Abstract
Transforming growth factor-β (TGFβ) conveys regulatory signals through multiple intracellular pathways, subsequently affecting various cellular functions. To identify new targets for TGFβ, we studied the changes in the proteome of Mv1Lu lung epithelial cells in response to TGFβ1 treatment. Thirty-eight non-abundant protein spots, affected by TGFβ1, were selected, and proteins were identified by peptide mass-fingerprinting (PMF). Among them, proteins involved in regulation of immune response, apoptosis, regulation of TGFβ signalling, metabolism and DNA repair were identified. Twenty-eight of the 38 proteins are new targets for TGFβ1, thus suggesting novel ways of integration of TGFβ signalling in intracellular regulatory processes. We show that TGFβ1-dependent decrease in expression of one of the new targets, Rad51, correlates with a decrease in DNA repair efficiency. This was evaluated by formation of nuclear Rad51-containing DNA repair complexes in response to DNA damage, by single cell gel electrophoresis and by cell survival assay. The TGFβ1-dependent inhibition of DNA repair was reversed by ectopic overexpression of Rad51. Therefore, TGFβ can promote DNA instability through down-regulation of Rad51 and inhibition of DNA repair.
Keywords: DNA repair/proteomics/Rad51/TGFβ
Introduction
Transforming growth factor-β (TGFβ) is a potent regulator of embryonal development, angiogenesis, immune response, bone remodelling, fibrosis and cancer (Roberts and Sporn, 1990; Massagué, 1998; de Caestecker et al., 2000). The first step in TGFβ signalling is binding of the ligand to specific serine/threonine kinase receptors. This leads to their activation, followed by phosphorylation of downstream substrates of the Smad family, which have been identified as major components of TGFβ intracellular signalling (Derynck et al., 1998; Piek et al., 1999; Miyazono et al., 2000; Moustakas et al., 2001). TGFβ is also known to regulate other pathways, among which are MAP kinases Erk, p38 and JNK, cAMP/cGMP-dependent kinases, phosphatidylinositol 3′-kinase and regulation of Ca2+ influx (Derynck et al., 1998; Massagué, 1998; Piek et al., 1999).
Components involved in TGFβ signalling and their targets have been extensively studied. Significant progress has been made in exploration of Smad-dependent signalling, with description of mechanisms of Smad activation and action (Derynck et al., 1998; Massagué, 1998; Piek et al., 1999; Miyazono et al., 2000). The molecular mechanisms of TGFβ-dependent regulation of MAP kinases Erk, p38 and JNK remain to be explored, and even less is known about the mechanisms that regulate other pathways. Although significant information is available regarding the effect of TGFβ on specific intracellular regulatory mechanisms, e.g. regulation of the cell cycle (CDK inhibitors, cyclins) or extracellular matrix formation (proteases, protease inhibitors, matrix proteins), an analysis of the global effect of TGFβ on cell proteome has not been reported. Since such an analysis is expected to provide more complete and unbiased insight into regulatory mechanisms, we studied changes in the proteome of Mv1Lu cells in response to treatment with TGFβ1. These cells express TGFβ receptors and Smad proteins, and are commonly used for studies of TGFβ signalling; their proliferation is potently inhibited by TGFβ, while extracellular matrix formation is induced (Tucker et al., 1984).
We identified TGFβ1-induced changes in 38 proteins that are involved in the regulation of cell proliferation, immune response, metabolism, transcriptional regulation or DNA integrity, or are components of the cytoskeleton. Twenty-eight of the proteins are previously undescribed targets of TGFβ. We show that the down-regulation of one of these new targets, Rad51, correlates with TGFβ1-dependent inhibition of DNA repair and cell survival, suggesting an effect of TGFβ signalling on the maintenance of genome stability, which is of importance in the prevention of cell transformation and tumour growth.
Results
Two-dimensional proteome maps of Mv1Lu cells
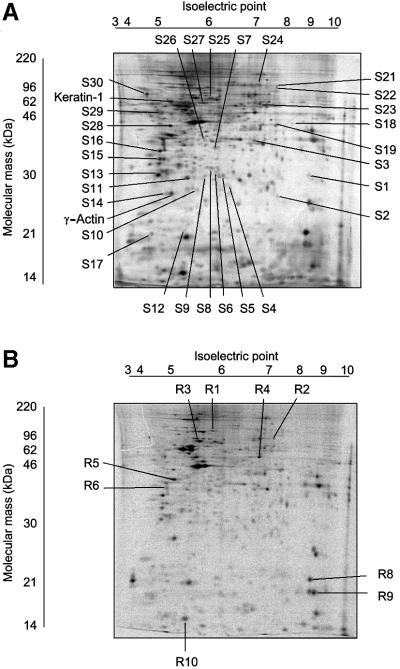
To identify changes in protein expression regulated by TGFβ1, we compared the proteome of Mv1Lu cells with that of the same cells stimulated with the ligand. To detect rapid and prolonged changes in protein expression, cells were treated with TGFβ1 for 2 and 24 h, respectively. Cell lysates were resolved by two-dimensional gel electrophoresis. We reproducibly detected ∼1000 protein spots on two-dimensional gels after silver staining (Figure 1A). The volumes of the spots were normalized to the volumes of landmark proteins used for internal standardization, and which did not change upon TGFβ1 treatment (keratin 1 and γ-actin). We analysed three to five gels for each experimental condition, and selected only spots whose normalized volumes were similar between the gels, as evaluated by the statistical analysis. We disregarded proteins that showed <30% changes between at least two of three experimental conditions (untreated cells, and cells treated for 2 or 24 h with TGFβ1). Twenty-nine protein spots that increased or decreased after TGFβ1 stimulation were selected for analysis by mass spectrometry.
Fig. 1. Two-dimensional electrophoresis gels, with annotations of spots of identified proteins. Images of a silver-stained gel (A) and a gel with [35S]methionine-labelled proteins (B), obtained from cells not treated with TGFβ1, are shown. Spots containing proteins that increased or decreased in amount in response to treatment of cells by TGFβ1 (5 ng/ml) and that were identified by PMF, are shown; S1–S30 represent annotated spots, identified by analysis of silver-stained gels, and R1–R10 represent annotated [35S]methionine-labelled spots. The pH gradient of the first-dimension electrophoresis is shown on top of the gels, and migration of molecular mass markers for SDS–PAGE in the second dimension is shown on the side of the gel. Migration positions for keratin-1 and γ-actin, used as internal landmarks, are shown.
To increase the sensitivity of detection of proteins with altered turnover, we also analysed cells labelled with [35S]methionine before TGFβ1 treatment. A similar number of labelled proteins was detected, compared with silver-stained gels (Figure 1B). However, TGFβ1-dependent changes were found for fewer [35S]methionine-labelled proteins, compared with silver-stained proteins. Moreover, many of the 35S-labelled proteins that were affected by TGFβ1 corresponded to or co-migrated with abundant silver-stained spots, and were therefore disregarded. Thus, only nine [35S]methionine-labelled proteins were subjected to identification by mass spectrometry in addition to the proteins selected from silver-stained gels (Table I).
Table I. List of proteins affected by TGFβ1 and identified by PMF.
| Spot | Protein | Probability | Est’d Z | Sequence coverage (%) | NCBI ID No. or accession No. | Theoretical value |
Experimental value |
Fold changes |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pI | Mr (kDa) | pH | Mr (kDa) | 2 h | 24 h | ||||||
| S1 | Tax interaction protein 43 | 8.6E–01 | 1.07 | 19 | AF028828 | 9.2 | 31.8 | 8.7 | 31 | 1.04 | 0.50 |
| S2 | elastase 1 precursor | 9.7E–01 | 0.62 | 22 | AF120493 | 9.2 | 27.8 | 7.7 | 26 | 1.56 | 0.85 |
| S3 | transcriptional factor TFEC | 6.0E–01 | 1.03 | 10 | AF077742 | 6.3 | 35.1 | 6.9 | 40 | 1.20 | 0.62 |
| S4 | isopropylmalate isomerase | 6.8E–01 | 0.29 | 22 | gi/3122365 | 7.0 | 22.5 | 6.5 | 29 | 1.46 | 0.72 |
| S5 | anti-oxidant protein 2 | 8.9E–01 | 0.57 | 45 | gi/6671549 | 6.0 | 24.8 | 6.4 | 31 | 0.88 | 0.66 |
| S6 | collagenase-3 precursor | 9.1E–01 | 0.47 | 18 | gi/116872 | 5.1 | 53.4 | 6.2 | 32 | 0.43 | 0.47 |
| S7 | cytosolic inorganic pyrophosphatase | 1.0E+00 | 1.55 | 29 | AF108211 | 5.4 | 31.8 | 6.2 | 35 | 0.99 | 0.65 |
| S8 | caspase 3 | 9.7E–01 | 0.63 | 19 | gi/2493528 | 6.2 | 32.1 | 6.1 | 32 | 0.98 | 0.47 |
| S9 | keratin 10, type 1, cytoskeleton | 1.0E+00 | 1.40 | 16 | gi/106849 | 4.7 | 39.7 | 5.9 | 30 | 1.37 | 0.81 |
| S10 | apolipoprotein-A1 | 9.5E–01 | 0.52 | 23 | U79574 | 5.5 | 30.5 | 5.7 | 26 | 1.13 | 0.75 |
| S11 | hypothetical protein | 1.0E+00 | 0.90 | 20 | gi/7705535 | 5.3 | 36.0 | 5.6 | 30 | 0.52 | 0.65 |
| S12 | GFAP | 6.9E–01 | 1.37 | 16 | gi/223869 | 6.0 | 24.9 | 5.5 | 21 | 1.59 | 0.58 |
| S13 | protein kinase C inhibitor | 1.0E+00 | 1.92 | 16 | gi/284655 | 4.8 | 27.8 | 5.0 | 31 | 1.33 | 0.84 |
| S14 | caspase 1 | 7.6E–01 | 0.53 | 20 | AF090119 | 6.1 | 45.3 | 5.2 | 26 | 0.38 | 0.44 |
| S15 | guanidine nucleotide binding protein, α-inhibiting 2 | 7.2E–01 | 1.37 | 21 | gi/6680037 | 5.4 | 40.5 | 5.1 | 32 | 1.14 | 0.72 |
| S16 | RAG1 | 7.6E–01 | 1.38 | 16 | AF203759 | 5.7 | 42.9 | 5.0 | 35 | 1.26 | 0.78 |
| S17 | Son of sevenless 1 | 4.7E–01 | 0.73 | 28 | D83014 | 4.9 | 23.6 | 4.7 | 20 | 1.12 | 0.69 |
| S18 | Musashi-1 homolog | 6.5E–01 | 0.28 | 11 | gi/6678940 | 7.9 | 39.1 | 8.2 | 43 | 1.13 | 2.41 |
| S19 | phosphoglycerate kinase-1 | 1.0E+00 | 2.43 | 15 | gi/104957 | 8.4 | 44.8 | 7.6 | 43 | 2.22 | 2.25 |
| S21 | ubiquitin-like fusion protein An1b | 5.7E–01 | 0.93 | 12 | gi/48443 | 8.6 | 78.6 | 7.6 | 100 | 1.10 | 2.54 |
| S22 | far upstream element-binding protein | 1.0E+00 | 1.33 | 12 | gi/4503801 | 7.2 | 67.5 | 7.6 | 98 | 0.98 | 2.23 |
| S23 | inosine-5′-monophosphate dehydro genase 2 | 1.0E+00 | 2.32 | 21 | gi/124419 | 6.4 | 55.8 | 7.2 | 55 | 0.76 | 0.68 |
| S24 | lamin A | 1.0E+00 | 1.05 | 16 | X76297 | 6.2 | 71.6 | 7.0 | 110 | 1.42 | 1.44 |
| S25 | indole-3-acetaldehyde dehydrogenase | 9.2E-01 | 0.57 | 17 | U74468 | 6.1 | 53.7 | 6.0 | 62 | 0.99 | 0.57 |
| S26 | caspase 10 | 1.0E+00 | 1.11 | 13 | AB038173 | 5.6 | 59.6 | 5.9 | 40 | 1.07 | 1.43 |
| S27 | p59 protein | 9.9E–01 | 0.68 | 17 | gi/122768 | 5.3 | 51.5 | 5.8 | 55 | 1.65 | 2.53 |
| S28 | type 2 keratin subunit protein | 1.0E+00 | 1.49 | 22 | M10938 | 5.3 | 52.8 | 5.4 | 42 | 1.22 | 1.50 |
| S29 | keratin 10 | 1.0E+00 | 2.43 | 15 | gi/4557697 | 5.0 | 57.2 | 5.4 | 48 | 2.39 | 1.19 |
| S30 | calreticulin | 9.6E–01 | 1.45 | 20 | gi/237420 | 4.3 | 46.3 | 4.6 | 70 | 0.81 | 1.34 |
| R1 | 67K cytoskeletal type 2 keratin | 9.9E–01 | 0.84 | 15 | gi/88054 | 6.0 | 65.5 | 5.9 | 100 | 0.89 | 0.41 |
| R2 | hnRNP L | 9.6E–01 | 0.72 | 10 | gi/4557645 | 6.7 | 60.2 | 7.2 | 70 | 1.17 | 0.59 |
| R3 | HSP60 | 1.0E+00 | 2.35 | 22 | X53585 | 5.3 | 57.9 | 5.6 | 65 | 1.10 | 0.68 |
| R4 | enolase-1 | 9.9E–01 | 1.55 | 20 | gi/4503571 | 7.0 | 47.2 | 6.8 | 50 | 1.54 | 1.01 |
| R5 | RAD51 | 1.0E+00 | 2.40 | 17 | gi/4506389 | 5.3 | 37.0 | 5.2 | 38 | 1.10 | 0.68 |
| R6 | tropomyocin α-chain | 1.0E+00 | 2.43 | 14 | gi/112448 | 4.9 | 32.1 | 5.0 | 35 | 0.95 | 1.58 |
| R8 | adducin | 9.9E–01 | 0.88 | 21 | gi/321247 | 8.8 | 20.3 | 8.4 | 22 | 1.16 | 0.83 |
| R9 | peptidylprolyl isomerase A | 1.0E+00 | 2.43 | 27 | gi/68401 | 8.8 | 17.7 | 8.6 | 18 | 1.31 | 0.76 |
| R10 | myosin regulatory light chain | 4.8E–01 | 0.53 | 43 | X54617 | 4.6 | 6.7 | 5.4 | 15 | 1.67 | 1.08 |
S1–30, silver-stained spots; R1–10, radioactively labelled spots; gi, NCBI sequence identification numbers.
S20 and R7 did not provide reliable results. Probability, Est’d Z, coverage and theoretical pI and Mr were obtained from the Pro Found search.
The calculation of experimental pI and Mr was based on migration of the protein on a 2D gel.
Fold changes presents the ratio compared with values in the absence of TGFβ1 stimulation, taken as 1.0.
The 38 spots selected were subjected to in-gel trypsin digestion and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). Peptide mass-fingerprinting (PMF) of the selected spots and database search with the peptide mass spectra obtained showed that eight of the 38 proteins identified were structural components of the cell, while others were proteins with functions primarily in regulatory pathways or processes (Table I).
Clustering of identified proteins
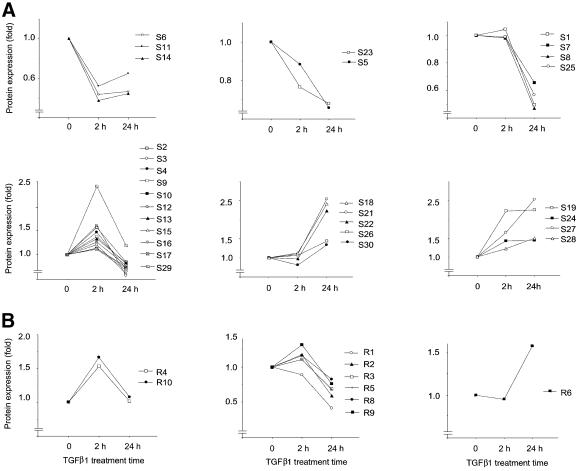
We found that the expression of 65% of the proteins identified (25 of 38) was decreased after 24 h treatment with TGFβ1 (Figure 2). However, after 2 h of incubation, the expression of only five proteins was decreased, compared with untreated cells. Moreover, 18 proteins showed transient or biphasic expression, with an increase after 2 h of TGFβ1 treatment, followed by a return to the level in untreated cells or to inhibition of expression by 24 h. Only 10 proteins were up-regulated after 24 h incubation with TGFβ1 (Figure 2). The TGFβ1-dependent inhibition of expression of some proteins and stimulation of expression of other proteins indicate that the changes described are specific and are not due to a non-selective effect of TGFβ1 on protein synthesis.
Fig. 2. Clustering of identified proteins according to time course of changes. Changes in protein expression level after treatment with TGFβ1 for 2 and 24 h, were calculated using normalized volumes of spots (for silver-stained gels; A), or of radioactivity (for [35S]methionine-labelled cells; B). For annotation of proteins, see Figure 1 and Table I; 1.0-fold refers to the volume value (A) or radioactivity value (B) at the zero time point.
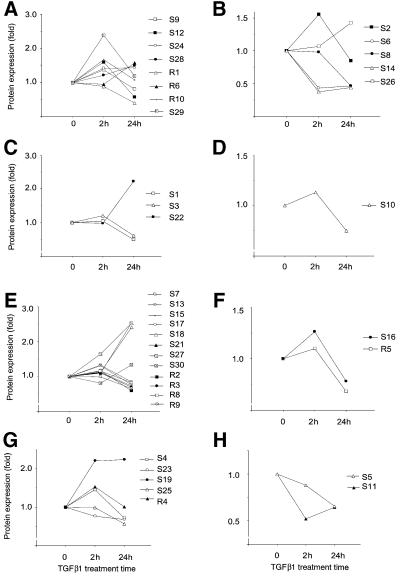
Analysis of TGFβ1-regulated proteins by PMF identified proteins involved in different cellular functions (Table I). Five proteases, three transcription factors, eight cytoskeletal components, two regulators of DNA repair and one protein involved in regulation of TGFβ signalling were identified (Figure 3). Among known TGFβ targets, we identified MMP13 (collagenase-3 precursor), glial fibrillary acidic protein (GFAP), elastase, caspases 1, 3 and 10, calreticulin, apolipoprotein-A1 and inorganic pyrophosphatase (Figures 3 and 4). In agreement with previous reports (Rydziel et al., 1997; Uria et al., 1998), expression of MMP13 was decreased after only 2 h of incubation with TGFβ1 (Figures 2 and 3). Stimulation of GFAP expression by TGFβ has been described previously, but with a delayed time response (Sakai et al., 1990). The time course of changes in expression of other known TGFβ targets was not always similar to that reported, which may depend on which type of cells were used in the experiments (Uria et al., 1998). Thus, several known targets of TGFβ were identified, proving the reliability of our approach.
Fig. 3. Clustering of identified proteins according to their function. Identified TGFβ1-regulated proteins, acting in the same signalling pathway, with similar activities or functions, are shown: (A) cytoskeletal components; (B) proteases; (C) transcription factors; (D) regulators of TGFβ signalling; (E) other components of regulatory pathways; (F) regulators of DNA recombination; (G) regulators of metabolism; (H) unclassified proteins. For annotation of proteins, see Figure 1 and Table I; 1.0-fold refers to the volume value (A) or radioactivity value (B) at the zero time point.
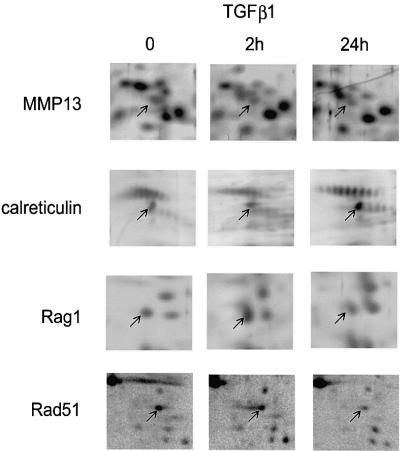
Fig. 4. Effect of TGFβ1 on expression of MMP13, calreticulin, Rag1 and Rad51 proteins. Representative parts of the 2D gels of the proteome of Mv1Lu cells, showing expression of MMP13 (S6), calreticulin (S30), Rag1 (S16) and Rad51 (R5) after treatment with TGFβ1 (5 ng/ml) for various lengths of times, are presented. Arrows show respective proteins.
Twenty-eight of the 38 proteins identified have not been described previously as targets of TGFβ (Table I). Among these targets, proteins were identified that are involved in DNA repair (R5, S16), transcriptional regulation (S22, S1), TGFβ signalling regulation (S10), regulation of RNA stability (R2), synthesis of ATP (S19) and other regulatory intracellular mechanisms (Figures 3 and 4; Table I). The effect of TGFβ1 on the expression of some of the new targets, e.g. Rad51, Rag1 and Sos1, was confirmed by western blotting with respective specific antibodies (Figure 5, for data not shown see Supplementary data at The EMBO Journal Online or http://www.licr.uu.se/groups/ is/index.html). Identification of these targets suggests new ways of integration of TGFβ signalling in the intracellular regulatory network.
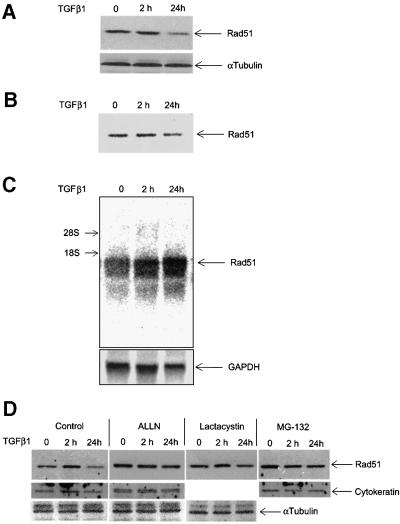
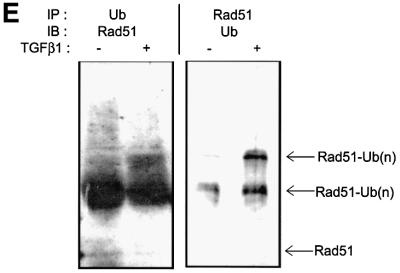
Fig. 5. TGFβ1 inhibits expression of Rad51 protein by affecting its degradation but not its mRNA expression. Expression of endogenous Rad51 protein was evaluated by western blotting (A) and by immunoprecipitation from 35S-labelled cells (B). Mv1Lu cells were treated with TGFβ1 (5 ng/ml) for 2 and 24 h, and total cell lysate was subjected to SDS–PAGE and western blotting (A), or to immunoprecipitation (B) with Rad51-specific antibodies. Migration of Rad51 is shown by arrows. For control of equal loading, expression of α-tubulin was determined using the same cell extracts (for western blotting; A). (C) Rad51 mRNA expression was evaluated by northern blotting of total RNA prepared from cells treated as in (A) and (B), with a human Rad51 probe. Equal loading of RNA was evaluated by re-probing the same membrane with GAPDH probe, as shown. Migration positions for 18S and 28S rRNA are shown. (D) Inhibition of proteasomal degradation by treatment of cells with MG-132 (10 µM), lactacystin (10 µM) or ALLN (12.5 µg/ml) for 6 h (for MG-132) or for 8 h (for lactacystin and ALLN) before cell lysis, blocks TGFβ1-dependent decrease in Rad51 protein expression. Cells were treated with TGFβ1 (5 ng/ml) as indicated. Rad51 protein expression was evaluated by western blotting. Cytokeratin expression was used for normalization of equal loading, as α-tubulin expression in Mv1Lu cells was found to be sensitive to the MG-132 treatment. Migration positions of Rad51, α-tubulin and cytokeratin are shown. (E) TGFβ1 stimulates ubiquitylation of Rad51. Mv1Lu cells were treated with TGFβ1 (5 ng/ml) or not, as indicated. After cell lysis, immunoprecipitation (IP) with anti-ubiquitin or anti-Rad51 antibodies was performed, followed by western blotting (IB) with anti-Rad51 or anti-ubiquitin antibodies, as indicated. Migration positions of unubiquitylated (Rad51) or ubiquitylated Rad51 [Rad51-Ub(n)] are shown. Representative experiments of four (for A, C and D) and three (for B and E) performed, are shown.
We observed increases in expression of certain cytoskeletal components after 2 h of treatment; of those, GFAP, keratins 2 and 10 were down-regulated after 24 h, while lamin A, keratin regulatory subunit 2 and tropomyocin α-chain were still up-regulated (Figure 3A). TGFβ is known to induce epithelial–mesenchymal transdifferentiation, including a potent rearrangement of the cytoskeleton (Cui et al., 1996). Whether the observed effect of TGFβ1 on expression of cytoskeletal components affects the cytoskeletal rearrangement seen in epithelial–mesen chymal transdifferentiation remains to be elucidated.
The effects of TGFβ1 observed on expression of various proteases can affect different cellular functions. Down-regulation of the protease MMP13 can contribute to extracellular matrix formation (Pendas et al., 2000), while down-regulation of caspase 1 could contribute to the effect of TGFβ on the inflammatory response, as caspase 1 regulates maturation of pro-inflammatory cytokines (Nicholson and Thornberry, 1997). It is possible that the regulation of caspase 10 and caspase 3 (Figure 3B) is linked to the effect of TGFβ on apoptosis.
TGFβ1 was found to affect the expression of markers of tumour progression [calreticulin, keratin 10, heat-shock protein-60 (HSP-60)], as well as proteins involved in regulation of tumour cell spreading (MMP13), DNA stability (Rad51, Rag1) and proliferation (FUSEBP) (Figures 3 and 4; Table I). Identification of these proteins is consistent with an active role of TGFβ in regulation of tumorigenesis.
TGFβ1 down-regulates Rad51 protein expression, inhibits DNA repair, and decreases the rate of cell survival after treatment with MMC
We selected one of the novel findings, that TGFβ1 down-regulates the Rad51 protein, for a functional analysis. Since Rad51 is an essential component in the repair of DNA double-strand breaks (Baumann and West, 1998), it is possible that TGFβ1-induced down-regulation of Rad51 expression results in an impairment of DNA repair. The effect of TGFβ1 on Rad51 expression was verified by immunoprecipitation and western blotting of endogenous Rad51 in TGFβ1-treated cells (Figure 5A and B). Moreover, quantification of radioactivity of immunoprecipitated Rad51 from metabolically labelled cells revealed a decrease after TGFβ1 stimulation for 24 h, consistent with the results obtained in the proteome analysis. However, northern blotting showed that TGFβ1 induced Rad51 mRNA expression (Figure 5C), suggesting that TGFβ1 inhibits Rad51 protein expression at the post-transcriptional level. The inhibitory effect of TGFβ1 on Rad51 protein expression was blocked by inhibitors of proteasomal degradation, MG-132, N-acetyl-leucinal-leucinal-norleucinal (ALLN) and lactacystin (Figure 5D). We also found that TGFβ1 stimulated ubiquitylation of Rad51 (Figure 5E). This suggests that TGFβ1 enhances proteasomal degradation of Rad51.
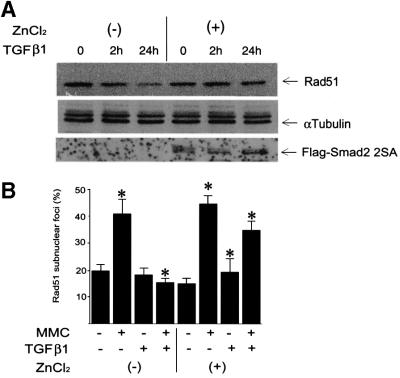
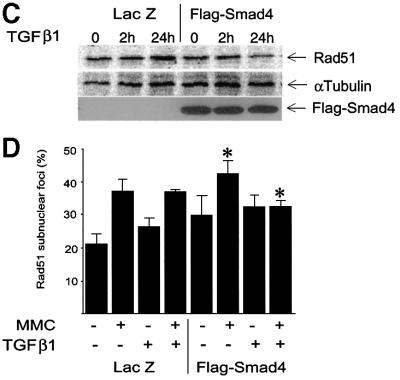
The effect of TGFβ1 on Rad51 expression was found to be Smad dependent, as TGFβ1 did not inhibit Rad51 expression in cells stably transfected with the Flag-Smad2 2SA mutant under a ZnCl2-inducible promoter (Figure 6A and B). The Flag-Smad2 2SA mutant has the two C-terminal serine residues, which are phosphorylated by the receptors, replaced with alanine residues, and has been shown to inhibit Smad-dependent signalling in a dominant-negative manner through inhibition of receptor-dependent phosphorylation of Smad2 and Smad3 (Souchelnytskyi et al., 1997; data not shown). To evaluate the possible involvement of different Smad proteins in TGFβ1-dependent inhibition of Rad51 expression, we performed assays with MDA-MB-468 cells, lacking Smad4, and with mouse embryonic fibroblasts, lacking the Smad3 or Smad2 gene (Figure 6C–F). We found that in the Smad4- or Smad3-deficient cells there was a complete abrogation of the TGFβ1 effect on Rad51 expression; overexpression of Smad4 in Smad4-deficient cells restored the TGFβ1 effect (Figure 6C and D). We observed that the deletion of Smad2 did not completely block TGFβ1-dependent decrease in Rad51 expression (Figure 6E). Thus, receptor-dependent activation of Smad3 and Smad4 is essential for the TGFβ1-dependent down-regulation of Rad51 level, while Smad2 is not. The observation that TGFβ1 inhibits the expression of Rad51 in cells of epithelial (Mv1Lu, MCF-7) as well as mesenchymal (mouse embryonic fibroblasts) origin from different species (human, mouse, mink), indicates that the effect of TGFβ1 on Rad51 is of general character in mammalian cells.
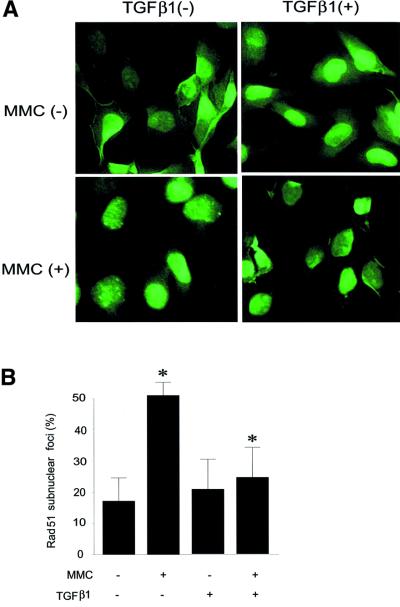
Fig. 6. Effect of TGFβ1 on Rad51 expression and formation of Rad51 nuclear complexes is Smad3 and Smad4 dependent. The effect of TGFβ1 on Rad51 protein expression and formation of Rad51 nuclear complexes in cells stably transfected with Flag-Smad2 2SA mutant (Mv1Lu; A and B), lacking Smad4 and infected with LacZ adenovirus (MDA-MB-468; LacZ; C and D) or with Flag-Smad4 adenovirus (MDA-MB-468; Flag-Smad4; C and D), and lacking Smad3 or Smad2 (mouse embryonic fibroblasts; E and F), was evaluated by western blotting with Rad51 antibodies (A, C and E), and by detection of Rad51-containing nuclear complexes (B, D and F). Equal loading of cell extract was monitored by immunoblotting of the same membrane with anti-α-tubulin antibodies. Expression of Flag-Smad2 2SA was induced by pre-treatment of cells with 50 µM ZnCl2, as evaluated by immunoblotting of cell extracts with anti-Flag antibodies and indicated in (A). Wild-type fibroblasts were used as a control, to show the effect of TGFβ1 on Rad51 expression in mouse embryonic fibroblasts (control; E). Relative expression of Rad51, normalized to α-tubulin expression in the same samples, is presented as indicated. MDA-MB-468 cells were infected with Flag-Smad4 or LacZ adenoviruses as indicated, and Flag-Smad4 expression was evaluated by immuno blotting of cell extracts with anti-Flag antibodies (C, lower panel). To evaluate the formation of Rad51-containing nuclear complexes, cells were treated with TGFβ1 (5 ng/ml) for 24 h, then MMC (500 ng/ml) was added, as indicated. After 1 h treatment with MMC, cells received fresh medium and were cultured for another 5 h, and immunofluores cence staining with Rad51-specific antibodies was then performed. Cells with more than five nuclear foci were scored as positive. Two hundred cells were analysed for each condition, and the percentage of positive cells is presented. *, p <0.05, cells treated with MMC and TGFβ1, compared with cells treated with MMC only (B, D and F), and with cells treated with TGFβ1 only (+ZnCl2, B; KO Smad2, E). MMC induced nuclear complex formation in all cell lines, including smad3-null embryonic fibroblasts (*), with a significance p <0.05.
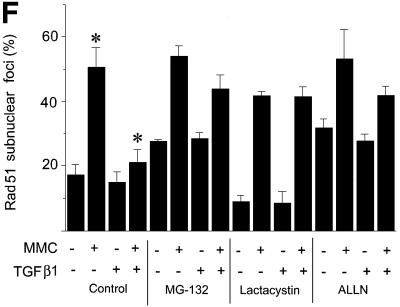
To explore the possibility that TGFβ modulates DNA repair via regulation of Rad51, we analysed the formation of Rad51-containing nuclear complexes after induction of DNA damage in Mv1Lu cells by treatment with mitomycin C (MMC). MMC is known to induce DNA double-strand breaks (Takata et al., 2000). Upon repair of DNA breaks, Rad51 forms nuclear complexes, detectable by immunofluorescence staining of cells (Raderschall et al., 1999), which is a prerequisite for DNA repair. We found that pre-treatment of Mv1Lu cells with TGFβ1 led to a decrease in the MMC-induced Rad51 nuclear complexes (Figure 7A and B), suggesting an impairment of the DNA repair mechanism. This effect of TGFβ1 was abrogated in cells lacking Smad3 or Smad4, and in cells overexpressing Flag-Smad2 2SA which inhibits receptor-dependent phosphorylation of Smads, while in Smad2-null cells the TGFβ1-dependent decrease in formation of Rad51 nuclear complexes is reversed only partially (Figure 6B, D and F), in agreement with the essential role of Smad3 and Smad4 in the TGFβ1-dependent decrease of Rad51 expression (Figure 6). In an attempt to clarify whether it is the variation of the level of Rad51 protein expression that is crucial for the effect of TGFβ1 on formation of DNA repair complexes, we generated Mv1Lu cells stably transfected with Rad51 (R51 cells). Despite the fact that the level of Rad51 protein in transfected cells was only moderately increased, compared with the level of endogenous protein (Figure 7D), it was sufficient to reverse the inhibitory effect of TGFβ1 on nuclear complex formation (Figure 7C). TGFβ1 also decreased Rad51 expression in R51 cells, but the Rad51 level, even after TGFβ1 treatment, was still significantly higher than in vector-transfected or wild-type cells (Figure 7D). We also found that MMC itself did not influence TGFβ1-dependent inhibition of Rad51 expression (Figure 7E). Proteasome inhibitors MG-132, lactacystin and ALLN blocked TGFβ1-dependent down-regulation of Rad51 nuclear complexes (Figure 7F). This is in an agreement with the TGFβ1-dependent stimulation of proteasomal degradation of Rad51 (Figure 5D and E). These results indicate that the TGFβ1-dependent inhibition of Rad51 expression affects DNA repair.
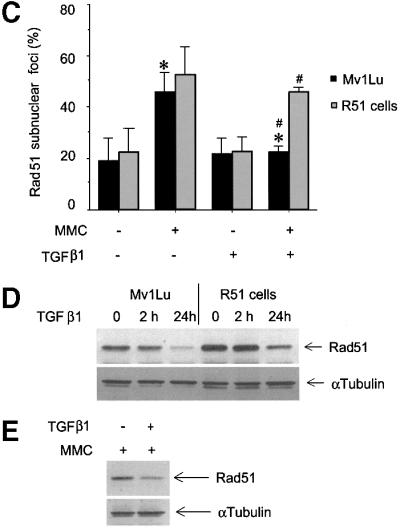
Fig. 7. TGFβ1 treatment inhibits the formation of Rad51-containing nuclear complexes upon repair of DNA damage. Rad51-containing nuclear complex formation in wild-type Mv1Lu cells (A and B) or in Mv1Lu cells stably transfected with Rad51 or empty vector (R51 or Mv1Lu cells, respectively) (C) was evaluated. Mv1Lu cells were treated with TGFβ1 (5 ng/ml) for 24 h, then MMC (500 ng/ml) was added, as indicated. After 1 h treatment with MMC, cells received fresh medium and were cultured for another 5 h, and immuno fluorescence staining with Rad51-specific antibodies was then performed. Cells with more than five nuclear foci were scored as positive (A). Quantification of positive wild-type Mv1Lu cells, Mv1Lu vector-transfected and R51 cells, are shown in (B) and (C). Two hundred cells were analysed for each condition, and the percentage of positive cells is presented. *, p <0.01, cells treated with TGFβ1 and MMC, compared with cells treated only with MMC. #, p <0.01, R51 cells compared with vector-transfected Mv1Lu cells treated with MMC and TGFβ1. (D) The expression level of Rad51 protein in Mv1Lu vector-transfected and R51 cells after TGFβ1 treatment was investigated by western blotting using Rad51 antibodies. (E) MMC does not affect inhibition of Rad51 protein expression after TGFβ1 treatment for 24 h, as evaluated by western blotting of Mv1Lu cell extracts. Cells were treated with MMC (500 ng/ml) for 1 h. To monitor equal sample loading, membrane was probed with anti-α-tubulin antibodies. The migration positions of Rad51 and α-tubulin are indicated. (F) Proteasome inhibitors MG-132, lactacystin and ALLN block TGFβ1-dependent inhibition of the formation of Rad51 nuclear complexes. Mv1Lu cells were treated with TGFβ1 and proteasome inhibitors, as indicated in Figure 5D. Treatment with MMC and detection of Rad51 nuclear complexes were performed as indicated in (A), (B) and (C) above. *, p <0.05, cells treated with MMC and TGFβ1 compared with cells treated with MMC only.
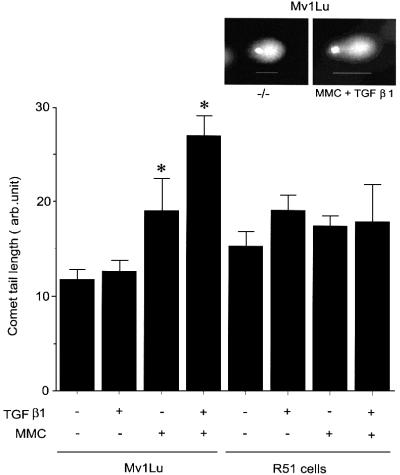
To investigate the effect of TGFβ on DNA repair in an alternative way, we measured the level of DNA fragmentation by single cell gel electrophoresis (Figure 8; SCGE or comet assay). This assay allows an evaluation of the degree of DNA fragmentation by measuring the size of genomic DNA ‘tail’ after electrophoresis of whole cells embedded in agarose, with larger size of the tail reflecting more fragmented DNA (Figure 8, insert). Treatment of Mv1Lu cells with MMC increased the size of the DNA tail, even if the cells were left to recover for 3 h to repair DNA after MMC treatment. Pre-treatment of cells with TGFβ1 resulted in longer comet tails, compared with untreated cells (Figure 8), indicating that TGFβ1 inhibits DNA repair. To evaluate the possible involvement of Rad51 in this DNA repair process, we performed a similar experiment with R51 cells. We found that R51 cells recovered from the MMC-induced DNA damage more efficiently than wild-type cells, with no significant difference between TGFβ1-treated and untreated cells (Figure 8). Therefore, the determination of DNA fragmentation shows that TGFβ1 inhibits the DNA repair process and that increased expression of Rad51 can counteract the TGFβ1 effect.
Fig. 8. TGFβ1 inhibits repair of DNA double-strand breaks after MMC treatment in a Rad51-dependent manner. The degree of fragmentation of genomic DNA was evaluated by single cell gel electrophoresis of wild-type Mv1Lu cells and of R51cells, after treatment with TGFβ1 (24 h; 5 ng/ml) and MMC (500 ng/ml), as indicated. Calculations of the average size of DNA tail of cells are presented. Insert shows representative wild-type Mv1Lu cells after SCGE of untreated or TGFβ1- and MMC-treated cells. *, p <0.05, TGFβ1- and MMC-treated cells compared with only MMC-treated cells.
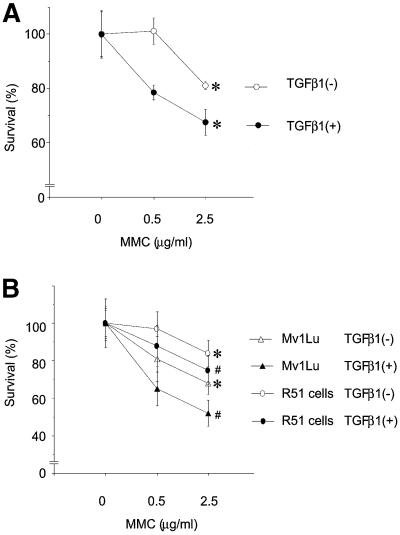
To test whether the inhibition of formation of Rad51 nuclear complexes and DNA repair affects cell survival, we performed a cell foci formation assay using Mv1Lu wild-type cells in order to evaluate the rate of cell recovery after DNA damage (Figure 9). Cells were treated or not with TGFβ1 for 24 h before, and for 5 h just after, induction of DNA damage by incubation with MMC, and were then incubated for up to 10 days in fresh medium without TGFβ1. We found that TGFβ1 significantly decreased the rate of cell survival after treatment with MMC (Figure 9A). Similar results were obtained with the MCF7 cell line (data not shown). In order to prove the involvement of Rad51 in TGFβ1-dependent decrease in cell survival, we performed the foci formation assay with Rad51 cells (Figure 9B). We found that increased expression of Rad51 significantly augmented the level of cell recovery after MMC treatment. The selection procedure during the establishment of the R51 cells did not affect the cells, as Mv1Lu cells stably transfected with empty vector behaved in a similar way to the wild-type cells (Figure 9B). Thus, the TGFβ1-dependent regulation of Rad51 suggests a mechanism whereby TGFβ affects the maintenance of genome stability.
Fig. 9. Pre-treatment of cells with TGFβ1 decreases cell survival after induction of DNA damage. (A) Wild-type Mv1Lu cells were pre-treated with TGFβ1 (5 ng/ml; closed circle) or not (open circle), before incubation with MMC. *, p <0.05, cells treated with TGFβ1 compared with untreated cells. (B) Mv1Lu cells stably transfected with empty vector (wild type, triangles) or Rad51 (R51, circles) were pre-treated with TGFβ1 (filled symbols) or not (open symbols). Cell survival was evaluated by measurement of the rate of colony formation. *, #, p <0.05, vector-transfected cells compared with R51 cells, with (#) or without (*) treatment with TGFβ1.
Discussion
In order to unravel new functional interactions in TGFβ signalling we investigated the changes in the proteome of Mv1Lu cells in response to treatment with TGFβ1. We identified 28 new targets of TGFβ1, providing insights into previously unknown effects of this growth factor, while the identification of several targets for TGFβ that were already known confirmed the effectiveness of our approach (Table I). Our data, and those of others (Soskic et al., 1999; Gerner et al., 2000; Lewis et al., 2000), illustrate the usefulness of proteomics for identification of new targets in signalling pathways in mammalian cells.
Many of the identified targets of TGFβ1 point to new aspects of TGFβ signalling in tumorigenesis. We found down-regulation of cytoskeletal components, notably keratin 10, which is known as a prognostic marker in certain human cancers. Keratin 10 has been shown to be down-regulated in low-differentiated human bladder tumours (Ostergaard et al., 1997), in human cervical carcinoma (Maddox et al., 1999) and mouse skin carcinoma (Santos et al., 1997). The effect of TGFβ on keratin 10 expression has not been reported previously. Another marker of aggressive human breast cancer, calreticulin (Franzen et al., 1996), was found to be up-regulated by TGFβ1 after 24 h (Figure 3E). In mammalian cells, no effect of TGFβ on calreticulin has been described previously, but in zebrafish, the TGFβ-related factor cyclops has been reported to induce expression of calreticulin (Rubinstein et al., 2000), supporting the possibility that calreticulin is also a target for TGFβ in other organisms. HSP60, which is down-regulated by TGFβ1 (Figure 3E), was found to correlate with poor survival prognosis for patients with ovarian carcinomas (Kimura et al., 1993). The consequences of the identified TGFβ1-dependent regulation of expression of keratin 10, HSP60 and calreticulin for the progression of human cancers remain to be explored.
The FUSEBP protein, down-regulated by TGFβ1 (Figures 2 and 3, S22), is an important transcription factor for the activation of the proto-oncogene c-myc (Duncan et al., 1996). Inhibition of c-Myc expression is important in the growth inhibitory action of TGFβ. The inability to inhibit c-Myc expression is often the cause of TGFβ resistance of tumour cells (Alexandrow et al., 1995; Chen et al., 2001). It is an interesting possibility that the inhibition of FUSEBP expression by TGFβ1 is linked to the shut-off of c-Myc expression.
Certain of the TGFβ1 targets identified suggest interesting possibilities for TGFβ cross-talks with regulation of mRNA transport (R2; heterogeneous nuclear ribonucleoprotein L), degradation by ubiquitylation (S21; ubiquitin-like fusion protein An1b), feed-forward regulation of TGFβ signalling (S10; apolipoprotein A-1), regulation of calcium homeostasis (S30; calreticulin) and glycolysis (S19; phosphoglycerate kinase-1). However, the precise molecular mechanisms and physiological significance of these effects remains to be determined.
Certain of the known targets of TGFβ in the regulation of the cell cycle were not detected, e.g. CDK inhibitors or cyclins. This suggests that more sensitive detection techniques need to be employed in order to cover all TGFβ-dependent changes in the cell proteome. 35S labelling allowed detection of additional proteins, compared with silver-stained gels (Figure 1), but the sensitivity of this method is still insufficient. One possible way to increase the sensitivity, albeit in a selective way, would be to use immunodetection. By probing a membrane with proteins transferred from two-dimensional gels with respective antibodies, we could detect expected changes in response to TGFβ1 stimulation in the expression of p21, c-Myc and PAI-1, and in pRb modification probably reflecting hypophosphorylation (data not shown).
Several important conclusions emerged from the comparison of our results with studies of the effect of TGFβ on the transcriptome of mammalian cells (Akiyoshi et al., 2001; Chen et al., 2001; Verrecchia et al., 2001; Zavadil et al., 2001). The pattern of protein turnover (35S-labelled proteins) showed a better correlation with the pattern of mRNA expression than with the pattern of protein presence (silver-stained proteins). Comparing inhibition with stimulation, a correlation of >90% was seen for 35S-labelled proteins, compared with only 47% for silver-stained proteins. However, there was poor correlation in amplitude of changes of 35S-labelled proteins and amplitude of changes of respective mRNAs, in such comparison. Another important conclusion is that the proteome was more inert and had a lower level of reactivity than the transcriptome, with regard to amplitude of changes. In transcriptome analyses, changes in the level of mRNA are often 2- to 10-fold. In our study, the majority of proteins changed their expression level by only ∼50%. Our study suggests that for a complete analysis of dynamic changes in a proteome, different protein detection techniques have to be used. These techniques should evaluate not only protein presence in a cell, but also its turnover, intracellular localization and post-transcriptional modifications.
Corruption of DNA repair is known to promote tumorigenesis (Hanahan and Weinberg, 2000). Here we show that TGFβ1 down-regulates expression of the Rad51 protein, which is involved in repair of DNA double-strand breaks (Baumann and West, 1998). Inactivation of the Rad51 gene results in early embryonal lethality (Lim and Hasty, 1996; Tsuzuki et al., 1996), suggesting that this gene is important for maintenance of genome stability. Rad51 protein or other proteins involved in the regulation of DNA repair have not previously been described as TGFβ targets. TGFβ has been proposed to affect cell sensitivity to DNA-damaging drugs indirectly, through induction of apoptosis, synergizing with cytotoxic effects of drugs (Raynal et al., 1997), or through inhibition of cell cycle progression, and therefore decreasing susceptibility of cells to DNA damage (Ohmori et al., 1998). TGFβ has been proposed also as an inhibitor of gene amplification, but the molecular mechanism behind this effect has not been explored (Glick et al., 1996). Here, we identified Rad51 as a TGFβ1 target involved in the regulation of DNA repair. We confirmed the TGFβ1-induced inhibition of Rad51 expression by immunoblotting and by immunoprecipitation from metabolically labelled cells. The TGFβ1 effect was blocked by overexpression of the dominant-negative Smad2 2SA mutant, and was not observed in cells lacking Smad4 or Smad3, indicating an involvement of Smad3 and Smad4 in the inhibition of Rad51 expression. In contrast, Smad2 is not essential for the TGFβ1 effect, as its deletion did not interfere with the down-regulation of Rad51 (Figure 6). We propose that TGFβ1 down-regulates Rad51 by enhancing its degradation, as inhibitors of proteasomal degradation, MG-132, ALLN and lactacystin, blocked the effect of TGFβ1 (Figure 5D), and since TGFβ1 stimulated ubiquitylation of Rad51 (Figure 5E).
TGFβ1 did not completely block the expression of Rad51 protein, but decreased it to a sufficiently low level to inhibit formation of intracellular Rad51 nuclear complexes in wild-type Mv1Lu cells after MMC-induced DNA double-strand breaks (Figure 7A and B). Moreover, an increase in the Rad51 level by ectopic expression in stably transfected cells reversed the TGFβ1-dependent inhibition of nuclear complex formation (Figure 7C and D). Formation of Rad51-containing complexes in regions of DNA damage is the first step in repair of DNA double-strand breaks. Thus, the inability to form such complexes indicates an impairment of DNA repair. An inhibitory role of TGFβ1 in DNA repair is also supported by the measurement of DNA fragmentation by electrophoresis of genomic DNA, which showed that the pre-treatment with TGFβ1 inhibited repair of DNA double-strand breaks induced by MMC (Figure 8). Absence of this TGFβ1 inhibitory effect in SCGE of Rad51-overexpressing cells, indicates strongly that a decrease in Rad51 protein is of key importance in TGFβ1-induced inhibition of DNA repair.
To test whether the down-regulation of Rad51 expression, inhibition of formation of nuclear complexes and DNA repair has an effect on cell survival, we performed cell foci formation assay. This assay evaluates the ability of cells to recover from treatment with DNA-damaging agents. A significant decrease was seen in foci formation by wild-type Mv1Lu cells pre-treated with TGFβ1 before incubation with MMC, suggesting that TGFβ1 inhibited cell recovery (Figure 9A). However, ectopic expression of Rad51 significantly increased the rate of cell survival (Figure 9B), which is consistent with our finding of Rad51 involvement in TGFβ1-dependent inhibition of DNA repair (Figures 7 and 8).
Our observations indicate that TGFβ1 decreases Rad51 expression, which correlates with inhibition of formation of nuclear DNA repair complexes and inhibition of cell survival after treatment with a DNA-damaging agent. Thus, our proteome analysis has unravelled a novel function of TGFβ as a regulator of genome stability. A possibility that deserves further analysis, is that TGFβ1, by increasing DNA instability, contributes to tumour progression by increasing the rate of tumour-promoting mutations in malignant cells.
Materials and methods
Cells and reagents
Mv1Lu, MCF7 and MDA-MB-468 cells were obtained from ATCC (Manassas, VA), cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and penicillin and streptomycin. Mouse embryonic fibroblasts, wild-type and deficient in Smad2 or Smad3, were obtained from Ester Piek (Piek et al., 2001). Cells were tested regularly for contamination, including Mycoplasma infection. High responsiveness of Mv1Lu cells to TGFβ, minimum 95% inhibition of DNA synthesis, was confirmed in [3H]thymidine incorporation tests. The intactness of Smad signalling was evaluated in assays with Smad-dependent luciferase reporters, i.e. (CAGA)12-luc and SBE4-luc.
For generation of stably transfected cells, Mv1Lu cells were transfected with pAneo/hRad51 plasmid (gift from Shunichi Takeda) and selected in the presence of neomycin/G418. Control Mv1Lu cells were transfected with empty pcDNA3 vector, and selected in the presence of neomycin/G418 according to the same procedure. Expression of Rad51 protein was monitored by western blotting with anti-Rad51 antibodies (H-92; Santa-Cruz). Anti-ubiquitin antibody (P4D1) was obtained from Santa-Cruz (CA). A clone that showed overexpression of Rad51 protein was used. Flag-Smad2 2SA stably transfected Mv1Lu cells were described earlier (Souchelnytskyi et al., 1997). Flag-Smad4 and LacZ adenoviruses were obtained from Aris Moustakas. Proteasome inhibitors MG-132, lactacystin and ALLN were obtained from Calbiochem (USA), Alexis Biochemicals (Switzerland) and Boehringer Mannheim (Germany), respectively. Human TGFβ1 was obtained from Napoleon Ferrara at Genentech.
Sample preparation
Cells were seeded in DMEM with 10% FBS, followed by a change to DMEM with 5% FBS. TGFβ1 (5 ng/ml) was added to the culture medium for 24 h or for the last 2 h of incubation. Cells were labelled with 30 µCi/ml ProMix (Amersham Life Sciences) for the last 6 h of incubation. After incubation, cells were washed twice with phosphate-buffered saline and collected directly in a sample buffer [8 M urea, 4% CHAPS, 0.5% dithiothreitol (DTT), IPG buffer pH 3–10]. The lysates were aliquotted and stored at –70°C. Protein concentration was determined by the Bradford assay.
Two-dimensional electrophoresis
Isoelectrofocusing was performed in strips with immobilized pH gradient (pH 3–10 non-linear gradient, 18 cm; Amersham Biosciences, Uppsala, Sweden). Samples (370 µl) were applied by the rehydration technique. First-dimension isoelectrophoresis was performed in IPGphor (Amersham Biosciences) according to the manufacturer’s recommendation and according to Görg et al. (2000). After the isoelectrofocusing, strips were equilibrated in 50 mM Tris–HCl pH 8.8, 6 M urea, 2.0% SDS, 30% glycerol, with 1% DTT; 10 min) and then in the same solution without DTT but with 4% iodoacetamide (10 min). Equilibrated strips were placed on top of 25 × 35 cm SDS-containing 12% polyacrylamide gels, and fixed with 0.5% agarose in concentrating buffer (62.5 mM Tris–HCl pH 6.8, 0.1% SDS). SDS–PAGE was performed for 14–18 h. Staining of IPG strips after the second-dimension SDS–PAGE showed complete transfer of proteins from the strips. After electrophoresis, gels were fixed in 7.5% acetic acid and 25% methanol for 1 h. Silver staining, compatible with subsequent in-gel digestion, was performed following a modified protocol (Shevchenko et al., 1996). Gels were vacuum-dried and exposed in a Fuji X2000 phosphoimager for 24 h. The pH gradient of the first-dimension electrophoresis was evaluated as proposed by the manufacturer of the strips. We did not observe concentration of proteins in any particular zone of the pH gradient, and we did not observe significant amount of proteins in zones of pH >10 or <3.
Gel analysis
For analysis of silver-stained proteins, gels were scanned in an ImageScanner with the MagicScan32 software and analysed with calculation of volumes of spots by ImageMaster 2D Elite software (Amersham Biosciences). Radioactively labelled gels were exposed in a Fuji X2000 phosphoimager. Incorporated radioactivities of spots were calculated and compared with other gels with the same experimental conditions. Average radioactivity and standard deviation were calculated.
Three to five gels corresponding to one time point, prepared with cell extracts from different experiments and which did not show deviations in the separation of abundant proteins, including internal standards (keratin-1, γ-actin), were used to generate a master gel corresponding to that time point. In total, 42 gels were used to generate master gels of the proteome of cells treated or not with TGFβ1 for 2 or 24 h. The average volume of silver-stained spots and standard deviation were calculated, with volumes of internal standards (keratin-1, γ-actin) used for normalization. Volume values (for silver-stained gels) or radioactivity values (for 35S-labelled gels) of the same spots from the master gels for different time points were compared. Spots with differences in expression for the various time points of TGFβ1 treatment were considered for identification by mass spectrometry.
Protein identification
Excised protein-containing spots were in-gel digested with trypsin essentially as described (Hellman, 2000). Briefly, spots were destained according to Gharahdaghi et al. (1999) and after introduction of trypsin (modified, sequence grade porcine, Promega, Madison, WI) into the dried gel pieces, incubated overnight. After peptide extraction, the sample was concentrated and desalted on a ‘nano-column’ (Gobom et al., 1999) and eluted with matrix directly onto the MALDI target. Spectra were generated on a Bruker Biflex III MALDI-TOF-MS (Bruker Daltonics, Bremen, Germany), using α-cyano-4-hydroxy cinnamic acid as matrix. The instrument was optimized for analytes with molecular masses up to 3500 Da. The spectra were internally calibrated using known internal tryptic peptides from trypsin and searches in the NCBInr sequence database using ProFound (http://prowl.rockfeller.edu/cgi-bin/ProFound) were performed with a tolerance of ± ∼0.1 Da. One miscut and partial oxidation of methionine were allowed. The search results were evaluated by considering the probability value, the Z parameter, peptide coverage, pattern of coverage and correspondence of predicted and experimental pI and molecular mass. Cluster analysis of proteins was performed by inspection of volume or radioactivity values of protein spots as function of time of TGFβ1 treatment.
Western blotting and immunoprecipitation
Mv1Lu cells were treated with TGFβ1 (5 ng/ml) for 2 and 24 h, and then cells were collected in lysis buffer [20 mM Tris–HCl pH 7.4, 1% Triton X-100, 0.5% deoxycholate, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM NaF, 20 mg/ml aprotinin]. For western blotting, total cell lysates were collected, clarified by centrifugation and subjected to SDS–PAGE. Proteins were transferred to a nitrocellulose membrane, Hybond-C extra (Amersham Life Science), blocked with 5% milk containing TBS-T buffer (10 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.1% Tween-20), and incubated with H-92 anti-Rad51 antibodies. Secondary antibodies and ECL detection were performed as described earlier (Yakymovych et al., 2001). To monitor equal sample loading, membranes were stripped and re-probed with anti-α-tubulin antibodies (TU-02; Santa Cruz) or with pan-cytokeratin antibodies (C11; Santa Cruz). For immunoprecipitation, cells were incubated with [35S]methionine (30 µCi/ml), treated with TGFβ1 (5 ng/ml) for 2 and 24 h, and lysed. Clarified lysates were incubated with anti-Rad51 or anti-ubiquitin antibodies and collected on protein A beads (MedicagoAB, Sweden). Immunoprecipitates were subjected to SDS–PAGE, gels were dried and exposed in a Fuji X2000 phosphoimager for 12–18 h. Values of radioactivity in specific and background bands were evaluated using the AIDA 2.11 software (Raytest, Germany).
Northern blotting
Total RNA was extracted using Trizol (Gibco BRL). Total RNA (20 µg/lane) was separated in a formaldehyde-containing 1.0% agarose gel, transferred to Hybond-N membrane (Amersham Biosciences), and then hybridized with a 32P-labelled human Rad51 cDNA probe. After hybridization, the same membrane was re-probed with a 32P-labelled GAPDH cDNA.
Immunofluorescence
Paraformaldehyde-fixed cells were incubated with rabbit anti-human Rad51 antibody H-92. Staining was visualized by incubation with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit antibody (Dako), and slides were mounted in Fluoromount G (Southern Biotechnology Associates, Inc.).
Single cell gel electrophoresis
Mv1Lu cells, wild type or stably transfected with Rad51, were pre-treated with TGFβ1 (5 ng/ml) for 24 h, followed by incubation with MMC (500 µg/ml) for 1 h. Cells were left to recover in fresh medium for 3 h. Then, cells were embedded in agarose in culture medium in Lab-Tek chamber slides (Nalge Nunc, USA). Cells were lysed for 4 h in 1% Triton X-100, 2.5 M NaCl, 20 mM Tris pH 10, 30 mM EDTA, with four changes of the lysis buffer. Agarose blocks were incubated in 300 mM NaOH, 10 mM EDTA for 1 h, and subjected to electrophoresis for 25 min in 300 mM NaOH, 10 mM EDTA solution. After electrophoresis, agarose blocks were incubated in 0.5 M Tris–HCl pH 7.2, for 30 min, and stained with ethidium bromide in water for 30 min. Cells were examined with an Axioplan2 microscope (Zeiss, Germany) connected to a C.4742-95 digital camera (Hamamatsu, Japan). Five different fields were photographed and the size of comet tail of at least 50 cells was measured for each experimental condition.
Cell survival assay
Mv1Lu cells were seeded in 10 cm dishes. To assess the effect of TGFβ1, cells were treated with TGFβ1 (5 ng/ml) for 24 h, then MMC (0.5 or 2.5 µg/ml) was added and incubation continued for 1 h. Thereafter, cells were incubated for 5 h in fresh culture medium with TGFβ1, and then medium was changed to culture medium without TGFβ1. As a control, cells were not treated with TGFβ1 at any step, but only with MMC. After incubation for 5–10 days, the number of colonies was counted. Colonies containing >100 cells were scored as positive.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online or at http://www.licr.uu.se/groups/is/index.html.
Acknowledgments
Acknowledgements
We are grateful to N.Ferrara (Genentech) for TGFβ1, E.Piek (NIH) for mouse embryonic fibroblasts, S.Takeda (Kyoto University) for Rad51 construct and A.Moustakas (Ludwig Institute for Cancer Research) for Flag-Smad4 adenovirus. This work was supported in part by a grant from the Royal Swedish Academy of Sciences to S.S.
References
- Alexandrow M.G., Kawabata,M., Aakre,M. and Moses,H.L. (1995) Overexpression of the c-Myc oncoprotein blocks the growth-inhibitory response but is required for the mitogenic effects of transforming growth factor β1. Proc. Natl Acad. Sci. USA, 92, 3239–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi S., Ishii,M., Nemoto,N., Kawabata,M., Aburatani,H. and Miyazono,K. (2001) Targets of transcriptional regulation by transforming growth factor-β: expression profile analysis using oligonucleotide arrays. Jpn J. Cancer. Res., 92, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. and West,S.C. (1998) Role of human Rad51 protein in homologous recombination and double-stranded break repair. Trends Biochem. Sci., 23, 247–251. [DOI] [PubMed] [Google Scholar]
- Chen C.-R., Kang,Y. and Massagué,J. (2001) Defective repression of c-myc in breast cancer cells: A loss at the core of the transforming growth factor β growth arrest program. Proc. Natl Acad. Sci. USA, 98, 992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W., Fowlis,D.J., Bryson,S., Duffie,E., Ireland,H., Balmain,A. and Akhurst,R.J. (1996) TGFβ1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinoma in transgenic mice. Cell, 86, 531–542. [DOI] [PubMed] [Google Scholar]
- de Caestecker M.P., Piek,E. and Roberts,A.B. (2000) The role of TGF-β signaling in cancer. J. Natl Cancer Inst., 92, 1388–1402. [DOI] [PubMed] [Google Scholar]
- Derynck R., Zhang,Y. and Feng,X.-H. (1998) Smads: transcriptional activators of TGF-β responses. Cell, 95, 737–740. [DOI] [PubMed] [Google Scholar]
- Duncan R., Collins,I., Tomonaga,T., Zhang,T. and Levens,D. (1996) A unique transactivation sequence motif is found in the carboxyl-terminal domain of the single-strand-binding protein FBP. Mol. Cell. Biol., 16, 2274–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen B., Linder,S., Alaiya,A.A., Eriksson,E., Uruy,K., Hirano,T., Okuzawa,K. and Auer,G. (1996) Analysis of polypeptide expression in benign and malignant human breast lesions: down-regulation of cytokeratins. Br. J. Cancer, 74, 1632–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner C., Fröhwein,U., Gotzmann,J., Bayer,E., Gelbmann,D., Bursh,W. and Schulte-Hermann,R. (2000) The Fas-induced apoptosis analyzed by high throughput proteome analysis. J. Biol. Chem., 275, 39018–39026. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F., Weinberg,C.R., Meagher,D.A., Imai,B.S. and Mische,S.M. (1999) Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis, 20, 601–605. [DOI] [PubMed] [Google Scholar]
- Glick A.B., Weinberg,W.C., Wu,I.-H., Quan,W. and Yuspa,S.H. (1996) Transforming growth factor β1 suppresses genomic instability independent of a G1 arrest, p53, and Rb. Cancer Res., 56, 3645–3650. [PubMed] [Google Scholar]
- Gobom J., Nordhoff,E., Mirgorodskaya,E., Ekman,R. and Roepstorff,P. (1999) Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom., 34, 105–116. [DOI] [PubMed] [Google Scholar]
- Görg A., Obermaier,Ch., Boguth,G., Harder,A., Sheibe,B., Wildgruber,R. and Weiss,W. (2000) The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis, 21, 1037–1053. [DOI] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg,R.A. (2000) The hallmarks of cancer. Cell, 100, 57–70. [DOI] [PubMed] [Google Scholar]
- Heldin C.-H., Miyazono,K. and ten Dijke,P. (1997) TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature, 390, 465–471. [DOI] [PubMed] [Google Scholar]
- Hellman U. (2000) Sample preparation by SDS/PAGE and in-gel digestions. In Jollès,P. and Jörnvall,H. (eds), Proteomics in Functional Genomics. Protein Structure Analysis. Birkhauser Verlag AG, Basel, Switzerland, pp. 88:43–88:54.
- Kimura E., Enns,R.E., Thiebaut,F. and Howell,S.B. (1993) Regulation of HSP60 mRNA expression in human ovarian carcinoma cell line. Cancer Chemother. Pharmacol., 32, 279–285. [DOI] [PubMed] [Google Scholar]
- Lewis T.S., Hunt,J.B., Aveline,L.D., Jonscher,K.R., Louie,D.F., Yeh,J.M., Nahreini,T.S., Resing,K.A. and Ahn,N.G. (2000) Identification of novel MAP kinase pathway signal targets by functional proteomics and mass spectrometry. Mol. Cell, 6, 1343–1354. [DOI] [PubMed] [Google Scholar]
- Lim D.S. and Hasty,P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol., 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P., Sasieni,P., Szarewski,A., Anderson,M. and Hanby,A. (1999) Differential expression of keratins 10, 17, and 17 in normal cervical epithelium, cervical intraepithelial neoplasia, and cervical carcinoma. J. Clin. Pathol., 52, 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem., 67, 753–791. [DOI] [PubMed] [Google Scholar]
- Miyazono K., ten Dijke,P. and Heldin,C.H. (2000) TGF-β signaling by Smad proteins. Adv. Immunol., 75, 115–157. [DOI] [PubMed] [Google Scholar]
- Moustakas A., Souchelnytskyi,S. and Heldin,C.H. (2001) Smad regulation in TGF-β signal transduction. J. Cell Sci., 114, 4359–4369. [DOI] [PubMed] [Google Scholar]
- Nicholson D.W. and Thornberry,N.A. (1997) Caspases: killer proteases. Trends Biochem. Sci., 22, 299–306. [DOI] [PubMed] [Google Scholar]
- Ohmori T., Yang,J.-L., Price,J.O. and Arteaga,C. (1998) Blockade of tumor cell transforming growth factor-βs enhances cell cycle progression and sensitizes human breast carcinoma cells to cytotoxic chemotherapy. Exp. Cell Res., 245, 350–359. [DOI] [PubMed] [Google Scholar]
- Ostergaard M., Rasmussen,H.H., Nielsen,H.V., Vorum,H., Orntoft,T.F., Wolf,H. and Celis,J.E. (1997) Proteome profiling of bladder squamous cell carcinomas: identification of markers that define their degree of differentiation. Cancer Res., 57, 4111–4117. [PubMed] [Google Scholar]
- Pendas A.M., Uria,J.A., Jimenez,M.G., Balbin,M., Freije,J.P. and Lopez-Otin,C. (2000) An overview of collagenase-3 expression in malignant tumors and analysis of its potential value as a target in antitumor therapies. Clin. Chim. Acta, 291, 137–155. [DOI] [PubMed] [Google Scholar]
- Piek E., Heldin,C.-H. and ten Dijke,P. (1999) Specificity, diversity, and regulation in TGF-β superfamily signaling. FASEB J., 13, 2105–2124. [PubMed] [Google Scholar]
- Piek E. et al. (2001) Functional characterization of transforming growth factor β signaling in Smad2- and Smad3-deficient fibroblasts. J. Biol. Chem., 276, 19945–19953. [DOI] [PubMed] [Google Scholar]
- Raderschall E., Golub,E.I. and Haaf,T. (1999) Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl Acad. Sci. USA, 96, 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynal S., Nocentini,S., Croisy,A., Lawrence,D.A. and Jullien,P. (1997) Transforming growth factor-β1 enhances the lethal effects of DNA-damaging agents in a human lung-cancer cell line. Int. J. Cancer, 72, 356–361. [DOI] [PubMed] [Google Scholar]
- Roberts A.B. and Sporn,M.B. (1990) The transforming growth factor-βs. In Sporn,M.B. and Roberts,A.B. (eds), Handbook of Experimental Pharmacology: Peptide Growth Factors and their Receptors. 95th edn. Springer-Verlag, New York, NY, pp. 419–472.
- Rubinstein A.L., Lee,D., Luo,R., Henion,P.D. and Halpern,M.E. (2000) Genes dependent on zebrafish cyclops function identified by AFLP differential gene expression screen. Genesis, 26, 86–97. [DOI] [PubMed] [Google Scholar]
- Rydziel S., Varghese,S. and Canalis,E. (1997) Transforming growth factor β1 inhibits collagenase 3 expression by transcriptional and post-transcriptional mechanisms in osteoblast cultures. J. Cell. Physiol., 170, 145–152. [DOI] [PubMed] [Google Scholar]
- Santos M., Ballestin,C., Garcia-Martin,R. and Jorcano,J.L. (1997) Delays in malignant tumor development in transgenic mice by forced epidermal keratin 10 expression in mouse skin carcinomas. Mol. Carcinog., 20, 3–9. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Rawson,C., Lindburg,K. and Barnes,D. (1990) Serum and transforming growth factor β regulate glial fibrillary acidic protein in serum-free-derived mouse embryo cells. Proc. Natl Acad. Sci. USA, 87, 8378–8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm,M., Vorm,O. and Mann,M. (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Soskic V., Gorlach,M., Poznanovic,S., Boehmer,F.D. and Godovac-Zimmermann,J. (1999) Functional proteomics analysis of signal transduction pathways of the platelet-derived growth factor β receptor. Biochemistry, 38, 1757–1764. [DOI] [PubMed] [Google Scholar]
- Souchelnytskyi S., Tamaki,K., Engström,U., Wernstedt,Ch., ten Dijke,P. and Heldin,C.H. (1997) Phosphorylation of Ser456 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J. Biol. Chem., 272, 28107–28115. [DOI] [PubMed] [Google Scholar]
- Takata M., Sasaki,M.S., Sonoda,E., Fukushima,T., Morrison,C., Albala,J.S., Swagemakers,S.M.A., Kanaar,R., Thompson,L.H. and Takeda,S. (2000) The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol., 20, 6476–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T., Fujii,Y., Sakumi,K., Tominaga,Y., Nakao,K., Sekiguchi,M., Matsushiro,A., Yoshimura,Y. and Morita,T. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA, 93, 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker R.F., Shipley,G.D., Moses,H.L. and Holley,R.W. (1984) Growth inhibitor from BSC-1 cells closely related to the platelet type β transforming growth factor. Science, 226, 705–707. [DOI] [PubMed] [Google Scholar]
- Uria J.A., Jimenez,M.G., Balbin,M., Freje,J.M.P. and Lopez-Otin,C. (1998) Differential effects of transforming growth factor-β on the expression of collagenase-1 and collagenase-3 in human fibroblasts. J. Biol. Chem., 273, 9769–9777. [DOI] [PubMed] [Google Scholar]
- Verrecchia F., Chu,M.L. and Mauviel,A. (2001) Identification of novel TGF-β/Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J. Biol. Chem., 276, 17058–17062. [DOI] [PubMed] [Google Scholar]
- Yakymovych I., ten Dijke,P., Heldin,C.-H. and Souchelnytskyi,S. (2001) Regulation of Smad signaling by protein kinase C. FASEB J., 15, 553–555. [DOI] [PubMed] [Google Scholar]
- Zavadil J., Bitzer,M., Liang,D., Yang,Y.C., Massimi,A., Kneitz,S., Piek,E. and Bottinger,E.P. (2001) Genetic programs of epithelial cell plasticity directed by transforming growth factor-β. Proc. Natl Acad. Sci. USA, 98, 6686–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]