Abstract
Antisense-mediated gene silencing (ASGS) and posttranscriptional gene silencing (PTGS) with sense transgenes markedly reduce the steady-state mRNA levels of endogenous genes similar in transcribed sequence. RNase protection assays established that silencing in tobacco plants transformed with plant-defense-related class I sense and antisense chitinase (CHN) transgenes is at the posttranscriptional level. Infection of tobacco plants with cucumber mosaic virus strain FN and a necrotizing strain of potato virus Y, but not with potato virus X, effectively suppressed PTGS and ASGS of both the transgenes and homologous endogenes. This suggests that ASGS and PTGS share components associated with initiation and maintenance of the silent state. Small, ca. 25-nt RNAs (smRNA) of both polarities were associated with PTGS and ASGS in CHN transformants as reported for PTGS in other transgenic plants and for RNA interference in Drosophila. Similar results were obtained with an antisense class I β-1,3-glucanase transformant showing that viral suppression and smRNAs are a more general feature of ASGS. Several current models hold that diverse signals lead to production of double-stranded RNAs, which are processed to smRNAs that then trigger PTGS. Our results provide direct evidence for mechanistic links between ASGS and PTGS and suggest that ASGS could join a common PTGS pathway at the double-stranded RNA step.
Posttranscriptional gene silencing (PTGS) refers to the trans-inactivation of homologous genes caused by increased RNA degradation (1, 2). This is a stable, reversible, epigenetic modification triggered by sequence-specific signals that, in some cases, can spread systemically. First described for transgenic plants, it is now recognized that very similar phenomena occur in a wide variety of organisms. Examples include quelling in Neurospora crassa (3) and RNA interference (RNAi) mediated by double-stranded RNA (dsRNA) in Caenorhabditis elegans (4), Drosophila melanogaster (5), and mouse (6). The finding that RNAi can spread systemically; the identification of small, ca. 25-nt sense- and antisense-RNAs (smRNAs) associated with PTGS in plants and RNAi in Drosophila (7, 8); and the strong genetic links between quelling, RNAi in C. elegans, and PTGS in Arabidopsis provide compelling evidence for a common, highly conserved molecular mechanism (9–12). Although the function of PTGS in plants is still unclear, the findings that PTGS can be suppressed by virus-encoded proteins that also suppress anti-viral defense in the host (13–16) and that PTGS mutants also affect virus infection (10, 11) strongly support a role in virus resistance. PTGS-deficient mutants (12) and plants overexpressing viral suppressor proteins (17) can show striking abnormalities, suggesting that PTGS may also be important in normal development.
Expression of endogenous plant genes can also be posttranscriptionally silenced by antisense transformation. Although little is known about the mechanism for antisense-mediated gene silencing (ASGS) (18–20), the involvement of dsRNAs and smRNAs in PTGS (1, 2, 7, 21) supports the early hypothesis proposed by Grierson et al. (22) and Mol et al. (23) that ASGS and PTGS are mechanistically linked and that antisense RNAs (asRNA) have a role in sequence-specific degradation of target RNAs in both processes (24).
We have obtained direct evidence linking ASGS and PTGS by comparing the silencing of genes encoding class I chitinases (CHN) and β-1,3-glucanases (GLU) in tobacco plants transformed with the homologous cDNAs in sense and antisense orientation. Earlier we showed that all members of the CHN and GLU gene families can be posttranscriptionally silenced in sense Nicotiana sylvestris transformants (25–27). Here we report that posttranscriptional sense- and antisense-silencing of these genes in tobacco is suppressed by plant viruses known to suppress PTGS and that smRNAs diagnostic for PTGS (7) are associated with both forms of silencing. These findings suggest that PTGS and ASGS share, at least in part, a common mechanism.
Materials and Methods
Plant Material.
The transgenic Nicotiana tabacum cv. Havana 425 plants used were single-locus homozygous lines obtained by Agrobacterium-mediated transformation with Ti-plasmid vectors carrying chimeric transgenes regulated by cauliflower mosaic virus 35S RNA expression signals (28). Sense-chitinase line TSC3.63 (TSC) carries the tobacco chitinase CHN48 cDNA (29) and a bacterial hptII gene conferring hygromycin resistance, and shows stochastic silencing starting at the two-leaf stage of seedling development. Antisense-chitinase line TAC11.7 (TAC) carries a tobacco chitinase CHN50 cDNA (29) in inverted orientation and a bacterial nptII gene conferring kanamycin resistance. Antisense-β-1,3-glucanase line TAG4.4 (TAG) carries a tobacco GLU cDNA in inverted orientation and has been described (30).
Virus Infection.
Plants at the six-leaf stage were assayed for expression of the silent and high-expressing phenotypes as described (25). A fully expanded leaf of plants with ca. nine leaves was dusted with carborundum (400 mesh) and inoculated with sap extracts prepared from Havana 425 plants infected with potato virus X (PVX), a satellite-free cucumber mosaic virus strain FN (CMV), or a necrotic strain of potato virus Y (PVY). Infection was verified by RNA blot hybridization using 32P-labeled cDNA probes specific for the coat protein genes of each virus kindly provided by D. Gallitelli (University of Bari, Italy). DNA fragments were labeled by using the Rediprime II random prime system (Amersham Pharmacia) according to the manufacturers instructions.
Hybridization Probes.
Unless indicated otherwise, standard molecular procedures were used (31). The cDNA clones pCHN48 and pGL43 have been described (29, 32). Intron/exon border sequences were cloned by PCR from N. sylvestris genomic DNA based on homology to the tobacco CHN50 genomic clone (positions 2011–2256, GenBank accession no. x64519) and the tobacco Gla genomic clone (positions 2236–2457, GenBank accession no. m60402), and then subcloned into pBSIIKS+. 32P-labeled RNA probes were synthesized by in vitro transcription from suitably linearized plasmids by using a Stratagene RNA transcription kit according to the manufacturer's instructions.
Total RNA Analysis.
Total RNA was purified from plant tissues by using TRIZOL reagent (GIBCO/BRL), followed by RQ1 DNase (Promega) digestion and two phenol extractions. RNA blot hybridization was performed by using the indicated probes. Membranes were rehybridized with a cDNA probe corresponding to 18S rRNA to confirm equal loading of the gels. RNase A/T1 protection assays with a U2 RNA probe as a constitutive control were performed as described (33). Hybridization signals were quantified with a PhosphorImager using imagequant software (Molecular Dynamics).
Small RNA Analysis.
To enrich for small RNAs, total RNA (100 μg) was applied to the column from the RNeasy plant mini kit 74904 (Qiagen, Chatsworth, CA) following the manufacturer's instructions for RNA clean-up until step 3. Small RNAs, which are present in the flow-through fraction, were precipitated by adding 70 μl of 3 M Na acetate (pH 4.8) and 540 μl of 2-propanol, separated by 15% PAGE, and electrotransferred to Hybond-N+ membranes. 32P-labeled CHN and GLU full-length RNA transcripts were gel-purified and hydrolyzed to fragments of ca. 150 nt (34). RNA blot hybridizations were carried out at 35°C as described by Hamilton et al. (7).
DNA Analysis.
DNA was prepared from plants and analyzed by Southern blot hybridization using 32P-labeled cDNA probes prepared from pCHN48 and pGL43 as described (29, 32).
Protein Analysis.
Immunoblot analysis of chitinase and β-1,3-glucanase proteins was performed as described (35), except that the second antibody was coupled to alkaline phosphatase. Under the conditions used, the antibodies detect class I and II chitinases and class I-III β-1,3-glucanases.
Results
Posttranscriptional Silencing in Sense and Antisense Transformants.
The endogenous tobacco genes targeted for silencing were CHN genes CHN48 and CHN50 encoding mRNAs 94.5% identical in sequence (29), and GLU genes Gla and Glb encoding mRNAs 97.5% identical in sequence (32). The homozygous, single-locus transformants used were sense (TSC) and antisense (TAC) lines carrying tobacco CHN transgenes with transcribed regions >94.5% identical to those of CHN48 and CHN50, and the antisense (TAG) line carrying a tobacco GLU transgene with a transcribed region >97.9% identical to those of Gla and Glb. The T-DNA loci were partially characterized by Southern blot analyses (see Figs. 6–8, which are published as supplemental data on the PNAS web site, www.pnas.org). The TAC locus consists of an intact T-DNA and a partial direct repeat with an intact antisense CHN gene. The TSC locus gives rise to a complex restriction pattern. It consists of three closely linked T-DNA inserts. One contains a partial inverted repeat with a deletion extending from the left border into the 3′ end of the sense CHN gene. The TAG locus consists of a single T-DNA with an intact antisense GLU gene. The fact that no inverted repeats were detected in the antisense lines supports our view that reduction of target gene expression in these lines is due to ASGS.
The plants were treated with ethylene to ensure that the endogenous CHN and GLU genes were transcriptionally active (36). An RNase A/T1 protection assay (RPA) using CHN50 and Gla intron/exon probes (see Fig. 9, which is published as supplemental data) was used to measure the contents of CHN50 and Gla pre-mRNAs as well as CHN48, CHN50, Gla, and Glb mRNAs in RNA prepared from mature leaves. Posttranscriptional inactivation was detected by comparing the content of unspliced, primary transcripts, which are not decreased in PTGS (37), with that of the mature mRNAs in sibling silent and high-expressing plants. Leaves of transformants were judged to express the silent phenotype if the protein and mRNA contents after ethylene treatment were appreciably lower than that of wild-type plants under comparable conditions in the same experiment (25).
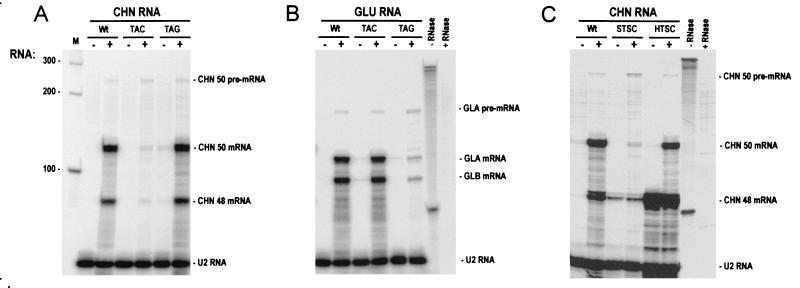
RPA confirmed that ethylene treatment induced the accumulation of CHN and GLU endogene transcripts in wild-type plants (Fig. 1 A and B). Accumulation of CHN but not GLU mRNAs was inhibited in TAC (Fig. 1A); whereas, accumulation of GLU but not CHN mRNAs was inhibited in TAG (Fig. 1B), indicating that ASGS acts in a sequence-specific fashion. In contrast, PhosphorImager quantitation of signals revealed that CHN50 and Gla pre-mRNAs were induced by ethylene treatment to comparable levels in TAC, TAG, and wild type (see Fig. 10, which is published as supplemental data). The fact that accumulation of the homologous mRNAs, but not the pre-mRNAs, was inhibited in the antisense lines leads us to conclude that ASGS acts at the posttranscriptional level and does not inhibit transcriptional induction by ethylene. Similarly, comparison of CHN50 endogene and CHN48 transgene expression in leaves of silent (STSC) and high-expressing (HTSC) siblings of the TSC line (Fig. 1C) confirmed PTGS in the CHN48 sense transformant.
Figure 1.
Pre-mRNA accumulation induced by ethylene treatment is unaffected by PTGS and ASGS. RNase protection assays of total RNA (25 μg) extracted from leaves of untransformed tobacco (Wt), TAC, TAG, sibling silent (STSC), and high-expressing (HTSC) TSC plants before (−) and after (+) treatment with 20 ppm ethylene for 2 days. Intron/exon RNA probes for CHN50 (A and C) and Gla (B) mixed with a control U2 RNA probe were used. The positions of the RNA-species protected and probes before (−RNase) and after (+RNase) RNase digestion protected with tRNA are indicated.
PVY and CMV Suppress PTGS and ASGS.
PVY and CMV, but not PVX, inhibit PTGS in Nicotiana benthamiana (14). CMV has also been shown to be effective in tobacco and Arabidopsis (38). We investigated the effect of CMV, PVY, and PVX infection on PTGS of chitinase in STSC plants, and on ASGS of CHN and GLU genes in the antisense lines TAC and TAG, respectively. Infection was confirmed by RNA-blot hybridization with RNA probes specific for each virus (data not shown). All lines tested, including control wild-type and HTSC plants, showed typical systemic symptoms 10–15 days after virus inoculation. Expression of GLU and CHN genes in wild-type leaves was not induced by systemic infection with CMV and PVX, but as expected (36) was induced in leaves showing a necrotic response to PVY infection (see Fig. 11, which is published as supplemental data).
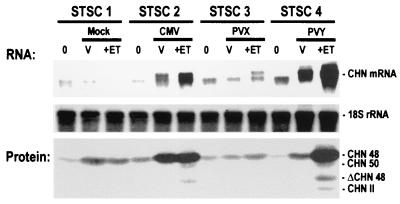
Strong accumulation of CHN mRNA and CHN protein was observed in STSC plants infected with CMV or PVY (Fig. 2), whereas low levels of CHN mRNA and weak expression of CHN protein were detected before as well as after mock or PVX inoculations. Because of posttranslational prolylhydroxylation, CHN48 and CHN50 proteins can be distinguished by immunoblotting (39), which revealed that the CHN50 endogene is either not expressed or expressed at low levels in plants before ethylene treatment. In contrast, following ethylene treatment the strong CHN mRNA signals obtained for CMV- and PVY-inoculated plants were accompanied by the appearance of both CHN48 and CHN50 proteins. Taken together, these results show that systemic infection with CMV and PVY, but not with PVX, suppresses PTGS of both endogenous CHN genes and CHN transgenes.
Figure 2.
CMV and PVY, but not PVX, suppress PTGS in TSC. RNA blot hybridization with a CHN cDNA probe and immunoblot analyses of leaves from sibling STSC plants (STSC1–STSC4). Tissues were harvested from the inoculated leaf just before inoculation (0), from leaves showing systemic virus infection 10–15 DPI (V), and from leaves showing systemic virus infection 2 days later after treatment of plants with 20-ppm ethylene (+ET). Total RNA (10 μg) was hybridized with a CHN48 cDNA probe, which detects CHN48 and CHN50 RNA (CHN RNA). The double bands detected with the probe have been described earlier and may result from alternative polyadenylation (26). Equal loading was confirmed by rehybridization with a probe for 18S rRNA. Immunoblot analyses of equal volumes of protein extracts of the same tissues used for RNA analyses are shown at the bottom. The positions of the class I chitinases CHN48, its truncated form ΔCHN48 and CHN50, and the class II chitinases (CHN II) are indicated.
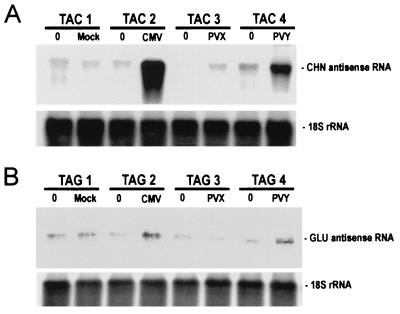
In parallel experiments, we examined the effect of virus infection on the accumulation of antisense CHN and GLU transcripts in TAC and TAG antisense transformants, respectively. Very low levels of antisense CHN50 RNA were detected in TAC plants before inoculation and following mock inoculation and inoculation with PVX (Fig. 3A). In contrast, strong signals were obtained with the CMV- and PVY-inoculated plants showing that TAC has the potential to express high levels of antisense CHN50 RNA and that infection with CMV and PVY suppressed degradation of the antisense transcripts. Similar results were obtained for antisense Gla RNA in the TAG plants (Fig. 3B), although for unknown reasons levels of expression in this transformed line were very low.
Figure 3.
CMV and PVY, but not PVX, suppress accumulation of transgene-encoded antisense transcripts in TAC and TAG. Tissues were harvested from the inoculated leaf just before inoculation (0) and from leaves showing systemic virus infection 10–15 DPI of sibling TAC (TAC1–TAC4) and TAG (TAG1–TAG4) plants. RNA blot hybridization of total RNA (10 μg) by using RNA probes to detect antisense CHN RNA (A) or antisense GLU RNA (B). Equal loading was confirmed by rehybridization with a probe for 18S rRNA.
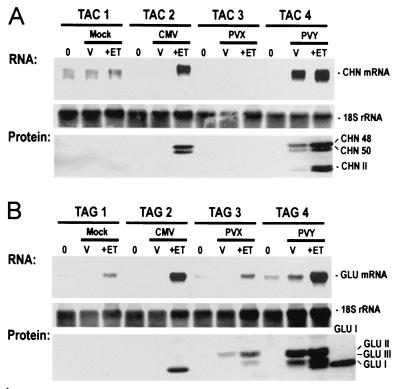
We also examined the effect of viruses on the expression of endogenes transcriptionally induced by ethylene treatment. In the case of TAC, strong CHN RNA signals were detected in CMV-infected plants after ethylene treatment and in PVY-infected plants both before and after ethylene treatment (Fig. 4A). On the other hand, low-level signals or no signals for CHN RNA were detected in mock-inoculated plants and in PVX-inoculated plants before inoculation and before and after ethylene treatment. Similar patterns of GLU RNA were found for TAG plants (Fig. 4B). Immunoblotting showed that accumulation of both CHN48 and CHN50 proteins was effectively inhibited by antisense transformation and that CMV and PVY infection suppressed this inhibition. Because the GLU encoded by Gla and Glb have the same apparent molecular masses in SDS gels, it was not possible to distinguish between the two isoforms. The additional bands detected on immunoblots correspond to the ca. 28-kDa class II chitinases, the ca. 36-kDa class II β-1,3-glucanases, and the ca. 33-kDa class III β-1,3-glucanase. These isoforms are induced as part of the necrotic response to infection by pathogens, but only weakly induced by ethylene treatment (36). Taken together, our results show that infection with suppressing viruses can restore both the expression and normal regulation by ethylene and pathogen infection of endogenous target genes in the antisense TAC and TAG transformants.
Figure 4.
CMV and PVY, but not PVX, restore ethylene-induction of endogenous-gene expression in antisense transformants. RNA blot and immunoblot analyses of leaf tissues harvested as indicated in Fig. 2 from sibling TAC plants (A) and TAG (B) plants. Total RNA (10 μg) was hybridized with RNA probes for detecting sense CHN RNA (A) or sense GLU RNA (B). Equal loading was confirmed by rehybridization with a probe for 18S rRNA. Protein extracts representing equal amounts of the same tissues were immunoblotted by using probes for CHN antigens and GLU antigens indicated in Fig. 2. Purified GLU I protein was used as size marker (B, GLU I). Positions of GLU I and the class II (GLU II) and class III (GLU III) β-1,3-glucanases are indicated. Note that induction of GLU I antigen is suppressed in mock- and PVX-infected plants, but not in CMV- and PVY-infected plants.
Sense and Antisense smRNAs Accumulate in Association with PTGS and ASGS.
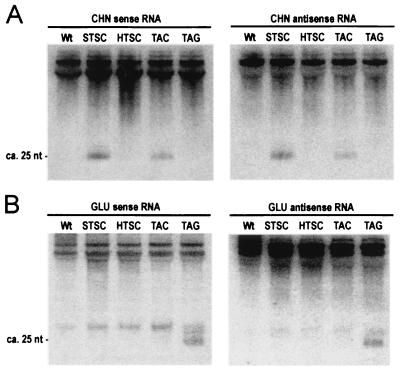
The association of smRNAs with PTGS in transgenic plants (7) and RNAi in Drosophila (8) prompted us to look for similar RNAs representing the transcribed sequences of CHN and GLU genes. RNA-blot hybridization (Fig. 5) and RNase A/T1 protection assays (data not shown) were used with probes for sense and antisense sequences. The CHN probes detected ca. 25-nt RNAs in STSC and TAC leaves, but not in HTSC, TAG, or wild-type leaves (Fig. 5A); whereas, the GLU probes detected ca. 25-nt RNAs in TAG leaves, but not in STSC, HTSC, TAC, or wild-type leaves (Fig. 5B). Taken together, these results indicate that accumulation of sense and antisense smRNAs representing the silenced gene is a feature of both PTGS and ASGS.
Figure 5.
Small, ca. 25-nt sense- and antisense-RNAs are associated with both PTGS and ASGS. RNA blot hybridization of 20 μg of the low-molecular-weight RNA fraction prepared from total RNA of untreated plants. Membranes were hybridized with RNA probes for sense and antisense CHN (A) and GLU (B) RNA. The position of the ca. 25-nt RNAs was determined by using single-stranded DNA primers as size markers. Unspecific cross-hybridization with tRNAs (top of each gel) was used as the control for equal loading. Note that an additional ca. 30-nt RNA was detected with sense and antisense GLU probes. This RNA was present at roughly the same abundance in wild type and transformed lines and was not correlated with ASGS.
Discussion
It is unclear whether ASGS is a form of PTGS or involves distinctly different mechanisms (20, 24). Our study shows for the first time that ASGS, like PTGS, is associated with the production of smRNAs and can be suppressed by infection with CMV and PVY that suppress PTGS. These results provide direct evidence for mechanistic links between the two forms of silencing. The suppressing viruses we used act at different steps in the PTGS pathway (14, 38, 40). Expression of the HC-Pro protein of PVY inhibits initiation and maintenance of PTGS; whereas expression of the 2b protein of CMV only inhibits initiation of PTGS and acts at sites in the cell nucleus (41). Both viruses suppressed ASGS of the homologous endogenes as well as expression of the antisense transgene indicating that ASGS and PTGS share at least some components associated with initiation and maintenance of the silent state.
Infection with CMV markedly increased the antisense RNA content of antisense transformants under conditions in which transcription of endogenes was very low (see Fig. 1). This indicates that high expression of sense genes is not required for posttranscriptional silencing of the antisense transgenes. The untreated antisense transformants, but not wild-type plants, also accumulated smRNAs of both polarities. Taken together, these results show that smRNA accumulation is associated with posttranscriptional silencing of antisense transgenes and does not depend on high-level expression of the sense endogenes. It seems likely, therefore, that the smRNAs are derived from the antisense transgene. Experiments using gene-specific probes are needed to confirm this hypothesis and to examine the possibility that smRNAs can also arise from silenced sense endogenes in the antisense transformants. Quantitation of hybridization signals corrected for RNA loading suggests that sense- and antisense-RNA species are present in roughly equal amounts. This raises the possibility that the smRNAs are duplex structures, or are derived from a dsRNA intermediate as reported for RNAi in Drosophila (8), which could also explain the observation that both sense and antisense RNAs can be targets of PTGS (42).
Several current models for PTGS hold that sequence-specific signals from several sources can feed into a common pathway for targeting degradation of RNAs (2, 43). Silencing signals can be generated by high-level transcription of single-copy sense loci, by transcription of transgenes with inverted repeats, by complex loci showing low transcriptional activity, and by cytoplasmically replicating viruses (44). Although the chemical nature of these signals is not known, it is often assumed that asRNAs provide sequence specificity (22–24, 44) and that dsRNAs act as primary signals or further downstream in the common pathway (1, 2).
Dalmay et al. (11) have proposed a general model to account for both virus-induced gene silencing (VIGS) and PTGS of sense transgenes. They envisage that dsRNAs generated by virus-encoded replicases in VIGS and by RNA-dependent RNA polymerases (RdRP) in PTGS are processed to smRNAs, which then trigger RNA degradation. We propose that ASGS could join the common PTGS pathway at the dsRNA step. Antisense transgenes could generate dsRNAs in at least two ways. When transcription of sense endogenes is low, dsRNAs could arise from highly expressed antisense transgenes by a mechanism similar to that of sense transgenes in a process, which may depend on RdRP. When transcription of sense endogenes is high, dsRNA could also arise by annealing of sense and antisense transcripts as reported for hybrids simultaneously expressing sense and antisense RNAs (45). According to our model, distinctive features of PTGS and ASGS (20), such as the patterns generated by sense and antisense alleles at the same transgene locus (46), could reflect differences upstream of the dsRNA step.
Based on studies of ASGS affecting chalcone synthase-gene expression in petunia flowers, Stam et al. (24) identified high-expressing, single-copy antisense loci, which presumably generate dsRNA by pairing with endogene transcripts, and inverted-repeat loci that trigger silencing by a mechanism similar to that of inverted sense repeats. These findings, which emphasize possible mechanistic links between ASGS and PTGS, and our studies, which provide direct evidence for common steps in the two processes, strongly support the hypothesis that ASGS is simply an antisense form of PTGS. More detailed studies employing informative mutants are needed to establish common pathways for silencing-related RNA metabolism and distinctive features of ASGS.
Supplementary Material
Acknowledgments
We thank D. Gallitelli for virus probes and CMV; J.-M. Neuhaus, V. Suarez, and P. Crété for transgenic plants; and our colleagues W. Filipowicz, O. Mittelsten Scheid, and U. Klahre for helpful comments.
Abbreviations
- ASGS
antisense-mediated gene silencing
- CHN
class I chitinase
- CMV
cucumber mosaic virus strain FN
- GLU
class I β-1,3-glucanase
- PTGS
posttranscriptional gene silencing
- PVX
potato virus X
- PVY
a necrotizing strain of potato virus Y
- RNAi
RNA interference
- smRNA
small sense and antisense RNA
- TAC
tobacco antisense CHN transformant
- TAG
tobacco antisense GLU transformant
- TSC
tobacco sense CHN transformant
- STSC
silent TSC
- HTSC
high-expressing TSC
- dsRNA
double-stranded RNA
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fire A. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- 2.Kooter J M, Matzke M A, Meyer P. Trends Plant Sci. 1999;4:340–347. doi: 10.1016/s1360-1385(99)01467-3. [DOI] [PubMed] [Google Scholar]
- 3.Cogoni C, Macino G. Proc Natl Acad Sci USA. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 5.Misquitta L, Paterson B M. Proc Natl Acad Sci USA. 1999;96:1451–1456. doi: 10.1073/pnas.96.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wianny F, Zernicka-Goetz M. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton A J, Baulcombe D C. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 8.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 9.Catalanotto C, Azzalin G, Macino G, Cogoni C. Nature (London) 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- 10.Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Jouette D, Lacombe A-M, Nikic S, Picault N, et al. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 11.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe D C. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 12.Fagard M, Boutet S, Morel J-B, Bellini C, Vaucheret H. Proc Natl Acad Sci USA. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. . (First Published October 3, 2000; 10.1073/pnas.200217597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anandalakshmi R, Pruss G J, Ge X, Marathe R, Mallory A C, Smith T H, Vance V B. Proc Natl Acad Sci USA. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brigneti G, Voinnet O, Li W-X, Ji L-H, Ding S-W, Baulcombe D C. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Kasschau K D, Carrington J C. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 16.Voinnet O, Pinto Y M, Baulcombe D C. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anandalakshmi R, Marathe R, Ge X, Herr J M, Jr, Mau C, Mallory A, Pruss G, Bowman L, Vance V B. Science. 2000;290:142–144. doi: 10.1126/science.290.5489.142. [DOI] [PubMed] [Google Scholar]
- 18.Mol J N M, van Blokland R, de Lange P, Stam M, Kooter J M. In: Gene Inactivation and Homologous Recombination in Plants. Paszkowski J, editor. Dordrecht, The Netherlands: Kluwer; 1994. pp. 309–334. [Google Scholar]
- 19.Bourque J E. Plant Sci. 1995;105:125–149. [Google Scholar]
- 20.Fagard M, Vaucheret H. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:168–221. doi: 10.1146/annurev.arplant.51.1.167. [DOI] [PubMed] [Google Scholar]
- 21.Chuang C-F, Meyerowitz E M. Proc Natl Acad Sci USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. . (First Published April 18, 2000; 10.1073/pnas.060034297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grierson D, Fray R G, Hamilton A J, Smith C J S, Watson C F. Trends Biotechnol. 1991;9:122–123. [Google Scholar]
- 23.Mol J, van Blokland R, Kooter J. Trends Biotechnol. 1991;9:182–183. [Google Scholar]
- 24.Stam M, de Bruin R, van Blokland R, Van der Hoorn R A L, Mol J N M, Kooter J M. Plant J. 2000;21:27–42. doi: 10.1046/j.1365-313x.2000.00650.x. [DOI] [PubMed] [Google Scholar]
- 25.Kunz C, Schöb H, Stam M, Kooter J M, Meins F., Jr Plant J. 1996;10:437–450. [Google Scholar]
- 26.Holtorf H, Schöb H, Kunz C, Waldvogel R, Meins F., Jr Plant Cell. 1999;11:471–484. doi: 10.1105/tpc.11.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunz C, Schöb H, Leubner-Metzger G, Glazov E, Meins F., Jr Planta. 2001;212:243–249. doi: 10.1007/s004250000383. [DOI] [PubMed] [Google Scholar]
- 28.Neuhaus J-M, Ahl-Goy P, Hinz U, Flores S, Meins F., Jr Plant Mol Biol. 1991;16:141–151. doi: 10.1007/BF00017924. [DOI] [PubMed] [Google Scholar]
- 29.van Buuren M, Neuhaus J-M, Shinshi H, Ryals J, Meins F., Jr Mol Gen Genet. 1992;232:460–469. doi: 10.1007/BF00266251. [DOI] [PubMed] [Google Scholar]
- 30.Beffa R S, Neuhaus J-M, Meins F., Jr Proc Natl Acad Sci USA. 1993;90:8792–8796. doi: 10.1073/pnas.90.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 32.Sperisen C, Ryals J, Meins F., Jr Proc Natl Acad Sci USA. 1991;88:1820–1824. doi: 10.1073/pnas.88.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambermon M H, Simpson G G, Wieczorek-Kirk D A, Hemmings-Mieszczak M, Klahre U, Filipowicz W. EMBO J. 2000;19:1638–1649. doi: 10.1093/emboj/19.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox K H, Goldberg R B. In: Plant Molecular Biology: A Practical Approach. Shaw C H, editor. Oxford: IRL; 1988. pp. 1–34. [Google Scholar]
- 35.Keefe D, Hinz U, Meins F., Jr Planta. 1990;182:43–51. doi: 10.1007/BF00239982. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J-M. In: Pathogenesis-Related Proteins in Plants. Datta S K, Muthukrishnan S, editors. Boca Raton, FL: CRC; 1999. pp. 195–206. [Google Scholar]
- 37.Hamilton A J, Brown S, Yuanhai H, Ishizuka M, Lowe A, Solis A-G A, Grierson D. Plant J. 1998;15:737–746. doi: 10.1046/j.1365-313X.1998.00251.x. [DOI] [PubMed] [Google Scholar]
- 38.Beclin C, Berthome R, Palauqui J C, Tepfer M, Vaucheret H. Virology. 1998;252:313–317. doi: 10.1006/viro.1998.9457. [DOI] [PubMed] [Google Scholar]
- 39.Sticher L, Hofsteenge J, Milani A, Neuhaus J-M, Meins F., Jr Science. 1992;257:655–657. doi: 10.1126/science.1496378. [DOI] [PubMed] [Google Scholar]
- 40.Dalmay T, Hamilton A, Mueller E, Baulcombe D C. Plant Cell. 2000;12:369–379. doi: 10.1105/tpc.12.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucy A P, Guo H-S, Li W-X, Ding S-W. EMBO J. 2000;19:1672–1680. doi: 10.1093/emboj/19.7.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Houdt H, Van Montagu M, Depicker A. Mol Gen Genet. 2000;263:995–1002. doi: 10.1007/pl00008700. [DOI] [PubMed] [Google Scholar]
- 43.Meins F., Jr Plant Mol Biol. 2000;43:261–273. doi: 10.1023/a:1006443731515. [DOI] [PubMed] [Google Scholar]
- 44.Baulcombe D C. Plant Cell. 1996;8:1833–1844. doi: 10.1105/tpc.8.10.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waterhouse P M, Graham M W, Wang M-B. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Que Q, Wang H-Y, Jorgensen R A. Plant J. 1998;13:401–409. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.