Abstract
Prion protein (PrP) plays a crucial role in prion disease, but its physiological function remains unclear. Mice with gene deletions restricted to the coding region of PrP have only minor phenotypic deficits, but are resistant to prion disease. We generated double transgenic mice using the Cre–loxP system to examine the effects of PrP depletion on neuronal survival and function in adult brain. Cre-mediated ablation of PrP in neurons occurred after 9 weeks. We found that the mice remained healthy without evidence of neurodegeneration or other histopathological changes for up to 15 months post-knockout. However, on neurophysiological evaluation, they showed significant reduction of afterhyperpolarization potentials (AHPs) in hippocampal CA1 cells, suggesting a direct role for PrP in the modulation of neuronal excitability. These data provide new insights into PrP function. Furthermore, they show that acute depletion of PrP does not affect neuronal survival in this model, ruling out loss of PrP function as a pathogenic mechanism in prion disease and validating therapeutic approaches targeting PrP.
Keywords: afterhyperpolarization potential/Cre–loxP system/prion disease/prion protein
Introduction
The normal cellular form of the prion protein (PrPc) is a ubiquitously expressed glycosylphosphatidylinositol (GPI)-anchored glycoprotein that plays a central role in the pathogenesis of the transmissible spongiform encephalopathies or prion diseases, such as scrapie, bovine spongiform encephalopathy (BSE) and Creutzfeldt–Jakob disease (CJD). These are fatal neurodegenerative conditions in which there is accumulation of a protease-resistant isoform of PrPc, PrPSc, associated with infectivity but which may not be directly toxic to neurons (Brandner et al., 1996; Hill et al., 2000). Mice devoid of PrPc are resistant to scrapie and do not accumulate PrPSc, propagate infectivity or develop pathology (Bueler et al., 1993). However, while PrPc expression is essential for scrapie pathogenesis, the normal biological function of PrP remains enigmatic and the possibility that the loss of this function contributes to scrapie pathogenesis remains unresolved. This is of key significance to the development of potential therapeutic strategies in prion disease, where PrPc itself may be the major target.
PrP is encoded by the highly conserved single-copy gene Prnp (Oesch et al., 1985), which is expressed at high levels throughout adult life, predominantly in neurons (Kretzschmar et al., 1986; Manson et al., 1992). PrPc is mainly localized at synapses, in cholesterol-rich microdomains or caveolae (Vey et al., 1996; Naslavsky et al., 1997); yet two separate lines of PrP-null (Prnp0/0) mice in which the genetic ablation of Prnp is confined to its coding region reveal relatively little about PrP function. These mice appear developmentally and behaviourally normal (Bueler et al., 1992; Manson et al., 1994), although closer examination reveals defects in neurophysiological and biochemical function. These include defects in synaptic (Collinge et al., 1994; Manson et al., 1995) and hippocampal cell function (Colling et al., 1995) similar to those seen in mice and hamsters infected with scrapie (Jefferys et al., 1994; Johnston et al., 1998; Barrow et al., 1999) and reminiscent of the electroencephalographic abnormalities in human prion disease (Cathala and Baron, 1987). Changes in the membrane localization of neuronal nitric oxide synthase (nNOS) also occur in both Prnp0/0 and scrapie-infected animals (Ovadia et al., 1996; Keshet et al., 1999). PrP binds Cu2+ ions (Hornshaw et al., 1995; Brown et al., 1997a) with femtomolar affinity (Jackson et al., 2001), and reduced Cu2+/Zn2+-dependent SOD-1 activity has been found in Prnp0/0 mice (Brown et al., 1999) and Prnp0/0 neurons are more vulnerable to oxidative stress in culture than PrP-expressing wild-type neurons (Brown et al., 1997b). Increased neuronal excitability and aberrant nNOS function, as well as enhanced vulnerability to oxidative stress, are associated with neuronal damage and death. Therefore, the functional depletion of PrPc in prion disease may contribute to the process of neurodegeneration.
There are three further PrP-null lines of mice in which more extensive deletions of the Prnp gene result in ectopic expression in brain of the PrP-like protein Doppel (Dpl) due to up-regulation of the downstream gene Prnd (Moore et al., 1999; Li et al., 2000). These mice develop ataxia and cerebellar neurodegeneration (Moore et al., 1999; Li et al., 2000; Rossi et al., 2001), which is rescued by the co-expression of PrP, and transfection of cultured neuronal cells from one of these lines with a PrP-expressing plasmid rescues the cells from apoptosis (Kuwahara et al., 1999). Similarly, mice expressing the truncated PrP molecule Δ32–135 (but no wild-type PrP) also develop ataxia and neurodegeneration, but not if PrPc is co-expressed (Shmerling et al., 1998). Clearly, the neurotoxicity associated with Dpl or PrPΔ32–135 expression only occurs in the absence of PrPc.
The demonstration, or exclusion, of any neuroprotective role for PrPc is of central importance in understanding the pathological mechanisms involved in prion disease and in designing therapeutic strategies. In Prnp0/0 mice (without ectopic Doppel expression), loss of PrP function may be largely compensated for by adaptive mechanisms operative in the embryo masking such effects. Therefore, to address the key question of whether acute loss of PrP function is deleterious, we made transgenic mice using the Cre–loxP system (Sauer and Henderson, 1988) in which PrP knockout was targeted to neurons in the adult mouse, and the effects of acute PrP depletion on neuronal survival and function were analysed.
Results
Generation and characterization of MloxP transgenic mice expressing PrP
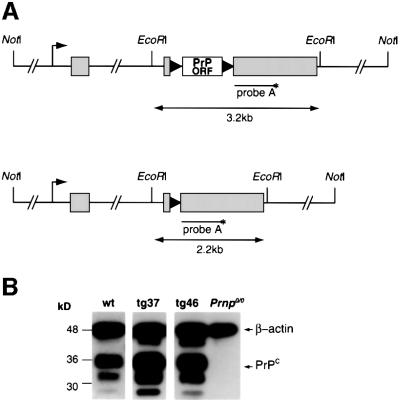
We microinjected FVB Prnp0/0 (Bueler et al., 1992) eggs with the MloxP transgene (Figure 1A) and produced five transmitting founders whose progeny expressed PrP. We performed all further phenotypic characterization in hemizygous mice, as this would reflect the zygosity of the MloxP transgene of double transgenic mice in the conditional knockout model. Hemizygous tg46 and tg37 mice derived from two founders expressed PrP in brain at or above wild-type levels, respectively, as determined by semi-quantitative western blotting (Figure 1B). tg46 mice also expressed wild-type levels of PrP in spinal cord, peripheral nerve and spleen (data not shown). We found the same regional variations in PrP immunostaining in brains of tg46 and tg37 mice as seen in wild-type mice (Salès et al., 1998; Figure 5A, a, f, k and p, wild-type mice; b, g, l and q, tg46 mice and tg37 mice, data not shown). tg46 mice succumbed to intracerebral inoculation of scrapie strain RML (Rocky Mountain Laboratories) with an incubation period of 122 ± 2 days, similar to that of wild-type FVB mice, and showed characteristic neuropathology and accumulation of PrPSc (data not shown). tg37 mice had a short incubation period of 77 ± 5 days, consistent with higher levels of PrP expression.
Fig. 1. The MloxP construct and PrP expression in tg37 and tg46 mice. (A) MloxP construct containing the floxed murine PrP coding region (PrP ORF, white box) within the CosSHaTet expression vector (exons represented by grey boxes) before (upper image) and after Cre-mediated recombination (lower image). Black triangles represent loxP sequences. The annealing site of DNA probe A used in Southern blotting is shown. (B) Western blotting showing PrP expression in wild-type, tg37 and tg46 mouse brains using antibody 6H4 (Prionics). β-actin was simultaneously detected to control for loading differences between the samples. After quantitation of relative signal intensities, expression levels of PrP by tg46 and wild-type mice are similar and ∼3-fold lower than in tg37 mice.
Fig. 5. Knockout of PrP expression in brains of tg46–Cre 22 mice. (A) Immunohistochemistry for PrPc in wild-type, tg46, tg46–Cre 22 and Prnp0/0 brains. Loss of PrP immunoreactivity to ICSM18 was seen in tg46–Cre 22 mouse brain sections, producing appearances equivalent to ICSM15-stained sections, which did not bind to mouse brain proteins and acted as a negative control. Serial frozen sections were stained for PrPc with ICSM18 and 6H4 and with anti-gangliocerebrosidase C (α-G) and ICSM15 antibodies. Counterstaining was with haematoxylin. (B) Macroscopic appearance of sections from tg46 and tg46–Cre 22 mouse brains immunostained as above. The staining patterns with α-G (positive control) and ICSM15 (negative control) were equivalent in the tg46 and conditional knockout mice, but the signal from PrP was undetectable with both 6H4 and ICSM18 in tg46–Cre 22 brain. (C) Section of tg46–Cre 22 mouse containing choroid plexus (CP) shows immunostaining of ependymal cells by 6H4 antibody, which does not occur in Prnp0/0 sections. Scale bar = 100 µm.
We also showed that expression of PrP from the MloxP transgene rescued a specific neurophysiological phenotype of Prnp0/0 mice: the abolition of the slow afterhyperpolarization (AHP) in CA1 pyramidal cells (Colling et al., 1996). tg46 and tg37 mice had slow AHP values in CA1 cells equivalent to those of wild-type FVB mice (Table I) and significantly different from values recorded in Prnp0/0 control mice (Barrow et al., 2000).
Table I. Characterization of MloxP transgenic lines.
| Mouse line | Prnp0/0 | tg37 | tg46 | FVB/wt |
|---|---|---|---|---|
| MloxP transgene copy number | n/a | 5–7 | 1 | n/a |
| PrP expression cf. wild type | 0 | ×3–4 | ×1 | 1 |
| Scrapie incubation period in days ± SD (RML, intracerebrally; n = number of mice) | n/a | 72 ± 5 (n = 7) | 122 ± 2 (n = 12) | 130 ± 1 (n = 6) |
| Slow AHP in CA1 cells (Barrow et al., 2000) | absent | rescued | rescued | present |
In summary, our results show that PrP expression in tg46 mice hemizygous for the MloxP transgene is quantitatively and qualitatively similar to that in wild-type mice (Table I).
Generation and characterization of NFH–Cre deleter lines
We produced NFH–Cre transgenic mice expressing the phage P1 enzyme Cre recombinase (Hamilton and Abremski, 1984) using the control elements of the murine neurofilament (NFH) gene (Julien et al., 1988; Figure 2A). We used Prnp0/0 FVB oocytes as before so that all PrP expression would be from the MloxP transgene in double transgenic NFH–Cre–MloxP animals.
Fig. 2. NFH–Cre transgene, temporal expression. (A) The NFH–Cre construct containing the Cre-coding region (grey box) inserted into exon 1 (E1) of the murine NFH gene upstream of the NFH start codon (ATG). Black box, SV40 polyadenylation signal; white boxes, NFH exons. NLS/ATG, nuclear localization signal and start codon of Cre gene. (B) Southern blotting of EcoRI-digested DNA from the brains of two tg46–Cre 22 (lanes 1 and 2), four tg46 (lanes 4–6 and 11) and two tg37–Cre 22 (lanes 8 and 9) mice. DNA from a Prnp0/0 mouse brain was loaded in lane 10. Hybridization of probe with unrecombined 3.2 kb and recombined 2.2 kb fragments is seen in double transgenic animals, the proportions of each fragment are shown beneath the image. There is no recombination in tg46 mice. (C) A histogram showing the time course of onset of Cre-mediated recombination of the MloxP locus. DNA from brains from three or more tg46–Cre 22 or tg37–Cre 22 mice or embryos were analysed for recombination by Southern blotting as above. E, embryonic day; P, post-natal day; w, weeks of age. Mean values for the number of mice indicated are depicted. For all time points up to 12 weeks, n = 3.
We detected Cre mRNA expression in brains of F1 progeny of four of five transmitting founders (Cre 7, 22, 27 and 29) by northern blotting (data not shown) and assessed functional expression of Cre by assessing the efficiency of in vivo Cre-mediated recombination of floxed transgenes. Using quantitative Southern blotting analysis on whole-brain DNA from double transgenic MloxP–Cre 22 mice, we found that the proportion of the MloxP transgene that had undergone Cre-mediated recombination was 29–38% (mean 33%; tg46–Cre 22 mice, n = 12; range 29–37%, Cre 22–tg37 mice, n = 5; range 32–38%; Figures 1A and 2B). This was consistent both within litters and also between unrelated double transgenic animals, and was independent of copy number of the transgene: single copy in tg46, five to seven copies in tg37. There were no double transgenic animals >12 weeks old in which recombination had not occurred or had occurred at a lower frequency. Neuron-derived DNA in whole-brain extracts is estimated at ∼20% (Ma et al., 1999), but may vary according to mouse strain, supporting the conclusion that Cre-mediated deletion of the MloxP transgene in Cre 22–MloxP double transgenic animals is occurring in the large majority of, if not all, neurons.
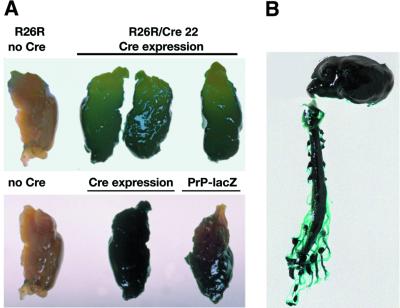
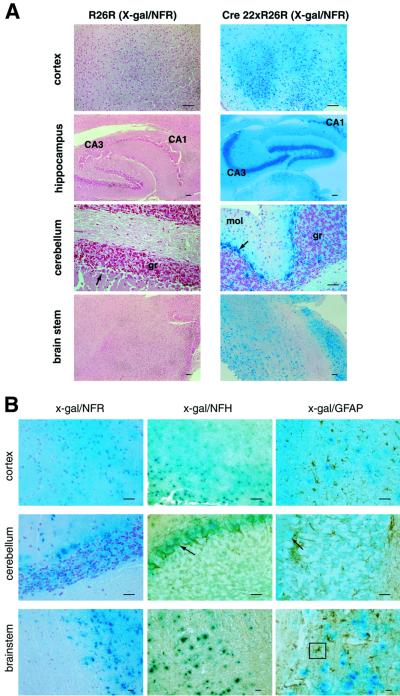
We confirmed this by demonstrating Cre-mediated activation of β-galactosidase (lacZ) transgene expression mice in neurons throughout the central and peripheral nervous systems using mice from the Cre reporter strain Rosa26R (R26R; Soriano, 1999) crossed with Cre 22 mice (Figures 3 and 4). Microscopically, lacZ expression was especially intense in neuron-rich regions such as the hippocampus (regions CA1–3 and dentate gyrus) and brain stem nuclei, as well as in cerebellar pyramidal cells (Figure 4A). In no sections did there appear to be subpopulations of cells that did not stain blue with 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (x-gal). In addition, we did not see leakiness of lacZ expression in any of the sections from control R26R animals. We confirmed neuronal specificity of Cre expression using counter-immunostaining techniques in x-gal-stained sections (Figure 4B). There is overlap of x-gal staining due to lacZ expression (blue) with neuron-specific NFH staining (brown) producing a greenish effect (Figure 4B, centre panel), but not with the astrocyte-specific marker glial fibrillary acidic protein (GFAP), which also stains brown (Figure 4B, right panel).
Fig. 3. Spatial features of NFH–Cre transgene expression in R26R–Cre 22 mice at the macroscopic level. (A) LacZ expression in R26R–Cre 22 mice and their R26R littermates (top panel) was determined by x-gal staining of brains. There is strong lacZ expression in whole brains of R26R–Cre 22 mice where Cre expression has allowed transcription of the lacZ gene. There is no lacZ expression in the absence of Cre, in the R26R brain. In the lower panel, a lacZ-expressing transgenic mouse, PrP-lacZ, which has high forebrain expression of lacZ (E.A.Asante, unpublished data), was used as a positive control; the R26R–Cre 22 mouse brain (centre) shows lacZ expression throughout fore and hind brain in comparison. (B) LacZ expression was seen throughout the central and peripheral nervous system in R26R–Cre 22 mice.
Fig. 4. Spatial features of NFH–Cre transgene expression in R26R–Cre 22 mice at the histological level. (A) The x-gal-stained brains of the R26R–Cre 22 and R26R mice above were sectioned and counterstained with Nuclear Fast Red (NFR). The strong expression of lacZ was seen throughout the brains of R26R–Cre 22 mice, particularly in neuron-rich regions such as the hippocampus. No lacZ expression was detected in the absence of Cre expression in R26R brains. (B) Sections from these mice were counter-immunostained using antibodies against NFH and GFAP to define cell types expressing lacZ. x-gal and NFH staining are superimposed (centre panel), whereas GFAP staining is distinct from the x-gal-stained cells and regions, confirming neuronal, but not astrocytic, distribution of Cre expression. Scale bar = 100 µm. CA1 and CA3 regions of hippocampus are indicated. An astrocyte is boxed. Arrows indicate pyramidal cells in cerebellum. mol, molecular layer; gr, granule cell layer of cerebellum.
We used data from Southern blotting analysis of recombination in brain DNA from MloxP–Cre 22 mice to detect the onset of activation of NFH–Cre expression and found that this occurred at ∼10 weeks postnatally. We analysed the brains of at least three embryos or mice daily from day E12 of embryonic development through to postnatal day 2, and then at intervals of 3 weeks up to 22 weeks of age. Quantitative DNA analysis on the Southern blotting did not reveal the presence of low amounts of the deleted fragment at earlier time points, consistent with genuine late activation of the NFH–Cre transgene in Cre 22 mice, rather than cumulative deletion of the MloxP locus over time.
In summary, these data show that in Cre 22 transgenic mice Cre recombinase is highly expressed from ∼10 weeks postnatally in post-mitotic neurons throughout the nervous system.
Adult-onset knockout of PrP in tg46–Cre 22 mice
We crossed Cre 22 mice with tg46 mice, in which PrP expression is both quantitatively and qualitatively very similar to wild-type mice, in order to knock out PrP in adult mice to study the uncompensated effects of acute PrP depletion.
Knockout of PrP expression in tg46–Cre 22 mice
We performed immunohistochemistry on frozen brain sections from three 12-week-old tg46–Cre 22 mice with confirmed deletion of the MloxP transgene by Southern blotting to examine the extent and distribution of PrP knockout over different regions and cell types. We found absence of detectable PrP expression using anti-PrP antibodies 6H4 and ICSM18 throughout all regions of the brain in these animals (Figure 5A, c, h, m and r, and B, lower panel), recapitulating the distribution of Cre expression inferred from x-gal staining of R26R–Cre 22 mice (Figure 4). Only ependymal cells of choroid plexus in tg46–Cre 22 mice, which are of glial origin, showed positive PrP immunostaining (Figure 5C). In contrast, tg46 littermates had a normal wild-type pattern of PrP expression (Figure 5A, a, f, k, p and b, g, l and q). Use of the avidin–biotin complex amplification system did not result in detection of PrP in double transgenic mice (data not shown). tg46–Cre 22 mice were equivalent to tg46 littermates with respect to α-galactocerebroside (α-G) antigen expression, used as a positive control of the immunohistochemical staining process (Figure 5B). On immunohistochemistry, tg46–Cre 22 mice pre-knockout (<10 weeks of age) had identical patterns of PrP expression to tg46 mice (data not shown).
Normal phenotype and brain morphology of tg46–Cre 22 mice
We screened tg46–Cre 22 mice and their non-double transgenic littermates for phenotypic abnormalities using the SHIRPA primary screening protocol (Rogers et al., 1997), using at least four males and four females of each genotype (tg46, Cre 22, Prnp0/0 and tg46–Cre 22) at two time points: aged 3–5 months, soon after loss of PrP expression; and later, 6–15 months post-knockout. PrP-deficient tg46–Cre 22 animals were indistinguishable from controls with respect to general morphology, development, gait, posture, motor control, co-ordination, autonomic function, general excitability and aggression at either time point post-knockout. All mice were equally healthy whether they were congenitally deficient in PrP (Prnp0/0 and Cre 22 mice), expressed physiological levels of PrP (tg46 mice) or for up to 15 months after the acute depletion of PrP expression (tg46–Cre 22 mice). We confirmed MloxP transgene recombination in two double transgenic mice from each group by Southern blotting (data not shown). We examined two 18-month-old double transgenic tg46–Cre 22 mice neuropathologically and found normal structure and appearance of all brain regions based on Nissl, NFH, synaptophysin, MAP2, calbindin and GFAP staining, indistinguishable from those of controls (data not shown). There was no evidence of neurodegeneration.
Conditional knockout models of PrP are resistant to scrapie
In order to confirm loss of PrP function with respect to scrapie susceptibility, five tg46–Cre 22 mice and four tg46 littermates were inoculated intracerebrally with RML prions at 13 weeks of age, after PrP knockout was estimated to have occurred. All tg46 mice developed scrapie symptoms in ∼122 days (range 120–125) and showed classical neuropathological features of scrapie (data not shown). None of the tg46–Cre 22 mice developed scrapie symptoms for up to 400 days post-inoculation, implying that susceptibility to scrapie was eliminated or the incubation period was significantly prolonged in these mice, at least up to this point. The brain of one animal culled due to intercurrent illness was examined neuropathologically at 370 days and no features of scrapie nor accumulation of PrPSc were seen on histopathological examination (data not shown).
Neurophysiological changes in hippocampal CA1 cells after conditional knockout of PrP
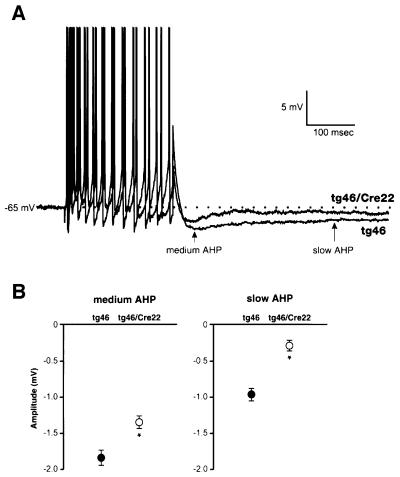
We looked at the AHP in our adult-onset PrP knockout model to see whether we could reproduce the AHP phenotype of Prnp0/0 mice (Colling et al., 1996) and whether this is directly due to the loss of PrP expression rather than reflecting complex secondary effects due to embryonic knockout. We found that both slow and medium AHPs resulting from trains of 1–15 action potentials in CA1 cells of tg46–Cre 22 mice were significantly reduced (slow AHP, –0.3 ± 0.05 mV; medium AHP, –1.35 ± 0.1 mV; n = 14 cells, six animals), in contrast to those of tg46 controls (slow AHP, –0.97 ± 0.13 mV; medium AHP, –1.8 ± 0.15 mV; n = 10 cells, five animals; P <0.001 for slow and medium AHP; Figure 6). Resting potentials, input resistances and action potential thresholds and amplitudes were not significantly different between the two groups (Table II).
Fig. 6. Abolition of AHP in hippocampal CA1 cells of conditional PrP-knockout mice. (A) Sample traces showing the AHPs following a burst of 12 action potentials in CA1 pyramidal cells (action potentials have been truncated). In tg46–Cre 22, amplitudes of both the medium and slow AHPs were significantly reduced compared with tg46. (B) Average medium and slow AHPs in the tg46 and tg46–Cre 22 groups. Results extracted from ANOVA test. Values were derived from all numbers of action potentials; SEMs represent the variance after that attributable to the different numbers of action potentials had been accounted for and removed (*two–way ANOVA, P <0.001 for both medium and slow AHPs; tg46, n = 10 cells; tg46–Cre 22, n = 14 cells).
Table II. Intrinsic properties of CA1 pyramidal cells in tg46 and tg46–Cre 22 mice.
| tg46 | tg46–Cre 22 | |
|---|---|---|
| Input resistance (MΩ) | 26.5 ± 2.8 (10) | 31.0 ± 3.5 (12) |
| Resting membrane potential (mV) | –61.5 ± 1.6 (10) | –62.3 ± 2.2 (12) |
| Action potential threshold (mV) | –52.2 ± 1.1 (10) | –53.4 ± 1.2 (14) |
| Action potential amplitude (mV) | 94.4 ± 1.6 (10) | 91.5 ± 2.2 (14) |
Numbers of cells are in parentheses.
In summary, we show the abolition of detectable neuronal PrP expression at ∼10 weeks in a transgenic mouse model in which its expression up to this point is qualitatively and quantitatively equivalent to that in wild-type mice. This knockout is without major detrimental effects for up to 15 months after its onset, but we have demonstrated neurophysiological abnormalities in hippocampal pyramidal cells that indicate a role for PrP in the proper activation of the AHP in these cells.
Discussion
The finding that the knockout of PrP in neurons of adult mice has no detrimental effect for up to 15 months post-knockout has significant implications. Until now, a major unresolved issue in prion biology has been whether the loss of PrP function contributes to the pathology of scrapie, which would also affect potential therapeutic strategies in prion disease.
Indeed, neurons from both Prnp0/0 mice and scrapie-infected animals show similar changes in neurophysiological function (Collinge et al., 1994; Jefferys et al., 1994; Manson et al., 1995; Colling et al., 1996; Johnston et al., 1997) and biochemical properties (Ovadia et al., 1996; Keshet et al., 1999). Altered neuronal excitability can predispose individuals to neuronal damage and death (Leist and Nicotera, 1998) and it is possible that loss of PrP function contributes to scrapie pathogenesis in this way. However, these abnormalities are non-pathogenic in Prnp0/0 mice, which are otherwise phenotypically normal (Bueler et al., 1992; Manson et al., 1994). It is possible that the viability of Prnp0/0 mice reflects the activation of mechanisms compensating for these deficits in the absence of PrP expression.
Our model affects PrP knockout in neurons of the fully developed mouse brain at ∼10 weeks of age, bypassing compensatory mechanisms during neurodevelopment and, for the first time, allows us to directly observe the effects of acute depletion of PrP after its normal expression throughout development. In a previous attempt at conditional knockout of PrP using the tet system (Kistner et al., 1996), expression of the PrP transgene prior to its induced repression was associated with embryonic and neonatal lethality in double transgenic mice. Thus, the authors had to repress PrP expression throughout gestation and weaning to obtain viable animals for interpretation of the effects of PrP knockout later in adult life (Tremblay et al., 1998). However, the fact that the acute knockout of PrP was found to be non-pathogenic when PrP was not physiologically expressed prior to knockout is uninterpretable. Indeed, significant levels of PrP persisted even after gene repression (Tremblay et al., 1998).
Critically, we established that PrP expression from the MloxP transgene in our model was quantitatively and qualitatively indistinguishable from that in wild-type mice up to the point of knockout, which is central to the interpretation of the effects of subsequent loss of expression. After PrP knockout in tg46–Cre 22 mice, we did not detect PrP expression in brain using immunohistochemical techniques, except in ependymal cells of choroid plexus. This must reflect loss of neuronal expression of PrP, recapitulating the pattern of Cre-mediated activation of lacZ expression in neurons seen in R26R–Cre 22 mice. Significantly, we were able to show functional consequences of PrP depletion, with reduction of medium and slow AHPs in hippocampal CA1 cells of tg46–Cre 22 mice.
Yet despite this demonstration of PrP knockout in adult mice at genomic, histological and neurophysiological levels after prior physiological expression, we found that these mice were morphologically and behaviourally normal and devoid of detectable neuropathological abnormalities for up to 15 months post-knockout. Therefore, our model confirms that the acute loss of PrP function in adult neurons is without detrimental sequelae.
PrP is also expressed at low levels in glia (Moser et al., 1995) and on the surface of blood cells (Cashman et al., 1990), and it has been noted that GPI-anchored proteins can transfer between cells in vivo by GPI painting (Kooyman et al., 1995). It is unlikely that such mechanisms are significant in our model, as our neurophysiological data strongly suggested that PrP knockout in neurons has not been functionally compensated.
The survival of conditional knockout mice inoculated with RML prions for up to 400 days without scrapie symptoms suggests that loss of neuronal PrPc has indeed significantly delayed or possibly even abrogated susceptibility to scrapie in this model. Interestingly, transgenic mice in which hamster PrP is only expressed in glia using the GFAP promoter are susceptible to the hamster scrapie strain 237K, although a few mice did not develop symptoms, but had PrPSc accumulation in brain (Raeber et al., 1999). It may be that scrapie resistance observed to date in our model reflects lower levels of PrP in glia, than when this is expressed using the GFAP promoter.
These data effectively eliminate any persisting doubt as to whether any crucial function of PrP is compensated for in previous Prnp0/0 models (Bueler et al., 1992; Manson et al., 1994). In fact, they confirm a specific phenotype (Colling et al., 1996) and the limited effects of loss of PrP function already reported in these models.
Significantly, our model provides new insights into the possible physiological function of PrP. The fact that the slow AHP in hippocampal CA1 cells was effectively abolished by the conditional knockout of PrP in tg46–Cre 22 mice as in Prnp0/0 mice (Colling et al., 1996), strongly suggests that this is specifically due to the absence of PrP, reflecting loss of a differentiated neuronal function rather than a developmental deficit. The slow AHP results from at least two Ca2+-dependent K+ currents and controls spike frequency adaptation, which modulates neuronal and synaptic activity in CA1 cells (Sah, 1996; Sah and Davies, 2000). Reduced Ca2+-gated K+ curents are also seen in cerebellar Purkinje cells of Prnp0/0 mice (Herms et al., 2001). Such channels are modulated by phosphorylation by non-receptor protein Src tyrosine kinases (PTKs), which, like PrP, are located in caveolae, as a means of modulating synaptic transmission. Recently, it was proposed that PrP is a neuronal signalling protein with the demonstration of caveolin-1-dependent activation of Fyn tyrosine kinase by cross-linking of PrPc on the surface of neuronal IC11 cells (Mouillet-Richard et al., 2000). Our data suggest that Ca2+-dependent K+ channels contributing to the AHP could be a potential target for modulation by Fyn-mediated PrP signalling.
In conclusion, it appears that the loss of the normal function of PrPc is unlikely in itself to be significant in the pathogenesis of prion disease. These findings are of critical importance in the context of potential therapeutic strategies in prion disease. It will be of great interest to observe the effects of Cre-mediated PrP depletion during the course of scrapie infection. The resolution of symptoms and pathology—a cure—in inoculated animals in which PrPc knockout occurs during scrapie infection would validate PrPc depletion as a target in the treatment of prion disease.
Materials and methods
Generation of transgenic mice
Primers S (5′-CCCGGGGTCGACATAACTTCGTATAGCATACATTATACGAAGTTATAGGAGAGCCAAGCAGACTAT-3′) and AS (5′-CGCGCGCTCGAGATAACTTCGTATAATGTATGCTATACGAAGTTATCTCATCCCACGATCAGGAAG-3′) were used to flank Prnp ORF [bases 100–862 of Prnp cDNA (Locht et al., 1986)] with loxP sites (bold). Thirty cycles (45 s at 94°C, 60 s at 57°C and 60 s at 72°C) were performed using Pfu DNA polymerase (Stratagene). SalI–XhoI-digested PCR product was ligated to CosSHaTet vector (Scott et al., 1992) and transgenic mice were generated by microinjection of a 43 kb NotI fragment (MloxP construct) into eggs from mice of mixed Sv129/C57Bl/FVB background obtained by crossing original Prnp0/0 mice (Bueler et al., 1992) with FVB mice.
The NFH–Cre construct was generated by cloning a 1.6 kb XbaI fragment of plasmid pNLSCre into the NotI site of the murine NFH gene. Transgenic mice were generated by microinjection of 14 kb NFH–Cre construct into Prnp0/0 eggs as above.
Southern blotting
Founder mice for both constructs were identified by PCR and Southern blotting using the capillary transfer method (Sambrook et al., 1989), Hybond N+ nylon membrane (Amersham) and alkaline blotting conditions. Transgene sequences were detected using DNA probes: a 949 bp SalI–HindIII fragment of the Syrian hamster PrP gene 3′UTR for MloxP sequences (probe A) and a 1.2 kb BamHI fragment of the Cre recombinase gene (probe B) for detection of NFH–Cre sequence. Probes were labelled with [α-32P]dCTP (Amersham) with the Prime-it II Random Primer Labelling Kit (Stratagene) and used at a concentration of 1 × 106 c.p.m./ml in ULTRAhyb hybridization solution (Ambion) at 42°C. Membranes were exposed to X-ray film or scanned with a STORM 840 phosphoimager (Amersham Pharmacia). Image analysis and signal quantitation were performed using ImageQuant and Fragment Analysis software programmes (Amersham Pharmacia). For transgene copy number estimation, membranes were stripped by boiling in 0.5% SDS, 0.1× SSC then re-probed with a 1 kb EcoRI–BamHI fragment of the neomycin resistance gene, neo (Bueler et al., 1992), present in two copies in all cells and the relative signals quantitated.
Northern blotting
Poly(A)+ mRNA [3 µg; obtained from total brain RNA using the Oligotex mRNA mini kit (Qiagen)] was electrophoresed on a 1% denaturing agarose midi-gel (1 M boric acid, 0.5 µM EDTA, 16.7% v/v formaldehyde). Capillary transfer and hybridization were performed as for Southern blotting, as were washing and image development. Filters were stripped by boiling in 0.5% SDS and checked as above, then probed with an 800 bp NotI–NruI fragment of the NFH gene and a 1.2 kb cDNA of the GAPDH gene for detection of NFH and GAPDH mRNAs, respectively.
Western blotting
Aliquots (20 µl) of 10% (w/v) tissue homogenates of tissue in 2× SDS–PAGE sample buffer (125 mM Tris–HCl pH 6.8, 4% SDS, 20% glycerol, 0.05% bromophenol blue) were electrophoresed through a 16% polyacrylamide 1 mm gel (NOVEX). Proteins were blotted onto Immobilon-P membrane (Millipore) by wet blotting. PrP was detected using primary monoclonal antibody 6H4 (Prionics AG) at 1:5000 dilution and secondary antibody goat anti-mouse IgG–alkaline phosphatase conjugate (Sigma) at 1:10 000 dilution. The signal was developed using enhanced chemifluorescence reagents (Pierce) and quantitated using software programs as above. Membranes were simultaneously probed with anti-β actin antibody (Sigma) at 1:25 000 dilution for quantitation of PrP expression. Homogenates were digested with 50 µg/ml proteinase K at 37°C prior to electrophoresis for detection of PrPSc.
Immunohistochemistry in frozen sections
Animals were killed by cranio-cervical dislocation. The brain was rapidly removed, immersed in Tissue-Tek OCT embedding medium (Bright Instruments Ltd) and placed on dry ice. Sections (8 µm) were cut from frozen tissue using a cryostat (Leica) and electrostatically attached to Super-frost glass slides (BDH), then left to dry overnight at room temperature. Sections were fixed in acetone for 10 min and dried. Endogenous peroxidase activity was blocked by incubation in 0.3% H2O2 in 70% methanol in phosphate-buffered saline (PBS) for 30 min at room temperature and a blocking step was performed by incubation in 5% goat serum in PBS for 30 min. Primary antibodies 6H4 (Prionics) or ICSM18 (Hill et al., 2000), which detect similar PrP epitopes (A.Khalili-Shirazi, unpublished data), were both used at 1:100 dilution added for 1 h. The slides were washed three times in PBS for 5 min each, followed by 45 min incubation in horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Sigma; 1:200 dilution), followed by washing as above. They were then incubated in 3,3′-diaminobenzidine tetrahydrochloride (DAB) solution (Sigma; 50% w/v DAB, 0.03% v/v H2O2 in PBS) for 10 min. After rinsing, counterstaining was with haematoxylin (Harris) for 5 min. Excess stain was removed by rinsing in ddH2O, followed by differentiation in acid alcohol (1% HCl in absolute alcohol). Stained sections were cleared in xylene and mounted. ICSM 15 was used as a negative isotype control at 1:100 dilution and anti-gangliocerebrosidase C (Sigma; 1:100 dilution) was used as a positive control for myelin detection.
Immunohistochemistry in fixed tissue
Tissues were fixed in 10% buffered formal saline overnight, processed and paraffin embedded. Sections (4 µm) were stuck to Super-frost slides and allowed to dry overnight at room temperature and then for 2 h at 60°C. Slides were de-waxed in increasing concentrations of alcohol. Endogenous peroxidase activity was blocked in 2.5% H2O2 in methanol for 30 min at room temperature, followed by two 5 min washes in distilled water, after which the sections were placed in 10 mM sodium citrate pH 6 and heated at 900 W in a microwave oven for 5 min. The sections were washed in running tap water for 5 min and placed in Tris-buffered saline pH 7.6 (TBS; 10 mM Tris, 140 mM NaCl). Non-specific binding was blocked with 10% swine serum (or rabbit serum in TBS depending on origin of secondary antibody). After primary antibody binding, sections were incubated in biotinylated porcine secondary antibody (Dako) for 45 min, washed twice in TBS for 2 min and then incubated with the ABC complex (Dako) for 30 min. Signal development was with DAB as described above.
The following antibodies and dilutions were used: anti-GFAP polyclonal antiserum (Dako; 1:500 dilution), anti-NFH 200 (1:200 dilution), anti-MAP2 clone HM-2 (1:500 dilution), anti-calbindin clone D28K (1:200 dilution) and anti-synaptophysin clone SVP-38 (1:200 dilution; Sigma).
β-galacotosidase staining
Mice were terminally anaesthetized and transcardially perfused with 2% paraformaldehyde, 0.2% gluteraldehyde, 2 mM MgCl2 and 5 mM EGTA in PBS pH 7.3. Brains and spinal cords were removed and fixed in the same solution for 30 min at room temperature with gentle agitation, then washed three times in rinse solution (2 mM MgCl2, 0.1% sodium deoxycholate, 0.1% NP-40 in PBS). Staining was performed overnight at 37°C in the rinse solution containing x-gal (Sigma) at 1 mg/ml, 4 mM potassium ferricyanide and 4 mM potassium ferrocyanide. Samples were washed three times in 1× PBS, then dehydrated in ethanol and glycerol 2:1 v/v for 15 min, followed by clearing in a 1:1 and then a 1:9 mix of 1× PBS and glycerol. Samples were paraffin embedded, cut into 4 µm sections and counterstained in 1% Nuclear Fast Red solution.
Scrapie transmissions to MloxP and tg46–Cre 22 mice
Mice were inoculated intracerebrally with 30 µl of 1% brain homogenate of scrapie strain PDG 586 (Rocky Mountain Laboratories). Detection of PrPSc after denaturation of tissue in formic acid was performed as described previously (Hill et al., 1997).
Phenotypic screening using SHIRPA primary screen
The SHIRPA primary screen (Rogers et al., 1997) was performed on mice. All assessments were undertaken by the same observer and performed blind to genotype.
Electrophysiology
Recordings from CA1 pyramidal cells of hippocampal slices from adult male mice, 12–18 weeks old, of tg46 or tg46–Cre 22 genotype were performed as described previously (Colling et al., 1996). Experiments and analysis were carried out in a double-blinded manner with regard to the identity of the mice, which were randomized using a Latin square design. Results were analysed statistically using ANOVAs and unpaired t-tests on a SPSS Systat software package. All results are expressed as mean ± SEM unless stated otherwise.
Acknowledgments
Acknowledgements
We thank M.Burley, A.Dickinson and S.Whitehead for technical assistance. We are also grateful to K.Hawkins, D.Moore and R.Bond for care of mice and to Ray Young for help with graphics. We are grateful to Professor C.Weissmann for the original Prnp0/0 mice, Dr P.Soriano and Dr R.Lovell Badge for R26R mice, and Dr V.Episkopou for helpful discussion. We thank Dr M.Scott for the CosSHaTet expression vector and Dr J.P.Julien for the NFH genomic clone. This work was funded by the Wellcome Trust, the Medical Research Council and the European Commission (Biomed 2 programme).
References
- Barrow P.A., Holmgren,C.D., Tapper,A.J. and Jeffreys,J.G.R. (1999) Intrinsic physiological and morphological properties of principal cells of the hippocampus and neocortex in hamsters infected with scrapie. Neurobiol. Dis., 6, 406–423. [DOI] [PubMed] [Google Scholar]
- Barrow P.A., Mallucci,G.R., Collinge,J. and Jefferys,J.G.R. (2000) Rescue of a physiological phenotype in mice with ablated PrP gene but expressing a transgene cassette for PrP. J. Physiol., 523P, 203P (Abstract). [Google Scholar]
- Brandner S., Isenmann,S., Raeber,A., Fischer,M., Sailer,A., Kobayashi,Y., Marino,S., Weissmann,C. and Aguzzi,A. (1996) Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature, 379, 339–343. [DOI] [PubMed] [Google Scholar]
- Brown D.R. et al. (1997a) The cellular prion protein binds copper in vivo. Nature, 390, 684–687. [DOI] [PubMed] [Google Scholar]
- Brown D.R., Schulz-Schaeffer,W.J., Schmidt,B. and Kretzschmar,H.A. (1997b) Prion protein-deficient cells show altered response to oxidative stress due to decreased SOD-1 activity. Exp. Neurol., 146, 104–112. [DOI] [PubMed] [Google Scholar]
- Brown D.R., Wong,B.S., Hafiz,F., Clive,C., Haswell,S.J. and Jones,I.M. (1999) Normal prion protein has an activity like that of superoxide dismutase. Biochem. J., 344, 1–5. [PMC free article] [PubMed] [Google Scholar]
- Bueler H., Fischer,M., Lang,Y., Bluethmann,H., Lipp,H.-P., DeArmond,S.J., Prusiner,S.B., Aguet,M. and Weissmann,C. (1992) Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature, 356, 577–582. [DOI] [PubMed] [Google Scholar]
- Bueler H., Aguzzi,A., Sailer,A., Greiner,R.A., Autenried,P., Aguet,M. and Weissmann,C. (1993) Mice devoid of PrP are resistant to scrapie. Cell, 73, 1339–1347. [DOI] [PubMed] [Google Scholar]
- Cashman N.R., Loertscher,R., Nalbantoglu,J., Shaw,I., Kascsak,R.J., Bolton,D.C. and Bendheim,P.E. (1990) Cellular isoform of the scrapie agent protein participates in lymphocyte activation. Cell, 61, 185–192. [DOI] [PubMed] [Google Scholar]
- Cathala F. and Baron,H. (1987) Clinical aspects of Creutzfeldt–Jakob Disease. In Prusiner,S.B. and McKinley,M.P. (eds), Prions: Novel Infectious Pathogens Causing Scrapie and Creutzfeldt–Jakob Disease. Academic Press, San Diego, CA, pp. 467–509.
- Colling S.B., King,T.M., Collinge,J. and Jefferys,J.G.R. (1995) Prion protein null mice: abnormal intrinsic properties of hippocampal CA1 pyramidal cells. Brain Res. Assoc. Abstr., 12 (Abstract). [Google Scholar]
- Colling S.B., Collinge,J. and Jefferys,J.G.R. (1996) Hippocampal slices from prion protein null mice: disrupted CA2+-activated K+ currents. Neurosci. Lett., 209, 49–52. [DOI] [PubMed] [Google Scholar]
- Collinge J., Whittington,M.A., Sidle,K.C.L., Smith,C.J., Palmer,M.S., Clarke,A.R. and Jefferys,J.G.R. (1994) Prion protein is necessary for normal synaptic function. Nature, 370, 295–297. [DOI] [PubMed] [Google Scholar]
- Hamilton D.L. and Abremski,K. (1984) Site-specific recombination by the bacteriophage P1 lox–Cre system. Cre-mediated synapsis of two lox sites. J. Mol. Biol., 178, 481–486. [DOI] [PubMed] [Google Scholar]
- Herms J.W., Tings,T., Dunker,S. and Kretzschmar,H.A. (2001) Prion protein affects Ca2+-activated K+ currents in cerebellar Purkinje cells. Neurobiol. Dis., 8, 324–330. [DOI] [PubMed] [Google Scholar]
- Hill A.F., Desbruslais,M., Joiner,S., Sidle,K.C.L., Gowland,I. and Collinge,J. (1997) The same prion strain causes vCJD and BSE. Nature, 389, 448–450. [DOI] [PubMed] [Google Scholar]
- Hill A.F., Joiner,S., Linehan,J., Desbruslais,M., Lantos,P.L. and Collinge,J. (2000) Species barrier independent prion replication in apparently resistant species. Proc. Natl Acad. Sci. USA, 97, 10248–10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornshaw M.P., McDermott,J.R., Candy,J.M. and Lakey,J.H. (1995) Copper binding to the N-terminal tandem repeat region of mammalian and avian prion protein: structural studies using synthetic peptides. Biochem. Biophys. Res. Commun., 214, 993–999. [DOI] [PubMed] [Google Scholar]
- Jackson G.S., Murray,I., Hosszu,L.L.P., Gibbs,N., Waltho,J.P., Clarke,A.R. and Collinge,J. (2001) Location and properties of metal-binding sites on the human prion protein. Proc. Natl Acad. Sci. USA, 98, 8531–8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferys J.G.R., Empson,R.M., Whittington,M.A. and Prusiner,S.B. (1994) Scrapie infection of transgenic mice leads to network and intrinsic dysfunction of cortical and hippocampal neurons. Neurobiol. Dis., 1, 3–15. [DOI] [PubMed] [Google Scholar]
- Johnston A.R., Black,C., Fraser,J. and MacLeod,N. (1997) Scrapie infection alters the membrane and synaptic properties of mouse hippocampal CA1 pyramidal neurones. J. Physiol., 500, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston A.R., Fraser,J.R., Jeffrey,M. and MacLeod,N. (1998) Alterations in potassium currents may trigger neurodegeneration in murine scrapie. Exp. Neurol., 151, 326–333. [DOI] [PubMed] [Google Scholar]
- Julien J.P., Cote,F., Beaudet,L., Sidky,M., Flavell,D., Grosveld,F. and Mushynski,W. (1988) Sequence and structure of the mouse gene coding for the largest neurofilament subunit. Gene, 68, 307–314. [DOI] [PubMed] [Google Scholar]
- Keshet G.I., Ovadia,H., Taraboulos,A. and Gabizon,R. (1999) Scrapie-infected mice and PrP knockout mice share abnormal localization and activity of neuronal nitric oxide synthase. J. Neurochem., 72, 1224–1231. [DOI] [PubMed] [Google Scholar]
- Kistner A., Gossen,M., Zimmermann,F., Jerecic,J., Ullmer,C., Lubbert,H. and Bujard,H. (1996) Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc. Natl Acad. Sci. USA, 93, 10933–10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooyman D.L. et al. (1995) In vivo transfer of GPI-linked complement restriction factors from erythrocytes to the endothelium. Science, 269, 89–92. [DOI] [PubMed] [Google Scholar]
- Kretzschmar H.A., Stowring,L.E., Westaway,D., Stubblebine,W.H., Prusiner,S.B. and DeArmond,S.J. (1986) Molecular cloning of a human prion protein cDNA. DNA, 5, 315–324. [DOI] [PubMed] [Google Scholar]
- Kuwahara C. et al. (1999) Prions prevent neuronal cell-line death. Nature, 400, 225–226. [DOI] [PubMed] [Google Scholar]
- Leist M. and Nicotera,P. (1998) Apoptosis versus necrosis: the shape of neuronal cell death. Results Probl. Cell Differ., 24, 105–135. [DOI] [PubMed] [Google Scholar]
- Li A. et al. (2000) Physiological expression of the gene for PrP-like protein, PrPLP/Dpl, by brain endothelial cells and its ectopic expression in neurons of PrP-deficient mice ataxic due to Purkinje cell degeneration. Am. J. Pathol., 157, 1447–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locht C., Chesebro,B., Race,R. and Keith,J.M. (1986) Molecular cloning and complete sequence of prion protein cDNA from mouse brain infected with the scrapie agent. Proc. Natl Acad. Sci. USA, 83, 6372–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Reis,G., Parada,L.F. and Schuman,E.M. (1999) Neuronal NT-3 is not required for synaptic transmission or long-term potentiation in area CA1 of the adult rat hippocampus. Learn. Mem., 6, 267–275. [PMC free article] [PubMed] [Google Scholar]
- Manson J.C., West,J.D., Thomson,V., McBride,P., Kaufman,M.H. and Hope,J. (1992) The prion protein gene: a role in mouse embryogenesis? Development, 115, 117–122. [DOI] [PubMed] [Google Scholar]
- Manson J.C., Clarke,A.R., Hooper,M.L., Aitchison,L., McConnell,I. and Hope,J. (1994) 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol. Neurobiol., 8, 121–127. [DOI] [PubMed] [Google Scholar]
- Manson J.C., Hope,J., Clarke,A.R., Johnston,A., Black,C. and MacLeod,N. (1995) PrP gene dosage and long term potentiation. Neurodegeneration, 4, 113–114. [DOI] [PubMed] [Google Scholar]
- Moore R.C. et al. (1999) Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein Doppel. J. Mol. Biol., 292, 797–817. [DOI] [PubMed] [Google Scholar]
- Moser M., Colello,R.J., Pott,U. and Oesch,B. (1995) Developmental expression of the prion protein gene in glial cells. Neuron, 14, 509–517. [DOI] [PubMed] [Google Scholar]
- Mouillet-Richard S., Ermonval,M., Chebassier,C., Laplanche,J.L., Lehmann,S., Launay,J.M. and Kellermann,O. (2000) Signal transduction through prion protein. Science, 289, 1925–1928. [DOI] [PubMed] [Google Scholar]
- Naslavsky N., Stein,R., Yanai,A., Friedlander,G. and Taraboulos,A. (1997) Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J. Biol. Chem., 272, 6324–6331. [DOI] [PubMed] [Google Scholar]
- Oesch B. et al. (1985) A cellular gene encodes scrapie PrP 27–30 protein. Cell, 40, 735–746. [DOI] [PubMed] [Google Scholar]
- Ovadia H., Rosenmann,H., Shezen,E., Halimi,M., Ofran,I. and Gabizon,R. (1996) Effect of scrapie infection on the activity of neuronal nitric-oxide synthase in brain and neuroblastoma cells. J. Biol. Chem., 271, 16856–16861. [DOI] [PubMed] [Google Scholar]
- Raeber A.J., Klein,M.A., Frigg,R., Flechsig,E., Aguzzi,A. and Weissmann,C. (1999) PrP-dependent association of prions with splenic but not circulating lymphocytes of scrapie-infected mice. EMBO J., 18, 2702–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D.C., Fisher,E.M., Brown,S.D., Peters,J., Hunter,A.J. and Martin,J.E. (1997) Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm. Genome, 8, 711–713. [DOI] [PubMed] [Google Scholar]
- Rossi D., Cozzio,A., Flechsig,E., Klein,M.A., Rülicke,T., Aguzzi,A. and Weissmann,C. (2001) Onset of ataxia and Purkinje cell loss in PrP null mice inversely correlated with Dpl level in brain. EMBO J., 20, 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah P. (1996) Ca2+-activated K+ currents in neurones: types, physiological roles and modulation. Trends Neurosci., 19, 150–154. [DOI] [PubMed] [Google Scholar]
- Sah P. and Davies,P. (2000) Calcium-activated potassium currents in mammalian neurons. Clin. Exp. Pharmacol. Physiol., 27, 657–663. [DOI] [PubMed] [Google Scholar]
- Salès N., Rodolfo,K., Hässig,R., Faucheux,B., Di Giamberardino,L. and Moya,K.L. (1998) Cellular prion protein localization in rodent and primate brain. Eur. J. Neurosci., 10, 2464–2471. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fisher,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sauer B. and Henderson,N. (1988) Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl Acad. Sci. USA, 85, 5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M.R., Kohler,R., Foster,D. and Prusiner,S.B. (1992) Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci., 1, 986–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmerling D. et al. (1998) Expression of amino-terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell, 93, 203–214. [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet., 21, 70–71. [DOI] [PubMed] [Google Scholar]
- Tremblay P. et al. (1998) Doxycycline control of prion protein transgene expression modulates prion disease in mice. Proc. Natl Acad. Sci. USA, 95, 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vey M., Pilkuhn,S., Wille,H., Nixon,R., DeArmond,S.J., Smart,E.J., Anderson,R.G.W., Taraboulos,A. and Prusiner,S.B. (1996) Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc. Natl Acad. Sci. USA, 93, 14945–14949. [DOI] [PMC free article] [PubMed] [Google Scholar]