Abstract
High efficiencies of recombination between LoxP elements were initially recorded when the Cre recombinase was expressed in meiotic spermatocytes. However, it was unexpectedly found that LoxP recombination fell to very low values at the second generation of mice expressing Cre during meiosis. The inability of the LoxP elements to serve as recombination substrates was correlated with cytosine methylation, initially in LoxP and transgene sequences, but later extending for distances of at least several kilobases into chromosomal sequences. It also affected the allelic locus, implying a transfer of structural information between alleles similar to the transvection phenomenon described in Drosophila. Once initiated following Cre–LoxP interaction, neither cis-extension nor transvection of the methylated state required the continuous expression of Cre, as they occurred both in germinal and somatic cells and in the fraction of the offspring that had not inherited the Sycp1-Cre transgene. Therefore, these processes depend on a physiological mechanism of establishment and extension of an epigenetic state, for which they provide an experimental model.
Keywords: Cre–LoxP/meiosis/methylation/recombination/transvection
Introduction
Epigenetic modifications are changes in the chromatin structure which result either in the activation or repression of defined regions of the genome without modifying (mutating) the primary nucleotide sequence. An ever increasing number of such modifications are under study in various organisms. In mammals, the two best studied cases, namely the inactivation of one of the X chromosomes in the female and the differential expression of imprinted loci in the paternal and maternal genomes, are both of critical importance in development (for reviews see Henikoff and Comai, 1998; Surani, 1998; Tilghman, 1999; Wolffe and Matzke, 1999). However, our knowledge of the mechanisms by which an epigenetic change is created remains rudimentary. An interesting situation has been described in Drosophila, fungi and higher plants in which an epigenetic modification is induced in one allele when the other one is itself modified. Several instances of this type of interallelic communication have been reported under the denominations of transvection and paramutation. Initially identified in studies of the bithorax complex (Lewis, 1954), transvection effects leading to modifications of gene expression and regulation were subsequently demonstrated in a number of Drosophila genes (for review see Pirrotta, 1999) and in the fungus Neurospora crassa (Aramayo and Metzenberg, 1996). The term paramutation has been used to describe situations in plants in which the phenotype corresponding to a ‘paramutable’ allele is similarly modified due to an interaction with a ‘paramutagenic’ one, the change being maintained after meiotic segregation (Hollick et al., 1997). These effects imply a physical interaction between either adjacent genes or between synapsed alleles at the pachytene stage of meiosis and, when it is the case, in somatic cells (Henikoff and Comai, 1998). A tantalizing question has been whether the same mechanisms are also at work in mammalian genomes, but no clear answer has yet been given.
Our results demonstrate a transvection effect in the mouse. It involves the spreading through the genome of an epigenetic modification: the 5′ methylation of cytosine residues in DNA. Cytosine methylation is one of the most extensively documented modifications in mammalian genomes. Frequently, although not in all cases, it corresponds to a state of transcriptional repression. A complex family of DNA methyltransferases has been identified, whose prototype is the group of proteins encoded at the Dnmt1 locus of the mouse (Mertineit et al., 1998). The fact that homozygous Dnmt1–/– mutants are early lethals is one of the most striking pieces of evidence for an important role in mammalian development (Li et al., 1992). The methylation state of a gene is often modified during development and during gametogenesis, but little is known of the controls responsible for the selective de novo addition (or removal) of methyl groups. Whether or not the possibility of transvection has to be taken into account therefore appears as a relevant question.
In the course of our analysis into the site-specific recombination between LoxP sequences catalyzed by the Cre recombinase expressed at the pachytene stage of meiosis from the stage-specific Sycp1 promoter (Vidal et al., 1998), we demonstrated methylation of cytosines within the LoxP sequences, dependent on Cre expression and inhibiting further recombination. This effect appears to be unique to the meiotic cells, as it has never been described in the numerous studies using Cre–LoxP recombination as a genetic engineering tool either in the somatic tissues of a transgenic mouse or in embryonic stem (ES) cells (for review see Porter, 1998). We then observed that, once initiated during gametogenesis, the methylated state was extended in the following generations from the LoxP sequence into the surrounding chromosomal sequences. These studies were conducted in parallel on two independent target loci: a LoxP insert in the Rxrα locus and one of the widely used ‘Cre monitor’ mouse strains carrying a stop cassette surrounded by LoxP elements (‘floxed’; Mao et al., 1999; Soriano, 1999) in the ROSA26 locus. While the initiation of methylation clearly resulted from a non-physiological situation created in the Sycp1-Cre transgenic males, its extension subsequently occurred in somatic tissues, which do not express the Cre transgene. Moreover, it followed the same pattern in the Cre+ and Cre– progenies of hemizygous parents. Therefore, the spreading of this epigenetic mark appears to result from a cellular mechanism and determining its rules should be informative. One striking observation in this respect was the induction of a methylated state in allelic loci that have not been exposed to the recombinase. Transvection thus appears to occur in the mouse, possibly dependent on chromosome synapsis during meiosis.
Results
Expression of the Cre recombinase in pachytene spermatocytes: efficient recombination of LoxP sites during the first meiosis followed by complete inhibition in the following generations
Recombination between LoxP sequences mediated by the Cre recombinase is of general use as a tool to delete floxed regions in mammalian cells (Sauer and Henderson, 1989). Targeted mutations can in this way be created in vivo, in animals where Cre expression is directed by the appropriate promoter–enhancer systems and/or by hormonal induction of a recombinase–receptor fusion protein (Brocard et al., 1997 and references therein). In a number of the transgenic mouse strains expressing Cre in various somatic tissues described in the literature, recombination frequencies were stably maintained generation after generation.
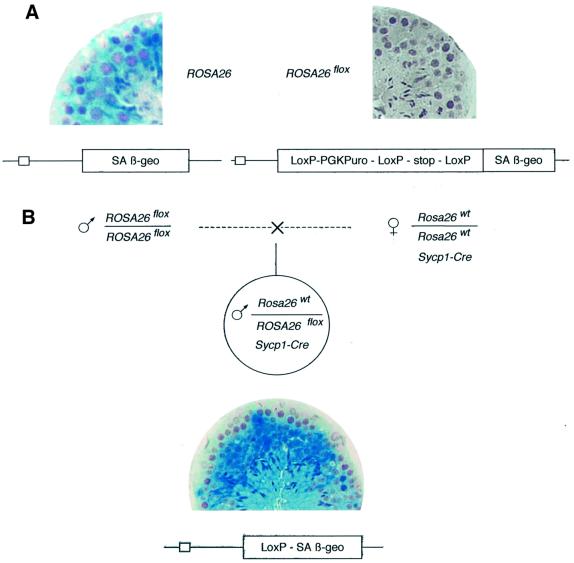
In order to extend this technology to the differentiating germ cells, we generated transgenic animals in which Cre expression was directed by a promoter fragment from the Sycp1 gene exclusively active at the leptotene to early pachytene stages of the male meiosis (Sage et al., 1999). Double transgenic mice were generated that carried both the Sycp1-Cre transgene and the Cre reporter locus ROSA26flox (Mao et al., 1999). Expression of the β-galactosidase coding sequence in ROSA26flox (βgeo fused gene) depends on the prior excision of a floxed stop cassette and can be easily detected, even in a minor fraction of the germ cells, by x-gal assays on testis sections. As shown in Figure 1, results appeared in complete agreement with data previously obtained with other floxed loci (Vidal et al., 1998). In the testes of Cre+ males generated by Sycp1-Cre;Rosa26wt/wt females mated with ROSA26flox/flox males, the stop cassette was excised with high efficiency in pachytene spermatocytes and their derivatives (meiosis II and haploid stages), thus generating the ROSA26del genotype and a β-galactosidase-positive phenotype. As expected from the strict specificity of the promoter, all somatic tissues tested were negative (not shown).
Fig. 1. Efficient excision of a floxed cassette in the testis of a male who inherited the Sycp1-Cre transgene. (A) Left: x-gal staining of a section of an adult (4 months) ROSA26 male mouse showing β-galactosidase expression at all stages of germinal differentiation. Right: no expression is detectable in a male of the same age carrying the floxed locus (ROSA26flox). (B) The indicated cross generated heterozygous ROSA26flox/Rosa26wt;Sycp1-Cre males. x-gal staining shows extensive β-galactosidase accumulation in testicular germ cells starting at the pachytene stage (note that the earliest stages at the periphery of the tubule remain negative), demonstrating excision of the floxed cassette generating the ROSA26del recombined allele.
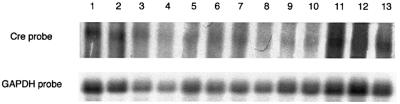
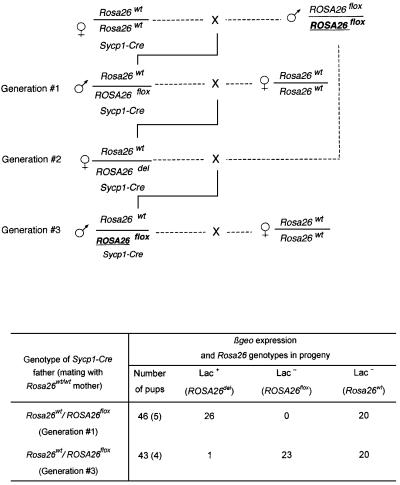
Therefore, it was surprising to observe a sharp decrease in efficiency in the progeny of these Sycp1-Cre males (Figure 2), contrasting with the high frequency of recombination in the progenitors. In these experiments, the locus deleted at the previous generation, with only one remaining LoxP site, had been associated by the appropriate crosses with the original floxed allele (ROSA26del/flox genotype). In spite of the fact that the Sycp1-Cre transgene was maintained, the newly introduced floxed region did not undergo excision during spermatogenesis. Several possible explanations were taken into consideration. We considered the possibility of a loss of Cre expression during successive germ line passages. Northern blot analysis was performed on testis RNA in a series of Sycp1-Cre males, in which excision of the floxed region had taken place, and in the recombination-deficient animals (‘Generation#3’ in Figure 2). Low and variable levels of Cre RNA were detected, as expected from the relatively low strength of the Sycp1 promoter and the small number of expressing cells in the testis, but without any significant difference between males who had or had not recombined (Figure 3). As the levels of recombinase expression appeared to be unchanged in males in which recombination did not take place, we considered the alternative possibility of an epigenetic modification of the LoxP sites, with methylation of cytosine residues as an obvious candidate.
Fig. 2. The floxed cassette is not excised after propagation for more than one generation of the Sycp1-Cre transgene. Same experiment as in Figure 1, but the floxed ROSA26 locus was transferred to a third generation progeny in one of the Sycp1-Cre;ROSA26 families. Indicated in the table are the numbers of β-galactosidase-expressing mice in the male progeny (ROSA26del) and, among the negative phenotypes, the wild-type and floxed genotypes distinguished by PCR analysis. Note that meiosis in the male, indicated as Generation #1, was the first exposure of the floxed locus to the recombinase. The total number of pups analyzed is indicated, with the number of litters shown in parentheses.
Fig. 3. Expression of the Cre transgene in males that have not excised the floxed cassette. Northern blot analysis of testis RNA of males designated as generation #1 (lanes 1–6) and generation #3 (lanes 7–13) in the experiment described in Figure 2. Hybridization was performed with a Cre probe amplified from pBS185 DNA with primers Cre1 and Cre2 and a GAPDH probe as a control (Lopez et al., 1999).
LoxP sites undergo methylation after exposure to the Cre recombinase during meiosis
Although the effect of cytosine methylation on the recombination of LoxP elements has not been previously documented, mutation of two cytosines in the sequence has been found to decrease, to a large extent, recombinase binding (Hartung and Kisters-Woike, 1998). Cytosine methylation was demonstrated at these two CpG sites in the LoxP elements of Sycp1-Cre;ROSA26del males by using bisulfite modification and PCR amplification for detection of modified cytosine residues (Figure 4). Methylation affected the two CpG residues in the LoxP sequence, but, interestingly, was also found in the surrounding chromosomal sequences and not only in CpG but also in CpT and CpA dinucleotides (see also Figure 7). Although recently described in ES cells (Ramsahoye et al., 2000), non-CpG methylation is not considered as usual. In this case, it was clearly confirmed on a total of 24 amplified sequences after bisulfite treatment performed on two males of the same generation. At the first generation in which LoxP recombination has been efficient (Gener ation #1 in Figure 2), distinct methylation patterns were observed in PCR clones derived from testis DNA in several animals of the same generation [Figure 4, 2.1(b) and 2.2(b)]. In the following generation, as one would have expected, a homogeneous pattern was observed in the somatic (tail) DNA of the progeny [in a total of 36 independent sequences established on six animals; Figure 4, 3(b) and 4(b)].
Fig. 4. Cytosine methylation in the LoxP elements of the ROSA26flox locus. Bisulfite treatment and subsequent PCR amplification and sequencing (Olek et al., 1996) were performed to determine the extent of cytosine methylation. Methylated cytosines are detected as cytosines (indicated as bold underlined characters) and the unmethylated bases as thymine nucleotides. u, untreated DNA; b, bisulfite-treated DNA. Sequence 1 shows amplification from testis DNA of a ROSA26 mouse without the Cre transgene (sequences identical in 12 amplified clones per animal and in two animals); sequences 2.1 and 2.2 are different sequences obtained from the ROSA26del testis of Rosa26wt/ROSA26flox male (generation #1 in Figure 2, the two sequences being found among 12 amplified clones for three animals of the same generation), and sequences 3 and 4 show amplification from the somatic (tail) DNA of two groups of three males (six strands sequenced for each animal all showing the same pattern). Amplification was performed with primers Rosa26–1S, 2S, 1R and Rosa26–1S, 2S, ch3R (see Materials and methods).
Fig. 7. Cytosine methylation in the LoxP element of RxrαAF1 and surrounding sequences. Bisulfite treatment and subsequent PCR amplification and sequencing were performed as shown in Figure 4 (u, untreated; b, bisulfite treated). Amplification was performed with primers 285S, 370S and 491R. (A) Testis DNA of heterozygous AF1/+ animals. Sequences 1 and 2, found in equivalent numbers, correspond, respectively, to the wild-type and mutant allele. Upper case, chromosomal sequence; lower case, transgene sequence; LoxP sequence in bold. The results shown were identical in 12 clones sequenced per animal (five animals). (B) Sequence 3 was established from the testis DNA of first generation heterozygotes (Figure 8, top) and sequence 4 from the tail DNA at the following generation. In each case, sequences were identical in 12 clones independently amplified from four distinct males of the same generation. (C) Same analysis performed on Sycp1-Cre;RxrαAF1/AF1 homozygotes at the following generation. Sequence 5, tail DNA; sequence 6, testis DNA. Identical sequences in 12 clones amplified from each one of three males of the same generation. Note that methylation extends into chromosomal sequences.
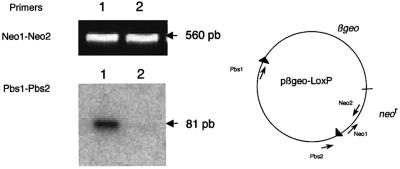
We directly confirmed that CpG methylation in LoxP inhibits Cre recombination. The recombinase was transiently expressed in cultured fibroblasts co-transfected with a Cre expression vector driven by the cytomegalovirus (CMV) early promoter and with a floxed target either unmethylated or methylated in vitro by the SssI CpG methyltransferase. As shown in Figure 5, 72 h after transfection, excision of the floxed DNA was observed only in cells transfected with the unmethylated DNA samples.
Fig. 5. Cytosine methylation of LoxP inhibits Cre recombination. Actively growing STO cells were transfected with a mixture of 1 µg of pBS185 (Cre expression vector) and 0.2 µg of pβgeo–LoxP plasmid DNA. DNA was prepared 72 h after transfection. PCR amplification of a 560 bp fragment with primers Neo1 and Neo2 (1 min at 94°C, 1 min at 60°C, 15 s at 72°C, 25 cycles) was performed as a control on undiluted extracts. PCR analysis with primers PBS1 and PBS2 (1 min at 94°C, 1 min at 56°C, 15 s at 72°C, 30 cycles) was then performed to amplify a fragment of 81 bp diagnostic of the excision of the floxed βgeo region, revealed by hybridization with LoxP oligonucleotide probe (labeled with T4 polynucleotide kinase and [γ-32P]ATP). Experiments were performed in parallel with βgeo-LoxP DNA methylated in vitro by SssI (lane 2) and with the unmethylated DNA (lane 1). Amplification of the diagnostic Pbs1–Pbs2 fragment was performed on a 1:100 dilution of the extract in the case of unmethylated DNA (lane 1) and on undiluted extract in the case of methylated DNA (lane 2). Note that the absence of the amplified Pbs1–Pbs2 fragment in lane 2 provides a control for the absence of the recombined structure in the starting construct.
LoxP methylation in meiotic cells expressing Cre is not unique to the ROSA26 genotype
Inhibition of the recombination process after a first exposure to the recombinase could have resulted from some abnormal structure of the target ROSA26 locus, created by the integration of an exon-trapping vector into an unknown locus (Friedrich and Soriano, 1991). We therefore checked whether similar observations could be made on a better defined genetic structure, with a minimal deviation from the wild-type locus, such as the AF1 mutant of the Rxrα locus (Figure 6). Besides a deletion of intron 2, RxrαAF1 contains a single LoxP site in the third intron of the gene, introduced by homologous recombination followed by excision of a LoxP–neo–LoxP cassette by Cre recombination in ES cells (D.Metzger and P.Chambon, personal communication). Both heterozygous and homozygous AF1 mutants develop normally and are fully viable.
Fig. 6. The AF1 region of Rxrα. Top: an out-of-scale map of the regions encompassing introns 1–4 and 7–10. The two regions are ∼16 kb apart. The 5′ region is mutated in RxrαAF1 by a complete deletion of intron 2 fusing exons 2 and 3, and the insertion of a single LoxP site in intron 3. E, EcoRI restriction sites. Bottom: Southern blot analysis after EcoRI cleavage and hybridization with the Rxrα5′ probe. Lane 1, wild type; lane 2, RxrαAF1/+.
Sycp1-Cre;RxrαAF1 males were generated and mated with RxrαAF1/AF1 females to obtain successive generations of mutant males either with or without the Sycp1-Cre transgene. When the methylation status of the LoxP and surrounding sequences was analyzed by the bisulfite assay, it was again found that methylation of cytosines, initiated within the LoxP sequence, was extending at the second generation into the surrounding chromosomal region (Figure 7).
Spreading of the methylated state: resistance to EcoRI cleavage in RxrαAF1 sequences as a marker
Results described so far indicate that the LoxP sequences and the neighboring DNA sequences undergo methylation starting at the first generation at which the recombinase is expressed in the male germ line. At this stage, the extent of methylation is still limited (Figure 7). Extension of the modified state was then observed starting at the second generation. The finding that methylation-sensitive restriction sites, up to at least 2 kb away from the LoxP site in the surrounding chromosomal region, were inactivated provided useful markers to measure precisely how the methylated state is spreading.
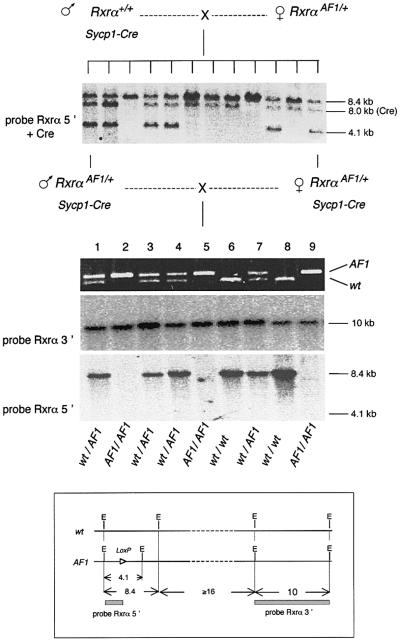
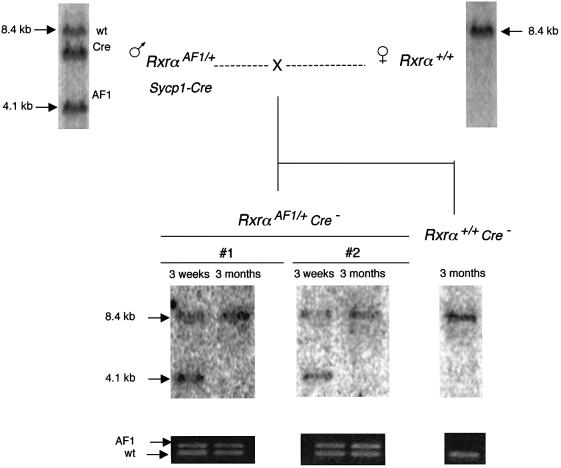
A major change in the Southern blot profile of the AF1 region was observed in routine checks performed at successive generations (Figure 8). EcoRI cleavage generated a 4.1 kb fragment from the region of RxrαAF1, which contains the LoxP element that was detected on Southern blots by the Rxrα5′ probe (Figure 6). Cleavage of the wild-type locus yields an 8.4 kb fragment that hybridizes with the same probe. When a transgenic Sycp1-Cre male was mated with a heterozygous Rxrα+/AF1 female, the distribution of the 4.1 and 8.4 kb fragments in the first generation, confirmed by PCR genotyping, showed the expected Mendelian distribution with the AF1 allele present in one half of the offspring. In the next generation (brother–sister crosses), the expected distribution of homozygous and heterozygous genotypes was again demonstrated by PCR analysis. However, the 4.1 kb EcoRI fragment could not be detected by Southern blot hybridization with the same probe. Only the wild-type-derived 8.4 kb fragment was detected from +/AF1 DNA and no band at all was seen in the lanes corresponding to the AF1/AF1 homozygotes, whereas the Rxrα3′ probe, covering the region from exon 8 to intron 10, 15 kb downstream of the LoxP site, still identified the expected 10 kb fragment on the same blots. The most likely interpretation of these results was that the EcoRI sites surrounding the LoxP element have been either lost or modified. In eukaryotic cells, as in bacteria, the endonuclease is sensitive to cytosine methylation (Brennan et al., 1986) and its inability to cut has been used as a marker to detect site-specific methylation (e.g. Kleymenova et al., 1998). In addition, restriction sites for the methylation-sensitive HpaII endonuclease in the region recognized by the Rxrα5′ probe were converted into a cleavage-resistant form, still recognized by the methylation-insensitive MspI isoschizomer (data not shown). And the conclusion that disappearance of the 4.1 kb EcoRI fragment results from an epigenetic modification was confirmed by experiments performed on primary fibroblast cultures. Starting from embryos in which EcoRI did not cut in the AF1 region, blot analysis showed a fragment of the correct size reappearing at the second to third transfer of the cultures (data not shown). Once established, the methylated state appeared, on the other hand, to be fairly stable in the mouse, at least during three successive generations of breeding in the absence of the Cre transgene.
Fig. 8. The 4.1 kb EcoRI fragment spanning the LoxP sequence of the RxrαAF1 region is not detectable at the second generation following meiotic exposure to the Cre recombinase. Analysis of a litter of nine offspring (middle) generated by two double transgenic first generation Sycp1-Cre;Rxrα+/AF1 parents, both from the same litter of 12 (top). Southern blot analysis of the parents and their littermates (top) showed the presence of the 4.1 kb fragment derived from the AF1 mutant, together with the 8.4 kb EcoRI fragment characteristic of the wild-type locus. Bacterial vector sequences are included in the probe to reveal in addition the Sycp1-Cre transgene (8.0 kb fragment). PCR analysis (middle) conducted with primers RxrAF1-1 and -2 (located on both sides of the AF1 LoxP site) demonstrated the expected distribution of homozygous (2, 5 and 9) and heterozygous (1, 3, 4 and 7) genotypes. Southern blot analysis (bottom) detected the 10 kb fragment hybridizing with the Rxrα3′ probe. The Rxrα5′ probe detected the 8.4 kb fragment from the wild-type locus, but not the 4.1 kb fragment from the AF1 allele. Hybridization of a probe for the Cre gene (not shown) indicated inheritance of the transgene only by animals 5, 8 and 9. Insert: simplified scheme showing the relative positions of the restriction sites and probes within the locus.
Spreading of the methylated state: cis extension in chromosomal sequences
Disappearance of the 4.1 kb EcoRI fragment provided a marker of the methylated state in the Rxrα locus, in all cases strictly correlated with inactivation of the LoxP sites. Like LoxP methylation itself, it was seen only after prior exposure of the locus to the recombinase in meiotic cells. AF1 mice, which had not been derived from Sycp1-Cre;RxrαAF1 males, always showed the complete EcoRI profile, as well as Sycp1-Cre;RxrαAF1 females, which do not express the transgene (Sage et al., 1999), and their progeny. The modification was not dependent on continuous exposure to the recombinase, since it occurred to the same extent in the Cre+ and Cre– fractions of the progeny (Figure 8). Furthermore, it was observed to progress in somatic tissues during the development of a young mouse to an adult. One example illustrated in Figure 9 is that of two young Sycp1-Cre;RxrαAF1/+ mice in which the 4.1 kb EcoRI fragment characteristic of the unmethylated site was present in tail DNA prepared at the age of 3 weeks. When DNA was prepared from a second tail biopsy 3 months later, the fragment was not detectable anymore in either of the two mice, thus showing that methylation had progressed in the somatic DNA in the time elapsed. In the 4-month-old male parent, the 4.1 kb fragment was still present in tail DNA, again indicating that the methylated state was spreading only after an initial exposure to the recombinase during meiosis and was not related to aging per se.
Fig. 9. Somatic propagation of the methylated state. A first generation Sycp1-Cre;Rxrα+/AF1 male was mated with a wild-type female. PCR analysis with primers RxrAF1-1 and -2 and Southern genotyping with the Rxrα5′ probe were performed at 2 weeks on the tail DNA of two offspring. It showed the complete EcoRI pattern, including the 4.1 kb fragment encompassing the LoxP site. The same analysis performed 3 months later showed the same pattern of amplified fragments, but the 4.1 kb EcoRI fragment is no longer detected.
Transfer of the methylated state to the allelic locus: a case of transvection in the mouse
Propagation in trans of the methylated state to the allelic region of the homologous chromosome was demonstrated in RxrαAF1 using resistance to EcoRI cleavage as an assay of the methylated state. In the experiment shown in Figure 10, a first generation Sycp1-Cre;RxrαAF1/+ male with unmethylated EcoRI sites was crossed with a homozygous RxrαAF1/AF1 female. In the progeny, as shown in previous sections, the AF1 locus inherited from the mother was sensitive to EcoRI cleavage. Only half of the progeny inherited the Sycp1-Cre transgene and a Cre–;RxrαAF1/+ male was mated with the parental homozygous RxrαAF1/AF1 female. Surprisingly, the 4.1 kb EcoRI fragment was not found in the RxrαAF1/AF1 progeny, implying that the unmethylated allele from the mother has been converted into the methylated state in the absence of Cre. In other words, the LoxP-containing region, which had been initially modified following its exposure to Cre during meiosis, was able to induce modification of the allelic locus even though the recombinase was no longer present.
Fig. 10. Transvection of the methylated state mediated by the RxrαAF1 allele. A first generation Sycp1-Cre;RxrαAF1/+ male was mated with a homozygous RxrαAF1/AF1 female. A back-cross was performed to re-introduce one of the AF1 alleles from the mother into one of the Cre–;RxrαAF1/+ offspring. The EcoRI pattern with the Rxrα5′ probe indicated the modification of the restriction site in the alleles underlined, which have not been exposed to the recombinase.
A similar instance of interallelic communication was demonstrated between ROSA26 loci. It can actually be noted that the result of the experiment shown in Figure 2, which first demonstrated a decrease in recombination efficiency, implies an effect of the ROSA26del allele in inducing methylation of ROSA26flox as early as the first generation in the presence of Sycp1-Cre. In other words, transvection was mediated by the del allele with one methylated LoxP site. It was demonstrated further in the same series of crosses that it may also be mediated by the wild-type Rosa26 locus with no LoxP element. In this case (Figure 11), the males at generation #3 received one copy of the ROSA26flox allele that has never been exposed to the recombinase, from the same progenitor as that extensively recombined at generation #1. The only difference was that it is present together with a copy of the wild-type allele that had been exposed to the recombinase for the previous three meiotic generations. In spite of the apparent identity of the generation #1 and #3 genotypes, the floxed allele in the latter was extensively resistant to Cre recombination, and bisulfite analysis confirmed CpG methylation in the LoxP sequence (data not shown). A signal resulting in the methylation of the floxed locus had therefore been transmitted by the wild-type allele.
Fig. 11. Transvection of the methylated state mediated by the Rosa26wt locus. In the same experiment as in Figure 2, the ROSA26del allele was replaced at generation #3 by a floxed allele (bold underlined) that had not been previously exposed to the recombinase. Note the apparently identical genotypes of the generation #1 and #3 males. Analysis of the progeny was performed as described in Figure 2.
Discussion
Studies on Cre–LoxP recombination in pachytene spermatocytes, the stage of meiosis at which chromosomal pairing (initiated at the zygotene stage) has been achieved, led us to the unexpected finding that, in this particular cell type, recombination was accompanied by methylation of cytosines. The change was made apparent by the fact that the methylated elements are no longer substrates for the recombinase. The initial establishment of this epigenetic mark is of interest by itself, but we do not have much indication on its mechanistic aspects, which will be the subject of further studies. With respect to these aspects, only a few remarks can be made at this stage.
One clear feature of the methylation process, in this case, is the minor, but clear, occurrence of modified cytosines 5′ of either A or T, in addition to the more frequent CpG doublets. The number of amplified clones sequenced (Figures 4 and 7) excludes a technical problem. The same observation has been made recently in ES cells, with the proposal that the Dnmt3a enzyme might be the methyltransferase involved (Ramsahoye et al., 2000).
Methylation of LoxP sites in cells that express the recombinase has not been reported so far. Our current hypothesis is that it is unique to the meiotic spermatocyte. In fact, because the progeny of a cellular genome in a somatic tissue cannot be followed in the successive generations in the same way as they can during gametogenesis, it is difficult to ascertain whether Cre expression in a somatic tissue induces methylation of LoxP sites and prevents further recognition by the recombinase. However, there is no indication that would suggest such a conclusion in the number of reports of Cre-induced deletion of a floxed sequence. There is even one instance in which we can safely conclude that this is not the case, namely the operation of excising a selection cassette in ES cells by the transient expression of Cre, one example being the construction of the RxrαAF1 mutant used in our experiments (D.Metzger and P.Chambon, personal communication).
The observed methylation of LoxP sites does not appear as a property of one specific mouse genotype, as is the case for the methylation of a V(D)J recombination site by the modifier Ssm1 allele of the C57BL/6 mouse (Engler et al., 1991). The present results were obtained with mice of such diverse genetic backgrounds as C57BL/6, DBA/2, 129/Sv and their hybrids, and we never observed segregation of methylated and unmethylated loci upon successive breeding with other supposedly non-methylating strains.
Inhibition of recombination by DNA methylation has been reported in several instances. It was described in Ascobolus, and a mechanistic relationship between methylation and recombination has been suggested (Colot et al., 1996; Maloisel and Rossignol, 1998). Another example is the inhibition by methylation of V(D)J recombination in immunoglobulin genes (Engler and Storb, 1999). Methylation has been more generally proposed to play a role in maintaining the integrity of eukaryotic genomes by preventing recombination within their large repetitious fractions (Colot and Rossignol, 1999). Establishing whether, in the case of the Cre–LoxP interaction, it is the binding of the protein that is prevented by methylation will require further studies. On the other hand, we already know that a complete recombination event is not required for methylation to be initiated since the single LoxP site present in RxrαAF1 is sufficient.
With all the uncertainties regarding the raison d’être and the mechanism of initiation of methylation within LoxP sequences, the main point of the present report is the possibility offered of following the spreading of an epigenetic modification through a mammalian genome. It must be noted that while inducing Cre expression in a mouse spermatocyte is clearly an artificial situation, the subsequent propagation of the methylated state occurs in somatic tissues that do not express the recombinase. Once initiated in Cre-positive meiotic cells, methylation proceeds in the Cre– as well as the Cre+ fractions of the progeny; therefore, it must reflect a normal cellular mechanism. Some elements can be already delineated from the present results. The most striking is that the modification does not extend only into the neighboring regions of the same chromosome, but also to the allelic region of the homologous chromosome. Transvection, paramutation or trans-sensing are all modulations in the state of the chromatin, in the levels and regulation of expression of one allele dictated by the other one in a paired structure. They have been identified first in Drosophila, then in lower eukaryotes, but although suggested by studies on imprinted domains (LaSalle and Lalande, 1996; Forne et al., 1997), never clearly demonstrated in a mammalian genome.
The mode of propagation of the methylated state, its possible directionality and boundaries, and the requirements for transvection to the allelic locus are all subjects for further studies. One important question is the nature of the modification of the inducing (‘paramutagenic’) wild-type allele in the transvection experiments shown in Figures 10 and 11. It may be methylation as much as it may be another ‘mark’ of unknown nature. In Rxrα, we have not so far observed extension of methylation to the EcoRI site(s) of the wild-type allele, which maintains the 8.4 kb EcoRI fragment. Establishing whether or not the inducing allele is itself methylated will require studies over long distances and, first, the establishment (in progress) of the complete genomic sequences of the Rxrα and ROSA26 loci.
An additional series of conclusions are of practical interest for the design of experiments using the meiotic expression of Cre as a tool for genomic engineering, for instance the ‘TAMERE’ system of targeted interchromosomal recombination (Hérault et al., 1998). Our results imply that one cannot propagate Sycp1-Cre males with genomic LoxP sites for successive generations without a rapid loss in recombination efficiency. It would now appear necessary to introduce the Sycp1-Cre transgene into the genome of a male with the required LoxP configuration. One round of breeding will then produce the recombinants without increasing the number of generations of Cre-expressing mice. Alternatively, one may consider maintaining the double transgenic Sycp1- Cre;LoxP genotypes through the female germ line. The Sycp1 promoter sequence driving the expression of the Cre transgene is a genomic fragment extending from –722 to +102 relative to the transcription start, which, unlike the endogenous promoter, is not active during the female meiosis (Sage et al., 1999). Accordingly, it was always found that the LoxP sites remain intact when transferred through the female germ line.
Materials and methods
Transgenic mice
The RxrαAF1 and RxrαAF2(LNL) (Mascrez et al., 1998) transgenic mouse families were kindly provided by P.Chambon, and the ROSA26flox transgenic strain by S.Orkin. The genetic backgrounds are B6D2F1 (Sycp1-Cre lines), C57BL/6 (Rxrα mutants) and 129/Sv (ROSA26 lines). The different alleles of the Rosa26 locus are designated Rosa26wt for the wild-type locus (no βgeo insert), ROSA26 for the locus with the βgeo proviral insertion, ROSA26flox for the locus in which a LoxP–stop–LoxP cassette prevents β-galactosidase expression and ROSA26del for the locus after excision of the cassette, leaving one LoxP site in front of the βgeo coding sequence (Mao et al., 1999).
Cell culture and transfection
STO cells (Hogan et al., 1994) were grown in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal calf serum (Hyclone). Transfection was performed with Fugene6 according to the manufacturer’s instructions (Boehringer Mannheim).
Plasmids
In the pβgeo–LoxP plasmid, the fusion gene βgeo (Friedrich and Soriano, 1991) was inserted between two LoxP sites. This plasmid was constructed by inserting a HindIII–NotI fragment corresponding to the coding sequence from the pβgeo plasmid (a gift from P.Soriano) into the HindIII–BamHI sites of plasmid pBS112 containing two LoxP sites (a gift from B.Sauer). In plasmid pBS185 (also a gift from B.Sauer), the Cre gene was expressed under the control of the CMV early promoter.
In vitro methylation of cytosine
The CpG methylase SssI (New England BioLabs) was used following the manufacturer’s instructions.
DNA sequencing
Purified PCR products were first subcloned in pGEM-T Easy vector System I (ref. A1360; Promega). The purified plasmids were then sequenced with the DNA sequencing system kit Big Dye Terminator (ref. 4303149) according to the manufacturer’s instructions. Sequences were analyzed with the PE Biosystems ABI PRISM 310 genetic analyzer.
Southern blot analysis
Southern blot analysis and hybridization were performed on 5 µg of DNA as described (Sambrook et al., 1989).
Northern blot analysis
Total RNA was prepared from decapsulated normal testes by the acid guanidinium thiocyanate method (Chomczynski and Sacchi, 1987). RNA concentrations were measured spectrophotometrically and the quality of the purified RNA preparations was checked by UV examination after agarose electrophoresis and ethidium bromide staining. Total RNA (20 µg) from testes or from cells in culture was electrophoresed in 1% agarose gels containing 1× TAE and 0.17 vol of 37% formaldehyde. RNAs were transferred to Hybond N+ (Amersham) membrane by capillary blotting with 20× SSC. The filters were rinsed with 2× SSC and RNAs were fixed on membranes by UV irradiation (Ultraviolet crosslinker; Amersham). The filters were prehybridized in 5× SSC, 50% formamide, 50 mM HEPES pH 6.8, 2 mM EDTA, 5× Denhart’s and 0.1% SDS for 4 h at 65°C, and hybridized overnight at 65°C in 10% dextran sulfate, 2× Denhart’s, 5× SSC, 50 mM EDTA, 50 mM HEPES pH 6.8, 40% formamide, 0.2% SDS and 50 µg/ml sheared thymus DNA, with random primed [α-32P]dCTP-labeled DNA. Filters were subsequently washed at 20°C in 2× SSPE, 0.1% SDS, and at 65°C in 1× SSPE, 0.1% SDS, and exposed to Kodak film for 2–5 days at –70°C with an intensifying screen.
Reverse transcription and PCR amplification
Total RNA (1 µg) was reverse transcribed using the MuLV reverse transcriptase and PCR amplification was performed with the Taq DNA polymerase according to the manufacturer’s instructions (Boehringer Mannheim). The pairs of oligonucleotide primers used, the polymerization conditions and expected product size are given in Table I.
Table I. Oligonucleotide primers, polymerization conditions and expected product size.
| Oligonucleotide primers | Polymerization conditions | Expected product size (bp) |
|---|---|---|
| Sa(5′-GATGATGTCATACTTATCCTG-3′)and Pgk(5′-GAAAAGCGCCTCCCCTACCCG-3′) | 94°C, 30 s, then 56°C, 30 s, 72°C, 30 s, 35 cycles | 288 |
| Rosa26-1S[5′-GA(CT)GGTAT(CT)GATAAGTTTTAGAG-3′], Rosa26-2S(5′-GGGGTTGTAGTTTAAGTAGT-3′)and Rosa26-1R(5′-CTTAAAATAACCCATCCCCTC-3′) | first PCR round: 1S+1R: 94°C, 1 min, 58°C, 1 min 72°C,1 min, 35 cycles; second round: 2S+1R: 94°C, 30 s, 56°C,30 s, 72°C, 30 s, 35 cycles | 158 |
| Sa and β-gal(5′-CGCTTGAGCAGCTCCTTG-3′) | 94°C, 30 s, 56°C, 30 s, 72°C 30 s, 35 cycles | 297 |
| Rosa26-ch3R[5′-CAAACTATCT(AT)TTCCCC(GA)AATC-3′] | first PCR reaction 1S+ch3R: 94°C, 1 min, 58°C, 1 min,72°C, 1 min, 35 cycles; second round of PCR, 2S+ch3R:94°C, 30 s, 56°C, 30 s, 72°C, 30 s, 35 cycles | 152 |
| RxrAF1-1(5′-GTGAGGTGGACATTAGGATGG-3′)and RxrAF1-2(5′-TCAGCCATCCATGCCTACATC-3′) | 94°C, 30 s, 62°C, 30 s, 72°C, 30 s, 35 cycles | 141 (if LoxP site present) and 88 (from wild type) |
| AF1-ch3-285S(5′-GAGAAGTAAGAGTTTAGGGTAG-3′),AF1-ch3-370S(5′-GTGAGGTGGATATTAGGATGG-3′)and AF1-ch3-491R(5′-CTCAACCATCCATACCTACATC-3′) | first round of PCR, 285S+491R: 94°C, 1 min, 58°C,1 min, 72°C, 1 min, 35 cycles; second round of PCR,370S/491R: 94°C, 30 s, 56°C, 30 s, 72°C, 30 s, 35 cycles | |
| Tkneo1(5′-GAAGGCGATGCGCTGCGAAT-3′)and Tkneo2(5′-GGTTCTCCGGCCGCTTGGGGT-3′) | 94°C, 1 min, 62°C, 1 min, 72°C, 1 min, 35 cycles | 700 |
| Cre1(5′-TGAGGACATGTTCAGGGATC-3′)and Cre2(5′-CAGCCACCAGCTTGCATGATCAG-3′) | 94°C, 1 min, 58°C, 1 min, 72°C, 1 min, 35 cycles | 800 |
| Neo1(5′-GCGATACCGTAAAGCACGAG-3′)and Neo2(5′-GAATGAACTGCAGGACGAGG-3′) | 94°C, 1 min, 60°C, 1 min, 72°C, 1 min, 35 cycles | 560 |
| PBS1(5′-GAATTCCGATCATTTCAATAAC-3′)and PBS2(5′-CCTCGAGGGGACCTATAAAC-3′) | 94°C, 1 min, 57°C, 1 min, 72°C, 1 min, 35 cycles | 81 |
| Gapdh1(5′-CACCACCAACTGCTTAGCC-3′)and Gapdh2(5′-CGGATACATTGGGGGTAGG-3′) | 94°C, 1 min, 58°C, 1 min, 72°C, 1 min, 35 cycles | 270 |
| LoxP(5′-ATAACTTCGTATAATGTATGCTATACGAAGT-3′), for hybridizationwith LoxP-containing fragment at 56°C |
Image analysis and densitometric measurements were performed using Adobe Photoshop 5.5 (Adobe Systems Inc.).
Bisulfite determination of methylcytosine
Levels of DNA methylation in chromosomal DNA of tail and testes were determined as described (Olek et al., 1996). For each determination, 2–5 animals of the same generation, but from independent crosses, were analyzed. PCR amplification was performed using the primers indicated in each case and 12 independent clones were sequenced. Methylated or unmethylated states of cytosines retained in the consensus sequences were identical in every one of the 24–60 sequencing reactions performed for each condition.
Acknowledgments
Acknowledgements
We thank Valérie Grandjean for critical reading of the manuscript, Luc Martin for expert help in the bisulfite determination of methylcytosine, and Daniel Metzger and Pierre Chambon for the gift of the RxrαAF1 mouse. The technical assistance of Eric Couchi, Mireille Cutajar, Yan Fantei, Max Radjkumar and Fariba Ranc is gratefully acknowledged. This work was made possible by grants from Association pour la Recherche sur le Cancer and GIP Aventis to M.R. and F.C.
References
- Aramayo R. and Metzenberg,R.L. (1996) Meiotic transvection in fungi. Cell, 86, 103–113. [DOI] [PubMed] [Google Scholar]
- Brennan C.A., Van Cleve,M.D. and Gumport,R.I. (1986) The effects of base analogue substitutions on the cleavage by the EcoRI restriction endonuclease of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J. Biol. Chem., 261, 7270–7278. [PubMed] [Google Scholar]
- Brocard J., Warot,X., Wendling,O., Messaddeq,N., Vonesch,J.L., Chambon,P. and Metzger,D. (1997) Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc. Natl Acad. Sci. USA, 94, 14559–14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. and Sacchi,S. (1987) Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Colot V. and Rossignol,J.L. (1999) Eukaryotic DNA methylation as an evolutionary device. BioEssays, 21, 402–411. [DOI] [PubMed] [Google Scholar]
- Colot V., Maloisel,L. and Rossignol,J.L. (1996) Interchromosomal transfer of epigenetic states in Ascobolus: transfer of DNA methylation is mechanistically related to homologous recombination. Cell, 86, 855–864. [DOI] [PubMed] [Google Scholar]
- Engler P. and Storb,U. (1999) Hypomethylation is necessary but not sufficient for V(D)J recombination within a transgenic substrate. Mol. Immunol., 36, 1169–1173. [DOI] [PubMed] [Google Scholar]
- Engler P., Haasch,D., Pinkert,C.A., Doglio,L., Glymour,M., Brinster,R. and Storb,U. (1991) A strain-specific modifier on mouse chromosome 4 controls the methylation of independent transgene loci. Cell, 65, 939–947. [DOI] [PubMed] [Google Scholar]
- Forne T., Oswald,J., Dean,W., Saam,J.R., Bailleul,B., Dandolo,L., Tilghman,S.M., Walter,J. and Reik,W. (1997) Loss of the maternal H19 gene induces changes in Igf2 methylation in both cis and trans. Proc. Natl Acad. Sci. USA, 94, 10243–10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G. and Soriano,P. (1991) Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev., 5, 1513–1523. [DOI] [PubMed] [Google Scholar]
- Hartung M. and Kisters-Woike,B. (1998) Cre mutants with altered DNA binding properties. J. Biol. Chem., 273, 22884–22891. [DOI] [PubMed] [Google Scholar]
- Henikoff S. and Comai,L. (1998) Trans-sensing effects: the ups and downs of being together. Cell, 93, 329–332. [DOI] [PubMed] [Google Scholar]
- Hérault Y., Rassoulzadegan,M., Cuzin,F. and Duboule,D. (1998) Engineering chromosomes in mice through targeted meiotic recombination (TAMERE). Nature Genet., 20, 381–384. [DOI] [PubMed] [Google Scholar]
- Hogan B., Costantini,F. and Lacy,L. (1994) Manipulating the Mouse Embryo—A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hollick J.B., Dorweiler,J.E. and Chandler,V.L. (1997) Paramutation and related allelic interactions. Trends Genet., 13, 302–308. [DOI] [PubMed] [Google Scholar]
- Kleymenova E.V., Yuan,X., LaBate,M.E. and Walker,C.L. (1998) Identification of a tumor-specific methylation site in the Wilms tumor suppressor gene. Oncogene, 16, 713–720. [DOI] [PubMed] [Google Scholar]
- LaSalle J.M. and Lalande,M. (1996) Homologous association of oppositely imprinted chromosomal domains. Science, 272, 725–728. [DOI] [PubMed] [Google Scholar]
- Lewis E.B. (1954) The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat., 88, 225–239. [Google Scholar]
- Li E., Bestor,T.H. and Jaenisch,R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915–926. [DOI] [PubMed] [Google Scholar]
- Lopez P., Vidal,F., Rassoulzadegan,M. and Cuzin,F. (1999) A role of inhibin as a tumor suppressor in Sertoli cells: down-regulation upon aging and repression by a viral oncogene. Oncogene, 18, 7303–7309. [DOI] [PubMed] [Google Scholar]
- Maloisel L. and Rossignol,J.L. (1998) Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev., 12, 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Fujiwara,Y. and Orkin,S.H. (1999) Improved reporter strain for monitoring cre recombinase-mediated DNA excisions in mice. Proc. Natl Acad. Sci. USA, 96, 5037–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascrez B., Mark,M., Dierich,A., Ghyselinck,N., Kastner,P. and Chambon,P. (1998) The RXRα ligand-dependent activation function 2 (AF-2) is important for mouse development. Development, 125, 4691–4707. [DOI] [PubMed] [Google Scholar]
- Mertineit C., Yoder,J.A., Taketo,T., Laird,D.W., Trasler,J.M. and Bestor,T.H. (1998) Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development, 125, 889–897. [DOI] [PubMed] [Google Scholar]
- Olek A., Oswald,J. and Walter,J. (1996) A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res., 24, 5064–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. (1999) Transvection and chromosomal trans-interaction effects. Biochim. Biophys. Acta, 1424, M1–M8. [DOI] [PubMed] [Google Scholar]
- Porter A. (1998) Controlling your losses: conditional gene silencing in mammals. Trends Genet., 14, 73–79. [DOI] [PubMed] [Google Scholar]
- Ramsahoye B.H., Biniszkiewicz,D., Lyko,F., Clark,V., Bird,A.P. and Jaenisch,R. (2000) Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA, 97, 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J., Martin,L., Meuwissen,R., Heyting,C., Cuzin,F. and Rassoulzadegan,M. (1999) Temporal and spatial control of the Sycp1 gene transcription in the mouse meiosis: regulatory elements active in the male are not sufficient for expression in the female gonad. Mech. Dev., 80, 29–39. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sauer B. and Henderson,N. (1989) Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res., 17, 147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet., 21, 70–71. [DOI] [PubMed] [Google Scholar]
- Surani M.A. (1998) Imprinting and the initiation of gene silencing in the germ line. Cell, 93, 309–312. [DOI] [PubMed] [Google Scholar]
- Tilghman S.M. (1999) The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell, 96, 185–193. [DOI] [PubMed] [Google Scholar]
- Vidal F., Sage,J., Cuzin,F. and Rassoulzadegan,M. (1998) Cre expression in primary spermatocytes: a tool for genetic engineering of the germ line. Mol. Reprod. Dev., 51, 274–280. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. and Matzke,M.A. (1999) Epigenetics: regulation through repression. Science, 286, 481–486. [DOI] [PubMed] [Google Scholar]