and C). This indicates that Spc34p is required in anaphase to ensure spindle stability.
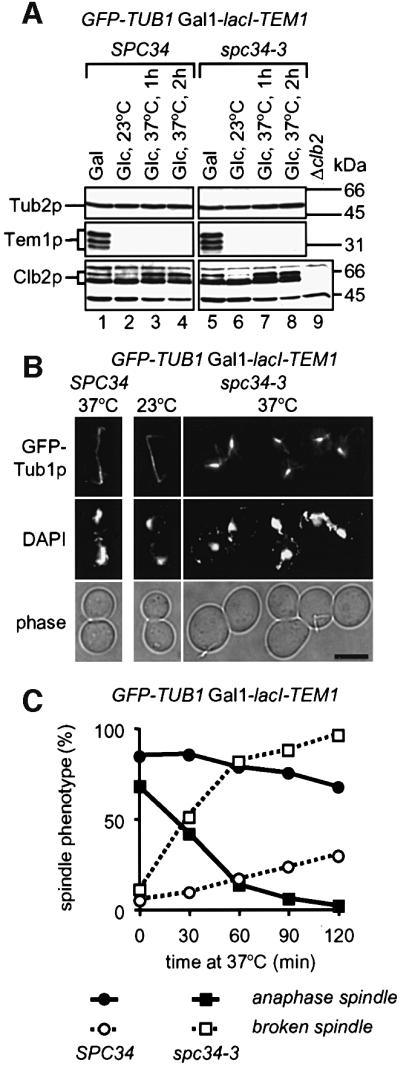
Fig. 8. Spc34p is required to maintain an anaphase spindle. (A) SPC34 (lanes 1–4) and spc34-3 cells (lanes 6–8) with Gal1-lacI-TEM1 GFP-TUB1 were grown in 2% galactose 3% raffinose medium at 23°C (lanes 1 and 5) followed by the addition of 2% glucose to repress Ga11-lacI-TEM1 expression (lanes 2 and 6). Anaphase cells were then incubated in glucose medium for up to 2 h at 37°C (lanes 3, 4, 7 and 8). Cells were tested by immunoblotting for Tem1p, Clb2p and Tub2p. Cells of Δclb2 in lane 9 were used as control for the anti-Clb2p antibodies. (B) Cells of (A), after incubation in glucose medium at 37°C, were fixed, stained with DAPI and analysed by fluorescence microscopy. Shown are cells after 1 h at 37°C. (C) Quantification of (B); n >100. Only the percentage of large-budded cells is indicated.
Discussion
We show here that the proteins Ask1p, Dad2p, Spc19p and Spc34p are components of the DDD complex. The fact that only Ask1p, Dad1p, Dam1p, Duo1p, Spc19p and Spc34p were purified with Dad2p–ProA without additional spindle or kinetochore components indicates that the DDD proteins form a relatively stable, defined complex that is distinct from other kinetochore subcomplexes. This notion is supported further by the association of all DDD complex proteins with kinetochores and nuclear MTs and by the common phenotype of conditional lethal DDD mutants, all affecting chromosome segregation and spindle stability (Figures 2 and 3; Hofmann et al., 1998; Adams and Kilmartin, 1999; Cheeseman et al., 2001; Enquist-Newman et al., 2001; He et al., 2001). Homologues of Ask1p, Dad1p, Dam1p, Duo1p and Spc34p are present in the Candida albicans and Schizosaccharomyces pombe genome database (Cheeseman et al., 2001; Enquist-Newman et al., 2001; see Supplementary figure 10), suggesting that the function of the DDD complex is conserved at least between fungi.
Since Dam1p is an MT-binding protein (Hofmann et al., 1998), the DDD complex may interact directly with MTs. Kinetochore binding of Dad2p was found to be dependent on MTs (Figure 3B). A similar dependency has been observed for Dad1p, Dam1p and Duo1p (Enquist-Newman et al., 2001). In summary, these data suggest a model in which the DDD complex is targeted from nuclear MTs to kinetochores upon MT binding of kinetochores. Consistently, kinetochore association of DDD complex components is also affected in ndc80-1 cells (Figure 3A), in which kinetochores fail to interact with MTs (Adams and Kilmartin, 1999).
Conditional lethal mutants of DAD1, DAM1 and DUO1 showed spindle elongation defects and chromosome missegregation of joined sister centromeres to one spindle pole (Hofmann et al., 1998; Cheeseman et al., 2001; Enquist-Newman et al., 2001; Jones et al., 2001). These phenotypes were also observed in dad1-9, dad2-9, spc19-4 and spc34-3 cells (Figure 4 and Supplementary figure 9). A closer inspection revealed that spc34-3 cells elongated the spindle although the securin Pds1p was not degraded and sister chromatid cohesion remained. This is in contrast to wild-type cells in which anaphase spindle elongation is triggered by Pds1p degradation followed by disjunction of sister chromatid cohesion (Uhlmann et al., 2000). Premature spindle elongation has been described for metaphase-arrested ipl1-321 cells, encoding a mutated form of the aurora-like kinase Ipl1p. In ipl1-321 cells, spindle extension is triggered by the frequent failure of sister kinetochore biorientation (Biggins et al., 1999). Thus, the spindle of spc34-3 cells may elongate because, in the absence of bipolar kinetochore attachments (Figure 4), poleward forces are not opposed.
Previous studies did not address the function of the DDD complex in establishing biorientation (Hofmann et al., 1998; Cheeseman et al., 2001; Enquist-Newman et al., 2001) nor whether the spindle defect in these mutants is a consequence of chromosome missegregation. Our analysis of spc34–3 cells now reveals three novel functions of the DDD complex. First, Spc34p is required to maintain biorientation of sister chromatids under conditions that do not affect the metaphase spindle. Secondly, Spc34p has an additional function in the establishment of biorientation. Thirdly, Spc34p is required after chromosome segregation to stabilize anaphase spindles.
Metaphase-arrested spc34-3 cells failed to maintain biorientation of sister kinetochores upon shifting cells to 37°C (Figure 5). Under this condition, the spindle structure was not visually affected. Thus, the failure to maintain biorientation is not caused by spindle collapse. Instead, it is likely that one of the two sister kinetochores of spc34-3 cells fails to maintain the interaction with MTs when tension is applied. Because sister kinetochores of spc34-3 cells always localize next to one spindle pole, the other kinetochore must maintain its MT connection. We have shown that all kinetochores of ΔScc1 spc34-3 cells are able to interact with MTs in the absence of tension (Figure 7), suggesting that Spc34p is not required for kinetochore binding to MTs. However, the requirement for Spc34p to maintain biorientation in the presence of tension implies that the DDD complex stabilizes kinetochore–MT interactions. We therefore propose that after an initial binding step of kinetochores to MTs, the DDD complex associates with kinetochores, thereby strengthening the interaction with MTs.
When metaphase-arrested ΔCdc20 spc34-3 cells were incubated at 37°C, biorientation failed and joined sister centromeres associated with equal likelihood to one of the two spindle poles (Figure 5C). This phenotype contrasts with the behaviour of spc34-3 cells shifted to the restrictive temperature before the establishment of biorientation. In this case, the majority of CEN5 DNA associated with MTs organized by the old SPB (Figure 6). This difference indicates that at least some sister kinetochores in spc34-3 cells do not go through a phase of bipolarity. Thus, Spc34p has a role not only in maintaining biorientation but also in a step leading to it. At present, it is unclear why spc34-3 cells fail to establish biorientation. It could simply be that Spc34p-stabilized MT–kinetochore interactions are required to resolve the monopolar attachment of sister centromeres.
DDD complex components are associated with nuclear MTs throughout the cell cycle (Figure 2; Hofmann et al., 1998; Adams and Kilmartin, 1999; Enquist-Newman et al., 2001). The DDD complex at MTs may only be a storage form that is targeted to kinetochores upon their binding to MTs. In such a case, the spindle defect of DDD mutants could be caused by premature spindle elongation in the presence of Pds1p (Figure 4F). Pds1p would then inhibit separase Esp1p (Ciosk et al., 1998), which has, besides degradation of the cohesin subunit Scc1p, an additional function in stabilizing the anaphase spindle (Sullivan et al., 2001). Alternatively, the DDD complex functions directly in MT stabilization. We found that the pre-formed anaphase spindle of ΔTem1 spc34-3 cells broke into two relatively stable half spindles when cells were incubated at the restrictive temperature (Figure 8B). This phenotype indicates that Spc34p helps to maintain the pole to pole MTs in anaphase, probably by stabilizing the spindle midzone.
In spc34-3, Scc1p-depleted (Figure 6) and ipl1-321 cells (T.U.Tanaka, unpublished), the monopolar attached sister CEN5 DNAs associate preferentially with the old SPB in the bud (Pereira et al., 2001). This result is explained most easily by assuming that at the time when centromere DNA is replicated in early S phase (McCarroll and Fangmann, 1988), the newly formed SPB is not fully functional. Due to this deficiency of the new SPB, sister kinetochores bind preferentially to the functional old SPB. When the centromere DNA is replicated later, as is the case with the late replicating plasmid pT334 (Figure 6D), the new SPB had time to mature such that both SPBs are equally active in binding to sister kinetochores. This result also excludes the possibility that a kinetochore defect is responsible for the preferential binding of sister kinetochores to the old SPB. We suggest that also in wild-type cells many of the joined sister centromeres bind first to MTs organized by the old SPB. Monopolar attachment of sister centromeres is then resolved in wild-type cells but persists in spc34-3 and ipl1-321 cells. Thus, our finding decisively contributes to the understanding of the steps leading to biorientation in budding yeast (Winey and O’Toole, 2001).
Materials and methods
Growth media, strains and plasmids
Yeast strains were grown in yeast extract, peptone, dextrose medium (YPD) or synthetic complete (SC) medium. For selective labelling of the old SPB, SPC42-RFP cells were grown on YPD plates for 6 days (Pereira et al., 2001). Yeast strains and plasmids are described in Table I. Yeast strains were constructed using PCR-amplified cassettes (Knop et al., 1999). To tag CEN5 DNA, plasmid pXH136 (He et al., 2000) was integrated next to CEN5 by homologous recombination. TetR–GFP of pCJ092 or TetR–YFP of pCJ097 were integrated into the ADE2 locus. Tub1p was labelled by integration of the plasmid pAFS125 (GFP-TUB1) into URA3 (Straight et al., 1997) or of pCJ141 (CFP-TUB1) into TRP1. Sites of TetO (112) were subcloned into the late and early replicating plasmids p13 and p13ARS (Friedmann et al., 1996), resulting in pT334 and pT331. Plasmid pWS103 (Shou et al., 1999) was used to construct Tem1p depletion strains. Conditional lethal mutants were constructed as described (Janke et al., 2001).
Table I. Yeast strains and plasmids.
| Name | Genotype | Reference |
|---|---|---|
| Yeast strains | ||
| CJY007 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD2-GFP::KanMX6 | this study |
| CJY008 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD1-3HA::KanMX6 | this study |
| CJY009 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD2-3HA::KanMX6 | this study |
| CJY011 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD2-TEV-ProA::KanMX6 | this study |
| CJY015 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 BIM1-3HA::KanMX6 | this study |
| CJY026 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 DAD1-3MYC::KanMX6 | this study |
| CJY067 | MATa Δdad1::KanMX6 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::pCJ045 | this study |
| CJY077 | MATa Δdad2::KanMX6 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::pCJ055 | this study |
| CJY092 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 NDC80-6HA::klTRP1 | Janke et al. (2001) |
| CJY094 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 SPC19-6HA::klTRP1 | this study |
| CJY097 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 SPC34-6HA::klTRP1 | this study |
| CJY114 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 Δspc19::HIS3MX4 leu2Δ1::pCJ074 | this study |
| CJY124 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 Δspc34::HIS3MX4 leu2Δ1::pCJ083 | this study |
| CJY138 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δsst1::URA3 PDS1-6HA::klTRP1 | Janke et al. (2001) |
| CJY178 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 Δspc34::HIS3MX4 leu2Δ1::pCJ083 Δsst1:: URA3 PDS1-6HA::klTRP1 | this study |
| CJY212 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ASK1-6HA::klTRP1 | this study |
| CJY295 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ097 trp1Δ63 his3Δ200 leu2Δ1 | this study |
| CJY299 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ097 trp1::pCJ141Δ63 his3Δ200 Δspc34:: HIS3MX4 leu2Δ1::pCJ083 | this study |
| CJY301 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 DAM1-6HA::klTRP1 | this study |
| CJY312 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ092 trp1Δ63 his3Δ200 leu2Δ1 PDS1-9MYC:: klTRP1 | this study |
| CJY316 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ092 trp1Δ63 his3Δ200 Δspc34::HIS3MX4 leu2Δ1:: pCJ083 PDS1-9MYC:: klTRP1 | this study |
| CJY332 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD2-9MYC::klTRP1 | this study |
| CJY333 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 ASK1-9MYC::klTRP1 | this study |
| CJY334 | MATa Δsst1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 Δspc34::HIS3MX4 leu2Δ1::pCJ083 DAD2- TEV-ProA::KanMX6 | this study |
| CJY352 | MATa cdc15-1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 DAD1-GFP::KanMX6 | this study |
| CJY353 | MATa cdc15-1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 DAD2-GFP::KanMX6 | this study |
| CJY354 | MATa cdc15-1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 SPC19-GFP::KanMX6 | this study |
| CJY355 | MATa cdc15-1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 SPC34-GFP::KanMX6 | this study |
| CJY356 | MATa cdc15-1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ASK1-GFP::KanMX6 | this study |
| CJY357 | MATa cdc15-1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 DUO1-GFP::KanMX6 | this study |
| CJY358 | MATa cdc15-1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 NDC80-GFP::KanMX6 | this study |
| CJY359 | MATa cdc15-1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 NUF2-GFP::KanMX6 | this study |
| CJY362 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 SPC34-GFP::HIS3MX4 | this study |
| CJY364 | MATa Δsst1 ura3-52::pAFS125 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 | this study |
| CJY365 | MATa Δdad1::KanMX6 Δsst1 ura3-52::pAFS125 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::pCJ045 | this study |
| CJY366 | MATa Δdad2::KanMX6 Δsst1 ura3-52::pAFS125 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::pCJ055 | this study |
| CJY367 | MATa Δsst1 ura3-52::pAFS125 lys2-801 ade2-101 trp1Δ63 his3Δ200 Δspc19::HIS3MX4 leu2Δ1::pCJ074 | this study |
| CJY368 | MATa Δsst1 ura3-52::pAFS125 lys2-801 ade2-101 trp1Δ63 his3Δ200 Δspc34::HIS3MX4 leu2Δ1::pCJ083 | this study |
| CJY371 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 SPC34-9MYC::klTRP1 | this study |
| CJY372 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DUO1-9MYC::klTRP1 | this study |
| CJY373 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD1-3HA::KanMX6 DAD2-9MYC::klTRP1 | this study |
| CJY374 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD1-3HA::KanMX6 ASK1-9MYC::klTRP1 | this study |
| CJY375 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD1-3HA::KanMX6 SPC34-9MYC::klTRP1 | this study |
| CJY376 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD1-3HA::KanMX6 DUO1-9MYC::klTRP1 | this study |
| CJY377 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD2-3HA::KanMX6 ASK1-9MYC::klTRP1 | this study |
| CJY378 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD2-3HA::KanMX6 SPC34-9MYC::klTRP1 | this study |
| CJY379 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 DAD2-3HA::KanMX6 DUO1-9MYC::klTRP1 | this study |
| CJY380 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ092 trp1Δ63 his3Δ200 leu2Δ1 PDS1-9MYC:: klTRP1 Gal1-3HA-CDC20::KanMX6 | this study |
| CJY381 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ092 trp1Δ63 his3Δ200 Δspc34::HIS3MX4 leu2Δ1:: pCJ083 PDS1-9MYC:: klTRP1 Gal1-3HA-CDC20::KanMX6 | this study |
| CJY388 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 SPC34-GFP::KanMX6 MCM21-9MYC::klTRP1 | this study |
| CJY399 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ092 trp1Δ63 his3Δ200 leu2Δ1 SPC42-RFP:: KanMX6 | this study |
| CJY400 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ092 trp1Δ63 his3Δ200 Δspc34::HIS3MX4 leu2Δ1:: pCJ083 SPC42-RFP::KanMX6 | this study |
| CJY405 | MATa Δsst1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 pAFS125::URA3Δmad2::KanMX6 | this study |
| CJY406 | MATa Δsst1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 Δspc34::HIS3MX4 leu2Δ1::pCJ083 pAFS125:: URA3Δmad2::KanMX6 | this study |
| CJY415 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ097 trp1Δ63 his3Δ200 leu2Δ1 Gal1-3HA-CDC20:: KanMX6 | this study |
| CJY416 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ097 trp1::pCJ141Δ63 his3Δ200 Δspc34:: HIS3MX4 leu2Δ1::pCJ083 Gal1-3HA-CDC20::KanMX6 | this study |
| CJY417 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ097 trp1Δ63 his3Δ200 leu2Δ1 Gal1-3HA-SCC1:: KanMX6 | this study |
| CJY418 | MATa Δsst1 ura3-52::pXH136 lys2-801 ade2-101::pCJ097 trp1::pCJ141Δ63 his3Δ200 Δspc34:: HIS3MX4 leu2Δ1::pCJ083 Gal1-3HA-SCC1::KanMX6 | this study |
| CJY440 | MATa Δsst1 pT331 lys2-801 ade2-101::pCJ097 trp1 Δ63 his3Δ200 leu2Δ1 | this study |
| CJY441 | MATa Δsst1 pT331 lys2-801 ade2-101::pCJ097 trp1::pCJ141Δ63 his3Δ200 Δspc34:: HIS3MX4 leu2Δ1::pCJ083 | this study |
| CJY442 | MATa Δsst1 pT334 lys2-801 ade2-101::pCJ097 trp1Δ63 his3Δ200 leu2Δ1 | this study |
| CJY443 | MATa Δsst1 pT334 lys2-801 ade2-101::pCJ097 trp1::pCJ141 Δ63 his3Δ200 Δspc34:: HIS3MX4 leu2Δ1::pCJ083 | this study |
| CJY446 | MATa ura3-52::pAFS125 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 ndc80-1 | this study |
| CJY448 | MATa Δsst1 ndc80-1 ura3-52::pAFS125 lys2-801 ade2-101 trp1Δ63 his3Δ200 Δspc34:: HIS3MX4 leu2Δ1::pCJ083 | this study |
| CJY460 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 ndc80-1 DAD2-GFP::KanMX6 | this study |
| CJY461 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 ndc80-1 SPC34-GFP::KanMX6 | this study |
| CJY462 | MATa Δsst1 ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 Δspc34::HIS3MX4 leu2Δ1:: pCJ083 SPC34-GFP::KanMX6 | this study |
| CJY486 | MATa Δsst1 ura3-52::pAFS125 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 pWS103::TEM1/TRP1 | this study |
| CJY487 | MATa Δsst1 ura3-52::pAFS125 lys2-801 ade2-101 trp1Δ63 his3Δ200 Δspc34::HIS3MX4 leu2Δ1:: pCJ083 pWS103::TEM1/TRP1 | this study |
| ESM477 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 SPC19-3HA::KanMX6 | this study |
| ESM482 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 SPC34-3HA::KanMX6 | this study |
| YPH499 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 | Sikorski and Hieter (1989) |
| Plasmids | ||
| pAFS125 | GFP-TUB1 in a URA3-based integration vector | Straight et al. (1997) |
| pCJ045 | dad1-9 in pRS305 | this study |
| pCJ055 | dad2-9 in pRS305 | this study |
| pCJ074 | spc19-4 in pRS305 | this study |
| pCJ083 | spc34-3 in pRS305 | this study |
| pCJ092 | tetR-GFP in pRS402 | this study |
| pCJ097 | tetR-YFP in pRS402 | this study |
| pCJ141 | CFP-TUB1 in pRS304 | this study |
| pGP103 | TEM1 in pET28c | this study |
| pJO02 | CLB2 in pGEX-5X-1 | this study |
| pRS305 | LEU2-based integration vector | Sikorski and Hieter (1989) |
| pSM700 | SPC19 in pGEX-5X-1 | this study |
| pT331 | p13-112×tetO: a minichromosome harbouring a short ARS (not late anymore) from a late ARS (c4037) marked by 112×tetO | this study |
| pT334 | p13-112×tetO: a minichromosome harbouring a late ARS (c4036) marked by 112×tetO | this study |
| pWS103 | Gal1-lacI-TEM1 in TRP1-based integration plasmid: TEM1 fused to the UPL degron of the ubiquitin fusion degradation pathway | Shou et al. (1999) |
| pXH136 | 112×tetO in pRS306-based integration vector contaning recombination signal for chromosome V | He et al. (2000) |
Purification of Dad2p-ProA and analysis by MALDI
Dad2p–ProA was purified from yeast extracts with IgG–Sepharose beads as described (Janke et al., 2001). The purified proteins were resolved by 6–12% Nupage gradient gels (Invitrogen) and stained with colloidal Coomassie Blue (Roth). Protein bands were analysed by MALDI analysis (Shevchenko et al., 1996).
Analysis of conditional lethal dad1, dad2, spc19 and spc34 cells
Yeast cells were incubated at 23°C for 3 h with α-factor (1 µg/ml) to arrest cells in G1 phase of the cell cycle. α-Factor was removed by washing the cells twice with pre-warmed (37°C) YPD medium (t = 0). Cells were then incubated in YPD at 37°C. At the indicated times, cells were fixed with formaldehyde. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). The DNA content of cells was analysed by flow cytometry (fluorescence-activated cell sorting) (Hutter and Eipel, 1979).
Immunological techniques, microscopy, immunoelectron microscopy and ChIP
Anti-Clb2p, anti-Spc19p and anti-Tem1p antibodies were raised in rabbits against purified, recombinant proteins. The polyclonal rabbit anti-Spc24p, anti-Nuf2p (Janke et al., 2001), anti-Spc72p (Knop and Schiebel, 1998) and anti-Mcm21p (Ortíz et al., 1999) antibodies have been described. Monoclonal mouse or rat anti-HA (12CA5, 3F10), mouse anti-Myc (9E10) or rabbit anti-GFP antibodies were obtained from Boehringer Mannheim or Boehringer Ingelheim. Indirect immunofluorescence and immunoelectron microscopy were performed as published (Knop and Schiebel, 1998). ChIPs of yeast cells and immunoprecipitation of HA-tagged proteins were as described (Hecht and Grunstein, 1999).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Drs W.Fangman, R.Deshaies, S.Jensen, J.V.Kilmartin, K.Nasmyth and P.Sorger for plasmids and yeast strains. We acknowledge the helpful comments of V.Cleghon and J.Grindlay on the manuscript and the generous help of P.Andrews and J.Swedlow with video microscopy. The work of E.S. is supported by the Cancer Research Campaign and Human Frontiers Science Program Organization (RG0319/1999) and that of T.T. by The Wellcome Trust.
References
- Adams I.R. and Kilmartin,J.V. (1999) Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol., 145, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin,F.F., Bhalla,N., Sassoon,I., Hyman,A.A. and Murray,A.W. (1999) The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev., 13, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B. and Goetsch,L. (1975) Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol., 124, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Enquist-Newman,M., Müller-Reichert,T., Drubin,D.G. and Barnes,G. (2001) Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol., 152, 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., Zachariae,W., Michaelis,C., Shevchenko,A., Mann,M. and Nasmyth,K. (1998) An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell, 93, 1967–1076. [DOI] [PubMed] [Google Scholar]
- Enquist-Newman M., Cheeseman,I.M., Van Goor,D., Drubin,D.G., Meluh,P.B. and Barnes,G. (2001) Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell, 12, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke,L. and Carbon,J. (1982) Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell, 29, 235–244. [DOI] [PubMed] [Google Scholar]
- Friedmann K.L., Diller,J.D., Ferguson,B.M., Nyland,S.V., Brewer,B.J. and Fangman,W.L. (1996) Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev., 10, 1595–1607. [DOI] [PubMed] [Google Scholar]
- Goh P.Y. and Kilmartin,J.V. (1993) NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol., 121, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Asthana,S. and Sorger,P.K. (2000) Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell, 101, 763–775. [DOI] [PubMed] [Google Scholar]
- He X., Rines,D.R., Espelin,C.W. and Sorger,P.K. (2001) Molecular analysis of kinetochore–microtubule attachment in budding yeast. Cell, 106, 195–206. [DOI] [PubMed] [Google Scholar]
- Hecht A. and Grunstein,M. (1999) Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol., 304, 399–414. [DOI] [PubMed] [Google Scholar]
- Hofmann C., Cheeseman,I.M., Goode,B.L., McDonald,K.L., Barnes,G. and Drubin,D.G. (1998) Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol., 143, 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter K.J. and Eipel,H.E. (1979) Microbial determination by flow cytometry. J. Gen. Microbiol., 113, 369–375. [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau,L.F., Smith,D.L., Mistrot,C.A., Hardwick,K.G., Hwang,E.S., Amon,A. and Murray,A.W. (1998) Budding yeast Cdc20: a target of the spindle checkpoint. Science, 279, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Ito T., Tashiro,K., Muta,S., Ozawa,R., Chiba,T., Nishizawa,M., Yamamoto,K., Kuhara,S. and Sakaki,Y. (2000) Toward a protein–protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl Acad. Sci. USA, 97, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortíz,J., Lechner,J., Shevchenko,A., Shevchenko,A., Magiera,M.M., Schramm,C. and Schiebel,E. (2001) The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J., 20, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q.-W., Fuchs,J. and Loidl,J. (2000) Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci., 113, 1903–1912. [DOI] [PubMed] [Google Scholar]
- Jones M.H., Bachant,J.B., Castillo,A.R., Giddings,T.H. and Winey,M. (1999) Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol. Biol. Cell, 10, 2377–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.H., He,X., Giddings,T. and Winey,M. (2001) Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc. Natl Acad. Sci. USA, 98, 13675–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M. and Schiebel,E. (1998) Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J., 17, 3952–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Lechner J. and Carbon,J. (1991) A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell, 64, 717–725. [DOI] [PubMed] [Google Scholar]
- McCarroll R.M. and Fangmann,W.L. (1988) Time of replication of yeast centromeres and telomeres. Cell, 54, 505–513. [DOI] [PubMed] [Google Scholar]
- Ortíz J., Stemmann,O., Rank,S. and Lechner,J. (1999) A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev., 13, 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Tanaka,T.U., Nasmyth,K. and Schiebel,E. (2001) Modes of spindle pole body inheritance and segregation of the Bfa1p/Bub2p checkpoint protein complex. EMBO J., 20, 6359–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L. and Allshire,R.C. (2000) Centromeres: getting a grip of chromosomes. Curr. Opin. Cell Biol., 12, 308–319. [DOI] [PubMed] [Google Scholar]
- Schwartz K., Richards,K. and Botstein,D. (1997) BIM1 encodes a microtubule-binding protein in yeast. Mol. Biol. Cell, 8, 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Jensen,O.N., Podtelejnikov,A.V., Sagliocco,F., Wilm,M., Vorm,O., Mortensen,P., Boucherie,H. and Mann,M. (1996) Linking genome and proteome by mass spectrometry: large scale identification of yeast proteins from two dimensional gels. Proc. Natl Acad. Sci. USA, 93, 14440–14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W. et al. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell, 97, 233–244. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Marschall,W.F., Sedat,J.W. and Murray,A.W. (1997) Mitosis in living budding yeast: anaphase A but no metaphase plate. Science, 277, 574–578. [DOI] [PubMed] [Google Scholar]
- Sullivan M., Lehane,C. and Uhlmann,F. (2001) Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nature Cell Biol., 3, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Fuchs,J., Loidl,J. and Nasmyth,K. (2000) Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nature Cell Biol., 2, 492–499. [DOI] [PubMed] [Google Scholar]
- Uetz P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Wernic,D., Poupart,M.-A., Kooin,E.V. and Nasmyth,K. (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell, 103, 375–386. [DOI] [PubMed] [Google Scholar]
- Wigge P.A. and Kilmartin,J.V. (2001) The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol., 152, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Jensen,O.N., Holmes,S., Souès,S., Mann,M. and Kilmartin,J.V. (1998) Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol., 141, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M. and O’Toole,E.T. (2001) The spindle cycle in budding yeast. Nature Cell Biol., 3, E23–E27. [DOI] [PubMed] [Google Scholar]