Abstract
Transcription of the major histocompatibility complex class II family of genes is regulated by conserved promoter elements and two gene-specific trans-activators, RFX and CIITA. RFX binds DNA and nucleates the assembly of an enhanceosome, which recruits CIITA through protein–protein interactions. Transcriptional activation is a complex, multi-step process involving chromatin modification and recruitment of the transcription apparatus. To examine the roles of the enhanceosome and CIITA in these processes, we analysed the level of promoter-associated hyperacetylated histones H3 and H4, TBP, TFIIB and RNA poly merase II in cells lacking RFX or CIITA. We compared four genes co-regulated by RFX and CIITA (HLA-DRA, HLA-DPB, HLA-DMB and Ii) and found that the enhanceosome and CIITA make variable, promoter-dependent contributions to histone acetylation and transcription apparatus recruitment. CIITA is generally implicated at multiple levels of the activation process, while the enhanceosome contributes in a CIITA-independent manner only at certain promoters. Our results support the general notion that the impact of a particular activator on transcription in vivo may vary depending on the promoter and the chromatin context.
Keywords: chromatin/gene expression regulation/promoter regions/trans-activators/transcription factors
Introduction
Transcriptional activity of eukaryotic genes can be regulated at multiple levels, including histone acetylation and chromatin remodelling, recruitment of the transcriptional machinery and assembly of the preinitiation complex, promoter clearance and elongation (Lee and Young, 2000; Lemon and Tjian, 2000). A wealth of studies has focused on the roles of individual activators in these processes. The general conclusion from these studies is that an activator can affect transcription at one or more of these steps. However, a question that remains unanswered is whether a given activator always performs the same function(s) at different promoters in vivo. To address this question we have turned to a well-defined model system, regulation of major histocompatibility complex class II (MHCII) expression, in which several related genes are co-regulated by the same transcriptional activators.
MHCII molecules are heterodimeric surface glycoproteins that present antigenic peptides to the T-cell receptor of CD4+ T lymphocytes and thereby play a central role in the adaptive immune response (Viret and Janeway, 1999). MHCII expression is constitutive in professional antigen-presenting cells, such as B cells, dendritic cells and macrophages. It is inducible by cytokines, particularly by interferon γ, in most other cell types (Reith and Mach, 2001). MHCII molecules are encoded by a family of genes that is coordinately regulated at the level of transcription (Reith and Mach, 2001). Transcription of all MHCII genes is controlled by a conserved regulatory region situated within the first 150 bp upstream of the transcription initiation site. This promoter-proximal regulatory region consists of four cis-acting elements, referred to as the S (also called W or Z), X, X2 and Y ‘boxes’ (Figure 1A), which function together as a single composite MHCII regulatory module (Reith and Mach, 2001). A similar arrangement of sequence elements has been conserved in the promoter regions of the MHCII-related Ii, HLA-DM and HLA-DO genes (Brown et al., 1991; Ting et al., 1997; Westerheide et al., 1997; Tai et al., 1999; Taxman et al., 2000), which code for proteins implicated in the intracellular traffic and peptide loading of MHCII molecules (Cresswell, 1996; Alfonso and Karlsson, 2000).
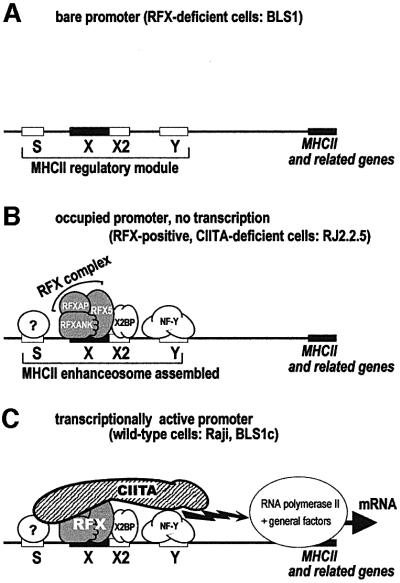
Fig. 1. Regulation of MHCII transcription and molecular defects in BLS. Transcription of MHCII and related genes is directed by a conserved regulatory module consisting of the S, X, X2 and Y ‘boxes’. This module is the target of four essential MHCII-specific trans-activators, RFX5, RFXAP, RFXANK (RFXB) and CIITA, identified by virtue of the fact that they are mutated in BLS. The first three factors assemble into a heterotrimeric complex called RFX. Binding of RFX to the X box nucleates the assembly of an enhanceosome complex containing RFX, X2BP, NF-Y and an as yet unidentified S box-binding factor. The enhanceosome serves as a landing pad for the coactivator CIITA. (A) In cells deficient in one of the RFX subunits (in this study, RFXANK–/– BLS1 cells), the RFX complex cannot assemble and the enhanceosome cannot form. MHCII promoters thus remain unoccupied. (B) In B cells deficient in CIITA (RJ2.2.5), RFX is functional and the promoters are stably occupied by the enhanceosome. However, they remain silent because CIITA is missing. (C) In wild-type cells (Raji and BLS1c), enhanceosome assembly and CIITA recruitment lead to promoter activation.
The regulation of MHCII expression is among the rare mammalian transcriptional control systems that have been dissected genetically in great detail. This has been facilitated greatly by the identification of genes that are mutated in the bare lymphocyte syndrome (BLS), a hereditary immunodeficiency disease resulting from deficient MHCII gene expression (Masternak et al., 2000a). The four genes mutated in BLS encode essential trans-activators of MHCII transcription, namely CIITA and the three subunits of the RFX complex (Masternak et al., 2000a).
Genetic and biochemical studies have led to the following model for the control of MHCII transcription (Harton and Ting, 2000; Waldburger et al., 2000). The MHCII regulatory module is recognized by three DNA-binding factors, RFX, X2BP and NF-Y, which bind in a highly cooperative manner to their respective X, X2 and Y box target sites (Reith et al., 1994a,b; Wright et al., 1994; Louis-Plence et al., 1997). These factors form a higher-order MHCII enhanceosome complex, which also contains an as yet unidentified S box-binding factor (Figure 1B). However, RFX, X2BP and NF-Y are expressed ubiquitously and in an apparently unregulated manner, implying that they do not provide the regulatory function necessary for cell-specific and inducible MHCII expression. It follows that the MHCII enhanceosome is not sufficient on its own to support transcription. This requires an additional factor, the MHCII-specific transcriptional co-activator CIITA, which is recruited to the enhanceosome via multiple protein–protein interactions (Figure 1C) (DeSandro et al., 2000; Hake et al., 2000; Masternak et al., 2000b; Zhu et al., 2000). In contrast to the ubiquitous expression of RFX and the other enhanceosome components, CIITA expression is highly regulated, imposing a tight qualitative and quantitative control over MHCII expression (Harton and Ting, 2000; Waldburger et al., 2000).
By virtue of its absolute control over MHCII genes, CIITA has become the primary focus of research in the field of MHCII regulation (Reith and Mach, 2001). Numerous in vitro and functional studies have implicated CIITA in multiple steps of the transcriptional activation process. First, CIITA may facilitate chromatin remodelling, as it interacts with histone acetyltransferases (Kretsovali et al., 1998; Fontes et al., 1999; Harton et al., 2001); moreover, CIITA has recently been shown to have intrinsic acetyltransferase activity (Raval et al., 2001). Secondly, CIITA interacts with the general transcription factors TFIIB, hTAFII32 and hTAFII70, implying that it may recruit the transcriptional apparatus directly (Fontes et al., 1997; Mahanta et al., 1997). Thirdly, CIITA interacts with TFIIH and P-TEFb (Mahanta et al., 1997; Kanazawa et al., 2000), and may therefore enhance promoter clearance and transcription elongation. An implicit picture emerging from these studies is that CIITA activates transcription single-handedly, while the role of the MHCII enhanceosome is relegated to specific DNA sequence recognition and CIITA recruitment. However, this division in labour has never been demonstrated formally.
The aim of the present work was to examine the roles of CIITA and the MHCII enhanceosome in the transcriptional activation process in vivo. In particular, we wanted to elucidate the involvement of CIITA and the enhanceosome in two key processes that precede the initiation of transcription—the acetylation of core histones and recruitment of the general transcription machinery (GTM). To achieve this goal, we determined the levels of histone acetylation and promoter association of the transcription apparatus in wild-type, RFX-deficient and CIITA-deficient B cells by quantitative chromatin immunoprecipitation (ChIP). We performed a comparative study of different members of the family of genes co-regulated through the MHCII regulatory module by RFX and CIITA. Four representative promoters sharing the MHCII regulatory module were studied: the prototypical MHCII promoter HLA-DRA, HLA-DPB, HLA-DMB, and the invariant chain (Ii) promoter. This led to the unanticipated finding that the respective contributions of CIITA and the enhanceosome to histone acetylation and GTM recruitment vary from one promoter to another. CIITA was indeed found to be the major player at HLA-DRA and HLA-DPB, but it was of lesser importance at the other two promoters (HLA-DMB and Ii). A dominant role of the enhanceosome in supporting histone acetylation and GTM recruitment at the HLA-DMB promoter was evident, clearly demonstrating that the MHCII enhanceosome is not exclusively a ‘CIITA landing pad’, but can contribute to transcription in a CIITA-independent manner. On the whole, our work provides evidence that the effects of activators on transcription in vivo are promoter specific and may differ significantly, even within a family of closely related genes.
Results
Genes sharing the MHCII regulatory module show differential dependence on MHCII trans-activators
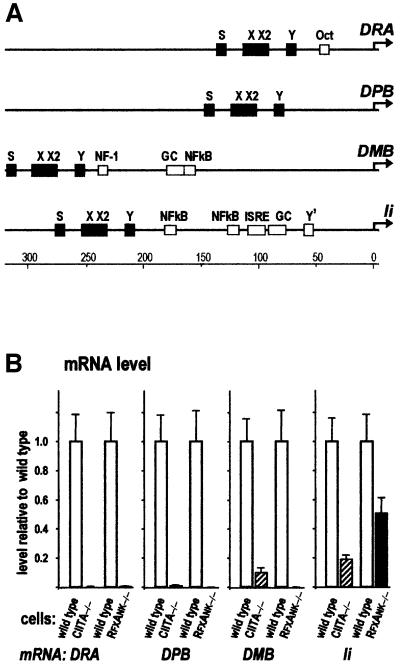
We chose for our study four representative genes controlled by RFX and CIITA: HLA-DRA (called hereafter DRA), HLA-DPB (DPB), HLA-DMB (DMB) and Ii. The promoters of these genes share the S–X–X2–Y boxes of the MHCII regulatory module. In addition, the individual promoters contain unique, potential or confirmed regulatory sequences (Figure 2A). The prototypical DRA promoter contains an octamer-binding site (Oct), which is required for maximal DRA promoter activity in B cells (Sherman et al., 1989). The DMB promoter contains putative NF-1, Sp1 (GC box) and NFκB sites located downstream of the MHCII regulatory module (Radley et al., 1994). These sequence elements are not found in the ‘classical’ MHCII genes and their importance for DMB expression has never been demonstrated. In the Ii pro moter, the extra sequence elements include two NFκB sites, a putative interferon-stimulated response element (ISRE), an Sp1-binding site (GC box) and a promoter-proximal imperfect Y box (Y′) (Figure 2A). Transient transfection experiments have suggested that all these sites, except the ISRE, are important for Ii expression in B cells (Brown et al., 1994; Wright et al., 1995; Tai et al., 1999).
Fig. 2. MHCII and related genes are co-regulated by RFX and CIITA through the MHCII regulatory module. (A) Schematic representation of the upstream promoter-proximal regions of HLA-DRA (DRA), HLA-DPB (DPB), HLA-DMB (DMB) and Ii. The S–X–X2–Y elements of the MHCII regulatory module are present in all promoters and are shown as filled boxes. The other potential or confirmed regulatory sequences are shown as empty boxes: Oct, octamer-binding site; NF-1, CTF/NF-1-binding site; GC, Sp1-binding site; NFkB, binding site for NFκB/Rel family members; ISRE, interferon-stimulated response element; Y′, an imperfect Y box (NF-Y-binding site). The promoters are drawn to scale and the distance (in bp) from the transcription initiation site (arrow) is shown at the bottom. (B) The effect of a deficiency in CIITA or RFXANK on DRA, DPB, DMB and Ii expression. Steady-state levels of the corresponding mRNAs were compared between isogenic pairs of B-cell lines (wild-type versus mutant). mRNA levels were determined by multiplex real-time RT–PCR and are represented relative to the wild type.
A wealth of previous reports has already proven the importance of CIITA and RFX for expression of the genes studied here. However, we wished to assess their CIITA- and RFX-dependence in a more precise, quantitative manner in order to demonstrate the functional relevance of our ChIP results. To achieve this goal, we compared expression of the endogenous genes in the presence and absence of either CIITA or RFXANK (also called RFXB). RFXANK is an integral subunit of the RFX complex, and is thus necessary for DNA binding and assembly of the MHCII enhanceosome (Kara and Glimcher, 1991; Masternak et al., 1998; Nagarajan et al., 1999). The assay was applied in parallel to two pairs of isogenic B-cell lines. The first pair consists of RJ2.2.5, a CIITA-deficient MHCII negative mutant, and the wild-type B-cell line Raji from which it was derived. The second pair consists of BLS1 (a cell line originating from an MHCII deficiency patient carrying an RFXANK mutation) and BLS1c, which was obtained by stable complementation of BLS1 with wild-type RFXANK. In BLS1c, RFXANK is expressed at physiological levels (data not shown). These two pairs of cell lines were used both for expression analysis and ChIP assays. The use of these isogenic cell lines permits a neat dissection of the respective contributions of CIITA and the enhanceosome in the transcriptional activation process.
Steady-state levels of specific mRNAs were quantified by multiplex real-time RT–PCR. Glyceraldehyde phosphate dehydrogenase (GADPH) mRNA was used as endogenous reference. The results plotted in Figure 2B show noticeable differences between the two classical MHCII genes (DRA and DPB) and the two MHCII-related genes (DMB and Ii) regarding CIITA- and RFXANK-dependent expression. In the mutant cells, both DRA and DPB mRNA levels were reduced by at least two orders of magnitude with respect to the wild type (Figure 2B). The dependence of the two classical MHCII genes on CIITA and RFXANK is therefore ‘strict’ or ‘absolute’, as there is no residual gene expression in the absence of either of these factors. The situation is somewhat different for DMB (Figure 2B). While DMB expression remains critically dependent on the enhanceosome, DMB mRNA was present in CIITA –/– cells at ∼10% of the wild-type level, which indicates that the enhanceosome supports some DMB expression independently of CIITA. This finding was rather unexpected, as others have shown previously that DMB mRNA is absent from CIITA-negative B-cell lines (Chang and Flavell, 1995; Kern et al., 1995; Westerheide et al., 1997; Quan et al., 1999) including the RJ2.2.5 B cells used here (Westerheide et al., 1997; Quan et al., 1999). This discrepancy reflects differences in accuracy and sensitivity between the traditional methods of nucleic acid quantification used previously and the real-time PCR analysis used here.
In the case of the invariant chain gene (Ii), we found that it is expressed in CIITA –/– and RFXANK –/– cells, albeit at a reduced level (Figure 2B). This is in line with earlier reports demonstrating that Ii expression is reduced but not completely abolished in B-cell lines that lack RFX or CIITA (de Preval et al., 1985; Chang and Flavell, 1995; Quan et al., 1999; Tai et al., 1999). A comparison between CIITA- and RFXANK-deficient cells reveals that Ii expression is actually less dependent on RFXANK (∼50% Ii mRNA levels in RFXANK –/– cells) than on CIITA (∼20% Ii mRNA levels in CIITA –/– cells). This is surprising because CIITA is believed to require RFX for function and RFXANK is an integral and essential subunit of the RFX complex (Masternak et al., 1998; Nagarajan et al., 1999; Nekrep et al., 2000). The fact that CIITA can at least partly activate Ii transcription in the absence of RFXANK points to a possible RFXANK-independent effect of CIITA on transcription.
Association of the enhanceosome and CIITA with MHCII promoters in vivo
We have demonstrated previously that both CIITA and RFX are associated physically with the promoters of various classical MHCII and related genes in wild-type MHCII-positive B cells (Masternak et al., 2000b). To extend our previous observations and define in more detail the respective contributions of CIITA and the enhanceosome in transcriptional activation in vivo, we developed a quantitative ChIP assay. Briefly, antibodies were used to pull down chromatin fragments cross-linked to specific proteins, and the immunoprecipitates were then analysed by real-time PCR for the presence of the promoters of interest.
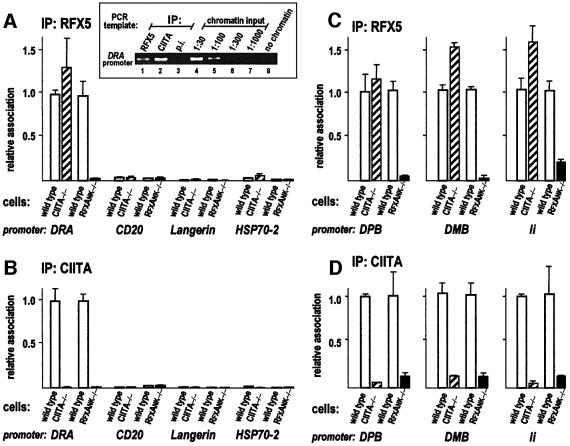
We used this quantitative ChIP assay to study the association of CIITA and RFX5 with the DRA, DPB, DMB and Ii promoters in wild-type and mutant cells (Figure 3). RFX5 is the largest, DNA-binding subunit of the RFX complex (Steimle et al., 1995), and RFX5 association with a promoter is diagnostic of MHCII enhanceosome assembly (Masternak et al., 2000b). The results indicate that CIITA and RFX are both associated specifically with the DRA, DPB, DMB and Ii promoters in wild-type cells (Figure 3). Specificity is demonstrated by the lack of association of either factor with the promoters of three unrelated genes, namely CD20 (encoding a cell surface molecule widely expressed in human B cells), Langerin (encoding the Langerhans cell-specific c-type lectin) and HSP70-2 (encoding a heat shock protein) (Figure 3A and B).
Fig. 3. Occupancy of MHCII and related promoters by the enhanceosome and CIITA. The association of RFX5 with promoter DNA is indicative of MHCII enhanceosome assembly. Antibodies specific for RFX5 or CIITA were used to immunoprecipitate crosslinked chromatin fragments. Immunoprecipitates were then analysed for the abundance of specific promoter sequences by PCR. The inset (top left) shows an example of a classical ChIP analysis; immunoprecipitated fractions (IP, lanes 1–3) and sequential dilutions of input chromatin from wild-type B cells (lanes 4–7) were amplified with DRA-specific primers and PCR products were analysed by gel electrophoresis. Pre-immune serum (p.i.) was used as negative control. In subsequent experiments, the levels of RFX5 and CIITA binding to different promoters in CIITA- and RFXANK-deficient cells and their matching wild-type counterparts were measured by quantitative real-time PCR. Association of RFX5 (A) and CIITA (B) with the DRA promoter. The unrelated CD20, Langerin and HSP70-2 promoters are included as controls. Results are shown relative to the values observed for the DRA promoter in wild-type cells. Association of RFX5 (C) and CIITA (D) with the DPB, DMB and Ii promoters. Results for the mutant cells (RJ2.2.5 and BLS1) are given relative to the values observed in the matching wild-type cells (Raji and BLS1c).
We find that binding of RFX5 to the DRA, DPB, DMB and Ii promoters is normal in the CIITA-deficient cells (Figure 3A and C), indicating that enhanceosome formation in these cells is independent of CIITA. This is in agreement with previous in vivo footprint experiments demonstrating that a deficiency in CIITA does not affect occupation of MHCII promoters by the enhanceosome in B cells (Kara and Glimcher, 1991, 1993). ChIP with the RFX5 antibody appears even more efficient in the CIITA-deficient cells as compared with wild-type cells (Figure 3A and C; data not shown). The reason for this difference is unknown. We speculate that binding of CIITA to the enhanceosome may somehow decrease the accessibility of the epitopes recognized by the RFX5-specific antiserum, resulting in reduced immunoprecipitation efficiency. As expected, association of both RFX5 and CIITA with the DRA, DPB, DMB and Ii promoters is lost in RFXANK-deficient cells (Figure 3). This confirms that RFXANK is essential for the assembly and binding of RFX (Masternak et al., 1998) and that enhanceosome formation is a pre-requisite for the recruitment of CIITA in vivo (Masternak et al., 2000b). Our ChIP results are thus entirely consistent with in vivo footprint experiments and in vitro recruitment assays showing that a deficiency in RFX abolishes both promoter occupation and the recruitment of CIITA (Kara and Glimcher, 1991, 1993; Masternak et al., 2000b).
A small fraction of RFX5 was found to be able to bind to the Ii promoter in the absence of RFXANK (Figure 3C). This may reflect the existence of a sub-complex composed of RFX5 and RFXAP (the third RFX subunit). Residual binding of RFX5–RFXAP sub-complexes has also been observed in vitro using promoter pull-down assays (K.Masternak and W.Reith, unpublished data).
Importance of the enhanceosome and CIITA in the induction of histone acetylation
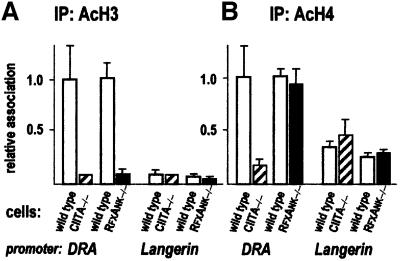
Histone hyperacetylation is now generally recognized as a characteristic property of transcriptionally competent chromatin (Struhl, 1998; Lee and Young, 2000; Strahl and Allis, 2000; Roth et al., 2001). To define the respective roles of CIITA and the enhanceosome in the induction of histone acetylation at the DRA, DPB, DMB and Ii promoters, we compared mutant and wild-type cells in quantitative ChIP experiments with antibodies specific for hyperacetylated histones H3 and H4. As negative control we included the promoter of the Langerin gene, which is inactive in B cells (Valladeau et al., 2000) and is thus hypoacetylated.
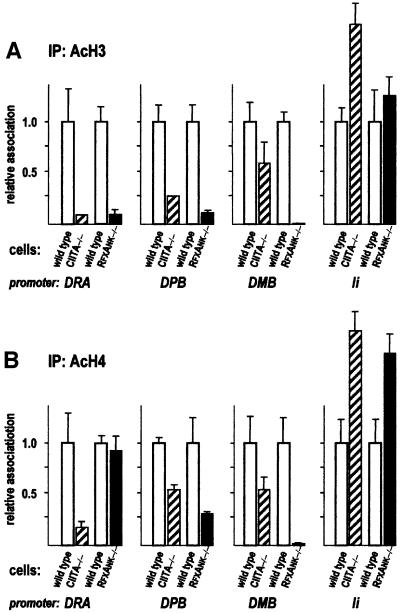
In CIITA –/– cells, H3 acetylation at the DRA promoter is decreased ∼10-fold, to the level observed at the negative control promoter (Figure 4A). The decrease is not stronger in RFXANK-deficient cells than in CIITA-deficient cells, indicating that the enhanceosome is unable to promote any H3 acetylation in the absence of CIITA. CIITA thus plays a major role in H3 hyperacetylation at the DRA promoter. Histone H4 acetylation levels were also clearly reduced in CIITA –/– cells. Curiously, however, no reduction in H4 acetylation was observed in RFXANK –/– cells (Figure 4B). This suggests that the basal level of H4 acetylation is already high at the bare DRA promoter, and that occupation by the enhanceosome in the absence of CIITA actually leads to histone H4 deacetylation. Similar observations have been made at the DRA and DQB promoters in ChIP experiments performed with cells lacking RFXAP, the third subunit of the RFX complex (M.Peretti, K.Masternak and W.Reith, unpublished data). Why binding of the enhanceosome can lead to a decrease in H4 acetylation at certain promoters is not clear. One possibility is that the enhanceosome recruits histone deacetylases. Alternatively, it could simply block access to acetyltransferases carrying out non-targeted H4 acetylation (Struhl, 1998; Roth et al., 2001). It is worth noting here that a specific decrease in H4 acetylation induced by binding of transcription factors has also been observed recently in yeast (Deckert and Struhl, 2001). As expected, the acetylation status of the unrelated Langerin promoter is not affected by a deficiency in either CIITA or RFX (Figure 4).
Fig. 4. Contribution of CIITA and the MHCII enhanceosome to histone acetylation at the DRA promoter. Histone acetylation at the DRA and Langerin promoters was analysed by quantitative ChIP using antibodies specific for acetylated histone H3 (A) or acetylated histone H4 (B). Results are normalized with respect to the values obtained with the internal control promoter HSP70-2, and are represented relative to the values observed for the DRA promoter in wild-type cells.
Although CIITA clearly plays a dominant role in the induction of histone acetylation at the DRA promoter, the situation is less clear-cut at other promoters. At the DPB and DMB promoters, H3 and H4 acetylation levels are lower in RFXANK –/– cells than in CIITA –/– cells (Figure 5). This difference is particularly striking for DMB, where only a 2-fold reduction is observed in the absence of CIITA, but a 50-fold reduction is evident in the absence of the enhanceosome. This implies that the enhanceosome directs efficient histone acetylation at the DMB promoter. At the DPB promoter, the contribution of the enhanceosome is also evident but clearly less important. Finally, at the Ii promoter, no reduction in histone acetylation is apparent in the absence of either CIITA or RFXANK. If anything, an increase in acetylation is observed.
Fig. 5. Contribution of CIITA and the MHCII enhanceosome to histone acetylation at different MHCII and related promoters. Histone acetylation at the DRA, DPB, DMB and Ii promoters was analysed as in Figure 4. Results are normalized and represented relative to the values observed in the matching wild-type cells.
Importance of the enhanceosome and CIITA for GTM recruitment
There is controversy concerning activator-dependent recruitment of GTM to promoters. At one extreme, the transcription apparatus may be recruited sequentially, factor by factor, as suggested by early in vitro studies. At the other extreme is the ‘two-step’ model, which predicts that all GTM components, except TFIID, reach the promoter simultaneously in the form of a single holoenzyme complex (Lee and Young, 2000; Lemon and Tjian, 2000). Activation at different promoters could actually involve a whole spectrum of mechanistic possibilities. To study GTM recruitment to MHCII and related promoters, we therefore targeted three key GTM components: (i) TATA-binding protein (TBP), the ‘core’ subunit of TFIID; (ii) TFIIB, a general transcription factor that is involved in start site selection and bridging of TFIID to the RNA polymerase II (Pol II) holoenzyme; and (iii) RNA Pol II itself. Quantitative ChIP analysis was performed with antibodies specific to these three proteins.
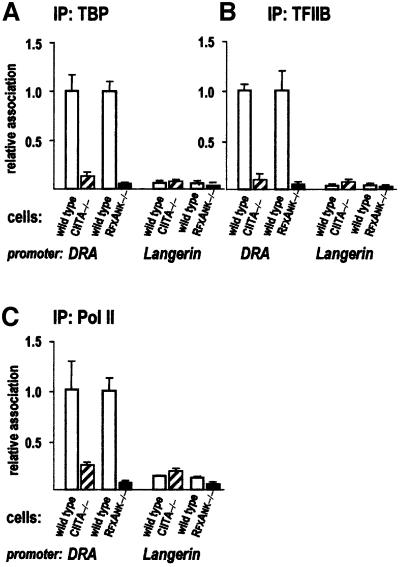
In wild-type cells, TFIID, TFIIB and Pol II are associated with the DRA promoter (Figure 6), which reflects ongoing DRA transcription (Figure 2A). In contrast, the association of these factors with the DRA promoter is reduced 5- to 20-fold in mutant cells, to the levels characteristic of the inactive Langerin promoter (Figure 6). A comparison between CIITA –/– and RFXANK –/– cells indicates that CIITA is required for efficient GTM recruitment to the DRA promoter: in the absence of CIITA, the enhanceosome does not suffice to tether TFIID or TFIIB, and makes at best a minor contribution to Pol II recruitment (Figure 6).
Fig. 6. Contribution of CIITA and the enhanceosome to GTM recruitment at the DRA promoter. GTM recruitment to the DRA and Langerin promoters was analysed by quantitative ChIP experiments using antibodies specific for TBP (A), TFIIB (B) and RNA Pol II (C). Results are represented as in Figure 4.
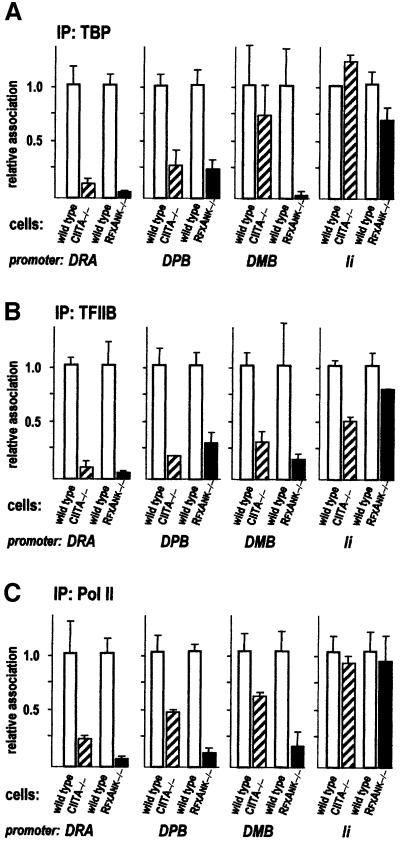
A more complex picture emerges when analysing DPB and DMB; CIITA is also implicated in GTM recruitment, but its contribution relative to the enhanceosome varies depending on the promoter and factor that is examined (Figure 7). At the DPB promoter, the decrease in TBP association appears to be the same in both mutant cells (Figure 7A), indicating that TFIID recruitment to DPB requires CIITA. On the other hand, TFIID recruitment to DMB is largely CIITA independent, as >70% of wild-type levels of TBP association are observed in CIITA –/– cells. Yet, TFIID recruitment is enhanceosome dependent, since only background levels of TBP association are detected in RFX –/– cells. TFIIB association with DPB and DMB is significantly reduced in both mutant cells (Figure 7B), indicating that the enhanceosome is inefficient at tethering this factor to the DPB and DMB promoters. On the contrary, its contribution to Pol II recruitment is evident for both DPB and DMB (Figure 7C).
Fig. 7. Contributions of CIITA and the enhanceosome to GTM recruitment at different MHCII and related promoters. Recruitment of TBP (A), TFIIB (B) and RNA Pol II (C) to the DRA, DPB, DMB and Ii promoters was analysed by quantitative ChIP and is represented as in Figure 5.
As already observed for histone acetylation, the situation at the Ii promoter is radically different from what is found at the other promoters. No major difference in GTM association with the Ii promoter is noticeable upon comparison of wild-type and mutant cells (Figure 7). The strongest effect is a 2-fold reduction in TFIIB association in CIITA –/– cells. This indicates that both the enhanceosome and CIITA play only a minor role in recruiting GTM components to the Ii promoter.
Discussion
The aim of the present work was to clarify the respective roles of CIITA and the MHCII enhanceosome in two key processes preceding transcription initiation: the acetylation of core histones and GTM recruitment. To achieve this goal, we compared the levels of histone acetylation and promoter-associated GTM components in CIITA-deficient and enhanceosome-deficient (RFXANK –/–) cells by quantitative ChIP. The functional relevance of our ChIP analysis was validated by an analysis of gene expression performed in parallel. We studied four representative genes, DRA, DPB, DMB and Ii, which share the MHCII regulatory module and are known to be co-regulated by RFX and CIITA (Harton and Ting, 2000; Reith and Mach, 2001). A comparison of the four genes demonstrated that the respective contributions of CIITA and the MHCII enhanceosome to histone acetylation and GTM recruitment vary considerably from one gene to another.
In the case of DRA, the most extensively studied human MHCII gene, CIITA is essential for both core histone acetylation and GTM recruitment, and the enhanceosome cannot independently support either task. Our results are in line with a recently published report showing that CIITA is required for histone acetylation at the DRA promoter (Beresford and Boss, 2001). CIITA has been proposed to enhance histone acetylation either by recruiting acetylases or via an intrinsic histone acetyltransferase activity (Kretsovali et al., 1998; Fontes et al., 1999; Raval et al., 2001). Transfection experiments performed with a CIITA mutant lacking the putative acetylation domain have suggested that both mechanisms could be involved (Beresford and Boss, 2001).
CIITA also plays a major role in histone acetylation and GTM recruitment at the DPB promoter. However, at this promoter, the MHCII enhanceosome provides a CIITA-independent contribution. This is apparent for both H3 and H4 acetylation, and for Pol II recruitment. Notwithstand ing, the enhanceosome contribution is not sufficient to activate transcription to detectable levels.
The example of the DMB promoter provides strong evidence for a CIITA-independent involvement of the MHCII enhanceosome in transcriptional activation. At this promoter, the dominant role in histone acetylation and GTM recruitment is clearly played by the enhanceosome rather than CIITA. While the loss of CIITA leads to at best a mere 2-fold reduction in both histone acetylation and TFIID and Pol II recruitment, the loss of the enhanceosome has a much stronger effect. Importantly, DMB mRNA is undetectable in RFXANK –/– cells, but remains present in CIITA –/– cells at ∼10% of the wild-type level. Thus, the enhanceosome is not just a ‘CIITA landing pad’, but can actually support CIITA-independent histone acetylation and GTM recruitment to levels that are sufficient for a partial DMB activation.
No major effect of a deficiency in CIITA or RFXANK on histone acetylation and GTM recruitment was observed at the Ii promoter. This is consistent with the expression analysis (Figure 2B) as well as earlier reports demonstrating that CIITA and RFX are not absolutely essential for Ii expression, at least in B cells (de Preval et al., 1985; Brown et al., 1991; Chang and Flavell, 1995; Quan et al., 1999; Tai et al., 1999).
At the Ii promoter, CIITA is clearly dispensable for histone acetylation, and its role in GTM recruitment is auxiliary, at best. CIITA is nevertheless necessary for full transcriptional activation, since Ii mRNA levels are reduced 5-fold in CIITA –/– cells (Figure 2B). This implies that CIITA may contribute to Ii transcription by promoting downstream, post-recruitment event(s), such as promoter escape or elongation. It may be relevant here that CIITA has been proposed to interact with factors implicated in promoter clearance (TFIIH) (Mahanta et al., 1997) and RNA Pol II processivity (pTEFb) (Kanazawa et al., 2000). A deficiency in RFXANK has virtually no effect on either histone acetylation or GTM recruitment at the Ii promoter (Figures 5 and 7), indicating that these processes are largely enhanceosome independent.
The MHCII enhanceosome incorporates factors that have been shown to interact directly with the transcriptional machinery. NF-Y, which may be implicated in the regulation of ∼20% of eukaryotic genes, can interact with TBP, the general coactivator PC4 and the histone acetyltransferases p300 and P/CAF (Mantovani, 1999). CREB, an X2BP component (Moreno et al., 1999), can interact with TAF II 130 and the Pol II holoenzyme-associated histone acetyltransferases CREB-binding protein (CBP) and p300 (Mayr and Montminy, 2001). Furthermore, the recruitment of CBP by CREB can be sufficient for transcriptional activation (Cardinaux et al., 2000). With this in mind, it is not surprising that the MHCII enhanceosome can actively sustain histone acetylation and GTM recruitment. What is actually more surprising is the finding that it is unable to perform the same functions at other, closely related promoters.
Whereas the mechanisms underlying the promoter-specific role of the MHCII enhanceosome remain to be determined, a number of possible explanations come to mind. First, the difference could reside in general properties of the chromatin environment and/or the core promoter. There is growing evidence that core promoters can function as regulatory elements by restricting the stimulatory capacity of upstream activators (Smale, 2001). Intrinsic nucleosome positioning, density and stability, as well as the accessibility of the locus to untargeted histone acetylation and deacetylation, would also be expected to affect activator efficiency in a gene-specific manner. A second plausible explanation is that the exact composition of the MHCII enhanceosome may vary at different promoters. The constituents of the enhanceosome have been defined mainly by reporter gene assays and biochemical studies performed with the DRA promoter. To the best of our knowledge, the enhanceosome contains RFX and NF-Y at all MHCII and related promoters. Concerning the identity of the X2 box-binding factor(s), CREB has been shown to be associated with the DRA promoter in vivo (Moreno et al., 1999), but several reports have suggested that the nature of the factor binding to the X2 box may be gene specific (Ono et al., 1991; Scholl et al., 1996). The S box-binding factor(s) also remains to be identified, and thus might not be the same at all genes. Another possibility is that the enhanceosome complex is invariant in its composition but can adopt subtly different conformations induced by minor differences in the sequence and spacing of the S, X, X2 and Y boxes constituting the MHCII regulatory module. Last but not least, at particular promoters the MHCII enhanceosome could be assisted by other activator(s) binding outside of the MHCII module. This possibility applies well to the DMB and Ii promoters, which harbour additional putative regulatory elements (Brown et al., 1994; Radley et al., 1994; Wright et al., 1995; Ting et al., 1997; Westerheide et al., 1997).
The studies performed here further our understanding of the function of CIITA and the MHCII enhanceosome in transcriptional activation of MHCII and related genes. We demonstrate that, depending on the promoter context, the MHCII enhanceosome can sustain CIITA-independent histone acetylation and GTM recruitment. This challenges a widespread belief, according to which CIITA controls virtually all steps of the transcriptional activation process, whereas the enhanceosome serves just as a ‘landing pad’ for CIITA. We also show that CIITA generally promotes hyperacetylation of core histones and GTM recruitment, but the relative importance of CIITA involvement may vary depending on the promoter. Taken together, our results suggest that CIITA may also be necessary for a downstream, post-recruitment event(s), such as promoter escape or elongation, which is in line with the model that CIITA can activate transcription at multiple levels in vivo.
From a broader point of view, our results demonstrate that the precise effect of a given activator on transcription in vivo may vary, depending on the promoter and chromatin context. Similar evidence for promoter-specific effects of activators in yeast and mammalian cells has also recently been provided by others (Deckert and Struhl, 2001; Frank et al., 2001). It is important to point out here that the notion of promoter-specific effects of activators is largely overlooked, because most research tends to be focused on the dissection of isolated model genes (in our case, DRA), whereas comparative studies including multiple genes co-regulated by the same activator(s) are rare.
Materials and methods
Cell lines and culture
The two pairs of isogenic B-cell lines used in this study are described as Supplementary data available at The EMBO Journal Online.
Quantification of mRNA levels by real-time PCR
Total RNA was prepared with Trizol (Life Technologies) according to the manufacturer’s instructions. cDNA was synthesized from total RNA using random hexamers and Superscript II reverse transcriptase (Life Technologies). cDNA was quantified by Multiplex real-time PCR with the TaqMan sequence detection system (Applied Biosystems). The TaqMan probes and primer sets were designed to detect spliced transcripts. For their sequences see Supplementary data available at The EMBO Journal Online. mRNA levels were quantified with respect to the endogenous reference (GAPDH mRNA) amplified in the same tube.
Antibodies
Polyclonal anti-CIITA and anti-RFX5 sera have been described previously (Steimle et al., 1995; Bontron et al., 1997). The anti-TFIIB antibody and the anti-Pol II antibody were generously provided by Nouria Hernandez and Jesper Svejstrup, respectively. Antibodies specific to acetylated histones were purchased from Upstate Biotechnology and the anti-TBP antibody from Santa Cruz Biotechnology.
ChIP
ChIP was essentially performed as described by Masternak et al. (2000b), with the following modifications: (i) chromatin input per immunoprecipitation reaction was scaled up to 10 µg (corresponding to ∼12 million cells); (ii) the amount of detergent was reduced in the TBP, TFIIB and Pol II IPs; (iii) SDS was eliminated from the IP and washing buffers; and (iv) the fifth and sixth washes contained only 0.25% sodium deoxycholate, instead of 0.5% sodium deoxycholate and 0.5% NP-40.
Quantification of ChIP results with real-time PCR
Two hundred nanograms of immunoprecipitated chromatin DNA and a series of standards containing 0.2–20 ng of input chromatin were analysed by real-time PCR using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems) and a kit for quantitative PCR (Eurogentec, Belgium) containing the intercalating fluorescent dye SYBR Green I. PCR reactions were performed with promoter-specific primers that amplify a region of 60–100 bp ranging from –120 to +30 with respect to the start of transcription. For their sequences see Supplementary data available at The EMBO Journal Online. Real-time PCR amplifications were repeated three to five times for each primer pair. The fraction of immunoprecipitated promoter DNA (the relative IP value) was calculated from a standard curve. The relative IP values obtained with different chromatin preparations and the antibodies for acetylated histones, TBP, TFIIB and Pol II were normalized to the values obtained for the reference promoter HSP70-2, which displays moderate to high levels of histone acetylation and high levels of in vivo occupation by TBP, TFIIB and Pol II (data not shown).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to thank Nouria Hernandez and Jesper Svejstrup for their generous gift of antibodies, Madeleine Zufferey and Salomé Landmann for help with cell culture and TaqMan experiments, Marie Peretti for sharing unpublished data, and Michel Strubin for critical reading of the manuscript and helpful discussions. This work was supported by grants from the Swiss National Science Foundation, the Novartis Foundation and NovImmune S.A.
References
- Alfonso C. and Karlsson,L. (2000) Nonclassical MHC class II molecules. Annu. Rev. Immunol., 18, 113–142. [DOI] [PubMed] [Google Scholar]
- Beresford G.W. and Boss,J.M. (2001) CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nature Immunol., 2, 652–657. [DOI] [PubMed] [Google Scholar]
- Bontron S., Ucla,C., Mach,B. and Steimle,V. (1997) Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol. Cell. Biol., 17, 4249–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.M., Barr,C.L. and Ting,J.P. (1991) Sequences homologous to class II MHC W, X and Y elements mediate constitutive and IFN-γ-induced expression of human class II-associated invariant chain gene. J. Immunol., 146, 3183–3189. [PubMed] [Google Scholar]
- Brown A.M., Linhoff,M.W., Stein,B., Wright,K.L., Baldwin,A.S.,Jr, Basta,P.V. and Ting,J.P. (1994) Function of NF-κB/Rel binding sites in the major histocompatibility complex class II invariant chain promoter is dependent on cell-specific binding of different NF-κB/Rel subunits. Mol. Cell. Biol., 14, 2926–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinaux J.R., Notis,J.C., Zhang,Q., Vo,N., Craig,J.C., Fass,D.M., Brennan,R.G. and Goodman,R.H. (2000) Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol. Cell. Biol., 20, 1546–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.H. and Flavell,R.A. (1995) Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J. Exp. Med., 181, 765–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P. (1996) Invariant chain structure and MHC class II function. Cell, 84, 505–507. [DOI] [PubMed] [Google Scholar]
- Deckert J. and Struhl,K. (2001) Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol., 21, 2726–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Preval C., Lisowska-Grospierre,B., Loche,M., Griscelli,C. and Mach,B. (1985) A trans-acting class II regulatory gene unlinked to the MHC controls expression of HLA class II genes. Nature, 318, 291–293. [DOI] [PubMed] [Google Scholar]
- DeSandro A.M., Nagarajan,U.M. and Boss,J.M. (2000) Associations and interactions between bare lymphocyte syndrome factors. Mol. Cell. Biol., 20, 6587–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes J.D., Jiang,B. and Peterlin,B.M. (1997) The class II trans-activator CIITA interacts with the TBP-associated factor TAF II 32. Nucleic Acids Res., 25, 2522–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes J.D., Kanazawa,S., Jean,D. and Peterlin,B.M. (1999) Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol. Cell. Biol., 19, 941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.R., Schroeder,M., Fernandez,P., Taubert,S. and Amati,B. (2001) Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev., 15, 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S.B., Masternak,K., Kammerbauer,C., Janzen,C., Reith,W. and Steimle,V. (2000) CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome and MHC class II gene transactivation. Mol. Cell. Biol., 20, 7716–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harton J.A. and Ting,J.P. (2000) Class II transactivator: mastering the art of major histocompatibility complex expression. Mol. Cell. Biol., 20, 6185–6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harton J.A., Zika,E. and Ting,J.P. (2001) The histone acetyltransferase domains of CBP and pCAF are not necessary for cooperativity with CIITA. J. Biol. Chem., 276, 38715–38720. [DOI] [PubMed] [Google Scholar]
- Kanazawa S., Okamoto,T. and Peterlin,B.M. (2000) Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity, 12, 61–70. [DOI] [PubMed] [Google Scholar]
- Kara C.J. and Glimcher,L.H. (1991) In vivo footprinting of MHC class II genes: bare promoters in the bare lymphocyte syndrome. Science, 252, 709–712. [DOI] [PubMed] [Google Scholar]
- Kara C.J. and Glimcher,L.H. (1993) Three in vivo promoter pheno types in MHC class II deficient combined immunodeficiency. Immunogenetics, 37, 227–230. [DOI] [PubMed] [Google Scholar]
- Kern I., Steimle,V., Siegrist,C.-A. and Mach,B. (1995) The two novel MHC class II transactivators RFX5 and CIITA both control expression of HLA-DM genes. Int. Immunol., 7, 1295–1299. [DOI] [PubMed] [Google Scholar]
- Kretsovali A., Agalioti,T., Spilianakis,C., Tzortzakaki,E., Merika,M. and Papamatheakis,J. (1998) Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol. Cell. Biol., 18, 6777–6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I. and Young,R.A. (2000) Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet., 34, 77–137. [DOI] [PubMed] [Google Scholar]
- Lemon B. and Tjian,R. (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev., 14, 2551–2569. [DOI] [PubMed] [Google Scholar]
- Louis-Plence P., Moreno,C.S. and Boss,J.M. (1997) Formation of a regulatory factor X/X2 box-binding protein/nuclear factor-Y multiprotein complex on the conserved regulatory regions of HLA class II genes. J. Immunol., 159, 3899–3909. [PubMed] [Google Scholar]
- Mahanta S.K., Scholl,T., Yang,F.C. and Strominger,J.L. (1997) Transactivation by CIITA, the type II bare lymphocyte syndrome-associated factor, requires participation of multiple regions of the TATA box binding protein. Proc. Natl Acad. Sci. USA, 94, 6324–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. (1999) The molecular biology of the CCAAT-binding factor NF-Y. Gene, 239, 15–27. [DOI] [PubMed] [Google Scholar]
- Masternak K. et al. (1998) A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nature Genet., 20, 273–277. [DOI] [PubMed] [Google Scholar]
- Masternak K., Muhlethaler-Mottet,A., Villard,J., Peretti,M. and Reith,W. (2000a) Molecular genetics of the bare lymphocyte syndrome. Rev. Immunogenet., 2, 267–282. [PubMed] [Google Scholar]
- Masternak K., Muhlethaler-Mottet,A., Villard,J., Zufferey,M., Steimle,V. and Reith,W. (2000b) CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev., 14, 1156–1166. [PMC free article] [PubMed] [Google Scholar]
- Mayr B. and Montminy,M. (2001) Transcriptional regulation by the phosphorylation-dependent factor CREB. Nature Rev. Mol. Cell Biol., 2, 599–609. [DOI] [PubMed] [Google Scholar]
- Moreno C.S., Beresford,G.W., Louis-Plence,P., Morris,A.C. and Boss,J.M. (1999) CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity, 10, 143–151. [DOI] [PubMed] [Google Scholar]
- Nagarajan U.M., Louis-Plence,P., DeSandro,A., Nilsen,R., Bushey,A. and Boss,J.M. (1999) RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity, 10, 153–162. [DOI] [PubMed] [Google Scholar]
- Nekrep N., Jabrane-Ferrat,N. and Peterlin,B.M. (2000) Mutations in the bare lymphocyte syndrome define critical steps in the assembly of the regulatory factor X complex. Mol. Cell. Biol., 20, 4455–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S.J., Liou,H.C., Davidon,R., Strominger,J.L. and Glimcher,L.H. (1991) Human X-box-binding protein 1 is required for the transcription of a subset of human class II major histocompatibility genes and forms a heterodimer with c-fos. Proc. Natl Acad. Sci. USA, 88, 4309–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan V., Towey,M., Sacks,S. and Kelly,A.P. (1999) Absence of MHC class II gene expression in a patient with a single amino acid substitution in the class II transactivator protein CIITA. Immunogenetics, 49, 957–963. [DOI] [PubMed] [Google Scholar]
- Radley E., Alderton,R.P., Kelly,A., Trowsdale,J. and Beck,S. (1994) Genomic organization of HLA-DMA and HLA-DMB. Comparison of the gene organization of all six class II families in the human major histocompatibility complex. J. Biol. Chem., 269, 18834–18838. [PubMed] [Google Scholar]
- Raval A., Howcroft,T.K., Weissman,J.D., Kirshner,S., Zhu,X.S., Yokoyama,K., Ting,J. and Singer,D.S. (2001) Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol. Cell, 7, 105–115. [DOI] [PubMed] [Google Scholar]
- Reith W. and Mach,B. (2001) The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol., 19, 331–373. [DOI] [PubMed] [Google Scholar]
- Reith W., Kobr,M., Emery,P., Durand,B., Siegrist,C.A. and Mach,B. (1994a) Cooperative binding between factors RFX and X2bp to the X and X2 boxes of MHC class II promoters. J. Biol. Chem., 269, 20020–20025. [PubMed] [Google Scholar]
- Reith W., Siegrist,C.A., Durand,B., Barras,E. and Mach,B. (1994b) Function of major histocompatibility complex class II promoters requires cooperative binding between factors RFX and NF-Y. Proc. Natl Acad. Sci. USA, 91, 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Scholl T., Stevens,M.B., Mahanta,S. and Strominger,J.L. (1996) A zinc finger protein that represses transcription of the human MHC class II gene, DPA. J. Immunol., 156, 1448–1457. [PubMed] [Google Scholar]
- Sherman P.A., Basta,P.V., Heguy,A., Wloch,M.K., Roeder,R.G. and Ting,J.P. (1989) The octamer motif is a B-lymphocyte-specific regulatory element of the HLA-DRα gene promoter. Proc. Natl Acad. Sci. USA, 86, 6739–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S.T. (2001) Core promoters: active contributors to combinatorial gene regulation. Genes Dev., 15, 2503–2508. [DOI] [PubMed] [Google Scholar]
- Steimle V., Durand,B., Barras,E., Zufferey,M., Hadam,M.R., Mach,B. and Reith,W. (1995) A novel DNA binding regulatory factor is mutated in primary MHC class II deficiency (Bare Lymphocyte Syndrome). Genes Dev., 9, 1021–1032. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Tai A.K., Zhou,G., Chau,K. and Ono,S.J. (1999) Cis-element depend ence and occupancy of the human invariant chain promoter in CIITA-dependent and -independent transcription. Mol. Immunol., 36, 447–460. [DOI] [PubMed] [Google Scholar]
- Taxman D.J., Cressman,D.E. and Ting,J.P. (2000) Identification of class II transcriptional activator-induced genes by representational difference analysis: discoordinate regulation of the DNα/DOβ heterodimer. J. Immunol., 165, 1410–1416. [DOI] [PubMed] [Google Scholar]
- Ting J.P., Wright,K.L., Chin,K.C., Brickey,W.J. and Li,G. (1997) The DMB promoter: delineation, in vivo footprint, trans-activation and trans-dominant suppression. J. Immunol., 159, 5457–5462. [PubMed] [Google Scholar]
- Valladeau J. et al. (2000) Langerin, a novel C-type lectin specific to Langerhans cells, is an endocytic receptor that induces the formation of Birbeck granules. Immunity, 12, 71–81. [DOI] [PubMed] [Google Scholar]
- Viret C. and Janeway,C.A.J. (1999) MHC and T cell development. Rev. Immunogenet., 1, 91–104. [PubMed] [Google Scholar]
- Waldburger J.M., Masternak,K., Muhlethaler-Mottet,A., Villard,J., Peretti,M., Landmann,S. and Reith,W. (2000) Lessons from the bare lymphocyte syndrome: molecular mechanisms regulating MHC class II expression. Immunol. Rev., 178, 148–165. [DOI] [PubMed] [Google Scholar]
- Westerheide S.D., Louis-Plence,P., Ping,D., He,X.F. and Boss,J.M. (1997) HLA-DMA and HLA-DMB gene expression functions through the conserved S-X-Y region. J. Immunol., 158, 4812–4821. [PubMed] [Google Scholar]
- Wright K.L., Vilen,B.J., Itoh-Lindstrom,Y., Moore,T.L., Li,G., Criscitiello,M., Cogswell,P., Clarke,J.B. and Ting,J.P. (1994) CCAAT box binding protein NF-Y facilitates in vivo recruitment of upstream DNA binding transcription factors. EMBO J., 13, 4042–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K.L., Moore,T.L., Vilen,B.J., Brown,A.M. and Ting,J.P. (1995) Major histocompatibility complex class II-associated invariant chain gene expression is up-regulated by cooperative interactions of Sp1 and NF-Y. J. Biol. Chem., 270, 20978–20986. [DOI] [PubMed] [Google Scholar]
- Zhu X.S., Linhoff,M.W., Li,G., Chin,K.C., Maity,S.N. and Ting,J.P. (2000) Transcriptional scaffold: CIITA interacts with NF-Y, RFX and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol. Cell. Biol., 20, 6051–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]