Abstract
We previously showed that the ANGUSTIFOLIA (AN) gene regulates the width of leaves of Arabidopsis thaliana, by controlling the polar elongation of leaf cells. In the present study, we found that the abnormal arrangement of cortical microtubules (MTs) in an leaf cells appeared to account entirely for the abnormal shape of the cells. It suggested that the AN gene might regulate the polarity of cell growth by controlling the arrangement of cortical MTs. We cloned the AN gene using a map-based strategy and identified it as the first member of the CtBP family to be found in plants. Wild-type AN cDNA reversed the narrow-leaved phenotype and the abnormal arrangement of cortical MTs of the an-1 mutation. In the animal kingdom, CtBPs self-associate and act as co-repressors of transcription. The AN protein can also self-associate in the yeast two-hybrid system. Furthermore, microarray analysis suggested that the AN gene might regulate the expression of certain genes, e.g. the gene involved in formation of cell walls, MERI5. A discussion of the molecular mechanisms involved in the leaf shape regulation is presented based on our observations.
Keywords: ANGUSTIFOLIA/cortical microtubules/CtBP/leaf morphogenesis/MERI5
Introduction
Plants are composed of three basic organs, namely, leaves, stems and roots. The floral organs, such as petals, stamens and carpels, can be considered to be modified leaves. Thus, the leaf is a key organ for a full understanding of plant morphogenesis. As is the case for animal embryos, the development of leaf primordia is dependent on the establishment of several polarities (for review see Tsukaya, 1998, 2002). Using the techniques of developmental genetics to unravel the details of leaf morphogenesis (Tsukaya, 1995) we previously showed that two genes are responsible in Arabidopsis for the polarity-specific expansion of leaves (Tsukaya et al., 1994; Tsuge et al., 1996). They are the ANGUSTIFOLIA (AN) gene, which regulates polar elongation in the leaf-width direction, and the ROTUNDIFOLIA3 (ROT3) gene, which regulates polar elongation in the leaf-length direction. We cloned the ROT3 gene by T-DNA tagging and showed that the ROT3 gene encodes a novel cytochrome P450 (Kim et al., 1998a, 1999). The phenotype of an mutant plants is similar to that of rot3 plants, but the defects in the polarity-dependent elongation of leaf cells affect leaf expansion in different directions (Tsuge et al., 1996). The defect in both cases is limited to leaves and to floral organs, which are modified leaves. The product and function of the AN gene remains to be determined.
The polarity-dependent expansion of plant cells is generally associated with the organization of cortical microtubules (MTs) that are arranged beneath the plasma membrane (Cyr, 1994; Shibaoka, 1994). In Arabidopsis, disturbance by mutation of the organization of MTs can cause dwarfism (Traas et al., 1995; McClinton and Sung, 1997), and the abnormal arrangement of cortical MTs has been noted in mutants with altered polar development of roots (Aeschbacker et al., 1995; Hauser et al., 1995) and shoots (Furutani et al., 2000). Several Arabidopsis mutants with abnormal amounts of cellulose microfibrils have also been reported (Potikha and Delmer, 1995). However, the role of the regulation of the arrangement of MTs in the genetic control of leaf-cell expansion remains to be clarified.
We show here that a defect in the arrangement of cortical MTs in leaf cells is associated with the an mutation and is reversed by AN cDNA, which we cloned in the present study. The abnormal pattern of cortical MTs in an leaf cells can provide a complete explanation for the polarity-dependent alteration in the shape of the leaf cells and, therefore, it can also explain the change in shape of the leaves. We discuss the results of our analysis in terms of possible roles of the AN gene, the first member of the family of CtBP genes to be identified in plants.
Results
The orientation of cortical MTs in the an mutant is abnormal
The regulation of polar changes in cell morphology, which is disrupted in leaf cells of Arabidopsis plants with the an mutation (Figure 1) should be closely related to the regulation of the orientation of cellulose microfibrils which is governed, in turn, by the arrangement of cortical MTs. Therefore, we analyzed the orientation of cortical MTs in leaf cells of an and rot3 mutant plants by immunohistochemical staining and by monitoring the expression of the TUBULIN6::Green Fluorescent Protein (TUB6::GFP) gene for the TUB6–GFP fusion protein. We analyzed the orientation of MTs in epidermal cells and in the subepidermal layer of cells (palisade cells) on the adaxial side of the leaf. As shown in Figure 2, we found that the cortical MTs in epidermal cells of the an mutant were more regularly aligned parallel to the leaf-width direction than in those of the wild type. This pattern of MTs can explain the morphological phenotype of the an epidermal cells, which protrude less extensively than wild-type cells in the leaf-width direction (Figure 2; Tsuge et al., 1996). We next focused on the quantitative characterization of the palisade cells because of the simple rod-like shape of normal cells. We considered that the palisade cells represented lamina cells since the shapes of epidermal cells and spongy cells are irregular, as a consequence of multidirectional expansion, and this irregularity hampers detection of polarity-specific changes. We found no difference in numbers of MTs between wild-type (wt) and mutant plants when we examined the MTs immunohistochemically and by monitoring expression of the TUB6::GFP fusion gene (data not shown). Analysis by RT–PCR on one of the endogenous genes for tubulin, TUB4, indicated that there were no significant differences among the levels of expression of the transcript of this gene also (see Figure 7).
Fig. 1. Cross sections and gross morphology of the fifth rosette leaves of a wild-type (wt) plant, an an-1 mutant plant and a transgenic an-1 mutant that expressed the wild-type AN gene included in the TAC clone 1A13. (A and B) Cross sections of the fifth rosette leaves of the wt (A) and the an-1 mutant (B). Bar, 100 µm. Note the abnormal shape of the palisade cells in the an-1 mutant leaf. (C and D) The fifth rosette leaves of wt, an-1 and a transgenic an-1 plant that harbored 1A13 (C), and magnified views of the same leaves (D). Bars, 5 mm (C) and 1 mm (D).
Fig. 2. Confocal images of the arrangement of cortical MTs in the leaf epidermis. The adaxial epidermis is shown of wt, an-1 mutant and rot3-1 mutant leaves, from left to right. MTs are visible as fluorescence from the GFP–TUB6 fusion protein. Bars, 50 µm.
Fig. 7. Intracellular localization of AN::GFP in plant cells. (A) Intracellular localization of AN::GFP in trichomes of transgenic Arabidopsis leaves. (B) Non-transgenic wild-type trichomes that failed to generate a signal under the same conditions for fluorescence microscopy as those that yielded (A). (C) Transient expression of AN::GFP in onion epidermis. Bar, 100 µm.
We examined the orientation of the cortical MTs by analyzing the angle (θ) between the cortical MTs and the plane that was parallel to the paradermal plane, on both transverse and longitudinal sections of the palisade layer, as shown in Figure 3A. The mean value of θ was close to 45° in wt plants at the developmental stage examined (just after stage I; Tsuge et al., 1996), regardless of the direction within the leaf (longitudinal sections, 43.2 ± 22.4°; transverse sections, 48.5 ± 18.6°; Figure 3). In contrast, θ was significantly reduced (level of significance, 1%, Student’s t-test) in transverse sections of the an mutant (an, 29.7 ± 20.6°; wt, 48.5 ± 18.6°), but it was not significantly reduced (level of significance, 1%) in longitudinal sections (an, 39.3 ± 22.5°; wt, 43.2 ± 22.4°; Figure 3). Thus, θ was significantly smaller in the transverse sections of the an mutant than in the wild type. In contrast, θ was very slightly larger in longitudinal sections of the rot3 mutant than in the wild type (rot3, 45.1 ± 22.7°; wt, 43.2 ± 22.4°). However, there was no significant difference in the orientation of the cortical MTs between the wild type and the rot3 mutant both in longitudinal sections and in transverse sections. Thus, the regulation of cell elongation by the ROT3 gene seemed to be independent of the regulation of the orientation of cortical MTs.
Fig. 3. Arrangement of cortical MTs in leaf palisade cells in the subepidermal layer of wt, an and rot3 leaves. (A) Strategy for measurement of the angle θ between cortical MTs in a palisade cell and the paradermal plane (plane of the epidermis). The orientation of the clearest array of MTs was measured for each palisade cell. (B and C) Distribution of θ on longitudinal sections (B) and on transverse sections (C). n, number of palisade cells examined. The numbers indicated by arrows are means ± SD of the value of θ of the indicated strains.
No abnormalities in the arrangement of cortical MTs were apparent in cells of internodal stems, leaf petioles and primary roots of the an mutant (Table I). The organ-specific abnormal arrangement of cortical MTs in the an mutant corresponded to the organ-specific defect in the polarity-specific elongation of cells in this mutant (Tsuge et al., 1996). In the rot3 mutant, no abnormalities were recognized in the orientation of cortical MTs in the cells of the internodal stems, the petioles or the roots (Table I).
Table I. Orientation of cortical MTs in cells in the leaf petiole, stem and root of wild-type and mutant plants.
| Stema | Leaf petioleb | Rootc | |
|---|---|---|---|
| wt | 4.9 ± 4.1 (n = 27) | 15.6 ± 10.8 (n = 38) | 6.6 ± 4.6 (n = 69) |
| an | 5.4 ± 3.6 (n = 70) | 20.5 ± 12.1 (n = 56) | 6.1 ± 4.2 (n = 60) |
| rot3 | 5.3 ± 3.6 (n = 74) | 14.0 ± 11.9 (n = 66) | 6.3 ± 5.5 (n = 50) |
Results shown are mean angles ± SD between cortical MTs and the radial axis (n, number of cells examined). All data for mutants shown here are not significantly different from those for the wild type (level of significance, 1%; Student’s t-test).
aThe first internodes of type II metamers of 5–7 mm in length were examined.
bThe epidermis of petioles of fully matured fifth leaves was examined.
cPrimary roots of seedlings 3–7 days after sowing were used for analysis, and cells at the growth stage prior to elongation were examined.
Molecular cloning of the AN gene
The AN locus was first identified by Rédei (1962) in a study of plants derived from mutagenized seeds of ecotype Landsberg and it was mapped to the upper end of the first chromosome (Lee-Chen and Steinitz-Sears, 1967; Koornneef et al., 1983). Using Cleaved Amplified Polymorphic Sequence (CAPS; Research Genetics, Huntsville, AL) markers from this region, we found that the AN locus was strongly linked to the marker PVV4. No recombination between AN and PVV4 was recognized after five back-crosses with the Columbia wild type. In addition, we calculated that the NCC1 locus, which has been mapped to a site ∼10.4 cM below the PVV4 locus, was located 13.6 ± 2.7 cM from the AN locus (data from 144 wt and 57 an plants). Thus, we estimated that the AN locus was located close to the PVV4 locus. Using this new information, we screened genomic clones using the PVV4-containing genomic clone E3-pART (a kind gift from Dr T.Golden, University of Rochester, NY) as the probe. We obtained three positive genomic TAC clones (TAC = transformation-competent artificial chromosome; Liu et al., 1999; kindly supplied by Dr D.Shibata, Mitsui Plant Biotechnology Research Institute, Tsukuba, Japan) that covered this region, namely, 1A13, 2E23 and 9N19, and we examined the ability of these clones to complement the an mutation. The 4.5 kb long E3-pART clone did not complement the an mutation, but one of the TAC clones, 1A13, clearly complemented the mutation in transgenic plants (Figure 1C and D). Not only narrow leaves but also trichomes with fewer branches (Tsuge et al., 1996) were restored in five independent primary transformants that harbored the 1A13 clone (Figure 1C and D). Thus, we concluded that the AN gene was included within the 45 kb long genomic fragment of the 1A13 clone.
To identify the open reading frame (ORF) that actually encodes the AN protein, we performed RFLP analysis of wild-type and an mutant genomes using the 1A13 clone as the probe. We found one RFLP fragment that differed between the wild-type genome and the an genome and contained an ORF (data not shown). The sequence of this ORF suggested that it encoded a dehydrogenase-like protein that was also encoded within a 40 kb genomic clone that had previously been sequenced by Terryn et al. (1998). Our sequence of this ORF matched that of EST clone YAY141 (DDBJ/EMBL/GenBank accession No. Z48386) which had been partially sequenced. Sequencing and northern blotting analysis of the YAY141 clone revealed that this EST clone included full-length cDNA. Comparison of genomic sequences with the cDNA revealed the presence of six introns and seven exons in the genomic sequence. Using this cDNA as a probe, we performed Southern blotting analysis, under high-stringency conditions, of genomic DNA digested with several restriction enzymes. In each case, we obtained only one genomic fragment (data not shown). The cDNA encoded a predicted protein of 636 amino acids with a molecular mass of 70 kDa.
To identify the mutation in the an allele, we constructed a genomic DNA library of the an-1 mutant, which had originally been isolated after X-ray mutagenesis. We isolated three independent genomic DNA clones that included the coding sequence of the putative AN gene and sequenced the clones derived from the an-1 mutant. In each of the three independent genomic clones, we identified an insertion caused by translocation of a genomic fragment at the putative AN locus. A genomic DNA fragment of ∼3 kbp, which was located at a site 6 kbp from the AN gene on the same chromosome, was translocated to a site 22 bp upstream of the termination codon in the 3′ sequence of the AN gene (Figure 4). Furthermore, sequence analysis of the an-2 mutant allele (a kind gift from Dr T.Hattori, Mie University, Tsu, Japan), which was isolated after mutagenesis with ethyl methanesulfonate (EMS), had a nucleotide transition from G to A, which resulted in the introduction of a termination codon at position 291 (which replaced tryptophan in the wt protein) in the conserved d-isomer-specific 2-hydroxyacid dehydrogenase (2-Hacid_DH motif) (see below and Figure 4). These results confirmed that the ORF that we had identified as the putative AN locus did indeed encode the AN protein. Complementation analysis of the an phenotype by AN cDNA is described below.
Fig. 4. Alignment of the amino acid sequences of the AN protein of Arabidopsis and various animal CtBPs (h, human; m, mouse; x, Xenopus; z, zebrafish; d, Drosophila). Identical amino acids in all sequences are indicated by black boxes and similar amino acids are indicated by gray boxes. The underlined regions are regions of homology between CtBPs and 2-Hacid_DH. Double-underlined regions are Rb-binding motifs (see text). The open and the filled triangle indicate the site of the translocation in the an-1 allele and the site of the mutation in the an-2 allele, respectively.
The AN gene encodes a homolog of human CtBP
A homology search using the sequence of the AN gene in the WHICH database revealed strong sequence similarity between the product of the AN gene and a carboxy-terminal binding protein (CtBP), a human protein named for its ability to interact with C-terminal sequences of adenovirus E1A protein (Schaeper et al., 1995; Figure 4). Members of the CtBP family have been found in several animal species. The AN protein exhibited strong homology at the amino acid level to zebrafish CtBP (32%), human CtBP (31%) and other animal CtBPs. There is a strongly conserved region in the N-terminal amino acid sequences of AN and animal CtBPs (∼47% homology). This common conserved sequence resembles the 2-Hacid_DH motif at its N-terminus (Boyd et al., 1993; Turner and Crossley, 1998) as indicated by the underlined amino acids at positions 96–306 in Figure 4. It has been suggested that this motif is involved not only in energy metabolism but also in interactions with various proteins (Arthur et al., 1991; Singh and Green, 1993). Furthermore, we found the retinoblastoma-binding (Rb-binding) motif LxCxE (Lee et al., 1998) at the N-terminus of AN (double underlining in Figure 4). A putative NLS (NLS = nuclear localization signal) motif, PEGRRSR and KKRH, was found in the region between positions 412–427 in the AN protein (Figure 4). No obvious similarities to animal CtBPs and other proteins were evident in the C-terminal region of AN.
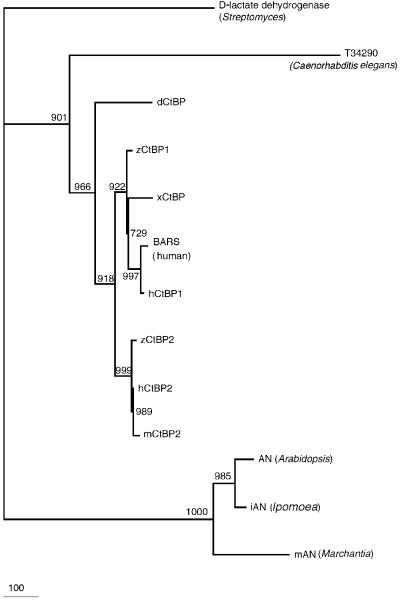
To examine the possible evolutionary history of CtBP genes, we compared the sequence of AN to available sequences of animal CtBPs. The phylogenetic tree revealed that common CtBPs can be divided into at least two subfamilies (Figure 5), i.e. one for plants and one for animals. We identified plant homologs of CtBPs by examining plant sequences similar to the sequence of the product of the AN gene, namely, sequences from Japanese morning glory (Ipomoea nil L., Convolvulaceae, Asteridae, Magnoliophyta; unpublished data from Dr E.Nitasata of Kyushu University, Fukuoka, Japan) and from liverwort (Marchantia polymorpha L., Marchantiaceae, Marchantiidae, Bryophyta; unpublished data from Dr H.Takano of Kumamoto University, Kumamoto, Japan). These polypeptides and the product of the AN gene clearly formed a monophyletic clade, suggesting the evolutionary conservation of AN homologs in the genomes of all land plants (Figure 5).
Fig. 5. Phylogenetic tree generated from the deduced amino acid sequences of animal CtBPs and other related proteins, as indicated. The tree was constructed with 1000 samplings of bootstrap values. The conserved region of the 2-Hacid_DH motif at the N-terminus, shown in Figure 4, was used to construct the tree. Branch lengths are proportional to the number of amino acid substitutions, and the scale bar indicates 100 substitutions.
Patterns of expression of the AN gene and cellular localization of the AN protein
We examined levels of AN mRNA by northern blotting and RT–PCR analysis. Northern blotting analysis showed that the AN gene was expressed throughout all organs examined in wild-type plants, namely, in cotyledons, leaves, roots, stems and floral buds (Figure 6A). We also examined the expression of two mutant an alleles by RT–PCR to determine the effects of these mutations on the accumulation of an mRNA. Wild-type levels of AN-type mRNA were detected in seedlings of both an-1 and an-2 mutant plants (Figure 6B), suggesting that these mutations alter the function of the AN protein and not the synthesis or stability of the AN transcript.
Fig. 6. Northern blotting and RT–PCR analysis of the AN transcript. (A) Expression of AN mRNA in various tissues of Arabidopsis. Aliquots of 20 µg of total RNA were loaded in each lane. Total RNA was isolated from roots (R), cotyledons (C), rosette leaves (L), stems (S) and floral buds (F). The intensity of the band of 25S rRNA served as an internal control (lower panel). (B) Levels of expression of AN mRNA in wild-type (wt, Columbia), an-1 and an-2 mutant plants, as determined by RT–PCR. Total RNA was isolated from aerial parts of 3-week-old seedlings and 0.5 µg of total RNA was used in each case. Results of the amplification by RT–PCR of the mRNA for β-tubulin 4 (TUB4) are shown as an internal control (lower panel).
The presence of a putative NLS motif in the AN protein suggested that AN might be localized in nuclei. To determine the localization of this protein, we fused the AN gene to a gene for green fluorescent protein (sGFP::S65T) under the control of the constitutively expressed 35S RNA promoter of cauliflower mosaic virus (CaMV). The fusion gene was expressed in both wild-type and an-1 mutant plants. Using a fluorescence microscope, we detected green fluorescence within the nuclei of cells in stable Arabidopsis transformants (Figure 7A). However, we detected the fusion protein not only within nuclei but also in the cytoplasm when the fusion gene was transiently expressed in onion epidermal cells (Figure 7C). Transgenic an-1 plants developed wild-type leaves and trichomes, suggesting that the expression of AN cDNA could complement the an mutation, with restoration of the normal arrangement of cortical MTs (Figure 9).

Fig. 9. Restoration of a normal arrangement of cortical MTs in the leaf epidermis by introduction of the AN gene into the an mutant. Confocal images of the adaxial epidermal cells of leaves are shown for the wt (A), the an-1 mutant (B) and a transgenic an-1 plant that expressed the wild-type AN gene (C). The direction perpendicular to the images in the figure corresponds to the longitudinal axis of the leaves (leaf-length direction), as shown in (C). Bars, 50 µm.
Control of cell shape in leaves by the AN gene
In an attempt to demonstrate genetic complementation of the an mutation by the ORF that we had identified and to clarify the function of AN, we transformed wild-type and an-1 mutant plants with the AN::GFP gene that was under the control of the CaMV 35S promoter. The an-1 phenotype was reversed by expression of the transgene (Figure 8A and B).
Fig. 8. Anatomical and molecular characterization of transgenic plants that expressed the AN::GFP transgene under control of the CaMV 35S promoter. (A) From left, rosette leaves of wt, an-1 mutant, and transgenic plants with the an-1 mutant background that expressed the AN::GFP fusion protein, respectively. (B) Analysis by RT–PCR of the levels of expression of AN mRNA in the transgenic plants shown in panel A. (C–E) Paradermal view of the cleared palisade layers of the fifth leaves of the wt (C), an-1 mutant (D) and transgenic an-1 plant that expressed AN::GFP (E). Bar, 0.1 mm. (F–H) Cross sections of the fifth leaves of the wt (F), of an-1 mutant plant (G) and of a transgenic an-1 that expressed AN::GFP plant (H). Bar, 0.1 mm.
To determine the cellular basis of the phenotypes of the transgenic plants, we performed an anatomical analysis of the fifth leaves from wild-type plants, an-1 mutant plants, and transgenic an-1 plants that expressed the AN gene and had a wild-type phenotype. The palisade cells of the an-1 mutant were narrower (Figures 1B and 8D) than those of the wild type (Figures 1A, 8C and F), but those of the transgenic an-1 plants that expressed the AN gene were similar to those of the wild type (Figure 8E and G). The arrangement of cortical MTs in epidermal cells of the transgenic an-1 plants was also normal, resembling that in the wild-type plants (Figure 9). The cortical MTs in epidermal cells of transgenic plants were not mostly oriented parallel to the leaf-width direction, unlike those in the epidermal cells of the an mutant. This result suggests that recovery from the an-type phenotype upon expression of the AN transgene might be attributable to normalization of the arrangement of cortical MTs.
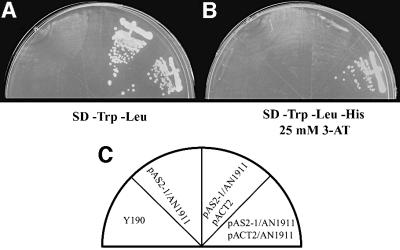
AN can self-associate in yeast cells
The CtBP of Drosophila (dCtBP; Poortinga et al., 1998) and of mouse (mCtBP2; Turner and Crossley, 1998) self-associate in a yeast two-hybrid system. It has been suggested that the dimerization of CtBP is important for linkage between a DNA-binding protein and other kinds of regulatory protein (for review see Turner and Crossley, 2001). Therefore, we examined the possible dimerization of AN in a yeast two-hybrid system. We found that AN was able to form dimers in yeast cells (Figure 10), as is the case for animal CtBPs. The proposed dimerization of AN was also supported by quantitation of the interaction in terms of the activity of β-galactosidase (host cells, 170 ± 8 µM/min/mg protein; cell lines with pAS2-1/AN1911 and pACT2 plasmids, 159 ± 10 µM/min/mg protein; and cell lines with pAS2-1/AN1911and pACT2/AN1911 plasmids, 1484 ± 61 µM/min/mg protein; three independent lines were examined in each case and results are means ± SD for each set of three lines).
Fig. 10. Self-association of AN in a yeast two-hybrid system. (A and B) Growth of yeast strain Y190 that harbored constructs as indicated in (C). (A) Growth on SD medium (–Trp –Leu) to confirm insertion of pAS2-1/AN and pACT2/AN into strain Y190. (B) Growth on selective SD medium (–Trp –Leu –His) that contained 25 mM 3-amino-1,2,4-triazole (3-AT) for the self-association test (see text for details).
The an mutation specifically affects the expression of the gene for a meristem-specific enzyme that is involved in cell-wall formation, MERI5
To examine the possible role of the AN protein as a regulator of the transcription of other genes, we performed microarray analysis under the auspices of the Monsanto Arabidopsis Microarray Program. Microarray analysis showed that the an mutant expressed some genes at higher levels than the wild type, suggesting that AN might function as a repressor of such genes. Possible candidates for target genes of the AN protein include a cell-wall modulating enzyme, a Zn-finger protein and a MAP kinase (Table II). To confirm the results of microarray analysis, we performed RT–PCR analysis of the expression of candidate genes. We confirmed that some genes were up-regulated in the an mutant but down-regulated in wild-type plants and transgenic an plants that expressed the wild-type AN gene (data not shown). In the present study, we focused on the expression of genes in the xyloglucan endotransglucosylase/hydrolase (XTH) family. These enzymes are thought to regulate loosening of cell walls, and one member of this family, MERI5 (Medford et al., 1991), was among the genes that were specifically up-regulated in the an mutant.
Table II. Genes up-regulated in the angustifolia leaves, as compared with wild-type leaves and identified by microarray analysisa.
| Accessionb | Encoded protein | Up-regulation (-fold; an/wt)c |
|---|---|---|
| AF08279 | Cys-3-His zinc finger protein | 11.3 |
| Monsanto | unknown | 8.3 |
| D21839 | MAP kinase 3 | 3.5 |
| Monsanto | putative mitochondrial uncoupling protein | 3.5 |
| Monsanto | putative protein | 3.4 |
| U40217 | homolog of eukaryotic release factor 1 | 3.4 |
| D63508 | MERI5 (xyloglucan endo-transglycosylase) | 3.3 |
| AF0(2109 | Hs1pro-1-related protein | 3.1 |
aGenes for which levels of transcripts were more than three times higher in angustifolia leaves than in wild-type leaves are shown.
bDesignations and accession numbers of registered genes that are identical to the identified cDNAs.
cRatio of the level of transcripts in the angustifolia mutant to that in wild type.
We analyzed the levels of expression of genes in the XTH family in rosette leaves by quantitative, real-time RT–PCR. We found that MERI5 was expressed at a 3-fold higher level in the an-1 mutant than in the wild type, as indicated by the microarray analysis (3.3-fold higher by microarray analysis; 3.27-fold higher by RT–PCR analysis). In contrast, levels of expression of two other genes for members of the XTH family, namely EXGT-A1 (EXT; Okazawa et al., 1993) and EXGT-A2 (Akamatsu et al., 1999; Yokoyama and Nishitani, 2001) did not show such differences (not significantly different from the wild type; level of significance, 5%) when we compared an-1 and wild-type plants (Figure 11). This result suggested that the level of expression of MERI5 was specifically affected by the an mutation, as compared to other members of the XTH family.
Fig. 11. Levels of expression of genes for three members of the XTH family in wild-type and angustifolia leaves, as detected by real-time RT–PCR. Average results from triplicate assays are shown with SD. An asterisk indicates a significant difference from the wild-type level (level of significance, 5%).
Discussion
In the present study, we showed that the AN gene, the first member of the family of CtBP genes to be identified in a plant genome, regulates the polarized growth of leaf cells via control of the arrangement of cortical MTs in epidermal cells (Figure 2) and in mesophyll cells (Figure 3). In the an mutant, the orientation of the cortical MTs was dramatically disrupted in leaves at an early stage of their development (Figure 2). The stage at which we examined palisade cells corresponded to the stage immediately after stage I, when the polar elongation of cells is initiated (Tsuge et al., 1996; Kim et al., 1998b). We proposed previously that the AN gene governs the polar elongation of leaf mesophyll cells after stage I (Tsuge et al., 1996). The mean value of the angle between cortical MTs and the paradermal plane (plane of the epidermis), was significantly smaller (61%; level of significance, 1%) in transverse sections of the an mutant than in those of the wild type. The smaller this angle, the greater is the extent to which rod-shaped cells elongate in a perpendicular direction relative to the paradermal plane. When cortical MTs (and the associated cellulose microfibrils) are arranged parallel to the paradermal plane and the volume of cells increases, the cells are forced to expand exclusively in the leaf-thickness direction and not in the leaf-width direction. We postulated previously (Tsuge et al., 1996) that the increase in cell length in the leaf-thickness direction in the an mutant might be a result of restricted elongation of cells in the leaf-width direction. The results in the present study now allow us to explain the unusually narrow and unusually thick leaves of an mutant plants in terms of a single phenomenon: the altered arrangement of cortical MTs in an mutant cells. Similarly, the reduced number of protrusions of epidermal cells that is specific to the leaf-width direction in the an mutant (Tsuge et al., 1996) can also be explained by the orientation of cortical MTs that we observed in the present study (Figure 3). Hogetsu (1989) reported that the pattern of the cortical MTs is the critical factor in the directional expansion of the leaf epidermis. Thus, the AN gene appears to play a role in the regulation of the orientation of cortical MTs in leaf morphogenesis.
Our analysis also revealed that the cortical MTs in the palisade cells in the subepidermal layer of the an mutant were abnormally oriented in different planes (in transverse sections and in longitudinal sections; Figure 3). The fact that the orientation was affected in a specific plane is, itself, of interest with respect to the regulation of cell polarity. The concept of the regulation of cell polarity in a specific plane is further supported by observations that changes in the orientation of cortical MTs, e.g. caused by treatment with a phytohormone, are also associated with changes in a specific plane (Yuan et al., 1995; Lloyd et al., 1996; Wymer and Lloyd, 1996).
The abnormal patterns of branching of the trichomes on an leaves (Hülskamp et al., 1994; Oppenheimer, 1998) can now also be readily explained. The branching of trichomes is controlled by cortical MTs in Arabidopsis (Mathur and Chua, 2000). Thus, as we speculated earlier (Tsuge et al., 1996), less extensive branching (two branches in an leaves and three or four in wt leaves) might be a result of the defect in the arrangement of cortical MTs in an plants that suppresses elongation of cell protrusions in the leaf-width direction.
AN as a regulator of the polar expansion of leaf cells
As discussed above, the AN gene appears to control the arrangement of cortical MTs and, in this way, AN is thought to influence the shape of leaf cells in a polarity-dependent manner. The leaf specificity of the mutant phenotype, with respect to gross morphology (Tsuge et al., 1996), was reflected by the leaf specificity of the abnormality in the arrangement of cortical MTs (Table I). In other words, the leaf-specific abnormality in the orientation of cortical MTs was consistent with the leaf-specific phenotype. Moreover, since the AN gene functions independently of the ROT3 gene (Tsuge et al., 1996), if the AN gene is involved in the control of the arrangement of cortical MTs, it would seem unlikely that the ROT3 gene would be involved in regulation of the same process in leaf cells. In the present study, rot3 mutant plants were found to have normally arranged cortical MTs in their leaf cells. In addition AN cDNA reversed the abnormality in the arrangement of cortical MTs in an leaf cells. Thus, all the available data indicate a parallelism between the arrangement of cortical MTs in leaf cells and the activity of the AN gene.
Previous studies have shown that disruption of the arrangement of cortical MTs can be responsible for a disturbance in the appropriate enlargement of cells (Aeschbacker et al., 1995; Hauser et al., 1995; Traas et al., 1995; McClinton and Sung, 1997). However, genetic correlations between the regulation of the orientation of cortical MTs and the direction of cell elongation have not been fully clarified, except in the case of the SPR genes of Arabidopsis (Furutani et al., 2000). As shown in the present study, the close relationship between the change in gross morphology and the abnormal arrangement of cortical MTs strongly suggests that the AN gene plays an important role in regulation of the orientation of cortical MTs in leaf cells. This is the first reported proof, to our knowledge, of the genetic regulation of the arrangement of cortical MTs in leaf cells.
AN is a member of the CtBP family and might act as a repressor of transcription
The AN gene is the first homolog of CtBP gene to be identified in a plant genome. The CtBP family has been identified as a new family of co-repressors from animal genomes and its members have been shown to play important roles in cell differentiation (for review see Turner and Crossley, 2001). Human CtBP (now hCtBP) was first isolated by yeast two-hybrid cloning, as the gene for a cellular phosphoprotein that interacts with a C-terminal domain of the E1A protein of adenovirus (Schaeper et al., 1995, 1998). It has been proposed that binding of hCtBP to E1A modulates E1A-mediated transformation (tumorigenesis). It has also been proposed that hCtBP might be a negative regulator of transcription (Schaeper et al., 1995; Sollerbrant et al., 1996; Molley et al., 1998). Schaeper et al. (1995) noted the sequence homology between hCtBP and d-2-hydroxy acid dehydrogenase but they did not detect any such enzymatic activity associated with hCtBP. Since several amino acids that are important for the enzymatic activity of d-lactate-dehydrogenase (Bernard et al., 1997) were not found in the amino acid sequence of AN, it is unlikely that AN has such dehydrogenase activity.
After the identification of hCtBP, many homologs of hCtBP were isolated from animal genomes. A homolog of hCtBP in mouse, mCtBP, was also isolated as a co-repressor that associates with regulators of transcription (Turner and Crossley, 1998) and a CtBP from Xenopus (xCtBP1) was identified as a transcriptional repressor that interacts with Polycomb which, itself, is involved in the repression of certain genes during the course of vertebrate development (Sewalt et al., 1999). Several extensive studies have revealed recently that a homolog of hCTBP from Drosophila, dCtBP, plays an important role in the repression of genes which is required for normal development of the embryo (Nibu et al., 1998, 2001; Poortinga et al., 1998; Mannervik and Levine, 1999; Rosée-Borggreve et al., 1999; Zhang and Levine, 1999; Phippen et al., 2000; Wen et al., 2000; Morel et al., 2001; Nibu and Levine, 2001). dCtBP is a key protein that regulates aspects of developmental processes via repression of various developmental genes, acting as a context-dependent transcriptional cofactor that interacts with several kinds of co-repressor. Thus, CtBPs in animals appear likely to be engaged in an evolutionarily conserved mechanism of transcriptional repression. It is also true, however, that brefeldin A ADP-ribosylated substrates (BARS; Spanfò et al., 1999; Weigert et al., 1999), which appear to play an important role in maintenance of the Golgi apparatus are also strongly homologous to products of CtBP genes (Figure 5) and the CtBPs are also known as CtBP/BARS.
Animal and plant CtBPs might share evolutionarily conserved functions as transcriptional co-repressors. The intracellular localization of the product of the AN::GFP gene (Figure 8) supports this hypothesis since it indicates that the AN protein is present in the nucleus. However, appreciable amounts of AN protein were also found in the cytoplasm. In animal cells, CtBPs are also abundant in nuclei but they are not exclusively nuclear (for review see Turner and Crossley, 2001).
Northern analysis of the AN transcript showed that the gene is expressed in all organs of Arabidopsis, even though the an phenotype is restricted to leaves and floral organs. The target gene(s) of the AN protein that is responsible for the regulation of the polar expansion of cells might be specifically expressed in leaf cells but the target gene(s) remains to be identified. The target genes of animal CtBPs are not conserved among species. In addition, as shown in Figure 5, the molecular phylogeny of the AN gene suggests that AN might have a unique function that is not shared by animal CtBPs. Our preliminary microarray analysis of cDNAs (performed in collaboration with Monsanto as part of their Arabidopsis Microarray Program) suggests that, in the an mutant plant, levels of transcripts of some genes (∼10 genes among 10 000 clones) might be more than 3-fold higher than in the wild type (Table II). We found no gene that was expressed at a lower level in the an mutant than in the wild type.
While we postulate that the shape of leaf cells might be regulated by the AN gene via control of the arrangement of MTs in leaf cells, some researchers have noted recently that, on occasion, a simple correlation between the orientation of MTs and cell shape cannot be observed (Wasteneys, 2000). Thus, it is also possible that the construction or composition of cell walls might be regulated by AN. It is of interest that one of the candidates for target genes identified by microarray analysis was MERI5, which encodes a member of the XTH family and is considered to play a role in the molecular grafting of components of cell walls (Nishitani, 1997). While levels of expression of genes for two other members of the XTH family, EXGT-A1 and EXGT-A2, were unaffected by the an mutation, the level of transcription of MERI5 in rosette leaves of the an mutant was three times higher than in the wild type (Figure 11). The MERI5 gene is preferentially expressed in apical meristems and leaf primordia in Arabidopsis (Medford et al., 1991), an observation that suggests that MERI5 might play a role at the early stages of leaf morphogenesis. Overexpression of MERI5 cDNA under control of constitutive CaMV 35S promoter caused the precocious elongation and/or abnormal expansion of leaf cells (Verica and Medford, 1997). Although the gross morphology of leaves of transgenic plants that carried CaMV 35S::MERI5 (Verica and Medford, 1997) was different from those of the an mutant, it is possible that the difference might have been due to a lack of tissue specificity of the overexpression in the transgenic plants. Since both MERI5-overexpressing transgenic plants and an mutant plants show alterations in the process of expansion of leaf cells, it is possible that the AN gene might control cell shape in leaves by regulating the expression of MERI5 directly or indirectly. In this case, the effect of AN gene on arrangement of MTs in leaf cells might be a secondary result of alteration of cell walls by activity of MERI5.
Our results support the hypothesis that the product of the AN gene might act as a repressor of transcription. The relationship between regulation of the arrangement of cortical MTs and such negative regulation of transcription is unclear. However, such a relationship is not implausible. Detailed analysis of the target(s) of the AN protein should reveal new aspects of gene regulation in plants and, in particular, of the regulation of the polarity-specific expansion of cells. As Folkers et al. (2002) strongly suggest, AN might interact with ZWICHEL (ZWI), a kinesin-like protein, in trichome cells. Since the morphological phenotype of zwi mutants is restricted to the shape of trichomes and since zwi leaves are normal in shape (Oppenheimer et al., 1997), AN should interact with some other component(s) in other leaf cells in the regulation of leaf width. It is noteworthy that AN self-associates in yeast cells (Figure 10). Homodimerization is thought to be necessary for linkage by animal CtBPs of two different proteins (for review see Turner and Crossley, 2001). The proteins interacting with animal CtBPs depends on circumstance. It is very possible that AN might interact with ZWI during the morphogenesis of trichomes and with other components in other types of cell or at other stages of leaf development.
Details of the functions of animal CtBPs remain to be determined. It is possible that CtBPs regulate gene expression directly via their enzymatic activities and/or by simply linking other proteins (Turner and Crossley, 2001). Our phylogenic analysis suggests that plant CtBPs, including the product of the AN gene, might have evolved to play a role that is specific to plants since plant CtBPs appear to be monophyletic and to form a unique subfamily among homologs of hCtBP. AN is the first member of the CtBP family to be identified in plants and future studies of AN should help us to understand how the functions of members of the CtBP family have been conserved in the control of the development of multicellular organisms.
Materials and methods
Plant culture
Seeds were sown on rockwool and/or vermiculite, moistened with MGRL medium, as described elsewhere (Tsukaya et al., 1991; Tsuge et al., 1996). Plants were grown at 23°C under continuous white fluorescent light (67.4 ± 14.0 µM/s/m2). For observations of root cells, plants were cultivated on agar plates, as described by Okada and Shimura (1992). The an-1 single-mutant lines were established as described by Tsukaya et al. (1994). Another allele, an-2, was isolated by Dr T.Hattori of Mie University, Tsu, Japan, from mutagenized seeds of the Columbia ecotype and kindly donated to the authors. The directions relating to the analysis of leaf blades are defined in an earlier report (Tsuge et al., 1996).
Anatomical and immunohistochemical analysis
The procedures for preparing sections of samples embedded in Technovit 7100 resin (Kulzer & Co. GmbH, Wehrheim, Germany) were described previously (Tsukaya et al., 1993; Tsuge et al., 1996). Immunohisto chemical observations of cortical MTs in cells of internodal stems and primary roots were made as described earlier (Tsukaya et al., 1995). Epidermal cells from leaf petioles were collected from the first set of foliage leaves at the fully mature stage. Stem samples were collected from the first internodal stem of type II metamers when they had reached 5–7 mm in length. Root samples were collected from primary roots of seedlings 3–7 days after sowing. Cells were examined at the growth stage prior to elongation.
The MTs in leaf cells were visualized by immunohistochemical staining, which was performed by a modified version of previously described methods (Ishida and Katsumi, 1991; Tsukaya et al., 1995) or by a freeze-shattering method (Wasteneys et al., 1997). Samples of mesophyll cells were prepared immediately after stage I (Tsuge et al., 1996), when the polar elongation of cells had just begun. We examined the arrangement of cortical MTs in cells in the subepidermal layer on the adaxial side of the leaf. We used monoclonal mouse antibodies against chicken α- and β-tubulin (Amersham Japan Co., Tokyo, Japan) diluted 500-fold in phosphate-buffered saline (PBS) plus 0.1% NaN3 and fluorescein isothiocyanate-labelled (FITC-labelled) sheep antibodies against mouse IgG (Amersham Japan Co.). Alternatively, we used an Alexa488-labelled Signal Amplification Kit for mouse antibodies (Molecular Probes, Leiden, The Netherlands). For observations of cellulose microfibrils, we treated samples with 0.01% Fluostain-I (Wako, Osaka, Japan) in 50 mM sodium phosphate buffer pH 7.0 for 5 min, with three subsequent washes with 50 mM sodium phosphate buffer. Samples on glass slides were set in mounting solution [1 mg/ml n-propyl gallate (Wako), dissolved in a 50% solution of glycerol in water plus 0.1% NaN3]. Samples were observed with a fluorescence microscope (DAS Mikroskop DMR; Leica, Wetzlar, Germany) or a confocal laser microscope (LEITZ DMIRB, with a True Confocal Scanner, LEICA TCS 4D; Leica).
For observations of cortical MTs in living cells, we also used transgenic Arabidopsis (ecotype Columbia) that harbored the TUB6::GFP fusion gene (a kind gift of Dr T.Hashimoto, NAIST, Nara, Japan; Ueda et al., 1999). This gene encodes GFP fused to a gene for tubulin, TUB6, under the control of the CaMV 35S promoter. We crossed this transgenic line with the an mutant to generate an mutant plants that expressed the fusion gene, in which we were able to examine the arrangement of cortical MTs with a confocal laser microscope (Olympus FLUoview/BX50; Olympus, Tokyo, Japan).
Molecular biological analysis
Routine procedures, including the isolation of DNA, Southern and northern blotting analysis, and nucleotide sequencing, were performed essentially as described elsewhere (Sambrook et al., 1989). The genomic DNA clone including the PVV4 fragment was a kind gift of Dr T.Golden (University of Rochester, Rochester, NY). TAC clones that gave positive signals in dot-blot analysis with the PVV4 fragment as a probe were identified and used to transform an-1 mutant plants. The Arabidopsis EST clone YAY141 was obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, OH) and it was sequenced in its entirety using primers derived from the previously determined DNA sequence. The deduced amino acid sequence encoded by the AN gene in the Columbia ecotype was identical to that in the Ler ecotype. The cDNA sequence of the AN gene was identical to that of the coding region of the genomic DNA sequence with the exception of a missense alteration at position 299 (Met to Ile) in the cDNA sequence. The cDNA sequence of the AN gene has been deposited in the DDBJ/EMBL/GenBank database (accession No. AB032060).
To identify the an-1 mutation, we constructed a genomic DNA library from an an-1 mutant plant by ligation into HindIII-digested lamda DASHII of fragments from a partial Sau3AI digest of the mutant genome using an In vitro Packaging Kit (Stratagene, La Jolla, CA). Southern blotting and plaque-hybridization analysis were performed with a Gene Images Random-Prime Labelling and Detection System and an AlkPhos Direct System (Amersham Pharmacia Biotech, Buckinghamshire, UK) according to the manufacturer’s instructions. We chose three independent genomic clones that gave positive signals for sequencing. To identify the mutation in an-2, we amplified the region that corresponded to the open reading frame (ORF) of the AN gene in the genome of the an-2 mutant using the total RNA of an an-2 mutant plant and a Superscript One-Step RT–PCR Kit (Gibco BRL, Rockville, MD) and subcloned the fragment into pBluescript SK (Stratagene). Three independent clones were chosen for sequencing.
Northern blotting analysis was performed with [32P]ATP-labelled AN cDNA as the probe. Total RNA was purified from various tissues as described previously (Kim et al., 1998a) and 20 µg aliquots of total RNA were used for northern analysis. Aliquots of 0.2–0.5 µg of total RNA were treated with DNase I at 37°C for 30 min before RT–PCR and each set of primers for PCR was designed such that a different exon sequence was included in each set of primers, to avoid amplification of any contaminating genomic DNA. The conditions for direct amplification by RT–PCR with a Superscript One-Step RT–PCR Kit (Gibco BRL) were one cycle at 50°C for 30 min and 94°C for 2 min, then 20, 25 or 30 cycles at 94°C for 15 s, 55°C for 15 s and 72°C for 90 s. The following oligonucleotides were used as primers for RT–PCR: AN-start, 5′-ATGAGCAAGATCCGTTC-3′; AN-stop, 5′-TTAATCGATCCAACGTGTA-3′; AN-RT-for1, 5′-TGAGACGGTGCCGTGGTATGG-3′; AN-RT-rev1, 5′-GTTGCCTACTGGTGGATTCC-3′ and AT-RT-rev2, 5′-CTTACCGGATCTGCTACGTC-3′. Amplification of β-TUB4 noncoding sequence was performed with two primers, as described previously (Kim et al., 1998a) as a positive control in the analysis by RT–PCR.
For microarray analysis, total RNA was extracted from 2-week-old rosette leaves (including shoot apical meristems) of plants that had been cultivated aseptically on MS0 plate, as described earlier (Tsukaya et al., 1991). The tissue was frozen and ground in liquid nitrogen and RNA was extracted with a Trizol™ reagent kit (Gibco BRL, Grand Island, NY). Microarray analysis of the RNA was carried out under the auspices of the Monsanto Arabidopsis Microarray Program (Monsanto, St Louis, MI).
Analysis of levels of expression of transcripts of EXGT-A1, EXGT-A2 and MERI5 in rosette leaves of 20-day-old seedlings of wild-type and an mutant plants was carried out by real-time RT–PCR (ABI Prism 5700 Sequence-Detection System; Perkin-Elmer Applied Biosystems, Foster City, CA) using specific primers as described earlier (Yokoyama and Nishitani, 2001).
Yeast two-hybrid assay
For two-hybrid analysis, we used the yeast Y190 [MATa, ura3-52, his3-200, lys2-801, ade2-101, trp1-901, leu2-3, 112, gal4–, gal80–, cyhr2, LYS2::GAL1UAS-HIS3TATA-HIS3, URA3::GAL1UAS-GAL1TATA-lacZ] as host strain. Two full-length AN cDNAs that differed in terms of a 5′ restriction site were constructed by PCR. AN cDNA with an NdeI–BamHI site inserted into pAS2-1 (Clontech Japan, Tokyo, Japan) that generated GAL4 DNA-binding domain (GAL4BD) was used as bait, and AN cDNA with an NcoI–BamHI site inserted into pACT2 (Clontech Japan, Tokyo, Japan) generating GAL4 activation domain (GAL4AD) was used as prey. Both clones were sequenced to confirm the nature of the products. SD medium (–Trp –Leu –His; prepared according to the manufacturer’s instructions) that contained 25 mM 3-amino-1,2,4-triazole (3-AT) was used for test of two hybrids.
Phylogenetic analysis
We generated phylogenetic trees using PROTPARS, a maximum-parsimony algorithm that is included in the PHYLIP version 3.5 software package (distributed by Dr J.Felsenstein, University of Washington, Seattle, WA). Topological robustness was assessed by bootstrap analysis with 1000 replicates using simple taxon addition (Felsenstein, 1985).
Sequences used for alignments were identified by BLAST searches of DDBJ/EMBL/GenBank. Sequences conserved in AN, homologs of AN, animal CtBPs and human BARS were aligned with the CLUSTAL_W program (Thompson et al., 1994) and alignments were refined manually. Several short sequences in the N-terminal region that could not be aligned unambiguously were excluded from the analysis.
Construction of transgenic Arabidopsis
For complementation tests, genomic TAC clones were introduced into an-1 mutant plants by a modified version of a previously published method for the Agrobacterium-mediated transformation of roots (Ueda et al., 1996). For construction of the AN::GFP fusion gene, we amplified a 1.9 kbp coding fragment of the AN gene by PCR, using oligonucleotides that introduced a SalI and an NcoI restriction site at the N- and C-terminal ends, respectively. Amplified fragments were sequenced and ligated into the SalI and NcoI sites of the sGFP vector pTH-2 (a kind gift of Dr Y.Niwa, Shizuoka Prefectural University, Japan; Chiu et al., 1996). The AN::GFP fusion gene was introduced into the pBI vector, which included a CaMV 35S promoter and a NOS terminator cassette. Lines of transgenic plants derived from the wild type (ecotype Columbia) and the an-1 mutant and harboring this construct were established by Agrobacterium-mediated transformation, which was performed by a simplified in planta infiltration method, as described by Kim et al. (1999). An experiment to examine the transient expression of the fusion gene in onion epidermal cells was performed using a particle delivery system (PDS-1000/He; Bio-Rad Japan, Tokyo, Japan) according to the manufacturer’s protocol.
Accession number
The DDBJ/EMBL/GenBank accession No. of the ORF in the AN gene is AB032060.
Acknowledgments
Acknowledgements
The authors thank Mr N.Inoue (Japan University, Tokyo, Japan) for his help with experiments. Drs I.Yabe (University of Tokyo) and S.Mano (NIBB, Okazaki, Japan) kindly taught H.T. to manipulate the confocal microscopes. The authors also thank Dr Y.Niwa (Shizuoka Prefectural University, Shizuoka, Japan) for the sGFP vector, Dr G.Wasteneys (Australia National University, Camberra, Australia) for kindly introducing them to his freeze-shattering method for staining MTs, Dr T.Hashimoto (NAIST, Nara, Japan) for the kind gift of transgenic Arabidopsis that expressed the TUB6::GFP fusion gene, Dr T.Golden (University of Rochester, Rochester, NY) and Dr D.Shibata (formerly of the Mitsui Plant Biotechnology Research Institute, Tsukuba, Japan and now at the Kazusa DNA Research Institute, Kazusa, Japan) for kind gifts of genomic clones and TAC clones of Arabidopsis. Dr T.Hattori (Mie University, Tsu, Japan) isolated the an-2 allele and kindly supplied the authors with seeds. Dr E.Nitasata (Kyushu University, Fukuoka, Japan) and Dr H.Takano (Kumamoto University, Kumamoto, Japan) kindly permitted the authors to use their unpublished sequence data for construction of the phylogenetic tree of AN homologs. The authors thank Dr C.W.Lloyd (John Innes Institute, Norwich, UK) for his critical reading of the MT-related parts of an early version of the manuscript. The authors also thank Dr H.Shibaoka (Osaka University, Osaka, Japan) for valuable discussions. Ms A.Shimotono of the University of Tokyo (Tokyo, Japan) helped with the sequencing of the ORF of the AN gene. Microarray analysis was carried out by Monsanto (St Louis, MI) under a Monsanto Arabidopsis Microarray Program Award. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture, Japan (to G.-T.K. and H.T.); by a grant from the Bio-Design Program of the Ministry of Agriculture, Forestry and Fishes of Japan (to H.T.) and by a grant from PRESTO, Japan Science and Technology Corporation, Japan (to H.T.).
References
- Aeschbacker R.A., Hauser,M.-T., Feldmann,K.A. and Benfey,P.N. (1995) The SABRE gene is required for normal cell expansion in Arabidopsis. Genes Dev., 9, 330–340. [DOI] [PubMed] [Google Scholar]
- Akamatsu T., Hanzawa,Y., Ohtake,Y., Takahashi,T., Nishitani,K. and Komeda,Y. (1999) Expression of endoxyloglucan transferase genes in acaulis mutants of Arabidopsis thaliana. Plant Physiol., 121, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Molinas,C., Dutka-Malen,S. and Courvalin,P. (1991) Structural relationship between the vancomycin-resistance protein VanH and 2-hydroxycarboxylic acid dehydrogenases. Gene, 103, 133–134. [DOI] [PubMed] [Google Scholar]
- Bernard N. et al. (1997) d-2-Hydroxy-4-methylvalerate dehydrogenase from Lactobacillus delbrueckii subsp bulgaricus. 2. Mutagenic analysis of catalytically important residues. Eur. J. Biochem., 244, 213–219. [DOI] [PubMed] [Google Scholar]
- Boyd J.M., Subramanian,T., Schaeper,U., La Regina,M., Bayley,S.T. and Chinnadurai,G. (1993) A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J., 12, 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W.-L., Niwa,Y., Zeng,W., Hirano,T., Kobayashi,H. and Sheen,J. (1996) Engineered GFP as a vital reporter in plants. Curr. Biol., 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Cyr R.J. (1994) Microtubules in plant morphogenesis: role of the cortical array. Annu. Rev. Cell. Biol., 10, 153–180. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Folkers U. et al. (2002) The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J., 21, 1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I., Watanabe,Y., Prieto,R., Masukawa,M., Suzuki,K., Naoi,K., Thitamabee,S., Shikanai,T. and Hashimoto,T. (2000) The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development, 127, 4443–4453. [DOI] [PubMed] [Google Scholar]
- Hauser M.-T., Morikami,A. and Benfey,P.N. (1995) Conditional root expansion mutants of Arabidopsis. Development, 121, 1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogetsu T. (1989) The arrangement of microtubules in leaves of monocotyledonous and dicotyledonous plants. Can. J. Bot., 67, 3506–3512. [Google Scholar]
- Hülskamp M., Misera,S. and Jürgens,G. (1994) Genetic dissection of trichome cell development in Arabidopsis. Cell, 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Ishida K. and Katsumi,M. (1991) Immunofluorescence microscopical observation of cortical microtubule arrangement as affected by gibberellin in d5 mutants of Zea mays L. Plant Cell Physiol., 32, 409–417. [Google Scholar]
- Kim G.-T., Tsukaya,H. and Uchimiya,H. (1998a) The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev., 12, 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.-T., Tsukaya,H. and Uchimiya,H. (1998b) The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana. Planta, 206, 175–183. [DOI] [PubMed] [Google Scholar]
- Kim G.-T., Tsukaya,H., Saito,Y. and Uchimiya,H. (1999) Changes in the shapes of leaves and flowers upon overexpression of the novel cytochrome P450 in Arabidopsis. Proc. Natl Acad. Sci. USA, 96, 9433–9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., van Eden,J., Hanhart,C.J., Stam,P., Braaksma,F.J. and Feenstra,W.J. (1983) Linkage map of Arabidopsis thaliana. J. Hered., 74, 265–272. [Google Scholar]
- Lee-Chen S. and Steinitz-Sears,L.M. (1967) The location of linkage groups in Arabidopsis thaliana. Can. J. Genet. Cytol., 9, 381–384. [Google Scholar]
- Lee J.-O., Alicia,A., Russo,A.A. and Pavletich,N.P. (1998) Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature, 391, 859–865. [DOI] [PubMed] [Google Scholar]
- Liu Y-G, Shirano,Y., Fukaki,H., Yanai,Y., Tasaka,M., Tabata,S. and Shibata,D. (1999) Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning Proc. Natl Acad. Sci. USA, 96, 6535–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C.W., Shaw,P.J., Warn,R.M. and Yuan,M. (1996) Gibberellic-acid-induced reorientation of cortical microtubules in living plant cells. J. Microscopy, 181, 140–144. [Google Scholar]
- Mannervik M. and Levine,M. (1999) The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc. Natl Acad. Sci. USA, 96, 6797–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J. and Chua,N.-H. (2000) Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell, 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClinton R.S. and Sung,Z.R. (1997) Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta, 201, 252–260. [DOI] [PubMed] [Google Scholar]
- Medford J., Elmer,J.S., Klee,H.J. (1991) Molecular cloning and characterization of genes expressed in shoot apical meristem. Plant Cell, 3, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molley D.P., Milner,A.E., Yakub,I.K., Chinnadurai,G., Gallimore,P.H. and Grant,R.J.A. (1998) Structural determinants present in the C-terminal binding protein binding site of adenovirus early region 1A proteins. J. Biol. Chem., 273, 20867–20876. [DOI] [PubMed] [Google Scholar]
- Morel V., Lecourtois,M., Massiani,O., Maier,D., Preiss,A. and Schweisguth,F. (2001) Transcriptional repression by Suppressor of Hairless involves the binding of a Hairless–dCtBP complex in Drosophila. Curr. Biol., 11, 789–792. [DOI] [PubMed] [Google Scholar]
- Nibu Y. and Levine,M.S. (2001) CtBP-dependent activities of the short-range Giant repressor in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 98, 6204–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y., Zhang,H. and Levine,M. (1998) Interaction of short-range repressors with Drosophila CtBP in the embryo. Science, 280, 101–104. [DOI] [PubMed] [Google Scholar]
- Nibu Y., Zhang,H. and Levine,M. (2001) Local action of long-range repressors in the Drosophila embryo. EMBO J., 20, 2246–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K. (1997) The role of endoxyloglucan transferase in the organization of plant cell walls. Int. Rev. Cytol., 173, 157–206. [DOI] [PubMed] [Google Scholar]
- Okada K. and Shimura,Y. (1992) Mutational analysis of root gravitropism and phototropism of Arabidopsis thaliana seedlings. Aust. J. Plant Physiol., 19, 439–448. [Google Scholar]
- Okazawa K., Sato,Y., Nakagawa,T., Asada,K., Kato,I., Tomita,E. and Nishitani,K. (1993) Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransferase that mediates molecular grafting between matrix polysaccharides in plant cell walls. J. Biol. Chem., 268, 25364–25368. [PubMed] [Google Scholar]
- Oppenheimer D.G. (1998) Genetics of plant cell shape. Curr. Opin. Plant Biol., 130, 630–650. [DOI] [PubMed] [Google Scholar]
- Oppenheimer D.G., Pollock,M.A., Vacik,J., Szymanski,D.B., Ericson,B., Feldmann,K. and Marks,M.D. (1997) Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl Acad. Sci. USA, 94, 6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phippen T.M., Sweigart,A.L., Moniwa,M., Krumm,A., Davie,J.R. and Parkhurst,S.M. (2000) Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both Mad and Groucho transcriptional repression. J. Biol. Chem., 275, 37628–37637. [DOI] [PubMed] [Google Scholar]
- Poortinga G., Watanabe,M. and Parkhurst,S.M. (1998) Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J., 17, 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potikha T. and Delmer,D.P. (1995) A mutant of Arabidopsis thaliana displaying altered patterns of cellulose deposition. Plant J., 2, 453–460. [Google Scholar]
- Rédei G.P. (1962) Single locus heterosis. Z. Vererbungs., 93, 164–170. [Google Scholar]
- Rosée-Borggreve A.L., Häder,T., Wainwright,D., Sauer,F. and Jäckle,H. (1999) hairy stripe 7 element mediates activation and repression in response to different domains and levels of Krüppel in the Drosophila embryo. Mech. Dev., 89, 133–140. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schaeper U., Boyd,J.M., Verma,S., Uhlmann,E., Subramanian,T. and Chinnadurai,G. (1995) Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl Acad. Sci. USA, 92, 10467–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeper U., Subramanian,T., Lim,L., Boyd,J.M. and Chinnadurai,G. (1998) Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem., 273, 8549–8552. [DOI] [PubMed] [Google Scholar]
- Sewalt R.G.A.B., Gunster,M.J., van der Vlag,J., Satijn,D.P.E. and Otte,A.P. (1999) C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol., 19, 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibaoka H. (1994) Plant hormone-induced changes in the orientation of cortical microtubules: alterations in the cross-linking between microtubules and the plasma membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol., 45, 527–544. [Google Scholar]
- Singh R. and Green,M.R. (1993) Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science, 259, 365–368. [DOI] [PubMed] [Google Scholar]
- Sollerbrant K., Chinnadurai,G. and Svensson,C. (1996) The CtBP binding domain in the adenovirus E1A protein controls CR1-dependent transactivation. Nucleic Acids Res., 24, 2578–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanfò S. et al. (1999) Molecular cloning and functional characterization of Brefeldin A-ADP-ribosylated substrate. J. Biol. Chem., 274, 17705–17710. [DOI] [PubMed] [Google Scholar]
- Terryn N. et al. (1998) Sequence analysis of a 40-kb Arabidopsis thaliana genomic region located at the top of chromosome 1. Gene, 215, 11–17. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL_W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas J., Bellini,C., Nacry,P., Kronenberger,J., Bouchez,D. and Caboche,M. (1995) Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature, 375, 676–677. [Google Scholar]
- Tsuge T., Tsukaya,H. and Uchimiya,H. (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development, 122, 1589–1600. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (1995) Developmental genetics of leaf morphogenesis in dicotyledonous plants. J. Plant Res., 108, 407–416. [Google Scholar]
- Tsukaya H. (1998) Genetic evidence for polarities that regulate leaf morphogenesis. J. Plant Res., 111, 113–119. [Google Scholar]
- Tsukaya H. (2002) Interpretation of mutants in leaf morphology: genetic evidence for a compensatory system in leaf morphogenesis that provides a new link between Cell and Organismal theory. Int. Rev. Cytol., 217, in press. [DOI] [PubMed] [Google Scholar]
- Tsukaya H., Ohshima,T., Naito,S., Chino,M. and Komeda,Y. (1991) Sugar-dependent expression of the CHS-A gene for chalcone synthase from petunia in transgenic Arabidopsis thaliana. Plant Physiol., 97, 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H., Naito,S., Rédei,G.P. and Komeda,Y. (1993) A new class of mutations in Arabidopsis thaliana, acaulis1, affecting the development of both inflorescences and leaves. Development, 118, 751–764. [Google Scholar]
- Tsukaya H., Tsuge,T. and Uchimiya,H. (1994) The cotyledon: a superior system for studies of leaf development. Planta, 195, 309–312. [Google Scholar]
- Tsukaya H., Inaba-Higano,K. and Komeda,Y. (1995) Phenotypic characterization and molecular mapping of acaulis2 mutant with flower stalks of much reduced length in Arabidopsis thaliana. Plant Cell Physiol., 36, 239–246. [Google Scholar]
- Turner J. and Crossley,M. (1998) Cloning and characterization of mCtBP2, a co-repressor that associates with basic Krüppel-like factor and other mammalian transcriptional regulators. EMBO J., 17, 5129–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. and Crossley,M. (2001) The CtBP family: enigmatic and enzymatic transcriptional co-repressors. BioEssays, 23, 683–690. [DOI] [PubMed] [Google Scholar]
- Ueda T., Anai,T., Tsukaya,H., Hirata,A. and Uchimiya,H. (1996) Characterization and subcellular localization of a small GTP-binding protein (Ara-4) from Arabidopsis: conditional expression under control of the promoter of the gene for heatshock protein HSP81-1. Mol. Gen. Genet., 250, 533–539. [DOI] [PubMed] [Google Scholar]
- Ueda K., Matsuyama,T. and Hashimoto,T. (1999) Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma, 206, 201–206. [Google Scholar]
- Verica J.A. and Medford,J. (1997) Modified MERI5 expression alters cell expansion in transgenic Arabidopsis plants. Plant Sci., 125, 201–210. [Google Scholar]
- Wasteneys G.O. (2000) The cytockeleton and growth polarity. Curr. Opin. Plant Biol., 3, 503–511. [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Willingale-Theun,J. and Menzel,D. (1997) Freeze shattering: a simple and effective method for permeabilizing higher plant cell walls. J. Microscopy, 188, 51–61. [DOI] [PubMed] [Google Scholar]
- Weigert R. et al. (1999) CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature, 402, 429–433. [DOI] [PubMed] [Google Scholar]
- Wen Y., Nguyen,D., Li,Y. and Lai,Z.-C. (2000) The N-terminal BTB/POZ domain and C-terminal sequences are essential for Tramtrack69 to specify cell fate in the developing Drosophila eye. Genetics, 156, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymer C. and Lloyd,C. (1996) Dynamic microtubules: implications for cell wall patterns. Trends Plant Sci., 1, 222–228. [Google Scholar]
- Yokoyama R. and Nishitani,K. (2001) A comprehensive expression-analysis of all members of a gene family encoding cell wall enzymes allows us to predict cis-regulatory regions involved in the cell wall construction in specific organs of Arabidopsis. Plant Cell Physiol., 42, 1025–1033. [DOI] [PubMed] [Google Scholar]
- Yuan M., Warn,R.M., Shaw,P.J. and Lloyd,C.W. (1995) Dynamic microtubules under the radial and outer tangential walls of microinjected pea epidermal cells observed by computer reconstruction. Plant J., 7, 17–23. [DOI] [PubMed] [Google Scholar]
- Zhang H. and Levine,M. (1999) Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 96, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]