Abstract
The distinction between stable (tRNA and rRNA) and unstable (mRNA) RNA has been considered an important feature of bacterial RNA metabolism. One factor thought to contribute to the difference between these RNA populations is polyadenylation, which promotes degradation of unstable RNA. However, the recent discovery that polyadenylation also occurs on stable RNA led us to examine whether poly(A) might serve as a signal for eliminating defective stable RNAs, and thus play a role in RNA quality control. Here we show that a readily denaturable, mutant tRNATrp does not accumulate to normal levels in Escherichia coli because its precursor is rapidly degraded. Degradation is largely dependent on polyadenylation of the precursor by poly(A) polymerase and on its removal by polynucleotide phosphorylase. Thus, in the absence of these two enzymes large amounts of tRNATrp precursor accumulate. We propose that defective stable RNA precursors that are poorly converted to their mature forms may be polyadenylated and subsequently degraded. These data indicate that quality control of stable RNA metabolism in many ways resembles normal turnover of unstable RNA.
Keywords: bacterial metabolism/polyadenylation/rRNA/tRNA/tRNA precursors
Introduction
Despite the high degree of accuracy in the synthesis of macromolecules in biological systems, a small number of molecules with sequences or structures different from those of the functional forms invariably are produced. These defective products, if they accumulate, can exert a deleterious effect on cells. Consequently, various mechanisms have developed to correct or eliminate them. These include the repair of DNA with incorrect or altered sequence, the degradation or refolding of denatured proteins and the degradation of prematurely terminated peptide chains. In the case of RNA biosynthesis, defective molecules can be generated by a number of mechanisms, including synthesis from mutant genes, misincorporation of incorrect nucleotides or premature termination during transcription, and inappropriate processing or folding. Since accumulation of such ‘bad’ RNAs may interfere with the translation machinery, resulting in lowered efficiency or non-functional proteins, it would be expected that RNA quality control mechanisms would be present in cells. For example, it is known that eukaryotic mRNAs with nonsense mutations are degraded by a nonsense-mediated decay pathway (Hilleren and Parker, 1999). However, despite its apparent importance, relatively little information about RNA quality control processes is available, particularly with regard to stable RNAs.
In bacteria, rapid turnover is a hallmark of the mRNA population, in contrast to the rRNAs and tRNAs, which are highly stable during exponential growth. One factor thought to contribute to the rapid decay of mRNA is its propensity to polyadenylation by poly(A) polymerase (Carpousis et al., 1999). Thus, removal of poly(A) polymerase by mutation largely eliminates polyadenylation and generally increases mRNA half-life, whereas overexpression of the enzyme leads to more rapid mRNA decay (O’Hara et al., 1995; Mohanty and Kushner, 1999). Likewise, polyadenylation of RNA I, an RNA that regulates replication of certain plasmids, targets this molecule for degradation (Xu et al., 1993). Consequently, it was quite surprising when, in earlier work, we observed that polyadenylated forms of many stable RNA precursors could be found in Escherichia coli (Li et al., 1998b). Such species are particularly prevalent in exoribonuclease-deficient strains, because the polyadenylated forms are able to accumulate. Based on these observations, we proposed that poly(A) polymerase adds adenylate residues to any RNA molecule with an exposed 3′ hydroxyl terminus, such as stable RNA precursors. As such, it competes with the 3′ to 5′ exoribonucleases, which convert these RNA precursors to their mature forms and which also can remove inappropriately added A residues. However, once a mature RNA is made, its 3′ terminus is protected and becomes relatively resistant to the action of either exoribonucleases or poly(A) polymerase (Li et al., 1998b).
We also raised the possibility that polyadenylation of stable RNA precursors might serve an important physiological role, i.e. a means to eliminate defective RNAs (Li et al., 1998b). Here, we test this idea using a temperature-sensitive mutant of tRNATrp (ts-tRNATrp). This tRNA has a G7→A7 substitution that disrupts the GC base pair at the bottom of the acceptor stem (Eisenberg et al., 1979), and as a consequence, it is more susceptible to denaturation in vivo than the wild type (Eisenberg and Yarus, 1980), making it a useful model for a ‘bad’ RNA.
The sole gene for tRNATrp is located at the distal end of the rrnC operon, eight nucleotides downstream from tRNAAsp. Downstream of tRNATrp is a stem–loop structure followed by eight U residues that presumably serves as a rho-independent terminator. A total of 35 residues are present in the 3′ trailer sequence. Although the maturation pathway for tRNATrp is not known, it would be expected, based on other transcripts, that the long 3′ trailer would first be cleaved by an endoribonuclease upstream of the stem–loop structure, followed by exonucleolytic trimming by RNases T and PH to generate the mature 3′ end (Li and Deutscher, 1996, 2002). The 5′ extra residues are presumably removed by RNase P.
In this paper we show, in agreement with Eisenberg et al. (1979), that the steady-state level of ts-tRNATrp is greatly reduced in vivo compared with its wild-type counterpart, and we present evidence that this is due to degradation of the ts-tRNATrp precursor. Moreover, we show that removal of poly(A) polymerase and/or polynucleotide phosphorylase (PNPase), two enzymes known to participate in the degradation of mRNA, leads to stabilization of the precursor to the defective tRNA. These data indicate that cells have quality control mechanisms for elimination of altered stable RNAs, and that these processes utilize enzymes already known to be involved in the normal turnover of unstable RNAs.
Results
ts-tRNATrp is present at a low amount
In order to test the hypothesis that cells eliminate defective stable RNAs to ensure RNA quality control, and that they do so by a mechanism that involves polyadenylation, we made use of strains carrying a mutant trpT gene that encodes a ts-tRNATrp (Eisenberg et al., 1979). The G7→A7 substitution in the tRNA removes the base pair at the bottom of the acceptor stem and renders the tRNA susceptible to denaturation. The structural and thermodynamic properties of the mutant tRNA have been described by Eisenberg and Yarus (1980). Inasmuch as there is only a single tRNATrp in E.coli, this mutation leads to lethality at 42°C and slows growth at 31°C, the temperature used for most of the studies described here. Based on these observations, ts-tRNATrp appeared to be a useful model RNA to evaluate the aforementioned hypothesis. Accordingly, to examine the metabolism of this mutant tRNA, strains were constructed in which a mutant trpT gene replaced the single wild-type copy on the chromosome (see Materials and methods).
Eisenberg et al. (1979) originally reported that the steady-state level of ts-tRNATrp, based on aminoacylation with tryptophan, reaches only 20–25% of that found when wild-type tRNATrp (wt-tRNATrp) is present. Although there may have been some uncertainty in measuring aminoacylation with a tRNA that has a high probability of denaturing to an inactive form, in fact, we have found a similar value of 15–20% based on northern analysis in multiple experiments using RNA preparations from strains containing either wt-tRNATrp or ts-tRNATrp. A typical example is shown in Figure 1 (compare lanes 1 and 5). In this experiment, which is quantitated in Table I, the strain with the mutant tRNA allele contains only 15% as much tRNATrp as the strain with its wild-type counterpart (compare wild-type lines for each tRNA group).
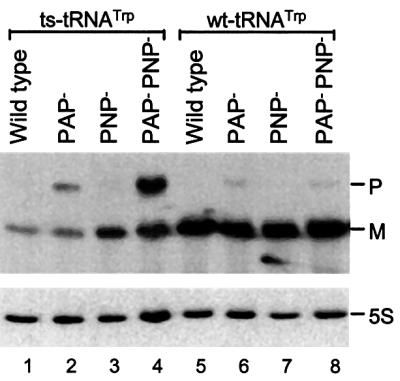
Fig. 1. Northern blot analysis of wt-tRNATrp and ts-tRNATrp in PAP– and PNP– strains. RNA was isolated and northern blot analysis performed as described in Materials and methods using a 16mer probe complementary to residues 39–54 of tRNATrp. Total RNA was isolated from strain CA244 and its derivatives grown at 31°C, and 8 µg were added to each lane. P and M indicate the migration positions of tRNATrp precursor and mature tRNATrp, respectively. 5S indicates the 5S RNA band determined after stripping the tRNATrp probe and reprobing with a probe specific for 5S RNA (Li and Deutscher, 1995).
Table I. Quantitation of northern blot analysis.
| Strain | Mature tRNATrp | Precursor tRNATrp | Total tRNATrp | Precursor form (%) | Total ts-tRNA/wt-tRNA (%) |
|---|---|---|---|---|---|
| wt-tRNATrp | |||||
| wild type | 310 | 9 | 319 | <5 | |
| PAP– | 263 | 17 | 280 | 6 | |
| PNP– | 404 | 5 | 409 | <5 | |
| PAP–PNP– | 374 | 14 | 388 | <5 | |
| ts-tRNATrp | |||||
| wild type | 44 | 3 | 47 | 7 | 15 |
| PAP– | 50 | 32 | 82 | 39 | 29 |
| PNP– | 121 | 12 | 133 | 9 | 33 |
| PAP–PNP– | 104 | 121 | 225 | 54 | 58 |
Individual bands from Figure 1 were analyzed with a PhosphorImager for quantitation. Data are expressed in arbitrary units that have been normalized to the 5S rRNA content of each lane. The experiment presented here is typical of multiple experiments with similar results. However, because the absolute numbers vary greatly from experiment to experiment due to the northern analysis and PhosphorImager quantitation, we have not presented standard errors.
The low amount of ts-tRNATrp could be due to either its decreased synthesis or its increased degradation. However, several pieces of evidence support the latter explanation. First, based on northern analysis, 5S RNA is present at normal levels in the mutant strain (Figure 1, compare lanes 1 and 5). This latter observation is particularly relevant, because the sole gene for tRNATrp is located at the distal end of the rrnC ribosomal operon and it is therefore co-transcribed with other members of the operon, namely the 5S RNA. Secondly, in separate experiments also based on northern analysis of the same strains, another tRNA, tRNASer3, was found to be synthesized in normal amounts (data not shown). Thus, these data make it unlikely that the decreased amount of ts-tRNATrp is due to its lower transcription or to an overall reduction in RNA synthesis in the mutant strain. Rather, one must consider the alternative possibility, that the mutant ts-tRNATrp and/or its precursor are unstable and are degraded, leading to a lower steady-state level of the mature form. Supporting this conclusion is the observation that in the absence of certain enzymes needed for its degradation, the total amount of ts-tRNATrp (mature plus precursor forms) increases dramatically (see below).
ts-tRNATrp precursor accumulates in the absence of poly(A) polymerase
As a first step to determine whether polyadenylation of ts-tRNATrp might play a role in its metabolism, we introduced a mutation which eliminates poly(A) polymerase, the primary enzyme responsible for polyadenylation (Mohanty and Kushner, 2000), into strain MY88, which harbors the ts-tRNATrp gene (Eisenberg et al., 1979). Northern analysis of RNA isolated from this strain lacking poly(A) polymerase (PAP–) grown at 31°C revealed the unexpected accumulation of a large amount of a second ts-tRNATrp product, ∼30 nucleotides longer than the mature tRNA (Figure 2). This product, which is not seen in the PAP+ strain, was also detected with a probe complementary to the 3′ trailer sequence of the tRNA (data not shown), indicating that it is a tRNATrp precursor containing an extended 3′ sequence. This conclusion was confirmed by the RT–PCR analysis that will be presented below. These data indicate that poly(A) polymerase, and presumably polyadenylation, plays an important role in the metabolism of the defective tRNATrp.
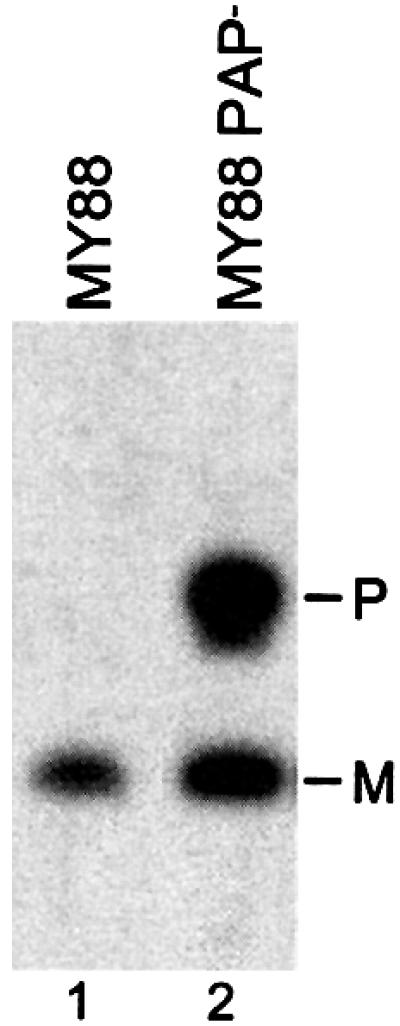
Fig. 2. Accumulation of ts-tRNATrp precursor in PAP– cells. Total RNA was isolated from strains MY88 and MY88 PAP– grown at 31°C, and 8 µg were added to each lane. Northern blot analysis was performed as described in Materials and methods.
Comparison of wt-tRNATrp and ts-tRNATrp metabolism
In order to compare directly the metabolism of ts-tRNATrp with its wild-type counterpart and to identify enzymes that might be responsible for its low level, two series of isogenic strains were constructed. These were made in the background of strain CA244, which has been used extensively in our laboratory (Reuven and Deutscher, 1993). One series contained wt-tRNATrp and the other ts-tRNATrp. The strains otherwise were either wild type, lacking poly(A) polymerase (PAP–), lacking PNPase (PNP–) or lacking both enzymes (PAP–PNP–). All eight strains were grown at 31°C, total RNA was isolated, and northern blot analysis was performed to determine the steady-state amounts of mature and precursor tRNATrp present in each strain. These data are presented in Figure 1, and their quantitation is shown in Table I. The values for tRNA in Table I have been normalized to the amount of 5S RNA in each lane, which remains relatively constant (Figure 1).
As noted earlier, wild-type strains contain much less total ts-tRNATrp than wt-tRNATrp (Figure 1, lanes 1 and 5), but as shown here, these amounts increase dramatically in the absence of enzymes known to participate in RNA degradation. Thus, in agreement with what was observed in the strain MY88 background (Figure 2), elimination of poly(A) polymerase leads to accumulation of the ts-tRNATrp precursor (Figure 1, lane 2), in this case to a level of ∼40% of the total ts-tRNATrp (Table I). In contrast, very little wt-tRNATrp precursor accumulates in a PAP– cell (Figure 1, lane 6), amounting to only 6% of the total tRNA (Table I). Despite the additional accumulation of the ts-tRNATrp precursor, the total amount of ts-tRNATrp (mature plus precursor) in the PAP– background still reaches only ∼30% that of total wt-tRNA (Table I), indicating that substantial degradation continues even in the absence of polyadenylation. Nevertheless, these data show significant differences in the metabolism of wt-tRNATrp and ts-tRNATrp in PAP– cells. While the wild-type precursor is effectively converted to its mature form, the ts-tRNATrp precursor accumulates to a high level. In addition, the low total amount of ts-tRNATrp compared with wt-tRNATrp indicates that much of it is degraded in this background. On the other hand, although the total amount of ts-tRNATrp remains low, it does almost double upon removal of poly(A) polymerase, suggesting that poly(A) polymerase-dependent degradation is a major process that cannot be fully compensated by other degradative pathways. Moreover, as what increases in the absence of poly(A) polymerase is the precursor of ts-tRNATrp, it is likely that this degradation occurs at the precursor stage.
Inasmuch as degradation appeared to be responsible for the low amount of ts-tRNATrp, we attempted to identify an RNase involved in the process. We focused initially on PNPase, because this enzyme had already been shown to play a role in the poly(A)-dependent decay of both mRNA and RNA I (Xu et al., 1993; Carpousis et al., 1999). In the absence of PNPase there is a 3-fold elevation in the total amount of ts-tRNATrp (Figure 1, lane 3 compared with lane 1; Table I). These data indicate that PNPase is one RNase that plays a role in the degradation of the defective tRNA.
The important roles of PNPase and poly(A) polymerase were even more evident when both of these enzymes were absent. In this mutant strain, there is both elevation of the mature form of ts-tRNATrp and massive accumulation of the precursor (Figure 1, lane 4). Quantitatively, the total amount of ts-tRNATrp (precursor plus mature) increases to close to 60% of that of total wt-tRNATrp (Table I). These data demonstrate that both poly(A) polymerase and PNPase are major contributors to the instability of the defective tRNA, and to its greatly decreased steady-state level in vivo. The fact that the total amount of ts-tRNATrp increases 4-fold upon removal of both poly(A) polymerase and PNPase (Table I) strongly supports the conclusion that its low level in wild-type cells is due to degradation. However, since the amount of ts-tRNA still is less than its wild-type counterpart, it is likely that additional enzymes also can participate in this process in the absence of these two proteins.
Polyadenylation of tRNATrp precursor
The data presented indicate an important role for poly(A) polymerase in the removal of defective tRNATrp precursors, suggesting that prior polyadenylation facilitates their degradation. To ensure that tRNATrp precursors can, in fact, be polyadenylated in vivo, we made use of an RT–PCR cloning procedure employed previously (Li et al., 1998b), and determined the 3′ terminal sequences of representative precursor molecules (Table II). As a control, we first examined the precursor to wild-type tRNATrp that accumulates in an RNase IIts PNP– D– BN– T– multiple exoribonuclease-deficient strain. In previous work we showed that precursors to many stable RNAs become polyadenylated when they cannot be converted to their mature forms due to the absence of processing exoribonucleases (Li et al., 1998b). In agreement with that study, we find that in this strain, wt-tRNATrp precursor also contains non-encoded 3′ terminal A residues (Table II). Of 19 clones sequenced, 10 had additional A residues that varied from one to three residues in length. Transcription of tRNATrp normally terminates at a rho-independent site, which results in multiple consecutive U residues in the tRNA precursor. Of the 19 clones examined, all had between five and eight of these U residues, and the additional A residues were found on molecules of each length.
Table II. RT–PCR analysis of clones derived from tRNATrp precursors.
| Strain |
tRNA |
Precursor |
Precursor +adenylates |
| IIts PNP– D– BN– T– | wt-tRNATrp | 9 | 10 (1–3) |
| PNP– | ts-tRNATrp | 3 | 6 (1–3) |
| PAP–PNP– | ts-tRNATrp | 10 | 0 |
The values presented are the number of RT–PCR clones derived from RNA with the indicated 3′ terminal sequence. Values in parenthesis are the lengths of non-encoded adenylate tails present.
A similar examination of the precursor to ts-tRNATrp was conducted (Table II). In the PNP– background, six of nine clones sequenced contained non-encoded stretches of A residues one to three residues in length. In contrast, none of the 10 clones derived from the PAP–PNP– strain contained additional A residues. These data show that the precursor to ts-tRNATrp can be polyadenylated in vivo, but only when poly(A) polymerase is present.
Stability of the precursor and mature forms of ts-tRNATrp
The data presented to this point are most consistent with the conclusion that the low level of ts-tRNATrp is due to degradation, and that this degradation occurs primarily at the precursor level. To prove this point directly, ideally it would be of interest to measure the decay rates of the precursor and mature forms of the defective tRNA directly. Unfortunately, it is not possible to carry out such an experiment satisfactorily, because in the wild-type background precursor does not accumulate, and its decay cannot be measured. In the PAP– strain, precursor does accumulate to levels at which it can be measured, but its rate of disappearance has been affected by the absence of poly(A) polymerase, which is the reason it accumulates in this background. Nevertheless, in order to obtain some information about the relative decay rates of the precursor and mature forms of ts-tRNATrp, their half-lives in the PAP– strain were measured after addition of rifampicin and nalidixic acid. In two experiments, the half-life of the precursor was 15–20 min, whereas that of the mature form was >1 h (data not shown). Thus, even in the absence of poly(A) polymerase, a condition under which the precursor is stabilized to some degree, the precursor remains a relatively unstable molecule.
In a second experiment, the stabilities of the precursor and mature forms of ts-tRNATrp were determined after cells were shifted from 31 to 42°C, a temperature which would be expected to accentuate the instability of the defective tRNA. However, as can be seen in Figure 3, in both wild-type MY88 cells and their PAP– derivative, the mature form of ts-tRNATrp is stable. On the other hand, the precursor to ts-tRNATrp essentially disappears by 7.5 min after the shift. This experiment demonstrates that even under extreme conditions, the mature form of ts-tRNATrp is quite stable. This observation, coupled with the instability of the precursor, supports the conclusion that at 31°C it is the degradation of the precursor that results in a low level of the defective tRNA. It should be noted that this experiment also shows that at 42°C degradation of the precursor can proceed rapidly even in the absence of polyadenylation. In contrast, at 31°C polyadenylation was demonstrated to play a significant role.
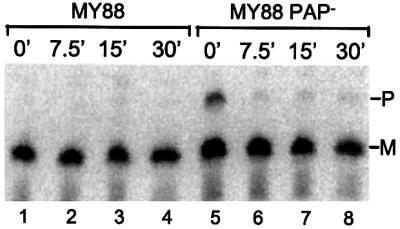
Fig. 3. Degradation of ts-tRNATrp precursor at 42°C. MY88 and MY88 PAP– cells were grown at 31°C and shifted to 42°C. Total RNA was isolated at time 0 (lanes 1 and 5) and at 7.5 min (lanes 2 and 6), 15 min (lanes 3 and 7) and 30 min (lanes 4 and 8) after the temperature shift. Five micrograms of RNA were added to each lane. Northern blot analysis was performed as described in Materials and methods.
Discussion
Using ts-tRNATrp as a model for a ‘bad’ RNA, we have investigated whether E.coli possesses a quality control system for eliminating defective, stable RNAs. In earlier work, we showed that stable RNA precursors are polyadenylated when they cannot be converted to their mature forms (Li et al., 1998b). In that artificial situation, maturation of RNA precursors was prevented because the necessary processing exoribonucleases had been removed by mutation. We suggested that such polyadenylation might normally serve as a physiological signal to promote RNA degradation when defective precursors are not matured or are processed slowly. The data presented here support that hypothesis. Thus, a mutant ts-tRNATrp, which is known to denature readily (Eisenberg and Yarus, 1980), was found to be present in cells at extremely low levels compared with the wt-tRNA, apparently because it is rapidly degraded at the precursor level. We also found that the total amount of ts-tRNATrp (mature plus precursor forms) was greatly increased in the absence of poly(A) polymerase and PNPase, implicating these two enzymes in the decay process. A large body of work has already shown that these enzymes play an important role in the degradation of mRNA and RNA I, which are unstable RNA molecules (Xu et al., 1993; Carpousis et al., 1999). Assuming that ts-tRNATrp would be representative of other defective, stable RNAs, these data suggest a commonality in the removal of all RNAs, and raise the question of whether the distinction made between stable and unstable RNAs is unwarranted.
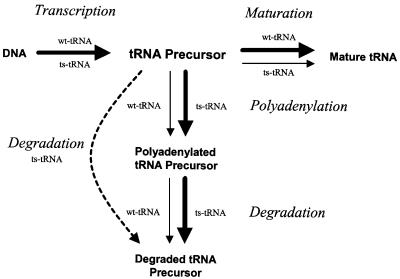
As illustrated in Figure 4, we propose that poly(A) polymerase is a scavenger enzyme that adds poly(A) tails to any exposed RNA 3′ terminus. We suggest that under normal circumstances such free ends would be found only on mRNAs and other unstable RNAs, whereas mature, stable RNAs would generally be protected either by aminoacylation, RNA secondary structure or by being buried within an RNP structure. In contrast, precursors to stable RNAs might have their extended 3′ termini exposed and accessible to poly(A) polymerase. Normally, stable RNA precursors are rapidly converted to their mature counterparts, precluding the possibility of polyadenylation. However, if this conversion were slowed due to an abnormal RNA structure, as would be the case for a defective RNA such as ts-tRNATrp, poly(A) polymerase would then be able to act. Once tagged by a poly(A) tail, the RNA would be subject to rapid degradation, often involving PNPase, as is already known to be the case for unstable RNAs (Xu et al., 1993; Carpousis et al., 1999). In such a scenario, as shown in Figure 4, RNA maturation and degradation are competitive processes that can be tilted in either direction by the structure of the RNA substrate, and the relative levels of the maturation and degradation enzymes.
Fig. 4. Model for quality control of stable RNA synthesis. Genes encoding normal (wt-tRNA) and defective (ts-tRNA) RNAs are transcribed with equal efficiency to generate tRNA precursors. However, whereas wt-tRNA precursor is rapidly converted to its mature form by the processing RNases, maturation of the defective precursor is greatly slowed. As a consequence, the defective tRNA precursor is first subject to polyadenylation by poly(A) polymerase, and is then degraded by PNPase and other RNases. Based on this model, there is competition at the tRNA precursor level between the processing RNases and poly(A) polymerase. Altered precursor structure or removal of processing RNases (Li et al., 1998b) makes the precursor available for polyadenylation and, in the case of the defective tRNA precursor, shifts the pathway from mature tRNA production to degradation. The thickness of the arrows indicates the relative rates of individual steps in the pathway with wt-tRNA or ts-tRNA. The dashed lines indicate the degradation of tRNA precursor that occurs in the absence of polyadenylation.
Thus, as shown here, removal of the degradative enzymes poly(A) polymerase and PNPase leads to massive accumulation of the ts-tRNATrp precursor, because in the absence of these enzymes the degradation pathway is dramatically slowed, and the abnormal structure of the precursor also limits the rate at which it is matured. In contrast, wt-tRNATrp maturation proceeds at a rapid rate and little precursor accumulates, independent of whether degradation enzymes are present. However, when the maturation machinery is disrupted, as was the case for the multiple exoribonuclease-deficient mutants studied previously, even precursors to wild-type, stable RNAs can accumulate and be polyadenylated (Li and Deutscher, 1996; Li et al., 1998a). We find this model attractive because it might provide an explanation for why RNA precursors exist at all, i.e. precursors serve as the substrates on which quality control of defective, stable RNA molecules is exercised, because their extended 3′ structure allows them to be polyadenylated.
Although this model explains most of our observations related to quality control of the defective tRNA, some details are not yet completely understood. For example, as shown in Table I, in the absence of PNPase alone, there is not only an increase in the amount of tRNA precursor, but also a >2-fold increase in the amount of mature ts-tRNATrp. Since mature ts-tRNATrp appears to be stable, even at an elevated temperature, we suspect that this increase is a secondary consequence of the decreased degradation of the precursor in the absence of PNPase, allowing more of it to be converted to the mature form, albeit slowly. However, we cannot eliminate the possibility that some degradation of the mature ts-tRNA may also occur. Secondly, it is evident that considerable degradation takes place even when poly(A) polymerase is absent, both at 42°C (Figure 3) and at 31°C (Figure 1; Table I), indicating that some RNases can act on the ts-tRNATrp precursor even in the absence of polyadenylation. Since polyadenylation assumes a larger role in the absence of PNPase, the action of other degradative enzymes may be even more dependent on the presence of a poly(A) tail on the ts-tRNATrp precursor than is PNPase. While we do not yet know which RNases might participate in the degradation of the ts-tRNATrp precursor in the absence of PNPase, two possible candidates for such a role include RNase II, which is already known to participate in mRNA decay (Coburn and Mackie, 1999; Regnier and Arraiano, 2000), and RNase R. Both of these enzymes have many properties in common with PNPase (Deutscher and Li, 2000). However, inasmuch as the removal of poly(A) polymerase and PNPase results in a major increase in the total amount of ts-tRNATrp (from 15 to 58% of wild type in Table I), we believe that the predominant degradation pathway is via polyadenylation and PNPase. Further work will be needed to evaluate the role of the various degrading activities.
The action of exoribonucleases may also account for why the poly(A) tails found on the accumulated RNA precursors are so short. First, RNA molecules with long tails may be degraded preferentially and thus will not be seen, and secondly, the RNases may shorten the poly(A) sequences. What we observe represents the steady state of poly(A) lengthening by poly(A) polymerase and its shortening by exoribonucleases. All of these aforementioned points suggest that the model presented above will require modification as more details about RNA quality control emerge.
Although the synthesis of wild-type tRNATrp is affected relatively little by the absence of poly(A) polymerase and/or PNPase (Figure 1; Table I), a few points deserve mention. We observed repeatedly that small amounts of precursor to wt-tRNATrp accumulate in PAP– cells (see Figure 1, lanes 6 and 8). In addition, strains lacking PNPase have slightly increased amounts of mature wt-tRNATrp (Table I). These findings suggest that even for wt-tRNATrp, a small number of defective molecules may be produced due to incorrect synthesis, processing or folding, and that they normally are degraded by the same pathways that remove the ts-tRNATrp. It remains to be determined whether this observation extends to the production of all stable tRNA molecules.
One interesting aside to this work is the question of how the altered structure of the ts-tRNATrp leads to a decreased rate of tRNA maturation. Do the processing exoribonucleases recognize tRNA structure? We suspect not. Rather, the tRNATrp precursor has a long 3′ tail of 35 nucleotides, with the possibility of formation of an extensive stem–loop structure. We have shown that for many tRNA precursors endonucleolytic cleavage by RNase E precedes exonucleolytic trimming to the mature 3′ terminus (Li and Deutscher, 2002). Since the precursors that accumulate here have essentially their complete 3′ tails, perhaps it is the initial 3′ endoribonucleolytic cleavage that is slowed down by the altered precursor structure. Additional studies are needed to resolve this question.
Materials and methods
Bacterial strains
Escherichia coli K12 strain CA244 (lacZ, trp, relA, spoT) (Reuven and Deutscher, 1993) was considered wild type for these studies. Strains (CA244) lacking poly(A) polymerase (PAP–), PNPase (PNP–) or both enzymes (PAP–PNP–) were as described previously (Li et al., 1998b). The mutation in pcnB, encoding PAP, is a kanamycin cassette interruption, and that in pnp is a Tn5 interruption. Each mutation leads to a null phenotype based on direct assay of the relevant enzyme. Strain MY88, carrying a G7→A7 temperature-sensitive mutation in trpT, the gene encoding tRNATrp, was provided by Dr Michael Yarus, University of Colorado (Eisenberg et al., 1979). The mutant trpT gene was transferred into the chromosome of the aforementioned strains by phage P1-mediated transduction, selecting initially for tetracycline resistance due to a nearby Tn10 insertion (ilv500::Tn10 from strain CAG18431, kindly provided by the Yale E.coli Genetic Stock Center). The tetracycline-resistant transductants were subsequently screened for temperature sensitivity at 42°C and affected colonies isolated and purified. The resulting strains CA244trpTts, CA244PAP–, trpTts and CA244PNP–, trpTts were found to be unable to grow at 42°C, whereas strain CA244PAP–, PNP–, trpTts grew slowly.
Materials
[3H]ATP was purchased from Amersham; [γ-32P]ATP was from Dupont/NEN; phage T4 RNA ligase was from New England Biolabs; phage T4 polynucleotide kinase and Moloney murine leukemia virus reverse transcriptase were from Gibco-BRL; RNasin was obtained from Promega; sequagel for northern analysis was purchased from National Diagnostics; the GeneAmp PCR Reagent kit was from Perkin-Elmer; and the TA cloning kit was from Invitrogen. Oligonucleotides were prepared at our local facility. The oligoribonucleotide used as a linker ligated to the 3′ end of cellular RNA was pUGGUGGUGGAUCCCGGGAUCp. The complementary oligodeoxynucleotide, GATCCCGGGATCCACCACCA, served as the primer for reverse transcription and as one of the primers for PCR. The oligodeoxynucleotide covering the 3′ terminus of the tRNA, CCGCCCCTGCCAGAAATCATCC, was used as the second PCR primer. Another oligodeoxynucleotide, ACTCCCAACACCCGGT, specific for tRNATrp and complementary to residues 39–54 of the tRNA, served as the probe for northern analysis.
RNA preparation and northern blot analysis
In order to avoid conversion of the slow-growing mutant cells to faster-growing forms, they were grown in yeast-tryptone medium to an A550 ∼0.5 from a colony without dilution. Total cellular RNA was isolated by phenol extraction as described previously (Li and Deutscher, 1995). The RNA was used for the experiments presented without further fractionation. Northern blot analysis was performed as reported previously (Li et al., 1998b) after electrophoresis on denaturing 6% acrylamide gels. Hybridization with the 16mer oligodeoxynucleotide complementary to tRNATrp, described above, and washing were at 37°C. Normalization of the amount of RNA loaded was by comparison with the amount of 5S RNA on the same membrane analyzed after stripping the tRNATrp probe, as described previously (Li and Deutscher, 1995, 1996). Comparison was also made with tRNASer3 to ensure normal synthesis of 5S RNA. Hybridization for 5S RNA was at 45°C. Quantitation was carried out by analysis on a Molecular Dynamics PhosphorImager.
RT–PCR cloning
The 3′ terminal sequences of tRNATrp accumulated in mutant cells were determined by RT–PCR, using one primer in the tRNA and one in the 3′ extension, as described previously (Li et al., 1998b).
Other assays
Poly(A) polymerase and PNPase activities were determined as described previously (Reuven et al., 1997). Protein concentration was determined using the method of Bradford (1976).
Acknowledgments
Acknowledgements
We thank Michael Yarus and the Yale E.coli Genetic Stock Center for strains, and Roy Parker for useful discussions. This work was supported by grant GM16317 from the National Institutes of Health.
References
- Bradford M.M. (1976) A rapid and sensitive method for the determination of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J., Vanzo,N.F. and Raynal,L.G. (1999) mRNA degradation. Trends Genet., 15, 24–28. [DOI] [PubMed] [Google Scholar]
- Coburn G.A. and Mackie,G.A. (1999) Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol., 62, 55–108. [DOI] [PubMed] [Google Scholar]
- Deutscher M.P. and Li,Z. (2000) Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acid Res. Mol. Biol., 66, 67–105. [DOI] [PubMed] [Google Scholar]
- Eisenberg S.P. and Yarus,M. (1980) The structure and aminoacylation of a temperature-sensitive tRNATrp (Escherichia coli). J. Biol. Chem., 255, 1128–1137. [PubMed] [Google Scholar]
- Eisenberg S.P., Soll,L. and Yarus,M. (1979) The purification and sequence of a temperature-sensitive tryptophan tRNA. J. Biol. Chem., 254, 5562–5566. [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (1999) Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet., 33, 229–260. [DOI] [PubMed] [Google Scholar]
- Li Z. and Deutscher,M.P. (1995) The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc. Natl Acad. Sci. USA, 92, 6883–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. and Deutscher,M.P. (1996) Maturation pathways for E.coli tRNA precursors: a random multienzyme process in vivo. Cell, 86, 503–512. [DOI] [PubMed] [Google Scholar]
- Li Z. and Deutscher,M.P. (2002) RNase plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA, 8, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pandit,S. and Deutscher,M.P. (1998a) 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 2856–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pandit,S.P. and Deutscher,M.P. (1998b) Polyadenylation of stable RNA precursors in vivo. Proc. Natl Acad. Sci. USA, 95, 12158–12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B.K. and Kushner,S.R. (1999) Analysis of the function of Escherichia coli poly(A) polymerase 1 in RNA metabolism. Mol. Microbiol., 34, 1094–1108. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K. and Kushner,S.R. (2000) Polynucleotide phosphorylase functions both as a 3′→5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 11966–11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara E.B., Chekanova,J.A., Ingle,C.A., Kushner,Z.R., Peters,E. and Kushner,S.R. (1995) Polyadenylation helps regulate mRNA decay in Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 1807–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier P. and Arraiano,C.M. (2000) Degradation of mRNA in bacteria: emergence of ubiquitous features. BioEssays, 22, 235–244. [DOI] [PubMed] [Google Scholar]
- Reuven N.B. and Deutscher,M.P. (1993) Multiple exoribonucleases are required for the 3′ processing of Escherichia coli tRNA precursors in vivo. FASEB J., 7, 143–148. [DOI] [PubMed] [Google Scholar]
- Reuven N.B., Zhou,Z. and Deutscher,M.P. (1997) Functional overlap of tRNA nucleotidyltransferase, poly(A) polymerase I and polynucleotide phosphorylase. J. Biol. Chem., 272, 33255–33259. [DOI] [PubMed] [Google Scholar]
- Xu F., Lin-Chao,S. and Cohen,S.N. (1993) The Escherichia coli pcnB gene promotes adenylation of antisense RNAI of ColE1 type plasmids in vivo and degradation of RNAI decay intermediate. Proc. Natl Acad. Sci. USA, 90, 6756–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]