Abstract
Post-transcriptional gene silencing (PTGS) provides protection against viruses in plants by homology-dependent RNA degradation. PTGS initiated locally produces a mobile signal that instructs specific RNA degradation at a distance. Here we show that this signal-mediated intercellular spread of PTGS does not occur after PTGS initiation in cells expressing cucumber mosaic virus 2b protein (Cmv2b), a nucleus-localized plant viral PTGS suppressor. Silencing spread via the signal was also effectively blocked in independent assays by expressing Cmv2b only in tissues through which the signal must travel to induce PTGS in the target cells. Furthermore, the signal imported externally into the Cmv2b-expressing cells was not active in triggering degradation of the target RNA and loss of signal activity in these cells was associated with a significantly reduced transgene DNA methylation. These findings indicate that Cmv2b inhibits the activity of the mobile signal and interferes with DNA methylation in the nucleus. Signal inactivation provides a mechanistic basis for the known role of Cmv2b in facilitating virus spread to tissues outside of the primarily infected sites.
Keywords: cucumber mosaic virus/DNA methylation/post-transcriptional gene silencing/silencing signal/viral silencing suppressor
Introduction
Post-transcriptional gene silencing (PTGS) destroys single-stranded RNAs in a homology-dependent manner and is conserved in a diverse group of eukaryotic organisms (Matzke et al., 2001; Waterhouse et al., 2001). In higher plants, PTGS functions as a natural anti-viral defense because plant viruses are both initiators and targets of PTGS, and encode proteins that are PTGS suppressors (Dougherty and Parks, 1995; Baulcombe, 1999a; Ding, 2000; Marathe et al., 2000; Carrington et al., 2001; Voinnet, 2001). In addition, a role for PTGS in development (Anandalakshmi et al., 2000; Fagard et al., 2000; Smardon et al., 2000; Grishok et al., 2001) and transposon restriction (Ketting et al., 1999; Wu-Scharf et al., 2000) has also been documented.
Three main approaches have been applied to the mechanistic studies of PTGS. Genetic screens have led to the identification of nearly a dozen genes that are required for silencing in Neurospora, Arabidopsis, Caenorhabditis elegans and Chlamydomonas (Vance and Vaucheret, 2001). The small RNAs (sRNAs) ∼21–25 nucleotides (nt) in length with sequence homology to the target RNAs represent the first component of the PTGS pathway, identified through the use of the biochemical approach (Hamilton and Baulcombe, 1999). These sRNAs accumulate in plants in which either a transgene or virus RNA is targeted by PTGS (Hamilton and Baulcombe, 1999; Llave et al., 2000; Serio et al., 2001; Sijen et al., 2001) and also in those silenced at the transcriptional level due to production of double-stranded (ds) RNA homologous to the promoter (Mette et al., 2000). Recent findings have confirmed an early prediction (Hamilton and Baulcombe, 1999) that sRNAs function as guide RNAs to prime specific degradation of homologous RNA targets (Hammond et al., 2000; Zamore et al., 2000; Elbashir et al., 2001a,b; Lipardi et al., 2001).
The third approach is to probe the PTGS pathway with suppressors encoded by plant viruses (Carrington et al., 2001; Li and Ding, 2001; Vance and Vaucheret, 2001). Suppression of RNA silencing by the potyviral helper component proteinase (HC-Pro) results in undetectable accumulation of sRNAs, but does not interfere with either the production/intercellular spread of the mobile silencing signal or DNA methylation associated with transgene RNA silencing (Llave et al., 2000; Mallory et al., 2001). The 25K protein (p25) of potato virus X (PVX) prevents the intercellular spread of PTGS, possibly by targeting either the production or transport of the silencing signal (Voinnet et al., 2000). However, when introduced after systemic silencing of a reporter transgene is established, p25 displays no detectable activity, unlike HC-Pro, which seems to reverse silencing wherever it is expressed (Brigneti et al., 1998). The 2b protein of cucumber mosaic virus (CMV) produces a unique pattern of silencing reversal in the same assay; while established silencing is not affected in older tissues, the signal-mediated spread of PTGS into the newly emerging tissues is suppressed (Brigneti et al., 1998). Thus, the CMV 2b protein (Cmv2b), which carries a functional nuclear localization signal (Lucy et al., 2000), may target a signaling step of PTGS that is distinct from p25.
Here we characterize further the suppressor activity of Cmv2b in two well characterized PTGS systems. We found that Cmv2b did not prevent PTGS initiation when co-expressed with either PTGS-inducing transgene. However, Cmv2b was able to block the silencing signal from leaving the site of production, passing through tissues expressing Cmv2b or activating RNA degradation once it arrived at target tissues also expressing Cmv2b. We conclude that Cmv2b suppresses PTGS by directly interfering with the activity of the mobile silencing signal. Furthermore, loss of the silencing signal activity was found to correlate with a significantly reduced DNA methylation of the target transgene in the Cmv2b-expressing tissues, suggesting that the silencing signal and the putative cytoplasmic signal that guides DNA methylation in the nucleus may share a key component that is the target of Cmv2b.
Results
Two PTGS systems were used for analyzing the mechanism of Cmv2b suppression of PTGS. In the first system, silencing of a green fluorescent protein (GFP) transgene in the Nicotiana benthamiana line 16c was induced de novo by a local Agrobacterium infiltration that initially directed a high level of transient GFP expression (Voinnet and Baulcombe, 1997). The second system is based on the autonomous PTGS induction of a β-glucuronidase (GUS) transgene in the Nicotiana tabacum line 6b5 (Elmayan and Vaucheret, 1996). PTGS of the GUS transgene in 6b5 plants, which occurs whether the transgene is in the homozygous or hemizygous state (Elmayan and Vaucheret, 1996; Palauqui et al., 1997; Mallory et al., 2001), shares many features with that of the GFP transgene in 16c plants (Voinnet et al., 1998; Jones et al., 1999). Silencing of both the GFP and GUS transgenes is associated with a specific degradation of the target mRNAs, accumulation of the target-specific sRNAs, cytosine methylation of the target transgene DNA in the transcribed region and production of the graft-transmissible silencing signal. However, the GUS transgene silencing and methylation in 6b5 plants were induced autonomously in each generation, whereas an artificial triggering step was required for the GFP transgene silencing and methylation in 16c plants.
Incomplete suppression of intracellular silencing by Cmv2b
We first examined the activity of Cmv2b in the Agrobacterium co-infiltration assay where PTGS initiation of the GFP transgene occurs in the presence of a silencing suppressor (Voinnet et al., 2000). In this assay, two A.tumefaciens strains, 35S-GFP (Brigneti et al., 1998) and 35S-Cmv2b, which encoded, respectively, a 35S promoter-controlled GFP transgene expression cassette and a similar cassette for Cmv2b in the transfer DNA of a binary plasmid, were co-infiltrated into the leaves of the 16c GFP plants, leading to a transient expression of both genes in the infiltrated regions.
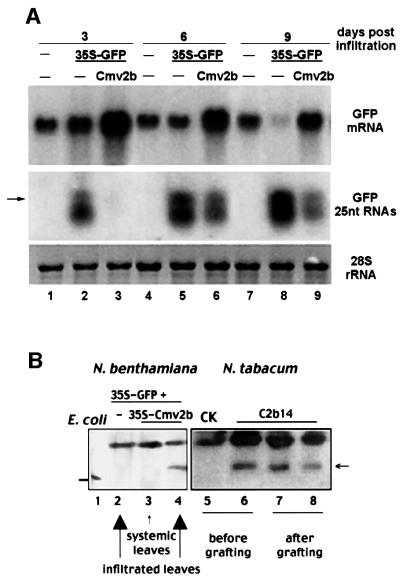
We found that co-infiltration with 35S-Cmv2b resulted in a significant increase in the GFP mRNA levels in the infiltrated leaves (Figure 1A, top, lanes 3, 6 and 9), as compared with infiltration with 35S-GFP alone (lanes 2, 5 and 8) or together with the untranslatable mutant 35S-CmvΔ2b (data not shown). Even at 9 days post-infiltration (p.i.), leaves co-infiltrated with 35S-Cmv2b maintained a higher level of GFP mRNA than that in the uninfiltrated 16c leaves (Figure 1A, compare lanes 7 and 9). At this point, the GFP mRNA in the leaves infiltrated with 35S-GFP alone was almost completely degraded (lane 8). Expression of Cmv2b was readily detectable by western blot analysis (Figure 1B) in the co-infiltrated leaves (lane 4) but not in the upper non-infiltrated leaves (lane 3) or in leaves infiltrated with 35S-GFP alone (lane 2). Thus, Cmv2b functions as a suppressor of PTGS in this co-infiltration assay, although this experiment alone does not exclude other mechanisms independent of silencing suppression, such as a Cmv2b-mediated enhanced T-DNA integration and/or transient expression. Furthermore, co-infiltration of 35S-GFP–35S-Cmv2b also resulted in significantly higher accumulation levels of GFP mRNA in wild-type (Wt) non-transgenic N.benthamiana plants than infiltration either with 35S-GFP alone or together with 35S-CmvΔ2b (data not shown). Thus, transient GFP expression by Agrobacterium infiltration induced GFP silencing in the absence of a stably integrated GFP transgene (Johansen and Carrington, 2001). This type of GFP silencing was also sensitive to Cmv2b suppression. This explains why higher levels of GFP mRNA were observed in leaves infiltrated with 35S-GFP/35S-Cmv2b (Figure 1A, lanes 3, 6 and 9) than in the uninfiltrated leaves of 16c plants (lanes 1, 4 and 7). In contrast, infiltrating 35S-Cmv2b into leaves of silenced 16c plants did not activate GFP expression in these leaves (data not shown), consistent with the early conclusion that Cmv2b does not reverse established PTGS (Brigneti et al., 1998).
Fig. 1. Incomplete suppression of GFP transgene silencing in the infiltrated leaves of the N.benthamiana plants. (A) Time course analyses of RNA accumulation in the infiltrated leaves. Total high (top, mRNA) and low (middle, 25 nt RNAs) molecular weight RNAs were extracted from the leaves 3, 6 and 9 days after infiltration with either 35S-GFP (lanes 2, 5 and 8) or 35S-GFP/35S-Cmv2b (lanes 3, 6 and 9). RNA samples were also obtained from non-infiltrated leaves of similar developmental stages and used as controls to show the levels of GFP mRNA from the stably integrated copy of the GFP transgene. The amount of total RNAs loaded was visualized by methylene blue staining of the 28S rRNA (bottom). A 32P-labeled DNA probe corresponding to the full-length GFP coding sequence was used for mRNA analysis (top), whereas 32P-labeled RNA transcripts corresponding to the minus-strand GFP mRNA were used for the detection of sRNAs (labeled as GFP 25 nt RNAs). The arrow indicates the position of a DNA oligo 25 bases long. A T7 RNA transcript 22 nt long was run immediately below the lower band of the GFP sRNAs (H.W.Li and S.W.Ding, unpublished data). (B) Western blotting detection of Cmv2b. Protein extracts were prepared from the 16c leaves infiltrated with 35S-GFP (lane 2), 35S-GFP+35S-Cmv2b (lane 4) and from upper non-infiltrated leaves (lane 3) of the plants infiltrated with 35S-GFP+35S-Cmv2b. Protein samples were also extracted from 8-week-old N.tabacum seedlings either non-transgenic (lane 5, CK) or transgenic for Cmv2b (line C2b14, lane 6) and from two middle C2b14 segments 2 weeks after the double-grafted tobacco plants were assembled (lanes 7 and 8, Figure 4E). Total plant proteins from 50 mg of leaf tissue were loaded in each lane. Lane 1 contained the purified Cmv2b expressed in Escherichia coli, which migrated faster than Cmv2b produced in plants (horizontal arrow; Ding et al., 1994).
Both fluorescence and RNA analyses indicate that there was PTGS initiation in the 16c leaves co-infiltrated with 35S-GFP–35S-Cmv2b. A red fluorescence zone surrounding the areas infiltrated with either 35S-GFP alone or together with 35S-CmvΔ2b were visible at 3 days p.i., due to the autofluorescence of chlorophyll in the absence of GFP accumulation. A similar red fluorescence zone of reduced intensity was also developed in the leaves co-infiltrated with 35S-Cmv2b, but was not visible until 5–6 days p.i. (Figure 2, left). An incomplete suppression of GFP silencing in these leaves was supported by the observed gradual decline in the GFP mRNA levels from 3 to 9 days p.i. (Figure 1A, top) and by the detection of the GFP sRNAs (Figure 1A, middle). While undetectable in the non-infiltrated 16c plants (Figure 1A, lanes 1, 4 and 7), the GFP-specific sRNAs, which appeared as two discrete bands ∼21–25 nt long (Voinnet et al., 2000), accumulated to high levels in the 35S-GFP infiltrated leaves at 3 days p.i. (Figure 1A, lane 2) and continued to increase for up to 9 days p.i. (Figure 1A, lanes 5 and 8). The sRNAs were not readily detectable in the 35S-Cmv2b/35S-GFP infiltrated leaves at 3 days p.i. (Figure 1A, lane 6), but accumulated to high levels at 6 and 9 days p.i., though lower than the levels detected in the 35S-GFP infiltrated leaves (Figure 1A, compare lanes 5 and 6 or 8 and 9).
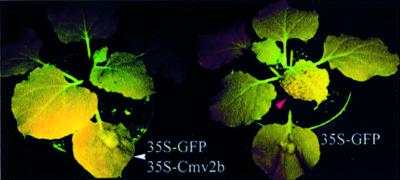
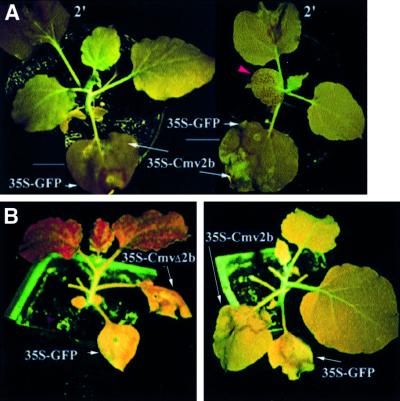
Fig. 2. Differential suppression of local and systemic silencing of the GFP transgene in the N.benthamiana GFP plants with or without Cmv2b expression. Six days after three leaves of each plant was infiltrated with 35S-GFP alone (right) or together with 35S-Cmv2b (left), the plants were photographed under UV light to show the local silencing as a red fluorescent zone surrounding the infiltrated area and emerging systemic silencing as red dots (red arrowhead) only in the 35S-GFP-infiltrated plant. Out of the 56 plants, 54 co-infiltrated with 35S-GFP and 35S-Cmv2b remained green fluorescent at least 2 months p.i. Note that only one infiltrated leaf from each plant is visible in this image and the fact that several leaves or leaf areas appear uniformly dark red is due to lack of exposure to UV light during photographing.
Cmv2b does not prevent silencing initiation of a GUS transgene
The observed incomplete suppression of GFP silencing in the co-infiltrated leaves may be a result of the transient expression of Cmv2b mediated by Agrobacterium infiltration. To address this concern, we made use of a transgenic tobacco (N.tabacum) line, C2b14, which carried a stably integrated 35S-Cmv2b transgene (Ji and Ding, 2001). Thus, in the progeny of a genetic cross between tobacco lines C2b14 and 6b5, referred to as C2bx6b5 plants, expression of Cmv2b and the PTGS-targeted GUS transgene were both from stably integrated transgenes and controlled under the same 35S promoter. The F1 progeny (Wtx6b5) of a cross between Wt tobacco and line 6b5 served as a control and was hemizygous at the GUS locus, as in C2bx6b5. As in the co-infiltrated 16c leaves, Cmv2b expression should occur during the autonomous initiation of GUS transgene silencing in the C2bx6b5 plants.
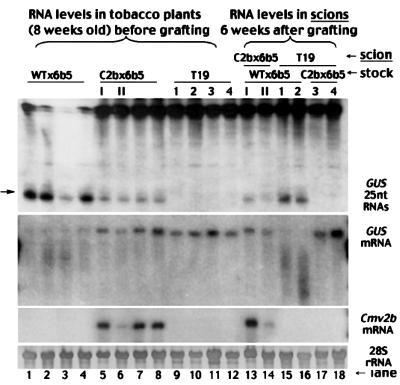
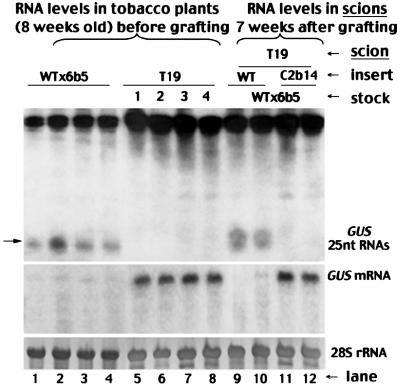
The GUS transgene was silenced as expected in Wtx6b5 plants (Figure 3, lanes 1–4) since there was degradation of the GUS mRNA (second panel) and accumulation of the GUS-specific sRNAs (top). In contrast, the full-length GUS mRNA (second panel) accumulated to high levels in individual C2bx6b5 plants (lanes 5–8) in which there was expression of the Cmv2b transgene (third panel). Similar levels of the full-length GUS mRNA (second panel) were found in tobacco line T19 (lanes 9–12), which carries a non-silenced GUS transgene (English et al., 1996). Histochemical staining further revealed significantly enhanced GUS enzymatic activity in the leaves of C2bx6b5 plants over the background level in WTx6b5 leaves (data not shown). However, there was also accumulation of the GUS-specific sRNAs in these C2bx6b5 plants (Figure 3, first panel, lanes 5–8), though at levels considerably lower than in Wtx6b5 plants (lanes 1–4). This is in contrast to the non-silenced T19 plants, in which these sRNAs were undetectable (lanes 9–12). Thus, there was suppression of the PTGS targeted against the GUS transgene by Cmv2b, but there were still lower levels of GUS transgene silencing in C2bx6b5 plants, as was also found in the co-infiltrated GFP-expressing 16c leaves. These results demonstrate that Cmv2b suppression of PTGS does not prevent PTGS initiation in two distinct PTGS systems, suggesting that Cmv2b targets a downstream step in the PTGS pathway.
Fig. 3. Suppression of the intercellular spread of GUS transgene silencing in N.tabacum plants. Total low (top) and high (middle) molecular weight RNAs were extracted either from individual 8-week-old tobacco plants of the genotypes WTx6b5 (lanes 1–4), C2bx6b5 (lanes 5–8) and T19 (lanes 9–12) or from scions of the genotypes T19 (lanes 15–18) and C2bx6b5 (lanes 13 and 14) 6 weeks after they were grafted (Figure 4A–C) on rootstocks of Wtx6b5 (lanes 13–16) or C2bx6b5 (lanes 17 and 18). Equal RNA loading was monitored by methylene blue staining of the 28S RNA and the blot was probed sequentially for the mRNA of GUS (second panel) and Cmv2b (third panel). The blot of sRNAs was hybridized with a 32P-labeled riboprobe corresponding to the minus strand of the full-length GUS mRNA. The position of a 25 nt oligonucleotide is indicated by an arrow. Note that RNA samples were prepared from the individually labeled T19 (1–4) and C2bx6b5 (I and II) plants before they were used as scions on Wtx6b5 or C2bx6b5 rootstocks, as indicated at the top of each lane. Hence, the accumulation level of Cmv2b mRNA was consistently lower in plant II of C2bx6b5 (lanes 6 and 14) than in plant I of C2bx6b5 (lanes 5 and 13) before and after grafting.
Cmv2b blocks the intercellular spread of PTGS
Despite the initiation of GFP silencing in all leaves infiltrated with 35S-GFP/35S-Cmv2b, 54 of the 56 co-infiltrated 16c plants did not become silenced for the GFP transgene outside of the infiltrated leaves (Figure 2, left) for as long as the observations were made (2 months p.i.). This is in contrast to the induction of systemic silencing in all of the 110 plants infiltrated with 35S-GFP alone (Figure 2, right) and in all of the 28 plants co-infiltrated with 35S-GFP and 35S-CmvΔ2b (data not shown). Thus, the signal-mediated intercellular spread of PTGS did not occur in 96% of the 16c plants co-infiltrated with 35S-GFP–35S-Cmv2b following GFP silencing initiation in the infiltrated leaves. The upper non-silenced tissues from these plants were fully responsive to silencing induction either by 35S-GFP infiltration or by grafting as scions onto silenced rootstocks (data not shown), suggesting that the restricted transient expression of Cmv2b in the infiltrated leaves had no effect on PTGS initiation in non-infiltrated tissues.
We further investigated whether there was intercellular spread of PTGS after PTGS initiation in the C2bx6b5 tobacco (Figure 3, lanes 5–8). We examined this possibility by grafting the non-silenced GUS-expressing T19 plants as the reporter scions onto the C2bx6b5 rootstocks as illustrated in Figure 4B. We also created grafted tobacco plants consisting of T19 scions and the GUS-silencing Wtx6b5 rootstocks (Figure 4A) as a positive control (Palauqui et al., 1997; Mallory et al., 2001). The T19 scions (Figure 3, lanes 9–12) contained high levels of GUS mRNA (second panel) but no GUS-specific sRNAs (first panel) at the time when the plants were grafted. Six weeks after grafting, the GUS transgene became silenced in the T19 scions grafted on the Wtx6b5 rootstocks (Figure 3, lanes 15 and 16), demonstrated by the abundant accumulation of the GUS sRNAs (first panel) and by the absence of the full-length GUS mRNA (second panel). This shows that the GUS transgene in the T19 scions was targeted for silencing by the signal exported from the Wtx6b5 rootstock, as expected (Palauqui et al., 1997; Mallory et al., 2001). However, silencing occurred in none of the individual T19 scions grafted on the C2bx6b5 rootstocks (Figure 3, lanes 17 and 18) in eight independent grafts because accumulation of the GUS mRNA remained high (second panel) and no GUS sRNAs were detected in these scions (first panel). Thus, there was no output of a functional GUS silencing signal from the C2bx6b5 rootstock, indicating the absence of signal-mediated intercellular PTGS spread in the C2bx6b5 tobacco after PTGS initiation as found in the 16c N.benthamiana plants infiltrated with 35S-GFP/35S-Cmv2b. However, it is not clear at this point whether Cmv2b inhibition of the intercellular PTGS spread occurred before or after the production of the silencing signal because in these experiments Cmv2b was co-expressed with the PTGS-inducing transgene in the same cells.
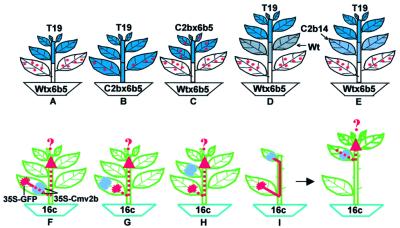
Fig. 4. Diagrams illustrating the grafting and infiltration experiments used to analyze the role of Cmv2b in blocking the signal-mediated spread of PTGS. Small red dots denote the GUS sRNAs, the red area indicates induction of GFP silencing in regions infiltrated with 35S-GFP, the red arrow indicates spread of GFP silencing in the 16c GFP plants, the green area represents GFP-expressing tissues, the blue area represents GUS-expressing tissues and the purple area is for Cmv2b-expressing tissues from the 35S-Cmv2b either stably integrated (E) or transiently delivered by Agrobacterium infiltration (F–I). Six or seven weeks after grafting, the GUS transgene in T19 scions became silenced in grafts (A) and (D), but not in grafts (B) and (E). The accumulation levels of both GUS mRNA and sRNAs in C2bx6b5 were unaltered after grafting on the Wtx6b5 rootstocks (C). There was normal induction of GFP transgene silencing in the 35S-GFP-infiltrated leaves of the 16c GFP plants (F–H), but systemic silencing outside of the infiltrated leaves was established in (G) and (H) but not in (F). GFP silencing was induced in the 35S-GFP-infiltrated leaf and spread to the remaining upper leaf outside of the 35S-Cmv2b-infiltrated area, but silencing did not spread into the new tissues that emerged from these 16c GFP plants.
Cmv2b prevents the signal-mediated induction of PTGS
There were similar levels of GUS mRNA (Figure 3) in the C2bx6b5 (lanes 5–8) and T19 (lanes 9–12) tobacco plants before they were grafted as scions on the GUS-silencing Wtx6b5 rootstocks (Figure 4A and C). As shown above, GUS mRNA in the T19 scions was degraded efficiently 6 weeks after grafting (Figure 3, second panel, lanes 15 and 16). However, the accumulation of GUS mRNA in the C2bx6b5 scions (lanes 13 and 14) was not affected, despite the predicted import of the silencing signal from the rootstock. In addition, there was no increase in accumulation of the GUS sRNAs in the C2bx6b5 scions 6 weeks after grafting (Figure 3, lanes 13 and 14 of the first panel) as compared with the levels before grafting (lanes 5 and 6). Thus, the silencing signal imported externally failed to trigger silencing of the target GUS mRNA in the C2bx6b5 scions, unlike in the T19 scions. This finding indicates that Cmv2b prevented the de novo induction of PTGS by the imported signal and the lack of intercellular PTGS spread in Cmv2b-expressing tissues (Figures 1, lanes 3, 6 and 9, and 3, lanes 5–8) is unlikely to be due to inhibition of the production of the silencing signal.
Cmv2b inhibits the activity of the silencing signal
It is possible that Cmv2b prevented the signal-mediated induction of PTGS by either a direct inhibition of the activity of the signal or a blockage in the signal transduction pathway. To resolve these alternatives, we performed three new experiments in which Cmv2b expression was targeted in tissues between the source and recipient cells of the silencing signal. Thus, they were different from the above experiments, where Cmv2b was co-expressed with either a PTGS-inducing transgene in the source cells (Figures 2, left, and 4B) or a PTGS target transgene in the recipient cells (Figure 4C). The first experimental design followed the three-way grafting strategy previously developed to demonstrate the long range transport of the silencing signal (Palauqui et al., 1997; Voinnet et al., 1998). We generated tobacco plants composed of three parts of distinct genotypes (Figure 4D and E). A segment from either Wt tobacco (Figure 4D) or C2b14 (Figure 4E) was used as the middle insert between a GUS-silencing Wtx6b5 rootstock and a GUS-expressing T19 reporter scion. As expected, the systemic silencing signal generated in the Wtx6b5 rootstocks was able to pass though the Wt intergraft to cause the degradation of GUS mRNA (Figure 5, second panel) and accumulation of the GUS sRNAs (first panel) in the T19 scions (lanes 9 and 10). However, silencing did not occur in the T19 scions from three double-grafted tobacco plants with C2b14 as the middle insert (two shown in lanes 11 and 12 of Figure 5), randomly selected from eight such independent grafts that survived. Similar levels of the GUS mRNA (second panel) were detected in the T19 scions both before grafting (lanes 7 and 8) and 7 weeks afterwards (lanes 11 and 12). Furthermore, the GUS-specific sRNAs (first panel) remained undetectable in these T19 scions (lanes 11 and 12) as in T19 plants before grafting (lanes 7 and 8). Cmv2b was only expressed in the C2b14 intergraft (Figure 1B, lanes 6–8) through which the silencing signal had to travel from the rootstock to induce silencing in the scion. Thus, the silencing signal became non-functional after spreading through the Cmv2b-expressing intergraft in these double-graft plants, suggesting that Cmv2b inhibits the activity of the silencing signal.
Fig. 5. Inhibition of the signal-mediated spread of PTGS in N.tabacum plants. The double-grafted plants were assembled as illustrated in Figure 4D and E, and the RNA analyses were carried out as in Figure 3. RNA samples were prepared from the 8-week-old individually labeled tobacco seedlings of genotype Wtx6b5 (lanes 1–4) and T19 (lanes 5–8) before they were assembled as rootstocks and scions, respectively, in the double-grafted plants with either line C2b14 (lanes 11 and 12) or Wt (lanes 9 and 10) tobacco plants as the intergrafts. RNA samples in lanes 9–12 were extracted from the T19 scions of these double-grafted plants 7 weeks after grafting.
Inhibition of the silencing signal activity in the presence of signal amplification
The C2b14 segment of the double-grafted tobacco plants does not carry the target GUS transgene so that signal spread through this intergraft occurred in the absence of signal amplification. Two experiments were thus conducted in the 16c GFP plants, as illustrated in Figure 4F–I, to determine whether Cmv2b was able to inhibit the activity of the silencing signal when signal spread was accompanied by signal amplification (Voinnet et al., 1998). In the first assay, 35S-Cmv2b was infiltrated to the basal region of a leaf while 35S-GFP was simultaneously infiltrated to the tip end of the same leaf (Figure 4F); two lower fully expanded leaves of each 16c plant were treated in this manner. There was efficient induction of GFP silencing in the tip end, visible under UV illumination as a broad red zone surrounding the infiltrated site (Figure 6A, left). However, GFP silencing did not occur outside of the infiltrated leaves in 26 of the 30 plants treated in this manner for at least 2 months after infiltration (Figure 6A, left). In contrast, when 35S-CmvΔ2b, instead of 35S-Cmv2b, was infiltrated to the leaf base, all of the 14 plants became silenced systemically (data not shown). Systemic silencing also occurred, as early as 6 days p.i., in all of the 14 plants (Figure 6A, right) in which 35S-Cmv2b was infiltrated in the leaf tip and 35S-GFP in the leaf base (Figure 4G). Thus, the GFP-specific silencing signal was also inactivated while spreading through the Cmv2b-expressing tissue of the 16c plants, which accumulates the GFP mRNA and thus has the potential to support signal amplification.
Fig. 6. Inhibition of the signal-mediated spread of PTGS in N.benthamiana plants. (A) The effect of Cmv2b expression on the spread of PTGS induced in the same leaf. Two leaves of each 16c plant were infiltrated with 35-GFP and 35S-Cmv2b according to the schemes illustrated in Figure 4F (left plant) and G (right plant), and photographed under UV light 6 days p.i. The second leaf (marked 2′) was similarly infiltrated. Note that red fluorescent dots were visible in an upper leaf (indicated by a red arrowhead) of the right plant but not in any non-infiltrated leaves of the left plant for as long as the observations were made (2 months). (B) Effect of Cmv2b expression on the spread of PTGS induced in a different leaf. As illustrated in Figure 4I, all leaves including the apical leaflets except two were removed from the 16c plants before 35S-GFP was infiltrated to the lower leaf and 35S-Cmv2b (right plant) or 35S-CmvΔ2b (left plant) was infiltrated to the upper leaf. The infiltrated plants were photographed 16 days p.i.
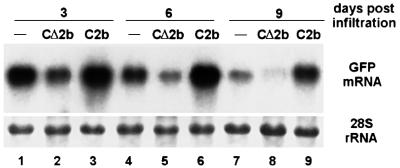
The second infiltration scheme was designed according to the finding that spread of the silencing signal follows the pathway of photoassimilate translocation from source (developed leaves) to sink (immature leaves) in the 16c GFP plants (Voinnet et al., 1998). In this assay, all leaves between an upper sink leaf and a lower source leaf, as well as the apical young leaflets, were removed before infiltrating 35S-GFP to the lower leaf and 35S-Cmv2b or 35S-CmvΔ2b to the upper leaf (Figure 4I). Signal translocation from the lower source leaf to the upper sink may occur within 2–3 days p.i. (Voinnet et al., 1998) and initiation of GFP silencing in this leaf and subsequently emerged tissues is dependent on the translocated signal (Figure 4I).
GFP silencing, as triggered by 35S-GFP infiltration to the lower leaf, became clearly visible in the upper leaf of plants non-infiltrated (44 plants, data not shown) or infiltrated with 35S-CmvΔ2b (30 plants) at 6 days p.i. and then spread to the new leaves that emerged subsequently in all of the plants treated (Figure 6B, left). Accordingly, northern blot hybridizations (Figure 7) detected a decline in the accumulation of the GFP mRNA from 3 to 9 days p.i. in these upper leaves, although it is not clear why this decline was more rapid in the leaves infiltrated with 35S-CmvΔ2b (lanes 2, 5 and 8) than in the uninfiltrated leaves (lanes 1, 4 and 7). Thus, there was induction of GFP RNA degradation by the silencing signal translocated from the lower silencing source leaf. Weak silencing was visible in the upper leaf infiltrated with 35S-Cmv2b, but occurred predominantly outside of the infiltrated area (Figure 6B, right). However, compared with leaves either non-infiltrated or infiltrated with 35S-CmvΔ2b, a much higher level of GFP mRNA was maintained in leaves at 3 and 6 days p.i. (Figure 7, lanes 3 and 6) and a decreased accumulation was only obvious at 9 days p.i. (lane 9). Significantly, silencing did not spread to any of the new leaves that emerged subsequently in 58 of the 66 plants infiltrated (Figure 6B, right). Although there were low levels of signal unloading and GFP silencing in the upper leaf expressing Cmv2b, further spread of PTGS out of this leaf to the newly emerging leaves was inhibited. This finding suggests that silencing induction in the emerging leaves is dependent on the signal translocated from the upper leaf rather than from the lower leaf, after the intervening leaves and the visible apical leaflets were removed. This is probably because the lower leaf was too old to be an active exporting source leaf by the time new leaves started to emerge. Notably, systemic silencing was established in all 16c plants either when 35S-GFP and 35S-Cmv2b were infiltrated simultaneously into a lower leaf and an upper leaf, respectively (Figure 4H) (14 plants), or 35S-Cmv2b was infiltrated into an upper leaf 3 days after the lower leaf was infiltrated with 35S-GFP (16 plants).
Fig. 7. Induction of GFP transgene silencing by the translocated silencing signal. As illustrated in Figure 4I, all leaves, including the apical leaflets except two were removed from the 16c plants before 35S-GFP was infiltrated to the lower leaf. The upper leaf was either non-infiltrated (lanes 1, 4 and 7) or infiltrated with 35S-CmvΔ2b (lanes 2, 5 and 8) or 35S-Cmv2b (lanes 3, 6 and 9). Total RNAs were isolated from this upper leaf 3, 6 and 9 days p.i., and analyzed for the accumulation of the GFP mRNA. Total RNAs loaded in each lane (4 µg) were visualized by methylene blue staining.
Cmv2b interferes with the restoration of transgene DNA methylation
The data presented above show that the silencing signal was unable to activate transgene RNA silencing in Cmv2b-expressing tissues. Thus, we next examined the methylation status of the GUS transgene DNA in C2bx6b5 plants, since a recent study suggested a correlation between production/export of the GUS silencing signal and DNA methylation at the two adjacent MluI sites towards the 3′ region of the GUS coding sequence at the same 6b5 locus in tobacco, which is restored in each generation in the first few days after germination (Mallory et al., 2001). The genomic DNA extracted from Wtx6b5, C2bx6b5 and T19 plants was digested with EcoRI and the methylation-sensitive restriction enzyme MluI, followed by DNA gel blotting analysis as described (English et al., 1996; Mallory et al., 2001). The MluI sites in the GUS coding sequence were not methylated in the GUS-expressing line T19 and were completely digested to give the two expected bands of 0.7 and 0.85 kb (Figure 8, lane 5; English et al., 1996; Mallory et al., 2001). Also as expected, these two bands were not detected in the gel blotting analysis of the genomic DNA from the silenced Wtx6b5 plants due to the methylation of these MluI sites; a new 1.5 kb band, as well as two larger bands, which resulted from cleavage upstream of the GUS coding region, were detected instead (Figure 8, lanes 1 and 2; Mallory et al., 2001). However, a large amount of the genomic DNA extracted from the C2bx6b5 plants was digested into the diagnostic 0.7 and 0.85 kb species (lanes 3 and 4). This result indicates that cytosine methylation at the MluI sites of a significant amount of the GUS transgene DNA was not restored in cells expressing Cmv2b. Thus, Cmv2b also interferes with the restoration of transgene DNA methylation in the nucleus.
Fig. 8. Cmv2b expression partially prevented restoration of transgene methylation in N.tabacum plants. Total genomic DNA was isolated from individual 8-week-old tobacco plants of the genotypes WTx6b5, C2bx6b5 and T19, digested with EcoRI and MluI, and DNA blotting was analyzed with a 32P-labeled DNA probe corresponding to the full-length GUS sequence (A). The positions of the 0.85 and 0.7 kb DNA fragments resulting from complete digestion at the MluI sites are indicated to the right. The ∼1.5 kb fragment is predicted if the MluI sites are protected by methylation. The blot was stripped and re-hybridized with a 32P-labeled probe specific for the 18S DNA sequence (B), and shows the amount of DNA loaded and the relative level of digestion in each lane.
Discussion
In this work, we demonstrate a novel activity of a viral silencing suppressor in targeting the mobile silencing signal for inactivation. We propose that Cmv2b interacts directly with the signal or a component of the signal complex so that the signal or signal complex becomes destabilized or inactivated. There is evidence for the long range transport of the silencing signal in plants through the phloem (Voinnet et al., 1998), which is composed of sieve elements that do not contain nuclei or support translation, surrounded by companion cells. Cmv2b expressed either constitutively in transgenic tobacco plants or transiently in 16c plants via Agrobacterium infiltration could also enter the phloem because of its small size (100 amino acids; Ding et al., 1994) and a relatively large size exclusion limit for plasmodesmata connecting sieve elements and companion cells (Oparka and Santa Cruz, 2000). The binding hypothesis is consistent with the required contact of Cmv2b with the signal for signal inactivation, as suggested by several experiments. For example, Cmv2b was active only when it was expressed between the source and recipient tissues of the signal (Figure 4E, F and I), but inactive when expressed outside of the tissue (Figure 4G) or leaf (Figure 4H) through which the signal must travel. It is of interest to note in this regard that Cmv2b binds ssRNA (H.S.Guo and S.W.Ding, unpublished data) and contains several positively charged regions with a predicted pI of 10.35 (Ding et al., 1994). Thus, it is possible that Cmv2b may bind the nucleic acid component of the signal, proposed to be an RNA template or product of the RNA-dependent RNA polymerase (Baulcombe, 1999b; Dalmay et al., 2000a; Fagard and Vaucheret, 2000; Mourrain et al., 2000; Waterhouse et al., 2001).
Alternatively, Cmv2b itself or its mRNA may translocate through the phloem (Ruiz-Medrano et al., 2001) and be unloaded in the upper leaves in a manner similar to the signal so that initiation of silencing by the signal in these leaves is blocked (Brigneti et al., 1998). While not mutually exclusive to the binding hypothesis, this idea would similarly explain the lack of silencing in the upper leaves of the double-grafted tobacco (Figure 4E) or the 16c plants infiltrated according to schemes illustrated in Figure 4F and I. Cmv2b unloading in cells that contain the target mRNA of the silencing signal would prevent initiation of PTGS in these cells. However, this suppressor translocation hypothesis does not explain why there was spread of GFP silencing into the upper tissues of the 16c plants when 35S-Cmv2b was infiltrated in the leaf tip (Figure 4G) or in an upper leaf (Figure 4H), because in both experiments Cmv2b should be similarly unloaded in the upper tissues as in Figure 4F. Regardless of the mechanism involved, the demonstrated inhibition of the signaling potential of the silencing signal by Cmv2b explains why silencing reversal in the silenced 16c plants after infection with CMV or a recombinant PVX expressing Cmv2b (PVX-C2b) occurs only in the newly emerging tissues (Brigneti et al., 1998). Arabidopsis PTGS mutants sde1/sgs1, sgs3 and sde3 display an enhanced susceptibility only to CMV among more than six viruses tested (Dalmay et al., 2000a, 2001; Mourrain et al., 2000; Voinnet, 2001). It is possible that unlike CMV, these viruses do not encode a suppressor capable of inhibiting the signal-mediated intercellular spread of PTGS, which may not have been disrupted in these Arabidopsis mutants.
Our previous work found that Cmv2b carries a functional arginine-rich nuclear localization signal and that Cmv2b nuclear targeting is required for its suppressor activity in the silencing reversal assay (Lucy et al., 2000). This led to the suggestions that Cmv2b may act either as a competitor in nuclear trafficking or as a transcriptional regulator (Lucy et al., 2000). However, results from several experiments presented in this work (Figure 4F–I) suggest that Cmv2b suppression of PTGS is unlikely to be due to an altered transcription of host genes. For example, Cmv2b expression either in the same leaf (Figure 4G) or plant (Figure 4H) was insufficient to ensure silencing suppression (Figure 4F). In addition, the GFP mRNA in the upper non-silencing leaves of the 16c plants co-infiltrated with 35S-Cmv2b and 35S-GFP (Figure 2, left) could be silenced readily, as demonstrated by 35S-GFP infiltration and grafting onto a GFP-silenced rootstock.
A possible nuclear function for Cmv2b is to prevent methylation of transgene DNA. Methylation of the GUS transgene in the Wtx6b5 plants, as measured at the two MluI sites of the GUS coding sequence, is reset in each generation in a manner similar to PTGS reset (Mallory et al., 2001). We found that unlike HC-Pro (Mallory et al., 2001), Cmv2b interferes with restoration of transgene methylation. The cytosine methylation at the same MluI sites was significantly reduced in C2bx6b5 plants as compared with that in Wtx6b5 plants (Figure 8). Methylation of the GFP transgene was also significantly reduced in the newly emerged green fluorescent tissues after silencing reversal in 16c plants by infection with a recombinant PVX expressing Cmv2b (H.W.Li and S.W.Ding, unpublished data), in contrast to the result obtained from silencing suppression with CMV (Jones et al., 1999). The association of DNA methylation with gene silencing has long been recognized (Wassenegger, 2000). Previous analyses have revealed a critical role of transgene methylation in the maintenance of PTGS (Dalmay et al., 2000b). Both PTGS and methylation of a GUS transgene were reversed in an Arabidopsis mutant impaired for the Met1 DNA methytransferase (Morel et al., 2000). The induction of homologous transgene DNA methylation by viral and subviral RNAs replicating in the cytoplasm strongly suggests nuclear import of a cytoplasmic RNA signal that initiates de novo DNA methylation in the nucleus (Wassenegger, 2000; Jones et al., 2001; Matzke et al., 2001; Wang et al., 2001). Thus, Cmv2b may interfere with either nuclear import of, or the methylation process mediated by, this methylation signal. However, no homologous nuclear DNA is involved in the natural host RNA silencing defense against CMV. Therefore, based on this, as well as the observed correlation between reduced levels of transgene methylation and loss of the activity of the silencing signal in C2bx6b5 plants, we propose that Cmv2b targets a signaling component of the silencing signal that also plays a key role in the RNA-induced DNA methylation. The hypothesis that the methylation and silencing signals share a key component is also consistent with the demonstrated link between transgene methylation and production/release of the silencing signal in HC-Prox6b5 tobacco, in which the GUS transgene is not silenced (Mallory et al., 2001).
In contrast to efficient inhibition of the intercellular silencing spread, Cmv2b suppression of intracellular silenc ing is incomplete. The incomplete suppression in the 35S-Cmv2b/35S-GFP co-infiltrated leaves can be due to (i) lack of Cmv2b expression in every silencing cell and/or (ii) the transient nature of Cmv2b expression in the 16c plants that support persistent GFP transgene silencing (Voinnet and Baulcombe, 1997; Voinnet et al., 1998). Indeed, an excess of agrobacteria carrying 35S-Cmv2b over those carrying 35S-GFP was required for an efficient suppression of systemic silencing in the co-infiltration experiments (see Materials and methods). However, incomplete suppression was also observed in C2bx6b5 tobacco where Cmv2b was expressed from a stably integrated transgene under the control of the same 35S promoter as the PTGS-inducing GUS transgene. There was also abundant accumulation of the virus-specific sRNAs in plants infected with Wt CMV that expresses Cmv2b (H.W.Li and S.W.Ding, unpublished data). Taken together, these data strongly indicate that Cmv2b is not able to prevent the signal-independent initiation of transgene and virus RNA silencing, both of which may be triggered by dsRNA (Sijen et al., 2001; Voinnet, 2001; Waterhouse et al., 2001).
The simultaneous initiation and suppression of transgene RNA silencing in C2bx6b5 tobacco and the co-infiltrated leaves in a way modulates what is predicted to occur in the leaves inoculated with CMV. Replication of the plus-strand RNA genome of CMV produces the dsRNA initiator of virus RNA silencing and RNA 4A, the viral mRNA for Cmv2b. Leaf-press blot hybridizations revealed defined viral RNA accumulation foci in the cucumber cotyledons 4 days after inoculation with either Wt CMV or a CMV mutant expressing no Cmv2b (CMV-Δ2b); these CMV-Δ2b foci decreased both in size and intensity subsequently, and systemic infection was not established, in contrast to Wt CMV (Ding et al., 1995). A similar requirement for Cmv2b in the CMV invasion of tissues outside of the initially infected leaves was recently confirmed in an alternative host species, Nicotiana glutinosa (Ji and Ding, 2001). We propose that the role of Cmv2b in facilitating the spread of CMV in infected plants is related to its ability to inactivate the silencing signal produced presumably in the primarily infected foci, as demonstrated in PVX-infected plants (Voinnet et al., 2000). Absence of the Cmv2b inactivation of the anti-viral signal in the CMV-Δ2b-infected foci would result in the inhibition of further virus spread due to immediate targeting of the viral RNAs spread to neighboring cells and distant tissues before replication is initiated. This active inhibition of the export of the anti-viral signal out of virus-infected tissues may also explain why Cmv2b is required in mixed infections to overcome the restricted infection of potato leafroll virus in the phloem tissues (Ryabov et al., 2001) and why CMV can alleviate a similar phloem restriction of a potyvirus in a resistant cultivar of Capsicum annuum (Guerini and Murphy, 1999).
Materials and methods
Agrobacterium strains and leaf infiltration
The binary plasmid 35S-GFP has been described (Brigneti et al., 1998). The Cmv2b coding sequence and its untranslatable mutant (CmvΔ2b), with the fourth codon replaced with a termination codon (Ding et al., 1995), were cloned between the 35S promoter and terminator in the binary plasmid pCAMBIA1300 (DDBJ/EMBL/GenBank accession No. AF234296) to generate 35S-Cmv2b and 35S-CmvΔ2b. These plasmids were transformed into A.tumefaciens strain EHA105 by electroporation and selected in Luria–Bertani medium containing kanamycin at 50 µg/ml and rifampicin at 10 µg/ml. The leaf infiltration of A.tumefaciens strains was as described (Brigneti et al., 1998). For the co-infiltration experiments, equal volumes of an Agrobacterium culture containing 35S-GFP (OD600 = 1.0) and an Agrobacterium culture containing 35S-Cmv2b (OD600 = 1.5) or 35S-CmvΔ2b (OD600 = 1.5) were mixed before infiltration. Similar results were obtained in co-infiltration experiments when the final Agrobacterium concentration in the mixture was 0.3 OD600 for 35S-GFP and 0.5 OD600 for 35S-Cmv2b (H.G.Xiao and S.W.Ding, unpublished data).
Transgenic plant lines and grafting procedure
The N.benthamiana line 16c, which expresses GFP (Brigneti et al., 1998), the N.tabacum line T19, which expresses GUS (English et al., 1996), and line 6b5, which contains a silenced GUS transgene (Elmayan and Vaucheret, 1996), have been described. Pollen grains from line 6b5 were used in crosses either with Wt N.tabacum plants or the N.tabacum line C2b14, which expresses Cmv2b (Ji and Ding, 2001), to generate progenies Wtx6b5 and C2bx6b5, respectively. GFP imaging (Brigneti et al., 1998) and GUS histochemical staining (Anandalakshmi et al., 1998) were performed as described. A wedge-grafting method (Palauqui et al., 1997; Guo et al., 1999) was used to generate single- and double-grafted tobacco plants. Tobacco plants used for grafting experiments were 8 weeks old. For triple grafting, the intergrafts were cut ∼10–15 cm long with at least two leaves retained (Palauqui et al., 1997). Leaf samples for RNA extraction were taken both from the individually labeled scion and stock plants immediately before grafting, and from the scions 6 (single grafts) or 7 weeks (double grafts) after grafting.
Analyses of RNA, DNA and protein
Total plant RNA extraction and northern blotting analysis were carried out as described previously (Li et al., 1999). Low molecular weight RNA analysis was as described (Hamilton and Baulcombe, 1999; Llave et al., 2000). The probes for GFP and GUS sRNAs were 32P-labeled full-length antisense RNAs transcribed by T7 RNA polymerase. The analysis of DNA methylation at the MluI sites of the GUS coding sequence was as described (Mallory et al., 2001). The DNA blots were first hybridized with a 32P-labeled full-length GUS coding sequence DNA probe, then stripped and re-hybridized with a 32P-labeled DNA probe specific for the 18S ribosomal DNA sequence (Li et al., 1999). The western blot detection of Cmv2b using a rabbit Cmv2b antiserum was essentially as described (Ding et al., 1994). The total plant proteins were extracted in an extraction buffer containing 10 mM KCl, 5 mM MgCl, 400 mM sucrose, 10 mM 2-mercaptoethanol, 10% glycerol, 10 mM Tris pH 8, 4 mM phenylmethylsulfonyl fluoride (PMSF; Sigma) and 1 mg/ml benzamidine (Sigma). The homogenates were then centrifuged for 10 min at 10 000 g. The resultant pellets were boiled for 5 min in a buffer containing 40 mM Tris pH 6.8, 8 M urea, 5% SDS, 0.1 mM EDTA, 10 mM 2-mercapto ethanol, 5 mM PMSF and 1 mg/ml benzamidine before loading. The Immun-Star Chemiluminescent Protein Detection Kit (Bio-Rad) and the manufacturer’s protocol were used in the detection of Cmv2b.
Acknowledgments
Acknowledgements
We wish to thank Wan-Xiang Li for the 35S-Cmv2b and 35S-CmvΔ2b constructs, Liang-Hui Ji for the C2bx6b5 and Wtx6b5 seeds, David Baulcombe for the 35S-GFP construct and 16c seeds, Herve Vaucheret for the 6b5 seeds, and Vicki Vance, Andrew Hamilton, James Carrington and Richard Jefferson for materials and protocols. This work was supported by grants to S.W.D. from the Singapore National Science and Technology Board and the USDA-NRICGP Plant Pathology Panel.
References
- Anandalakshmi R., Pruss,G.J., Ge,X., Marathe,R., Mallory,A.C., Smith,T.H. and Vance,V.B. (1998) A viral suppressor of gene silencing in plants. Proc. Natl Acad. Sci. USA, 95, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandalakshmi R., Marathe,R., Ge,X., Herr,J.M.,Jr, Mau,C., Mallory,A., Pruss,G., Bowman,L. and Vance,V.B. (2000) A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science, 290, 142–144. [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C. (1999a) Viruses and gene silencing in plants. Arch. Virol. Suppl., 15, 189–201. [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C. (1999b) Gene silencing: RNA makes RNA makes no protein. Curr. Biol., 9, 599–601. [DOI] [PubMed] [Google Scholar]
- Brigneti G., Voinnet,O., Li,W.X., Ji,L.H., Ding,S.W. and Baulcombe,D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J., 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Carrington J.C., Kasschau,K.D. and Johansen,L.K. (2001) Activation and suppression of RNA silencing by plant viruses. Virology, 281, 1–5. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Hamilton,A., Rudd,S., Angell,S. and Baulcombe,D.C. (2000a) An RNA-dependent RNA polymerase gene in Arabidopsis is required for PTGS mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Hamilton,A., Mueller,E. and Baulcombe,D.C. (2000b) Potato virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell, 12, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Horsefield,R., Braunstein,T.H. and Baulcombe,D.C. (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J., 20, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.W. (2000) RNA silencing. Curr. Opin. Biotech., 11, 152–156. [DOI] [PubMed] [Google Scholar]
- Ding S.W., Anderson,B.J., Haase,H.R. and Symons,R.H. (1994) New overlapping gene encoded by the cucumber mosaic virus genome. Virology, 198, 593–601. [DOI] [PubMed] [Google Scholar]
- Ding S.W., Li,W.X. and Symons,R.H. (1995) A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J., 14, 5762–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty W.G. and Parks,T.D. (1995) Transgenes and gene suppression: telling us something new? Curr. Opin. Cell Biol., 7, 399–405. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth.J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001a) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001b) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T. and Vaucheret,H. (1996). Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J., 9, 787–797. [Google Scholar]
- English J.J., Mueller,E. and Baulcombe,D.C. (1996) Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell, 8, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M. and Vaucheret,H. (2000) Systemic silencing signal(s). Plant Mol. Biol., 43, 285–293. [DOI] [PubMed] [Google Scholar]
- Fagard M., Boutet,S., Morel,J.B., Bellini,C. and Vaucheret,H. (2000) AGO1, QDE-2 and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi and RNA interference in animals. Proc. Natl Acad. Sci. USA, 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A. et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C.elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Guerini M.N. and Murphy,J.F. (1999) Resistance of Capsicum annuum ‘Avelar’ to pepper mottle potyvirus and alleviation of this resistance by co-infection with cucumber mosaic cucumovirus are associated with virus movement. J. Gen. Virol., 80, 2785–2792. [DOI] [PubMed] [Google Scholar]
- Guo H.S., Lopez-Moya,J.J. and Garcia,J.A. (1999) Mitotic stability of infection-induced resistance to plum pox potyvirus associated with transgene silencing and DNA methylation. Mol. Plant Microbe Interact., 12, 103–111. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe,D.C. (1999) A species of small antisense RNA in post-transcriptional gene silencing. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein,E., Beach,D. and Hannon,G. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cell extracts. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Ji L.H. and Ding,S.W. (2001) The suppressor of transgene RNA silencing encoded by cucumber mosaic virus interferes with salicylic acid-mediated virus resistance. Mol. Plant Microbe Interact., 14, 715–724. [DOI] [PubMed] [Google Scholar]
- Johansen L.K. and Carrington,J.C. (2001) Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol., 126, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Hamilton,A.J., Voinnet,O., Thomas,C.L., Maule,A.J. and Baulcombe,D.C. (1999) RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell, 11, 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Ratcliff,F. and Baulcombe,D.C. (2001) RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol., 11, 747–757. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Haverkamp,T.H., van Luenen,H.G. and Plasterk,R.H. (1999) Mut-7 of C.elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Li H.W., Lucy,A.P., Guo,H.S., Li,W.X., Ji,L.H., Wong,S.M. and Ding,S.W. (1999) Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J., 18, 2683–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.X. and Ding,S.W. (2001) Viral suppressors of RNA silencing. Curr. Opin. Biotech., 12, 150–154. [DOI] [PubMed] [Google Scholar]
- Lipardi C., Wei,Q. and Paterson,B.M. (2001) RNAi as random degradative PCR. siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell, 107, 297–307. [DOI] [PubMed] [Google Scholar]
- Llave C., Kasschau,K. and Carrington,J. (2000) Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl Acad. Sci. USA, 97, 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy A.P., Guo,H.S., Li,W.X. and Ding,S.W. (2000) Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J., 19, 1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A. et al. (2001) HC-Pro suppression of gene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell, 13, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe R., Anandalakshmi,R., Smith,T.H., Pruss,G.J. and Vance,V.B. (2000) RNA viruses as inducers, suppressors and targets of post-transcriptional gene silencing. Plant Mol. Biol., 43, 295–306. [DOI] [PubMed] [Google Scholar]
- Matzke M.A., Matzke,A.J.M. and Kooter,J.M. (2001) RNA: guiding gene silencing. Science, 293, 1080–1083. [DOI] [PubMed] [Google Scholar]
- Mette M.F., Aufsatz,W., van der Winden,J., Matzke,M.A. and Matzke,A.J. (2000) Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J., 19, 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel J., Mourrain,P., Beclin,C. and Vaucheret,H. (2000) DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol., 10, 1591–1594. [DOI] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Oparka K.J. and Santa Cruz,S. (2000) The great escape: phloem transport and unloading of macromolecules. Annu. Rev. Plant Physiol. Plant Mol. Biol., 51, 323–347. [DOI] [PubMed] [Google Scholar]
- Palauqui J.C., Elmayan,T., Pollien,J.-M. and Vaucheret,H. (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J., 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R., Xoconostle-Cazares,B. and Lucas,W.J. (2001) The phloem as a conduit for inter-organ communication. Curr. Opin. Plant Biol., 4, 202–209. [DOI] [PubMed] [Google Scholar]
- Ryabov E.V., Fraser,G., Mayo,M.A., Barker,H. and Taliansky,M. (2001) Umbravirus gene expression helps potato leafroll virus to invade mesophyll tissues and to be transmitted mechanically between plants. Virology, 286, 363–372. [DOI] [PubMed] [Google Scholar]
- Serio F., Schob,H., Iglesias,A., Tarina,C., Bouldoires,E. and Meins,F.,Jr (2001) Sense- and antisense-mediated gene silencing in tobacco is inhibited by the same viral suppressors and is associated with accumulation of small RNAs. Proc. Natl Acad. Sci. USA, 98, 6506–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Vijn,I., Rebocho,A., van Blokland,R., Roelofs,D., Mol,J.N. and Kooter,J.M. (2001) Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol., 11, 436–440. [DOI] [PubMed] [Google Scholar]
- Smardon A., Spoerke,J.M., Stacey,S.C., Klein,M.E., Mackin,N. and Maine,E.M. (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C.elegans. Curr. Biol., 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Vance V. and Vaucheret,H. (2001) RNA silencing in plants—defense and counterdefense. Science, 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2001) RNA silencing as a plant immune system against viruses. Trends Genet., 17, 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet O. and Baulcombe,D.C. (1997) Systemic signaling in gene silencing. Nature, 389, 553. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Vain,P., Angell,S. and Baulcombe,D.C. (1998) Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Lederer,C. and Baulcombe,D.C. (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell, 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Wang M.B., Wesley,S.V., Finnegan,E.J., Smith,N.A. and Waterhouse,P.M. (2001) Replicating satellite RNA induces sequence-specific DNA methylation and truncated transcripts in plants. RNA, 7, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M. (2000) RNA-directed DNA methylation. Plant Mol. Biol., 43, 203–220. [DOI] [PubMed] [Google Scholar]
- Waterhouse P.M., Wang,M. and Lough,T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Wu-Scharf D., Jeong,B., Zhang,C. and Cerutti,H. (2000) Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science, 290, 1159–1163. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]