Abstract
Class 1 peptide release factors (RFs) in Escherichia coli are N5-methylated on the glutamine residue of the universally conserved GGQ motif. One other protein alone has been shown to contain N5-methylglutamine: E.coli ribosomal protein L3. We identify the L3 methyltransferase as YfcB and show that it methylates ribosomes from a yfcB strain in vitro, but not RF1 or RF2. HemK, a close orthologue of YfcB, is shown to methylate RF1 and RF2 in vitro. hemK is immediately downstream of and co-expressed with prfA. Its deletion in E.coli K12 leads to very poor growth on rich media and abolishes methylation of RF1. The activity of unmethylated RF2 from K12 strains is extremely low due to the cumulative effects of threonine at position 246, in place of alanine or serine present in all other bacterial RFs, and the lack of N5-methylation of Gln252. Fast-growing spontaneous revertants in hemK K12 strains contain the mutations Thr246Ala or Thr246Ser in RF2. HemK and YfcB are the first identified methyltransferases modifying glutamine, and are widely distributed in nature.
Keywords: hemK/methyltransferase/peptide release factor/yfcB
Introduction
Translation termination occurs when the elongating ribosome encounters a stop signal on mRNA (for a review see Kisselev and Buckingham, 2000). Two class 1 protein release factors (RFs), with different but overlapping specificity of recognition, are required for termination in bacteria: RF1 recognizes UAG and UAA, and RF2 recognizes UAA and UGA. In eukarya and archaea, a single protein, eRF1 and aRF1, respectively, recognizes all three stop codons. How the stop codons are recognized in any of these systems remains poorly understood, although a tripeptide motif has been suggested to define the identity of class 1 peptide RFs from bacteria (Ito et al., 2000).
Eukaryotic and archaeal RFs are clearly homologous, but their primary and secondary structures differ radically from their bacterial counterparts. In fact, it is now recognized that the RFs from all kingdoms share only one sequence motif. This is the universal Gly–Gly–Gln (GGQ) tripeptide, flanked by sequences that are particularly rich in basic amino acids in eRF1 and aRF1 (Frolova et al., 1999). Substitutions affecting the glycine residues in GGQ lead to almost complete loss of release activity in both human (Seit Nebi et al., 2000) and Escherichia coli (L.Mora and A.Zavialov, unpublished results) RFs. In contrast, the glutamine residue can be changed to certain other amino acids with retention of partial release activity in vitro (Seit Nebi et al., 2000; L.Mora, unpublished results). However, none of these mutants substituted at the glutamine position is able to complement RF1 or RF2 themosensitive mutants in E.coli in vivo (L.Mora, unpublished results), or substitute for eRF1 in Saccharomyces cerevisiae (Song et al., 2000).
The glutamine residue in the GGQ motif is modified post-translationally to N5-methylglutamine in both RF1 and RF2 in E.coli (Dinçbas-Renqvist et al., 2000). This modification has an important stimulatory effect on the release activity of RF2, and explains earlier observations of a striking negative correlation between the specific activity of RF2 and the degree of overproduction of the factor (Tate et al., 1993). Overproduction of E.coli RF2 leads to a non-modified protein, presumably due to insufficient activity of the RF methyltransferase (MTase), and such overproduction of poorly active RF2 is highly inhibitory to growth in E.coli K12 strains (Uno et al., 1996; Dinçbas-Renqvist et al., 2000). These experiments have also revealed a striking functional interplay between the methylation of Gln252 in RF2 and the nature of the amino acid at position 246, four residues from the GGQ motif towards the N-terminus. The activity of RF2 in K12 strains is low compared with other E.coli strains due to the presence of a threonine residue at position 246, in place of an alanine or serine residue found in all other bacterial RFs. In vitro, the losses in activity due to the lack of glutamine methylation and to the presence of Thr246 instead of Ala246 are cumulative. Thus, overproduction of RF2 Ala246 has little inhibitory effect on cell growth (Uno et al., 1996; Dinçbas-Renqvist et al., 2000).
So far, both the extent of RF methylation among different organisms and the identity of the RF MTase are unknown. A single instance of glutamine transmethylation has been reported in the literature, involving ribosomal protein L3 in E.coli (Lhoest and Colson, 1977; Colson et al., 1979).
Here, we identify the MTases for both ribosomal protein L3 and the class 1 RF in E.coli. The L3 MTase is encoded by yfcB, and the RF MTase is encoded by hemK. The gene hemK, situated immediately downstream of and co-expressed with prfA (i.e. the gene for RF1), was suggested initially to encode a protoporphyrinogen oxidase (Nakayashiki et al., 1995). YfcB and HemK are the first N5-glutamine MTases to be identified.
Results
Identification of the gene encoding ribosomal protein L3 methyltransferase
An intensive search of the E.coli ribosome showed that at least 10 proteins of the 50S subunit become methylated (Chang and Chang, 1975). The modification occurs to varying extents and on several amino acids, with lysine being the most frequent target. In the case of ribosomal protein L3, methylation occurs on the nitrogen of the side chain amide group (Lhoest and Colson, 1977). Until the recent finding of the same modified amino acid in E.coli RFs (Dinçbas-Renqvist et al., 2000), this remained the only report in the literature of N5-methylglutamine. A mutant strain, unable to methylate L3, showing cryosensitive ribosome assembly and cell growth, allowed the putative L3 MTase gene prmB to be mapped to the aroC region at 50 min on the chromosome (Colson et al., 1979). A three-factor cross suggested the order aroC–prmB–purF.
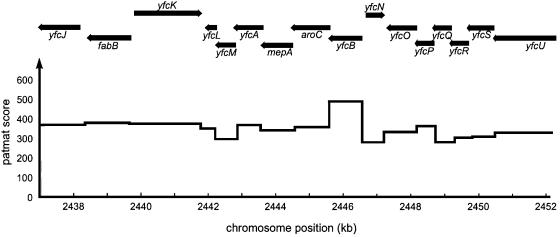
To identify the L3 MTase gene, we analysed coding sequences in the aroC region for possible S-adenosylmethionine (AdoMet)-binding motifs. Such sites are characterized by a series of three moderately conserved sequence motifs contained in a region of ∼100 amino acids (Liu et al., 2000). The first of these, a glycine-rich motif [vL(D/E)xgxGxg], is the most conserved and is therefore the best suited for searches with site-specific scoring matrices with the blocks technique (Wallace and Henikoff, 1992). A block covering motif 1 was constructed from a set of six protein MTase sequences (see Materials and methods) using Blockmaker (Henikoff et al., 1995) and employed to search coding sequences in the aroC region using the application PATMAT (Wallace and Henikoff, 1992). The results (Figure 1) show that the only reasonable candidate for an AdoMet-binding protein close to aroC is encoded by the gene yfcB, located in the same operon and immediately upstream of aroC, which possesses the partial match ILDMCTGSG to motif 1. This gene was therefore sequenced both in the mutant unable to methylate ribosomal protein L3 and in the parental wild-type strain. Sequencing revealed a single base change in the mutant that affected motif 1, altering the only completely conserved amino acid, the first glycine residue, to aspartate (mutant motif: ILDMCTDSG). Willcock et al. (1994) have shown that the same mutation in an adenine MTase abolishes its AdoMet binding. These data strongly suggested that, indeed, yfcB encodes the L3 MTase PrmB. The gene order in this case would be purF–aroC–prmB.
Fig. 1. Search for prmB in the vicinity of aroC. The pattern-matching application PATMAT (Wallace and Henikoff, 1992) was employed to locate coding sequences near aroC containing a potential glycine-rich AdoMet-binding motif 1 sequence. The pattern block used was derived from six protein MTases from E.coli. The highest score is associated with yfcB immediately upstream of aroC.
The cloned yfcB gene complements cryosensitive ribosome assembly in the yfcB mutant strain
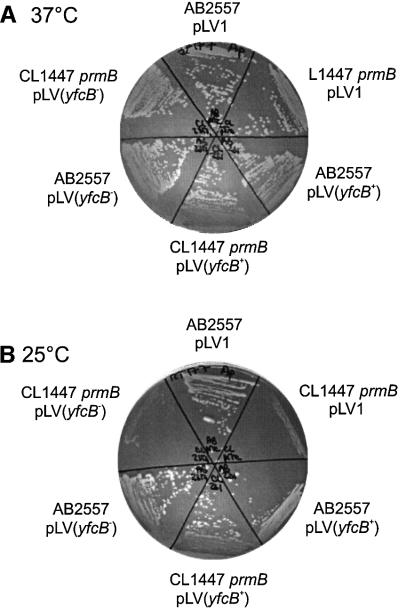
The cryosensitive growth observed by Colson et al. (1979) was ascribed to a prmB mutation leading to loss of L3 methylation. However, at the time, it could not be excluded that the nitrosoguanidine mutagenesis had created a secondary mutation close to prmB. To confirm that yfcB indeed encoded PrmB, the gene was first cloned from both the wild-type strain AB2557 and the prmB mutant strain CL1447. The genes were cloned into the plasmid pLV1 under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Ptrc promoter. Each of these plasmids was transformed into prmB mutant and wild-type strains. At 25°C, the growth defect of the prmB strain was complemented by the plasmid carrying wild-type yfcB but not by the plasmid carrying mutant yfcB or by the control plasmid without the insert (Figure 2). Induction by IPTG was not necessary to observe complementation, as sufficient leakiness of the Trc promoter occurs to express yfcB. This result confirms that expression of yfcB suppresses the growth defect and that the cryosensitivity was due to the absence of the L3 methylation.
Fig. 2. Complementation of prmB cryosensitive growth. The prmB strain CL1447 (Colson et al., 1979) and its wild-type parental strain AB2557 were transformed with plasmids carrying a wild-type yfcB gene [pLV(yfcB+)], a mutant yfcB gene [pLV(yfcB–)] or no insert (pLV1). The plate incubated at 25°C shows the cryosensitive growth of the prmB strain and complementation of the phenotype by plasmid pLV(yfcB+).
Methylation in vitro of ribosomal protein L3 by overexpressed YfcB
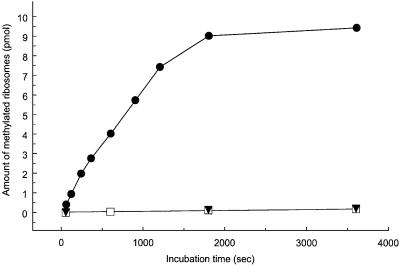
To show more directly that YfcB is the L3 MTase, methylation of ribosomal proteins was studied in vitro. The yfcB gene was recloned into the high level expression vector pET11a, allowing the preparation of cell-free extracts highly enriched in the putative MTase. Ribosomes were prepared from the prmB and wild-type parental strains and used as substrate in an MTase assay (Lhoest and Colson, 1977). As shown in Figure 3, mutant ribosomes, lacking the L3 modification, were stoichiometrically methylated by the YfcB-enriched extract. In contrast, no methylation occurred on wild-type ribosomes, showing that only non-methylated ribosomes from the prmB strain are substrates for the YfcB MTase. Similar cell extracts prepared from the prmB or wild-type strain (not shown) in the absence of YfcB overproduction did not exhibit MTase activity with undermethylated ribosomes as substrate. The results of these in vitro experiments support the conclusion that YfcB is responsible for methylation of ribosomal protein L3.
Fig. 3. Methylation of undermethylated ribosomal proteins by cell extracts enriched in YfcB. Ribosomes prepared from the prmB strain CL1447 (filled circles) or the wild-type parental strain AB2557 (open squares) were incubated with S-adenosyl-l-[methyl-3H]methionine and cell extract from cells overproducing YfcB. Control points (filled triangles) show the absence of methylation of undermethylated ribosomal proteins by a cell extract from the prmB strain CL1447. Each point corresponds to 12 pmol of ribosomes and 3 µg of cell extract protein (YfcB enriched) or 10 µg of cell extract (strain CL1447).
YfcB does not encode the RF MTase
Methylation of ribosomal protein L3 occurs at position N5 of Gln150 (Muranova et al., 1978). Since RF methylation also occurs on the same nitrogen atom of the glutamine residue in the GGQ motif, we considered the possibility that YfcB also catalyses peptide RF methylation. A priori, this seemed unlikely as the primary sequence surrounding Gln150 in L3 is quite different from that around the modified glutamine in RF.
Mass spectrometry analysis was performed on tryptic digests of RF1 prepared from the prmB and wild-type parental strains. Prior to RF1 preparation, a cassette adding a His6 tag to the C-terminus of RF1 was inserted in the chromosome by recombination (see below). These strains contained normal levels of RF1 and RF2, which was necessary as overproduction of the factors is known to produce non-methylated RF (Dinçbas-Renqvist et al., 2000). RF1 from both strains gave rise to peptides with a mass [M + H]+ of 1672, corresponding to that of the methylated peptide A-S-G-A-G-G-N5MeQ-H-V-N-R, and none with the mass of 1658, expected for the non-methylated peptide (Dinçbas-Renqvist et al., 2000). Attempts to methylate RF1 and RF2 using the in vitro methylation assay described also led us to conclude that RF methylation must be performed by an MTase other than YfcB.
HemK encodes the RF1 and RF2 MTase
As a known N5-glutamine MTase, YfcB could be expected to show sequence similarity to the putative peptide RF MTase. A search of the E.coli protein sequence databank showed that only one protein, encoded by the gene hemK, has significant sequence similarity to YfcB. When first cloned and sequenced, the product of hemK was believed to be a protoporphyrinogen oxidase involved in haem biosynthesis (Nakayashiki et al., 1995). The gene is immediately downstream of prfA, encoding RF1 at 27 min on the genome. The initiation codon of hemK overlaps the termination codon of prfA, suggesting that the synthesis of the two proteins is coupled. The tandem position of hemK and prfA in the genome was also in line with the hypothesis that hemK encodes the RF MTase.
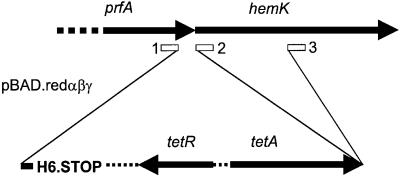
To test this idea, we attempted to inactivate hemK by recombination with the chromosome of a linear DNA fragment containing a tetracycline-resistant cassette and terminal sequences matching regions of prfA and hemK (Figure 4). Recombination was stimulated by the method of Zhang et al. (2000) depending on phage λ Redα/Redβ proteins. The cassette also added a C-terminal His6 tag to RF1 to facilitate purification of the factor when it was present at normal cellular abundance. Recombination was performed in two ways: (i) using terminal sequences 1 and 3 so as to truncate the hemK gene; and (ii) with terminal sequences 1 and 2, which maintains the integrity of hemK and creates a translational coupling between tetA and hemK similar to that which normally exists between prfA and hemK. Tetracycline-resistant recombinants were obtained in both cases, although in the first case they grew extremely poorly both on plates and in liquid media. It was verified that the poor growth resulted from the truncation of HemK, rather than from a polar effect that the truncation might have on any of the three downstream genes, two of which are of unknown function. To do this, hemK was first cloned into the vector pLV1. The poor growth of the partially deleted hemK recombinant strain was found to be totally complemented by the resulting plasmid.
Fig. 4. Insertion of the His6 tag and truncation of hemK by recombination with linear DNA. Redαβγ-stimulated recombination was used to insert a His6 tag-coding sequence and tetR-resistant cassette at the end of the prfA gene with or without truncation of hemK. The positions on the chromosome of the homologous terminal sequences allowing recombination are shown by boxes 1–3. PCR oligonucleotides used to make the inserts added the homologous sequences 1 and 3 so as to truncate hemK, or 1 and 2 so as to allow insertion without truncation and allow hemK expression by translation coupling to tetA.
Analysis by mass spectrometry was performed on tryptic digests of RF1 prepared from the two recombinant strains, with truncated or intact hemK. As described above, RF1 from the second strain yielded a normally modified peptide containing N5-methylglutamine. In contrast, RF1 from the hemK-truncated strain gave no equivalent peptide, but instead yielded a peptide of mass [M + H]+ of 1658, as expected for the non-modified peptide A-S-G-A-G-G-Q-H-V-N-R.
In vitro methylation of RF1 and RF2 by HemK
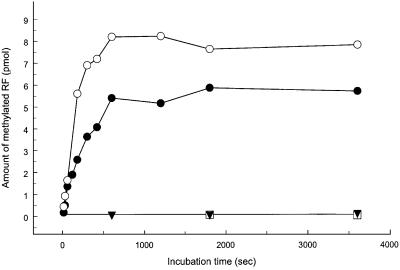
To confirm that HemK methylates both class 1 peptide RFs, hemK was cloned into the expression vector pET11a for overproduction. More than 15% of the total protein extracted from cells after 2 h of induction was HemK. Methylation of both RF1 and RF2 was observed in vitro using cell-free extracts enriched in HemK (Figure 5). No methylation could be detected using extracts from the hemK-truncated strain. Not all added RF could be methylated, particularly in the case of RF1. This observation may reflect the presence of an inactive fraction of RF, consistent with the findings that the percentage of RF molecules active in peptide chain release in vitro is significantly below 100% and that the specific activity of RF2 is higher than that of RF1. The observed methylation was specific for the glutamine residue located in the conserved GGQ motif, since it did not occur on mutant RF1 or RF2 with a GGE or GGA motif in place of GGQ (Figure 5).
Fig. 5. Methylation of RF1 and RF2 by HemK. Non-methylated RFs prepared by expression from overproducing vectors were incubated with S-adenosyl-l-[methyl-3H]methionine and cell extract from cells overproducing HemK: RF1 (filled circles), RF2Ala(His)6 (open circles), RF2AlaGGE(His)6 variant (open boxes). A control is shown with RF2Ala(His)6 and a cell extract prepared from the ΔhemK strain SC8 (filled triangles). Each point corresponds to 25 pmol of added RF and 0.5 µg of cell extract protein (HemK enriched) or 20 µg of cell extract (strain SC8).
Suppression of RF2 toxicity by HemK overexpression
Uno et al. (1996) showed that overexpression of RF2 from E.coli, but not from Salmonella typhimurium, inhibits cell growth. They also demonstrated that it is the identity of the residue at position 246 in RF2 and none of the other 15 differences between the two proteins that causes the toxicity. To explain this observation, Uno et al. (1996) postulated that overexpression of RF2 gives rise to one deficient and one fully active population of RF2 in the cell, and that these compete on the ribosome. They suggested, further, that normal activity of RF2 might require a post-translational modification of Thr246. Recently, it was shown that the change Thr246Ala and the methylation of Gln252 both gave positive and cumulative effects on RF2 activity in vitro (Dinçbas-Renqvist et al., 2000). We therefore proposed that overproduction of E.coli K12 RF2 is toxic because it abolishes methylation of Gln252 and that this, in combination with threonine at position 246, results in a termination activity that is too low to sustain growth (Dinçbas-Renqvist et al., 2000). Now that the RF MTase has been identified, this suggestion can be subjected to more precise experimental tests.
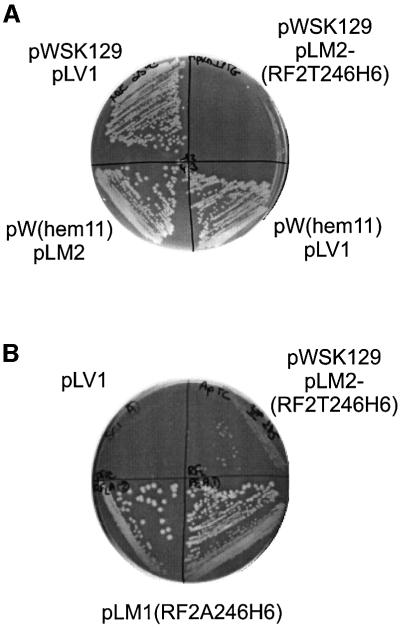
If true, it would imply that not only strains overproducing RFs, but also methylation-deficient strains producing RFs at the normal level, would show impaired growth. In line with this, we have found that strain SC5, in which the AdoMet-binding motif of hemK is deleted, grows poorly on rich media and not at all on poor media. Another prediction is that overexpression of RF2(Thr246) should be much less toxic in strains with increased HemK levels due to their enhanced capacity to methylate RF. To test this, the hemK gene was recloned into the low copy number plasmid vector pWSK129 (Wang and Kushner, 1991), which has an origin of replication compatible with that of pLV1. The growth inhibition resulting from the overproduction of RF2(Thr246) in a wild-type hemK strain is clearly shown when the factor is expressed from vector pLM2, a derivation of pLV1 (Figure 6A, top right segment). In contrast, the growth of cells overexpressing both RF2(Thr246) from pLM2 and HemK from pW(hem11) is comparable to that of cells transformed by control plasmids without inserts (pLV1 and pWSK129). These results are fully in line with the hypothesis. Further evidence came from the observation that RF2(Thr246) overproduction was not toxic in strain SC1, in which hemK is expressed from the comparatively strong tetA promoter instead of by transcription of the hemA–prfA–hemK operon. We conclude that the growth inhibition due to RF2(Thr246) overproduction results from the synthesis of non-methylated RF2 molecules and can be suppressed by increased levels of HemK.
Fig. 6. Growth inhibition due to undermethylation of RF2(Thr246). (A) Wild-type K12 strain Xac was transformed by plasmid pLM2(RF2T246H6) overexpressing His-tagged RF2Thr246 and co-transformed with plasmid pW(hem11) expressing hemK or control plasmid pWSK129 without insert, showing that growth inhibition due to RF2Thr246 overproduction is suppressed by hemK expression. IPTG (1 mM) was present to induce RF2 and HemK expression. Control constructions shown are the double transformant with pW(hem11) and pLV1 parent plasmid without insert, and the double transformant with both parent plasmids without inserts, pWSK129 and pLV1. Transformants were streaked on LB-agar, 1 mM IPTG and incubated at 25°C. (B) HemK-truncated mutant strain SC5 was transformed with plasmid pLM2(RF2T246H6) expressing His-tagged RF2Thr246, plasmid pLM1(RF2A246H6) expressing His-tagged RF2Ala246 (lower two streaks) or control plasmid pLV1 without insert. Transformants were streaked on LB-Amp-Tet and incubated at 37°C.
hemK revertant strains
Previous results show that non-methylated RF2(Ala246) is considerably more active in peptide release than non-methylated RF2(Thr246) and that methylation contributes little to the activity of RF1 (Dinçbas-Renqvist et al., 2000). It might therefore be anticipated that the poor growth of the truncated HemK strain SC5 would be suppressed by the expression of RF2(Ala246) but not RF2(Thr246) from a plasmid, and this expectation was confirmed experimentally (Figure 6B). Our results strongly suggest that the poor growth of strain SC5 was due to the presence of a threonine at position 246 of RF2 in combination with lack of methylation of the factor. Therefore, we expected that spontaneous revertants to faster growth could occur and that these would affect the prfB gene. Faster growing revertants of strain SC5 were found to arise in cultures in rich medium at frequencies of 10–9–10–8. Revertants were isolated from four independent cultures and prfB was sequenced. All sequences showed changes to the codon for residue 246, which was altered either to GCG, encoding alanine, or UGC, encoding serine. These two amino acids are ubiquitous at the corresponding position of all bacterial RFs with the exception of E.coli K12 strains, and both their codons are accessible from the threonine codon ACG by a single nucleotide change.
Homologues of HemK, YfcB and related proteins in other organisms
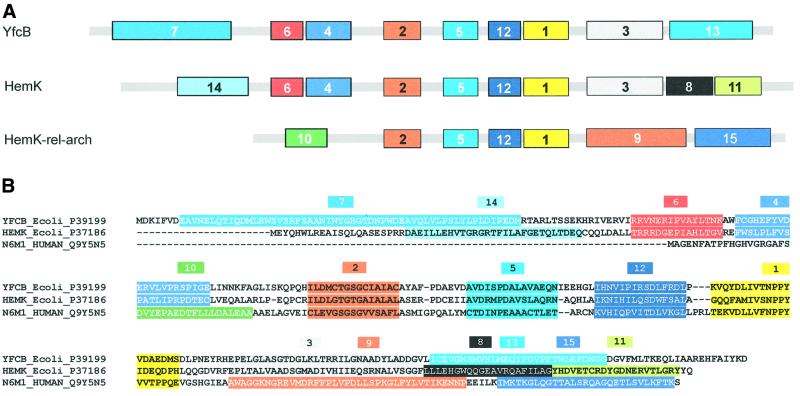
HemK and YfcB are classified in the InterPro and TIGR protein family protein databases as belonging to a class of enzymes of unknown function. This is referred to as the HemK family, and includes predicted proteins from bacteria, eukarya and archaea. The databases putatively class ∼60 proteins in the HemK family (InterPro signature IPR004556; TIGR signature TIGR00536). A smaller family of ∼20 proteins, referred to as the HemK-rel-arch family, contains mostly archaeal and eukaryotic proteins that are related in sequence to those of the HemK family but lack their typical N-terminal domain (InterPro signature IPR004557; TIGR signature TIGR00537). The 78 non-redundant sequences in these families were studied in more detail using the MEME motif discovery tool (Bailey and Elkan, 1994).
About one half of the sequences fall into three well-defined subfamilies, which show the pattern of motifs illustrated in Figure 7A. A set of four motifs, labelled 2, 5, 12 and 1, was common to all three families, and occupies the central part of the sequences. They include the motifs known to be involved in AdoMet binding, such as the glycine-rich motif referred to above, which is part of motif 2. Motif 1 includes the NPPY tetrapeptide, part of the AdoMet-binding site of DNA MTases (Nakayashiki et al., 1995). The N- and C-terminal parts of the sequences are specific to the three subfamilies. The precise locations of the motifs are shown in Figure 7B superimposed on sequences of typical members of the three subfamilies: E.coli YfcB, E.coli HemK and the human protein Q9Y5N5. The first subfamily comprises seven sequences and the second subfamily 19 sequences (Table I). Upstream of motif 2, the two adjacent motifs 6 and 4 are shared by both the YfcB and HemK subfamilies, but the motif closest to the N-terminus is distinct in each case. A similar situation is seen towards the C-terminus of the sequences, where motif 3 is common to the two subfamilies, whereas motif 13 in the YfcB subfamily is replaced by motifs 8 and 11 in the HemK subfamily. The third subfamily that emerged from the MEME analysis corresponds to the eukaryotic/archaeal HemK-related family, with the exclusion of four sequences, and comprises 16 sequences: eight from eukarya including two yeast sequences, and eight of archaeal origin. In this subfamily, the three N-terminal motifs of the other two subfamilies are replaced by a new motif 10 and the C-terminal motifs by two motifs, 9 and 15 (see Figure 7).
Fig. 7. Amino acid sequence motifs in putative proteins of the HemK family and HemK-rel-arch families (InterPro signatures IPR004556/7). A search for conserved motifs using MEME (Bailey and Elkan, 1994) showed three clearly defined subfamilies. The motifs found are numbered in the order of identification by MEME. (A) The proteins characteristic of the subfamilies are shown in Table II. (B) The localization of each of the motifs 1–15 identified by MEME is shown superimposed on a typical member of each subfamily. The regions of the sequences including the common motifs 2, 5, 12 and 1 were aligned using Clustal (Thompson et al., 1994) and manually adjusted to remove small gaps from the MEME motifs.
Table I. Putative proteins of the YfcB, HemK and HemK-rel-arch families.
| YfcB subfamily | HemK subfamily (continued) |
|
Eschericha coli YfcB (P39199) |
Rhizobium loti HemK (Q98G94) |
|
Haemophilus influenzae YfcB (P45106) |
Staphylococcus aureus SA19 (Q99SE1) |
|
Xylella fastidiosa YD68 (Q9PDL1) |
Caulobacter crescentus CC0875 (Q9A9T7) |
|
Vibrio cholera YL18 (Q9KQ83) |
Neisseria meningitidis A NMA0369 (Q9JWH6) |
|
Pasteurella multocida PM0390 (Q9CNN7) |
Bacillus halodurans BH3774 (Q9K6F5) |
|
Neisseria meningitidis NMA1912 (Q9JTA1) |
|
|
Pseudomonas aeruginosa PA1678 (Q9I347) |
HemK-rel-arch subfamily |
| |
Arabidopsis thaliana N6MT (Q9LJE6) |
| HemK subfamily |
Homo sapiens N6AMT1 (Q9Y5N5) |
|
Borrelia burgdorferi HemK (O51215) |
Caenorhabditis elegans C33C12.9 (O16582) |
|
Pasteurella multocida HemK (Q9CN82) |
Saccharomyces cerevisiae FYV9 (Q03920) |
|
Vibrio cholerae HemK (Q9KQ26) |
Schizosaccharomyces pombe SPAC323.05C (Q9UT94) |
|
Pseudomonas aeruginosa HemK (Q9HVC8) |
Drosophila melanogaster GC9960 (Q9VQF8) |
|
Haemophilus influenzae HemK (P45253) |
Methanococcus jannaschii Y928 (Q58338) |
|
Eschericha coli HemK (P37186) |
Archaeoglobus fulgidus YH84 (O28490) |
|
Xylella fastidiosa HemK (Q9PD67) |
Pyrococcus horikoshii PH1731 (O59386) |
|
Deinococcus radiodurans HemK (Q9RXR2) |
Thermoplasma volcanium TVG0197945 (Q97CB1) |
|
Bacillus subtilis HemK (P45873) |
Halobacterium sp. HemK (Q9HQH3) |
|
Clostridium acetobutylicum HemK (Q97F67) |
Pyrococcus abyssi PAB0284 (Q9V1J5) |
|
Streptomyces coelicolor 2SC6 (Q9K4E3) |
Fugu rubripes YDR140W (Q9YHV6) |
|
Lawsonia intracellularis pISI (O87889) |
Mus musculus 5830445C04RIK (Q9CST4) |
|
Treponema pallidum TP00 (O83091) |
Thermoplasma acidophilum TA0114 (Q9HLW1) |
| Buchnera aphidicola HemK (P57269) | Methanothermobacter thermautotrophicus MTH1329 (O27384). |
Subfamilies correspond to those in Figure 7. Accession numbers for protein sequences are shown in parentheses.
Other putative proteins bearing the InterPro signature IPR004556 are related to the HemK subfamily but are more divergent than the 19 most closely related sequences. Nevertheless, it is likely that some of these conserve HemK function since they show the same tandem organization of prfA and hemK as in E.coli and many other organisms. This is the case for HemK-like proteins in Chlamydia trachomatis (O84027), Mycobacterium tuberculosis (Q10602) and Streptococcus pneu moniae (Q97R19), for example. The mycoplasmas Mycoplasma pneumoniae, Mycoplasma genitalium and Ureaplasma urealyticum present further cases of tandem prfA–hemK genes of particular interest, as these proteins possess highly extended C-terminal regions in place of motifs 8 and 11. Proteins assigned from a group of six eukaryotic sequences, including two from yeast, are also less conserved than typical HemK proteins, particularly in the N-terminal region (motif 14). These putatively identified proteins, from man (Q9Y5RA), Drosophila (Q9VMD3), mouse (AAH11431), Arabidopsis (Q9FMI5), Schizosaccharomyces pombe (O14028) and S.cerevisiae (S58715), are predicted by the TargetP program (Emanuelsson et al., 2000) to be targeted to the mitochondrion. A functional analysis of the S.cerevisiae protein, called YNL063w, concluded that it did not possess protoporphyrinogen activity (Le Guen et al., 1999), thought to be the role of the E.coli HemK protein (Nakayashiki et al., 1995). No function has been determined for YNL063w. One related protein lacks motif 11 of the HemK subfamily and is unique amongst the putatively HemK-related proteins in that a function for it has been reported. This protein is PapM in Streptomyces pristinaespiralis, which catalyses two successive methylation steps of 4-amino-l-phenylalanine in the pathway to pristinamycin I synthesis (Blanc et al., 1997).
Discussion
GGQ is the only motif that is universally conserved among characterized class 1 peptide RFs in all organisms from eubacteria, eukarya and archaea (Frolova et al., 1999). The two glycine residues are essential for activity in both mammalian and bacterial factors. The glutamine residue can be changed to certain other amino acids with retention of partial peptide release activity in vitro. However, no change has been found at this position that allows RFs to fulfil their essential function in vivo in either yeast or E.coli (Frolova et al., 1999; Seit Nebi et al., 2000; L.Mora, unpublished results). The precise role of this region of class 1 RFs has remained unknown, in spite of its functional significance. The crystal structure of human eRF1 shows the GGQ motif to be at one extremity of the molecule (Song et al., 2000), offering some support for the hypothesis that it is a molecular mimic of the CCA terminus of a tRNA molecule (Frolova et al., 2000). We have shown previously that in E.coli the glutamine residue is N5-methylated, and that the modification is important for the activity of RF2 in vitro, for both RF2(Thr246) as found in K12 strains and the more active RF2(Ala246) present in other strains of E.coli. In contrast, in vitro peptide release experiments have not revealed any obvious role for glutamine methylation in the case of RF1 (Dinçbas-Renqvist et al., 2000).
In the experiments described here, two protein MTases that modify glutamine residues were identified. YfcB methylates ribosomal protein L3 and HemK targets peptide RFs RF1 and RF2. Our findings show that these enzymes are members of a new family of protein MTases, quite distinct from the family of adenine MTases to which they have been ascribed (Bujnicki and Radlinska, 1999). As in the case of HemK, the inactivation of YfcB (or PrmB) has a clear phenotype. Successful complementation of a prmB mutant with a plasmid-borne yfcB gene demonstrates that the cryosensitive growth phenotype of the prmB mutant is indeed caused by the mutation in the AdoMet-binding motif 1. In contrast, methylation-deficient mutants other than prmB and hemK often lack a clear phenotype. For example, the inactivation of PrmA, which can add nine methyl groups to L11, the most heavily modified ribosomal protein known in E.coli, has no apparent effect on cell growth (Vanet et al., 1994).
Experiments where hemK was inactivated or HemK overproduced were used here to establish the physiological importance of RF2 methylation in E.coli K12 strains. Our findings will greatly facilitate further studies of the role of RF methylation in vivo. Furthermore, large quantities of purified RFs for biochemical studies can now be obtained in both their unmethylated and methylated states.
This and previous (Dinçbas-Renqvist et al., 2000) work has shown that E.coli K12 peptide RF methylation is important or even necessary for cell growth, but in other organisms the physiological significance of HemK remains to be clarified. At this point, it cannot be excluded that HemK plays some role in the cell other than the methylation of RFs. It is also possible that RF methylation plays a role beyond that revealed by simple peptide RF assays in vitro. For example, one aspect of translation termination that has attracted attention is the possibility that aberrant RF activity during peptide chain elongation may contribute to errors in translational processivity. Particularly for large proteins, overall processivity errors may be considerable. Thus, ∼30% of ribosomes that initiate translation of a lacZ mRNA fail to reach the normal stop signal (Manley, 1978). The possible role of RFs in processivity errors was first studied by RF overproduction in vivo, necessarily producing non-methylated factors and accompanied by cell growth inhibition (Jørgensen et al., 1993). More recent studies in vitro, again with non-methylated factors, have shown that RFs function with remarkable accuracy, but that some sense codons represent hotspots for RF errors (Freistroffer et al., 2000). Since methylation of peptide RFs affects their interaction with the ribosome, the modification may influence their ability to induce abortive termination events at sense codons. The impact of peptide RF errors on the processivity of ribosomes may therefore be quite different from what was previously thought, and the present work offers the experimental tools to re-examine the question in vivo as well as in vitro.
AdoMet-dependent MTases comprise a large and complex family of enzymes, acting on a wide range of substrates, including DNA, RNA, proteins, lipids and numerous small molecules. The three-dimensional structures of 20 of these enzymes are known (Cheng and Roberts, 2001). All share a common core structure based on a mixed seven-stranded β-sheet, now referred to as the AdoMet-dependent MTase fold, which is poorly reflected in the conservation of primary structure. The classification of these enzymes is complicated further by the fact that different functional domains have become permutated circularly, leading to three observed subfamilies (Malone et al., 1995). In database annotations, HemK and YfcB have been recognized as likely AdoMet MTases and classed amongst the adenine-specific DNA MTases. To our knowledge, no authors or database annotations have placed either protein in a protein MTase family. One recent study suggested that HemK is a member of a missing subfamily of adenine MTases with the target recognition domain at the N-terminus of the protein (Bujnicki and Radlinska, 1999). This was motivated partly by the presence in HemK proteins of an NPPY motif, considered to be a hallmark [consensus sequence (S/N/D)PPY] of N6-adenine and N4-cytosine MTases (Malone et al., 1995; Roth et al., 1998). This motif, located in the loop region following the β4 strand of the AdoMet-dependent fold, contributes to the AdoMet-binding site and the active site of the enzyme. However, in protein MTases characterized up to now, or in C5-cytosine MTases such as the M·HhaI MTase (O’Gara et al., 1995), the NPPY motif is replaced by other amino acid residues. The presence of an NPPY motif in HemK and YfcB shows, therefore, that this motif cannot in general be used to identify DNA MTases. The only exception previously reported is the PapM MTase in Streptomyces pristinaespiralis, which transfers a methyl group from AdoMet to 4-aminophenylalanine and 4-methylaminophenylalanine (Blanc et al., 1997).
Protein sequence databases have recognized a distinct family of proteins related to HemK. Currently, ∼80 putative proteins from 56 different organisms have been classified thus, out of ∼2000 putative AdoMet-dependent MTases. PapM is the only HemK-related protein for which a function has been identified previously. Analysis of AdoMet-dependent MTases is clearly complicated by the fact that they share an AdoMet-dependent MTase fold and yet possess distinct domains that reflect the large variety characteristic of their target molecules. Our search for motifs within the group of HemK-related putative proteins identified three subfamilies. The smallest of these concerned seven organisms with genes encoding putative proteins closely similar to E.coli YfcB. In ribosomal protein L3 from these organisms, the Gln150 residue methylated by E.coli YfcB is conserved. In contrast, in organisms that do not appear to encode a YfcB-like protein, Gln150 has been replaced most often by another amino acid. This is consistent with the idea that the role of all members of the YfcB subfamily is L3 methylation.
The hemK gene is widespread in both Gram-negative and Gram-positive bacteria, and has been maintained in the small genomes of the obligate intracellular parasites Rickettsia prowazekii, Mycoplasma genitalium and Haemophilus influenzae. The hemK gene counts among the proposed minimal set of genes required for life (Koonin et al., 1996). Pairs of apparent homologues are present in yeast, human, mouse and fly cells, one of which may be targeted to the mitochondrion. In each case, the second putative protein is notably different in organization from typical HemK proteins. The function of the HemK-rel-arch subfamily to which they belong (Figure 7), and which is specific to eukarya and archaea, remains to be determined. The possibility that eRF1 and aRF1 are methylated by members of this subfamily needs to be examined experimentally.
The gene organization found in E.coli, with hemK immediately downstream of prfA within one operon, is widely but not universally conserved. In eubacteria, different cases are seen to occur: (i) the organization prfA–hemK is conserved, as for example in Pseudomonas aeruginosa, Bacillus subtilis, Streptococcus pneumoniae, Staphylococcus aureus and M.genitalium; (ii) the genes prfA and hemK are very close to each other but not adjacent, as in H.influenzae; and (iii) the genes are far from each other and the gene located just downstream of prfA is involved in another cellular function, as in R.prowazekii and Neisseria meningitidis. Such disruption of gene order is often observed in bacterial evolution and does not exclude the possibility that similar genes from two organisms are orthologues (Mushegian and Koonin, 1996). The maintenance of prfA–hemK gene organization supports the tentative functional identification of some hemK orthologues more distant in sequence similarity, such as hemK in mycoplasma.
In conclusion, two MTases in E.coli have been identified in this work: YfcB that methylates ribosomal protein L3 and HemK that methylates both RF1 and RF2 on the glutamine of the GGQ motif that is universally conserved among class 1 peptide RFs from all organisms. We propose that yfcB and hemK be renamed prmB and prmC. The present findings clarify the fundamental role played by methylation of RF2 for growth or survival of E.coli K12 strains. They suggest that RF methylation may be widespread in occurrence and importance, but it will require further work to determine whether HemK orthologues in higher organisms indeed target RFs.
Materials and methods
Bacterial strains, plasmids and bacteriophage
Bacterial strains are listed in Table II. SC1 and SC5 were constructed by ET recombination (Zhang et al., 2000). In both cases, plasmid pCP16 (Cherepanov and Wackernagel, 1995) carrying the tetracycline resistance genes tetR and tetA was used to prepare linear double-stranded DNA by PCR. For SC1, the upstream oligonucleotide contained a part complementary to prfA, nucleotides encoding His6, the prfA stop codon and a sequence complementary to pCP16. The downstream oligonucleotide contained a part complementary to the 5′ end of hemK and a sequence complementary to tetA from pCP16, giving a DNA fragment of 2303 bp. For SC5, the upstream oligonucleotide was similar, and the downstream oligo was complementary to a central part of hemK so that recombination deleted the first 123 amino acids of HemK; PCR yielded a DNA fragment of 2648 bp. PCR products were digested by BsaI to eliminate residual circular pCP16, and transformed into electrocompetent cells of strain Xac carrying pBAD-αβγ (Zhang et al., 2000). During cell preparation, the redα gene was induced by arabinose (0.1% final added at OD600 = 0.2) and growth continued until OD600 = 0.6. Recombinants were selected on LB-tetracycline plates and verified by PCR and sequencing.
Table II. Escherichia coli strains.
| Strain | Genotype | Reference/source |
|---|---|---|
| AB2557 | ilvD188, dsdA1, aroC4, purF1, mtl-2, xyl-7, malA1, lacY1 or lacZ4, rpsL8, 9 or 14, tonA2, A22 or 14, tsx-23 or 25, supE44, thi-1, F– | B.Bachmann, E.coli Genetic Stock center |
| BL21(DE3) | F–, ompT, hsdSB, dcm, λ prophage carrying T7 RNA polymerase | Studier and Moffatt (1986) |
| BL21(DE3) pLysS | BL21(DE3) carrying pLysS plasmid CmR | Studier and Moffatt (1986) |
| CL1447 | AB2557 purF+, prmB2 | Colson et al. (1979) |
| SC1 | Δ(lac-pro), argE, ara, gyrA, rpoB, thi, prfA(His)6-Tc | this work (ET recombination in Xac) |
| SC3 | AB2557, prfA(His)6-Tc | this work (AB2557 and P1 lysate on SC1) |
| SC4 | CL1447, prfA(His)6-Tc | this work (CL1447 and P1 lysate on SC1) |
| SC5 | Δ(lac-pro), argE, ara, gyrA, rpoB, thi, prfA(His)6-Tc, ΔhemK | this work (ET recombination in Xac) |
| SC6 | SC5 prfB(Ser246) | this work (a revertant of SC5) |
| SC8 | SC5 prfB(Ala246) | this work (a revertant of SC5) |
| Xac | Δ(lacpro), argE, ara, gyrA, rpoB, thi | Coulondre and Miller (1977) |
pET11a(hemK) and pET11a(yfcB) were constructed by PCR amplification of hemK and yfcB in DNA extracted from strain Xac with oligonucleotides specific for each gene and containing NdeI and BamHI sites. In vivo complementation and toxicity assays were carried out after subcloning genes (hemK, yfcB, prfB) from pET11a derivatives into inducible plasmid pLV1 between the NdeI and BamHI sites, giving, respectively, pLV(hemK), pLV(yfcB), pLM2(RF2Thr246) and pLM1(RF2Ala246). pLV1 is a derivative of pTrc99c (Amann et al., 1988) with substitution of an NdeI site for the NcoI site after deletion of the other NdeI site located outside of the polylinker (L.Mora, V.Heurgué-Hamard, S.Champ, M.Ehrenberg and R.H.Buckingham, in preparation). Plasmid pLV(yfcB–) was constructed similarly by PCR using chromosomal DNA from strain CL1447. pW(hem11) was constructed by subcloning the hemK gene from pLV(hemK). The DNA fragment containing hemK transcribed from the Ptrc promoter was obtained by digestion with ScaI and EcoRV and cloned into pWSK129 digested by EcoRV.
Bacterial growth
Luria broth (LB) medium was supplemented according to the requirements. Antibiotics were added at the following final concentration: tetracycline, 12.5 µg/ml; kanamycin, 50 µg/ml; ampicillin, 200 µg/ml. When induction was necessary to express hemK, yfcB, prfA or prfB genes, 1 M IPTG was added to LB plates or liquid medium to a final concentration of 1 mM. Growth comparisons were made by streaking the strains on plates under different conditions. For minimal medium plates, Vogel–Bonner medium (Vogel and Bonner, 1956) was used, supplemented either with 0.5% casamino acids or with the required amino acids and 0.2% glucose.
Recombinant DNA manipulations and genetic manipulations
General procedures for DNA recombinant techniques, plasmid extraction, etc. were performed as described by Sambrook et al. (1989). Purification of fragments on agarose gel was by Jetsorb gel extraction. Phage P1 lysates, transductions and transformations were performed as described by Miller (1992).
Protein purifications
RF2(Ala246) and RF1 proteins used as substrates for methylation were purified as described previously (Pavlov et al., 1998; Dinçbas et al., 1999) after expression from pET11a-derived plasmids in strain BL21(DE3) containing an inducible T7 polymerase gene. The construction and preparation of mutant RFs will be described in a forthcoming manuscript. RF1 with a His6 tag on its C-terminus, expressed from the modified chromosomal gene, was purified from strains SC3, SC4 and SC6 from 4 l saturated cultures. Cells were washed and resuspended in 30 mM Tris–HCl pH 8, 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and DNase I. After centrifugation at 13 000 g for 30 min, the supernantant was loaded on to an Ni-NTA superflow (Qiagen) column and RF1 was eluted with an imidazole gradient from 5 to 100 mM in 30 mM Tris–HCl pH 8, 1 mM DTT. Fractions containing RF1 were precipitated with 10% trichloroacetic acid (TCA) with tRNA carrier and the pellets washed with ether before purification on 12% SDS–PAGE. The band corresponding to RF1 was cut and and used for mass spectrometry analysis.
Preparation of ribosomes
Escherichia coli cells were grown in LB medium (400 ml) until late exponential phase (OD600 = 1), harvested by centrifugation, washed once with buffer 1 (10 mM Tris–HCl pH 7.6, 30 mM NH4Cl, 10 mM magnesium acetate, 6 mM β-mercaptoethanol) and opened with a French press. After removing the cell debris by centrifugation (20 000 g, 30 min), the supernantant was centrifuged at 100 000 g on a sucrose cushion (buffer 1 with 1.1 M sucrose) for 4 h to pellet the ribosomes. The ribosomal pellet was resuspended in 3 ml of buffer 1 and cleared by low-speed centrifugation (20 000 g, 30 min). The supernantant containing crude ribosomes was made 1 M in NH4Cl and stirred overnight at 4°C. Washed ribosomes were pelleted as before, resuspended in 250 µl of buffer 1 (final concentration of 10–20 pmol/µl) and kept at –20°C after low speed centrifugation.
Sources of methylating enzymes
HemK and YfcB proteins were overproduced in strain BL21(DE3)pLysS by IPTG induction from derivatives of pET11a. The cells were broken and ribosomes pelleted at 100 000 g as described above, and the S100 supernantant was dialysed against buffer 2 (10 mM Tris–HCl pH 8.3, 10 mM β-mercaptoethanol). Glycerol was added to a final concentration of 50% keeping the same final concentration in Tris–HCl and β-mercaptoethanol. Aliquots were kept at –20°C for methylation assays. Protein concentration was determined using the BCA protein assay reagent (Pierce).
Methylation in vitro
Methylation assays on ribosomal proteins were performed as described by Colson et al. (1979) in buffer 3 (50 mM Tris–HCl pH 8.3, 10 mM EDTA, 10 mM β-mercaptoethanol, 600 mM potassium acetate) with S-adenosyl-l-[methyl-3H]methionine (ICN, 78 Ci/mmol) diluted with unlabelled AdoMet to a final concentration of 2 µM (0.17 Ci/mmol). Each time point corresponds to 12 pmol of ribosomes added and amounts of S100 extract as indicated in the figure legends. Samples were withdrawn at different times and the reaction was stopped by cold TCA (5%) precipitation, followed by filtration on Whatman GF/C filters and measurement of radioactivity. RF methylation was performed in buffer 4 (10 mM Tris–HCl pH 7.6, 100 mM KCl, 10 mM magnesium acetate, 6 mM β-mercaptoethanol) as described above with 25 pmol of RF1 or RF2 and amounts of S100 extract as indicated in the figure legends.
Mass spectrometry analysis
The RF1-containing bands were excised from the SDS–polyacrylamide gel and digested with modified trypsin (Promega). The tryptic digests were analysed by MALDI-TOF in a Bruker Reflex IV instrument.
Protein sequence similarity analysis
A block 16 residues in width containing the sequence motif 1 for the AdoMet-binding site was obtained by applying the program Blockmaker (Henikoff et al., 1995) to the six E.coli MTase sequences (SWISS-PROT accession numbers in parentheses): TehB (EG11884), PimT (P22061), UbiG (P24206), BioC (P12999), Cfa (EG11531) and PrmA (EG11497). The small bank of protein sequences coded within the region of ∼15 kb around aroC was searched with this block using PATMAT (Wallace and Henikoff, 1992). Narrower blocks restricted to the apparently conserved motif were unsuccessful in identifying any MTase in the aroC region. Following identification of YfcB and HemK as N5-glutamine MTases, the non-redundant sequences of the InterPro protein families with signatures IPR004556 and IPR004557 were analysed using the MEME motif discovery tool (http://meme.sdsc.edu/meme/website/; Bailey and Elkan, 1994) with the maximum number of motifs set at 20 and maximum motif width at 80 amino acids.
Acknowledgments
Acknowledgements
We thank Glenn Bjork for the kind gift of the prmB mutant and parental strains. This work was supported by the Centre National de la Recherche Scientifique (UPR9073), l’Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Medicale, the Swedish Research Council and the Knut and Alice Wallenberg Foundation (WCN).
References
- Amann E., Ochs,B. and Abel,K.J. (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene, 69, 301–315. [DOI] [PubMed] [Google Scholar]
- Bailey T.L. and Elkan,C. (1994) Fitting a Mixture Model by Expectation Maximisation to Discover Motifs in Biopolymers. AAAI Press, Menlow Park, CA, pp. 28–36. [PubMed]
- Blanc V. et al. (1997) Identification and analysis of genes from Streptomyces pristinaespiralis encoding enzymes involved in the biosynthesis of the 4-dimethylamino-l-phenylalanine precursor of pristinamycin I. Mol. Microbiol., 23, 191–202. [DOI] [PubMed] [Google Scholar]
- Bujnicki J.M. and Radlinska,M. (1999) Is the HemK family of putative S-adenosylmethionine-dependent methyltransferases a ‘missing’ ζ subfamily of adenine methyltransferases? A hypothesis. IUBMB Life, 48, 247–249. [DOI] [PubMed] [Google Scholar]
- Chang C.N. and Chang,F.N. (1975) Methylation of the ribosomal proteins in Escherichia coli. Nature and stoichiometry of the methylated amino acids in 50S ribosomal proteins. Biochemistry, 14, 468–477. [DOI] [PubMed] [Google Scholar]
- Cheng X. and Roberts,R.J. (2001) AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res., 29, 3784–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P.P. and Wackernagel,W. (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalysed excision of the antibiotic-resistance determinant. Gene, 158, 9–14. [DOI] [PubMed] [Google Scholar]
- Colson C., Lhoest,J. and Urlings,C. (1979) Genetics of ribosomal protein methylation in Escherichia coli. III. Map position of two genes, prmA and prmB, governing methylation of proteins L11 and L3. Mol. Gen. Genet., 169, 245–250. [DOI] [PubMed] [Google Scholar]
- Coulondre C. and Miller,J.H. (1977) Genetic studies of the lac repressor III. Additional correlation of mutational sites with specific amino acid residues. J. Mol. Biol., 117, 525–567. [DOI] [PubMed] [Google Scholar]
- Dinçbas V., Heurgué-Hamard,V., Buckingham,R.H., Karimi,R. and Ehrenberg,M. (1999) Shutdown in protein synthesis due to the expression of mini-genes in bacteria. J. Mol. Biol., 291, 745–759. [DOI] [PubMed] [Google Scholar]
- Dinçbas-Renqvist V., Engström,Å., Mora,L., Heurgué-Hamard,V., Buckingham,R.H. and Ehrenberg,M. (2000) A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J., 19, 6900–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen,H., Brunak,S. and von Heijne,G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol., 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Freistroffer D.V., Kwiatkowski,M., Buckingham,R.H. and Ehrenberg,M. (2000) The accuracy of codon recognition by ribosome release factors. Proc. Natl Acad. Sci. USA, 97, 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L.Y., Tsivkovskii,R.Y., Sivolobova,G.F., Oparina,N.Y., Serpinsky,O.I., Blinov,V.M., Tatkov,S.I. and Kisselev,L.L. (1999) Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA, 5, 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L.Y., Merkulova,T.I. and Kisselev,L.L. (2000) Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA, 6, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff,J.G., Alford,W.J. and Pietrokovski,S. (1995) Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene, 163, 17–26. [DOI] [PubMed] [Google Scholar]
- Ito K., Uno,M. and Nakamura,Y. (2000) A tripepeptide anticodon deciphers stop codons in messenger RNA. Nature, 403, 680–684. [DOI] [PubMed] [Google Scholar]
- Jørgensen F., Adamski,F.M., Tate,W.P. and Kurland,C.G. (1993) Release factor-dependent false stops are infrequent in Escherichia coli. J. Mol. Biol., 230, 41–50. [DOI] [PubMed] [Google Scholar]
- Kisselev L.L. and Buckingham,R.H. (2000) Translational termination comes of age. Trends Biochem. Sci., 25, 561–566. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Mushegian,A.R. and Rudd,K.E. (1996) Sequencing and analysis of bacterial genomes. Curr. Biol., 6, 404–416. [DOI] [PubMed] [Google Scholar]
- Le Guen L., Santos,R. and Camadro,J.M. (1999) Functional analysis of the hemK gene product involvement in protoporphyrinogen oxidase activity in yeast. FEMS Microbiol. Lett., 173, 175–182. [DOI] [PubMed] [Google Scholar]
- Lhoest J. and Colson,C. (1977) Genetics of ribosomal protein methylation in Escherichia coli. II. A mutant lacking a new type of methylated amino acid, N5-methylglutamine, in protein L3. Mol. Gen. Genet., 154, 175–180. [DOI] [PubMed] [Google Scholar]
- Liu M. et al. (2000) Escherichia coli TehB requires S-adenosylmethionine as a cofactor to mediate tellurite resistance. J. Bacteriol., 182, 6509–6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone T., Blumenthal,R.M. and Cheng,X. (1995) Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases and suggests a catalytic mechanism for these enzymes. J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- Manley J.L. (1978) Synthesis and degradation of termination and premature termination fragments of β-galactosidase in vitro. J. Mol. Biol., 125, 407–432. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1992) A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Muranova T.A., Muranov,A.V., Markova,L.F. and Ovchinnikov,Y.A. (1978) The primary structure of ribosomal protein L3 from Escherichia coli 70S ribosomes. FEBS Lett., 96, 301–305. [DOI] [PubMed] [Google Scholar]
- Mushegian A.R. and Koonin,E.V. (1996) Gene order is not conserved in bacterial evolution. Trends Genet., 12, 289–290. [DOI] [PubMed] [Google Scholar]
- Nakayashiki T., Nishimura,K. and Inokuchi,H. (1995) Cloning and sequencing of a previously unidentified gene that is involved in the biosynthesis of heme in Escherichia coli. Gene, 153, 67–70. [DOI] [PubMed] [Google Scholar]
- O’Gara M., McCloy,K., Malone,T. and Cheng,X. (1995) Structure-based sequence alignment of three AdoMet-dependent DNA methyltransferases. Gene, 157, 135–138. [DOI] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer,D.V., Dincbas,V., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1998) A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol., 284, 579–590. [DOI] [PubMed] [Google Scholar]
- Roth M., Helm-Kruse,S., Friedrich,T. and Jeltsch,A. (1998) Functional roles of conserved amino acid residues in DNA methyltransferases investigated by site-directed mutagenesis of the EcoRV adenine-N6-methyltransferase. J. Biol. Chem., 273, 17333–17342. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Seit Nebi A., Frolova,L., Ivanova,N., Poltarus,A. and Kiselev,L. (2000) Mutation of a glutamine residue in the universal tripeptide GGQ in human eRF1 termination factor does not cause complete loss of its activity. Mol. Biol. (Mosk), 34, 899–900. [PubMed] [Google Scholar]
- Song H., Mugnier,P., Das,A.K., Webb,H.M., Evans,D.R., Tuite,M.F., Hemmings,B.A. and Barford,D. (2000) The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell, 100, 311–321. [DOI] [PubMed] [Google Scholar]
- Studier F.W. and Moffatt,B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- Tate W.P. et al. (1993) Translational stop signal: evolution, decoding for protein synthesis and recoding for alternative events. In Nierhaus,K.H., Franceshi,F., Subramanian,A.R., Erdmann,V.A. and Wittmann-Liebold,B. (eds), The Translational Apparatus: Structure, Function, Regulation, Evolution. Plenum Press, New York, NY, pp. 253–261.
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno M., Ito,K. and Nakamura,Y. (1996) Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie, 78, 935–944. [DOI] [PubMed] [Google Scholar]
- Vanet A., Plumbridge,J.A. and Alix,J.H. (1994) Ribosomal protein methylation in Escherichia coli: the gene prmA, encoding the ribosomal protein L11 methyltransferase is dispensable for normal growth. Mol. Microbiol., 14, 947–958. [DOI] [PubMed] [Google Scholar]
- Vogel H.J. and Bonner,D.M. (1956) Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem., 218, 97–106. [PubMed] [Google Scholar]
- Wallace J.C. and Henikoff,S. (1992) PATMAT: a searching and extraction program for sequence, pattern and block queries and databases. CABIOS, 8, 249–254. [DOI] [PubMed] [Google Scholar]
- Wang F.W. and Kushner,S.R. (1991) Construction of low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene, 100, 195–199. [PubMed] [Google Scholar]
- Willcock D.F., Dryden,D.T. and Murray,N.E. (1994) A mutational analysis of the two motifs common to adenine methyltransferases. EMBO J., 13, 3902–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Muyrers,J.P., Testa,G. and Stewart,A.F. (2000) DNA cloning by homologous recombination in Escherichia coli. Nature Biotechnol., 18, 1314–1317. [DOI] [PubMed] [Google Scholar]