Abstract
Recent work from several laboratories has demonstrated that proteolytic mechanisms significantly contribute to the molecular interplay between Streptococcus pyogenes, an important human pathogen, and its host. Here we describe the identification, purification and characterization of a novel extracellular cysteine proteinase produced by S.pyogenes. This enzyme, designated IdeS for Immunoglobulin G-degrading enzyme of S.pyogenes, is distinct from the well-characterized streptococcal cysteine proteinase, SpeB, and cleaves human IgG in the hinge region with a high degree of specificity. Thus, other human proteins, including immunoglobulins M, A, D and E, are not degraded by IdeS. The enzyme efficiently cleaves IgG antibodies bound to streptococcal surface structures, thereby inhibiting the killing of S.pyogenes by phagocytic cells. This and additional observations on the distribution and expression of the ideS gene indicate that IdeS represents a novel and significant bacterial virulence determinant, and a potential therapeutic target.
Keywords: cysteine proteinase/immunoglobulin/Streptococcus pyogenes/virulence
Introduction
Streptococcus pyogenes is one of the most common and significant human bacterial pathogens. Streptococcal infections vary in severity from relatively mild diseases, like impetigo and pharyngitis, to serious and life-threatening conditions such as septicemia, necrotizing fascitis and streptococcal toxic-shock syndrome (Bisno and Stevens, 1996; Cunningham, 2000). Clinically uncomplicated S.pyogenes infections still cause substantial morbidity and economic loss for society, and sequelae to skin and throat infections include important and serious clinical conditions such as acute rheumatic fever and post-streptococcal glomerulonephritis.
Immunoglobulins are a central part of the adaptive immune system that specifically recognize and mediate the elimination of invading microorganisms. Ig consists of antigen-recognizing Fab regions, linked through a flexible hinge region with the constant Fc effector part. The Fc region triggers the classical pathway of complement by binding C1q. In addition, opsonizing immunoglobulin G (IgG) antibodies bound to bacterial surface structures and exposing their Fc region mediate contact with phagocytic cells carrying receptors for IgGFc (FcγR) (Burton, 1985).
Streptococcus pyogenes expresses cell wall-anchored surface proteins with the ability to interact with abundant extracellular human proteins such as albumin, IgG, IgA, fibrinogen, fibronectin and α2-macroglobulin (for a review see Navarre and Schneewind, 1999). Many of these protein–protein interactions are mediated by members of the so-called M-protein family, proteins that contribute to the virulence of the bacterium (Fischetti, 1989; Cunningham, 2000). IgGFc-binding surface proteins are common among Gram-positive bacteria (Forsgren and Sjöquist, 1966; Björck and Kronvall, 1984; Reis et al., 1984; Heath and Cleary, 1987; Gomi et al., 1990). These proteins have evolved convergently, suggesting that IgGFc-binding adds selective advantages to the bacteria (Frick et al., 1992). This view is supported by the finding that repeated growth of S.pyogenes in human blood increases the level of IgGFc-binding proteins (Raeder and Boyle, 1993). Moreover, when bound to these surface proteins, the interaction between IgG and complement factor C1q is blocked, resulting in reduced surface deposition of opsonic C3b (Berge et al., 1997). In contrast to IgG bound to bacterial surface proteins via the Fc region, specific IgG antibodies directed against surface antigens expose their Fc region to Fcγ receptors present on phagocytes. Thus, S.pyogenes bacteria recognized by specific IgG antibodies are rapidly eliminated from human blood (Lancefield, 1962).
The S.pyogenes cysteine proteinase, SpeB, was the first cysteine proteinase isolated from a prokaryote (Elliott, 1945), and several investigations have suggested that SpeB might be an important virulence determinant. SpeB has broad proteolytic activity and degrades a number of different human proteins. In addition, the enzyme activates interleukin-1β (Kapur et al., 1993a) and the matrix metalloproteinase MMP-2 (Burns et al., 1996), and it releases active proinflammatory kinins from H-kininogen (Herwald et al., 1996). A role in virulence has also been suggested by in vivo experiments, as certain SpeB mutant strains are significantly less virulent in mice compared with isogenic wild-type strains, and are also less prone to disseminate than wild-type S.pyogenes (Lukomski et al., 1997, 1998; Svensson et al., 2000). However, contradictory results on the importance of SpeB in severe infections have been reported (Ashbaugh et al., 1998; Ashbaugh and Wessels, 2001), and the precise role of SpeB remains elusive. Thus, patients with severe and life-threatening S.pyogenes infections were reported to have low antibody titers against SpeB (Holm et al., 1992), suggesting that such antibodies are protective against severe disease, whereas another study reported an inverse correlation between disease severity and SpeB production in vitro (Kansal et al., 2000).
The crystal structure of the 40 kDa precursor of SpeB was recently determined, and revealed structural homology of SpeB to enzymes of the papain superfamily (Kagawa et al., 2000). Papain has frequently been used to cleave IgG in the flexible hinge region of the γ-heavy chain to generate Fab and Fc fragments. SpeB also cleaves IgG in this region, but at a different site (Collin and Olsén, 2001a). In addition, SpeB modulates the amount of IgGFc-binding surface proteins of S.pyogenes by cleaving and releasing these proteins from the bacterial surface. Released IgGFc-binding proteins in complex with IgG have the ability to activate and consume complement at a distance from the pathogen (Berge and Björck, 1995; Berge et al., 1997).
The starting point for the present investigation was the surprising observation that S.pyogenes, grown under conditions repressing the production of SpeB, still expressed proteolytic activity towards IgG. A previously unknown secreted cysteine proteinase was found to be responsible for this activity and was denoted IdeS (Immunoglobulin G-degrading enzyme of S.pyogenes). This novel and highly specific enzyme profoundly affects the host–microbe relationship, and provides S.pyogenes with means for dealing with specific opsonizing IgG antibodies.
Results
Streptococcus pyogenes secretes an IgG-cleaving enzyme distinct from SpeB, the classical streptococcal cysteine proteinase
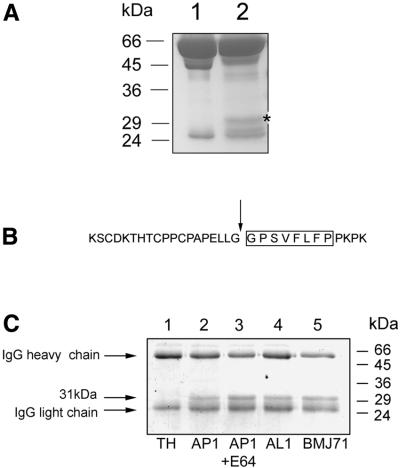
The proteolytic activity of extracellular enzymes of S.pyogenes strain AP1 was analyzed by growing AP1 bacteria in Todd–Hewitt (TH) medium supplemented with 10% human plasma. Following growth to stationary phase, bacteria were removed by centrifugation and the supernatant was subjected to SDS–PAGE (all SDS–PAGE experiments in this work were performed under reducing conditions). The band pattern was compared with the pattern of human plasma proteins that had not been in contact with bacteria (Figure 1A). Interestingly, the bacterial supernatant contained a protein band of ∼31 kDa, which was absent in the plasma control. The N-terminal sequence of this protein was determined to be GPSVFLFP, a sequence that corresponds to amino acids 237–244 of human IgG1, and which is located in the hinge region of the protein (Figure 1B). Recent work has shown that the streptococcal cysteine proteinase, SpeB, cleaves IgG at this site (Collin and Olsén, 2001a,b). However, most strains of S.pyogenes, including AP1, do not express the speB gene in TH medium (Elliott, 1945; J.C.Cooney and L.Björck, unpublished data). Moreover, the proteolytic activity of SpeB is efficiently blocked by the specific cysteine proteinase inhibitor E64 (Björck et al., 1989), but E64 did not inhibit the cleavage of IgG when added to the AP1 growth medium (Figure 1C, lanes 2 and 3). In addition, growth medium from the isogenic SpeB-deficient mutant strain AL1 (Collin and Olsén, 2001a) also contained IgG-cleaving activity (Figure 1C, lane 4). Taken together, these data demonstrate that SpeB is not responsible for the cleavage of IgG in TH medium.
Fig. 1. Cleavage of IgG in S.pyogenes growth medium. (A) Identification of an extra protein band in the culture supernatant of S.pyogenes grown in the presence of human plasma (see asterisk). Lane 1, 5% human plasma in TH. Lane 2, AP1 bacteria grown in TH medium supplemented with human plasma to 10%. Samples were separated on 12% SDS–PAGE and stained with Coomassie Blue. (B) The determined N-terminal amino acid sequence of the 31 kDa protein found in bacterial culture supernatant is boxed. This sequence was identified in the hinge region of human IgG1 and the cleavage site is indicated by an arrow. (C) Human polyclonal IgG was incubated with growth medium (TH) alone (lane 1), with growth medium from strain AP1 lacking (lane 2) or containing E64 (lane 3), with growth medium from the speB-deficient mutant strain AL1 (lane 4), or the mga mutant strain BMJ71 (lane 5). IgG cleavage was analyzed by 12% SDS–PAGE, and the gel was stained with Coomassie Blue. Molecular weight markers are indicated.
The streptococcal strain AP1 studied here expresses a surface-associated C5a peptidase (Wexler et al., 1985), and two IgGFc-binding proteins, H and M1 (Åkesson et al., 1990, 1994). The genes encoding these surface proteins are controlled by the transcriptional activator Mga (Caparon and Scott, 1987), and BMJ71 is an isogenic mutant of AP1, carrying a Tn916 insertion within the mga gene (Kihlberg et al., 1995). The 31 kDa IgG cleavage product is also generated in growth medium of BMJ71 (Figure 1C, lane 5), showing that the proteolytic activity is not under Mga control.
Purification and sequence characteristics of IdeS,a novel proteinase of S.pyogenes
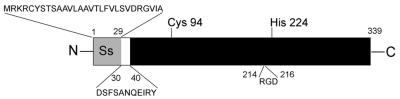
As the IgG-cleaving activity was found in the growth medium of strain AP1, we fractionated culture medium proteins after bacterial growth by adding increasing amounts of ammonium sulfate (10–80%). These initial experiments revealed that precipitates of 60–70% ammonium sulfate contained most of the IgG-cleaving activity. For purification, the growth medium was fractionated with 50% ammonium sulfate, the resulting pellet was discarded and the ammonium sulfate concentration in the supernatant was adjusted to 70%. Proteins pelleted by this second precipitation were subjected to ion-exchange chromatography, and peak fractions were tested for enzymatic activity. Maximum IgG-cleaving activity was eluted at 0.1 M NaCl, and the corresponding fractions contained a major band of ∼34 kDa as judged by SDS–PAGE. This protein band was excised and subjected to N-terminal sequence analysis. The sequence obtained, DSFSANQEIRY, was used to search the Streptococcal Genome Sequencing Project database (Ferretti et al., 2001; Roe et al., 2001). A perfect match was found in an open reading frame of 339 amino acids designated SPy0861. The N-terminal sequence obtained corresponds to amino acids 30–40 (Figure 2) and was preceded by a potential signal sequence of 29 amino acids as predicted by the SignalP algorithm (Nielsen et al., 1997). The protein does not contain a cell wall attachment signal (LPXTGX), a common feature of cell wall-anchored proteins of S.pyogenes (Navarre and Schneewind, 1999; Janulczyk and Rasmussen, 2001), and the predicted size of the protein, without the potential signal sequence, is 34.9 kDa, which is in accordance with the size of the purified protein estimated by SDS–PAGE. Apart from the putative signal sequence, the protein has an RGD motif at amino acids 214–216 (Figure 2). This motif is important for ligand recognition by integrins, and a variety of bacterial and viral pathogens have been shown to bind to host cell integrins (Isberg and Van Nhieu, 1994). The full-length putative protein sequence was used in a similarity search against the DDBJ/EMBL/GenBank database using a BLASTp algorithm (Altschul et al., 1990). This search revealed no similarities to any prokaryotic protein and a weak similarity (24% identity in a region of 204 amino acid residues) to human MAC-1 integrin α M precursor (Arnaout et al., 1988). Owing to the absence of any previously reported function, and based on the enzymatic activity against human IgG, the protein was denoted IdeS, as stated above.
Fig. 2. Schematic representation of the IdeS protein. The obtained N- terminal sequence (amino acids 30–40), the putative signal sequence (Ss) and an RGD motif (amino acids 214–216) are indicated. The potential catalytic cysteine and histidine residues are designated Cys94 and His224, respectively.
To confirm further that the identified IdeS protein has IgG-cleaving activity, the ideS gene was cloned in plasmid pGEX-5X-3 (Amersham Pharmacia Biotech), and expressed in Escherichia coli. Partially purified lysates were incubated with IgG and analyzed by SDS–PAGE. Lysates from E.coli carrying the ideS gene generated the 31 kDa IgG-derived band, whereas extracts from cells carrying only a plasmid control did not cleave IgG (data not shown).
IdeS is a novel cysteine proteinase highly specific for IgG
We noticed that the sequence of the IdeS protein contains a single cysteine residue at position 94 (Figure 2). Despite the lack of sequence homology to other cysteine proteinases, IdeS also contains a histidine residue at position 224. The distance between these two amino acids is similar to the distance found in other cysteine proteinases (Rawlings and Barrett, 1994). Thus, although the enzymatic activity was not inhibited by the cysteine proteinase inhibitor E64 (Figure 1C), sequence characteristics indicated that IdeS could still represent a member of the cysteine proteinase family. The peptide derivative Z-LVG-CHN2, structurally based on the inhibitory reactive site of cystatin C (Abrahamson et al., 1987), and carrying a diazomethyl ketone group to inactivate the sulfhydryl group of the catalytic cysteine (Green and Shaw, 1981), has been shown previously to irreversibly bind to and inhibit papain and SpeB (Björck et al., 1989). Moreover, cysteine proteinases are also inactivated by iodoacetic acid through an irreversible modification of the catalytic sulfhydryl group (Elliott, 1945). We therefore investigated whether treatment with these specific inhibitors would affect the enzymatic activity of IdeS. Analysis of IgG incubated with IdeS alone (Figure 3, lanes A and B), or with IdeS pre-incubated with inhibitors (lanes C–E), revealed that Z-LVG-CHN2 and iodoacetic acid efficiently inhibited the activity of IdeS (lanes C and D), whereas E64 had no effect on the enzyme (lane E). Equal amounts of active SpeB were used as a control (lanes F–I). As expected, Z-LVG-CHN2 and E64 efficiently inhibited the activity of SpeB (lanes H and I). The activity, sequence characteristics and inhibition profile of IdeS establish it as a new member of the cysteine proteinase family. A detailed characterization of the enzymatic properties of IdeS will be published elsewhere.
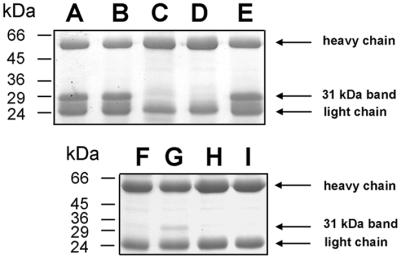
Fig. 3. IdeS is a novel streptococcal cysteine proteinase. Purified IdeS or activated SpeB as a control, were separately pre-incubated with proteinase inhibitors prior to incubation with IgG. Cleavage was analyzed by 12% SDS–PAGE. (A) Untreated IdeS. (B–E) IdeS in 1% DMSO (solvent for Z-LVG-CHN2) (B), iodoacetic acid (C), Z-LVG-CHN2 (D) or E64 (E). (F) IgG. (G–I) Untreated SpeB (G), Z-LVG-CHN2 (H) or E64 (I).
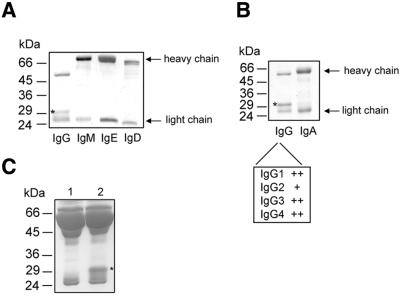
Recently, the streptococcal cysteine proteinase SpeB was shown to cleave the heavy chains of all classes of human immunoglobulins: IgG, IgM, IgA, IgD and IgE (Collin and Olsén, 2001b). In contrast, when human IgG, IgM, IgA, IgD or IgE were incubated with purified IdeS for 2 h at 37°C, only IgG was degraded (Figure 4A and B). We also analyzed the activity of IdeS against the different subclasses of IgG and found that all were susceptible to IdeS digestion, although compared with the other subclasses IgG2 was less efficiently digested (data not shown). The high specificity of IdeS is further emphasized by the observation that only the 31 kDa IgG-derived band and no additional degradation products could be identified following incubation of human plasma with higher concentrations of purified IdeS (1 mg/ml) (Figure 4C). Although it cannot be excluded that the enzyme has other substrates, these data show that IdeS has a higher degree of specificity for IgG than previously described proteinases with proteolytic activity towards immunoglobulins, e.g. papain, pepsin (Porter, 1973) or SpeB (Collin and Olsén, 2001b).
Fig. 4. IdeS is specific for IgG. (A and B) Human immunoglobulins of different classes were separately incubated with IdeS, followed by SDS–PAGE analysis. Only IgG was degraded by IdeS. All four IgG subclasses are cleaved by IdeS, but IgG2 is more resistant to IdeS cleavage (+). Results from different experiments have been combined. (C) Samples of 5% human plasma in TH (lane 1) or 5% human plasma in TH pre-incubated with IdeS (lane 2) were separated by 12% SDS–PAGE. The asterisk indicates the 31 kDa IgG-derived band.
Distribution and expression of the ideS gene in S.pyogenes strains
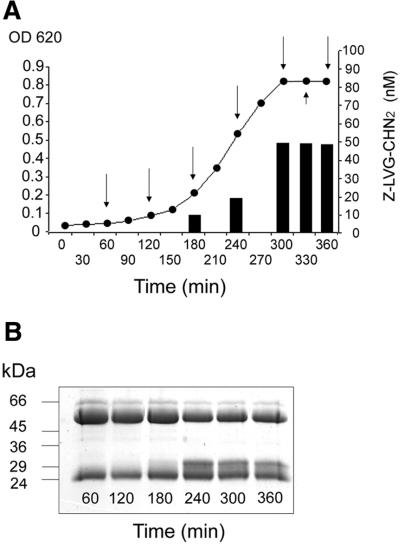
The distribution of ideS among S.pyogenes strains was investigated by PCR analysis using primers designed to amplify the internal coding region of ideS. We analyzed chromosomal DNA preparations from 11 S.pyogenes strains of nine different M serotypes, and were able to amplify identical PCR fragments of the expected size from all strains (Table I; data not shown). However, when analyzing the cleavage of IgG during bacterial growth in TH medium, only five of the tested strains expressed the IgG-degrading activity (AP1, KTL3, SF370, AP12 and AP55), and among these strains KTL3 and SF370 showed weak activity. Thus, although the ideS gene seems to be present in all S.pyogenes isolates, the expressed enzyme activity under the conditions used here is restricted to some strains and varies even within the same M serotype (Table I; U.von Pawel-Rammingen, B.P.Johansson and L.Björck, in preparation). The secretion pattern of the IgG-cleaving activity during growth of strain AP1 in TH medium (i.e. no SpeB expression) was also investigated. The peptide derivative Z-LVG-CHN2 irreversibly binds to and inhibits IdeS activity at stoichiometric concentrations (see Materials and methods; data not shown). Thus, the molar amount inhibitor needed to inhibit IdeS activity (>90%) is equal to the amount of active enzyme present in the sample. Samples were taken from the growth medium at different time points during bacterial growth, and tested for enzymatic activity against IgG. IgG-degrading activity started to appear in samples taken during early logarithmic growth phase (enzyme concentration ∼10 nM), and the activity increased initially during logarithmic growth as determined from the degree of IgG cleavage and the amount of active enzyme present in the culture supernatant (∼20 nM). The enzymatic activity did not further increase beyond OD600 ∼0.4, but appeared to be persistent at a constant level (∼50 nM) (Figure 5).
Table I. Distribution of the ideS gene and IdeS enzyme activity during S.pyogenes growth in TH medium supplemented with human IgG.
| Strain | M-serotype | Reference or source | IgG cleavage (in the presence of E64) | PCR product |
|---|---|---|---|---|
| SF370 | 1 | Suvorov and Feretti (1996); Ferretti et al. (2001) | (+) | + |
| AP1 | 1 | WHO Prague collectiona | + | + |
| AL1 | 1 | speB mutant of AP1; Collin and Olsén (2001a) | + | n.d.b |
| BMJ71 | 1 | mga mutant of AP1; Kihlberg et al. (1995) | + | n.d. |
| KTL3 | 1 | Finnish institute for health | (+) | + |
| AP4 | 4 | WHO Prague collection | – | + |
| M5 | 5 | sequencing in progessc | – | + |
| AP6 | 6 | WHO Prague collection | – | + |
| AP12 | 12 | WHO Prague collection | + | + |
| AP49 | 49 | WHO Prague collection | – | + |
| AP53 | 53 | WHO Prague collection | – | + |
| AP55 | 55 | WHO Prague collection | + | + |
| AP57 | 57 | WHO Prague collection | – | + |
aWHO Collaborating Center for Reference and Research on Streptococci, Institute of Hygiene and Epidemiology, Prague, Czech Republic.
bn.d., not determined.
cSanger Center in collaboration with Dr Michael Kehoe, University of Newcastle, UK.
+, indicates weak activity.
Fig. 5. IgG-cleaving activity generated during streptococcal growth. (A) Strain AP1 was grown in TH medium; supernatant samples were taken at indicated time points (arrows) and IdeS concentrations were determined by adding increasing amounts of Z-LVG-CHN2. Bars indicate the amount inhibitor required to inhibit >90% of enzyme activity. (B) IgG-cleaving activity at different time points as shown by SDS–PAGE.
Fc-mediated phagocytosis and killing of S.pyogenes is inhibited by IdeS
Opsonizing IgG antibodies bound to surface antigens of S.pyogenes will expose their Fc regions to complement factor C1q and Fcγ receptors of phagocytic cells, and thereby facilitate phagocytosis and killing of the bacteria. To test the hypothesis that IdeS, by proteolytic cleavage of IgG, could interfere with this defense mechanism, AP1 bacteria were incubated with either human immune or non-immune plasma. After incubation, bacteria were washed and incubated with IdeS or with a buffer control followed by another washing step to remove IdeS and degradation products. Confluent RAW264.7 macrophage-like cells or isolated human polymorphonuclear leukocytes (PMNs) were infected with these bacteria at ∼0.1–5 bacteria/cell. Infections were synchronized by gentle centrifugation and cells were lysed immediately to determine the number of c.f.u. at time zero. Incubations were continued for various periods of time, after which cells were lysed and the number of c.f.u. was determined. The ratios of c.f.u. at time 30 min or 1 h, divided by the number of c.f.u. at time zero were determined as survival factors, and are shown in Figure 6. The relatively short incubation time was chosen to minimize IgG-independent phagocytosis. While bacteria pre-incubated with non-immune plasma survived contact with macrophage-like cells or neutrophils, the number of living bacteria that had been exposed to opsonizing immunoglobulins in immune plasma, was significantly reduced (P <0.03 or <0.05, respectively). However, this effect was abolished when bacteria carrying opsonizing IgG were treated with IdeS prior to the incubation with phagocytes (Figure 6A and B).
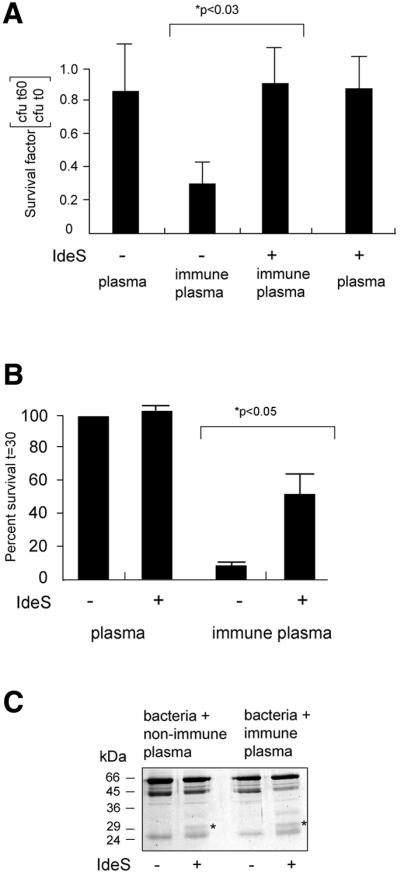
Fig. 6. IdeS cleaves IgG at the bacterial surface and confers resistance against killing of S.pyogenes by phagocytes. (A) AP1 bacteria were incubated with human non-immune or immune plasma, washed, and incubated with or without IdeS. Bacteria were washed and added to RAW264.7 macrophage-like cells. Bacteria were recovered either directly after centrifugation (t0) or after 1 h (t60). The survival rate is shown as number of c.f.u. at 1 h divided by the number of c.f.u. at time zero. Bars indicate the mean values +SEM of at least three different experiments. (B) Bacteria and neutrophils were mixed and incubated at 37°C for various periods of time. Samples were withdrawn and phagocytosis was stopped by putting samples on ice. The bacterial survival rate is shown as number of c.f.u. at 30 min divided by the number of c.f.u. at time zero. The ratio obtained with AP1 treated with non-immune plasma was arbitrarily set to 100%. All other ratios are correlated to this number. Bars indicate the mean values +SEM. (C) AP1 bacteria were incubated with human non-immune or immune plasma, and incubated with IdeS or buffer control. Plasma proteins bound to the bacterial surface and released by boiling in SDS–PAGE sample buffer were subjected to SDS–PAGE analysis. The 31 kDa IgG fragment is indicated by an asterisk.
AP1 bacteria express surface proteins that bind several abundant human plasma proteins. As reported previously (Åkesson et al., 1994; Frick et al., 1994), the major protein bands eluted from AP1 bacteria following plasma incubation represent albumin, fibrinogen, and IgG heavy and light chains. The same protein pattern was obtained following absorption of non-immune or immune plasma, and in both cases IgG was cleaved by IdeS, generating IgGFc fragments, which, under the reducing conditions used, gave rise to the 31 kDa band (Figure 6C). These Fc fragments are associated with IgGFc-binding proteins, interactions that efficiently block their capacity to bind complementation factor C1q (Berge et al., 1997). However, as shown in Figure 6A and B, IdeS protects bacteria pre-incubated with plasma containing specific IgG antibodies. These antibodies are bound to the streptococcal surface via their antigen-binding Fab regions, suggesting that cleavage of this IgG population by IdeS will result in the removal of exposed Fc fragments from the bacterial surface. These data demonstrate that cleavage of IgG by IdeS can occur at the bacterial surface and that IgG cleavage by IdeS increases the capacity of S.pyogenes to evade phagocytic cells.
Discussion
Since the late 1980s, a worldwide increase of severe S.pyogenes infections has attracted considerable attention and concern (Stevens, 1995, 2000; Curtis, 1996). Despite the fact that S.pyogenes continues to be exquisitely susceptible to β-lactam antibiotics, these hyperacute and toxic infections are still connected with high mortality. Streptococcus pyogenes virulence is based on multiple molecular host–microbe interactions, including several proteolytic mechanisms. The pathogen not only produces and secretes proteolytic enzymes, but also utilizes and modulates host proteolytic cascades, such as the complement, coagulation and contact systems. In addition, during infection, damaged host cells and tissues will leak intracellular proteinases at the site of infection. As a result, an intense inflammatory state is induced, which is a typical feature of severe S.pyogenes infections (Rasmussen and Björck, 2002).
Several investigations have suggested that the streptococcal cysteine proteinase SpeB is a virulence determinant, and SpeB was recently shown to cleave the hinge region of IgG and to degrade the heavy chains of all classes of human immunoglobulins (Collin and Olsén, 2001a,b). Therefore, the discovery of an additional extracellular cysteine proteinase in S.pyogenes was unexpected. However, at least under laboratory conditions, SpeB is not expressed until S.pyogenes reaches stationary growth phase (Chaussee et al., 1997), which makes a possible function of SpeB as an enzyme cleaving opsonizing IgG antibodies questionable. Thus, it should be important for such a proteinase to be present continuously during infection. IdeS production starts during early logarithmic growth and continues into late stationary growth phase, which makes the enzyme more suitable to remove opsonizing IgG from the bacterial surface. However, the actions of IdeS and SpeB could well be complementary. In fact, the identification and characterization of IdeS might explain some previous and puzzling observations concerning SpeB. IdeS is not affected by the cysteine proteinase inhibitor E64, but is inhibited by a synthetic peptide derivative (Z-LVG-CHN2) structurally based on the proteinase-binding center of cystatin C, a human cysteine proteinase inhibitor. Z-LVG-CHN2 and E64 both irreversibly block the proteolytic activity of SpeB, but only Z-LVG-CHN2 inhibited streptococcal growth in vitro and in vivo (Björck et al., 1989). However, the observation that IdeS is inhibited by Z-LVG-CHN2 but not by E64 suggests that the previously observed effect of Z-LVG-CHN2 on S.pyogenes growth and virulence, could be due to interference with both SpeB and IdeS.
In severe invasive S.pyogenes infections, strains of the M1 serotype are the most common (Holm et al., 1992; Musser et al., 1995), and the AP1 strain, which was the most studied here and produced IdeS, is of this serotype. Strains of serotypes M12 and M55, also producing proteolytically active IdeS under the growth conditions used, are phylogenetically closely related (Whatmore et al., 1994), and represent clinically relevant strains often connected with post-streptococcal glomerulonephritis (Hartas and Sriprakash, 1999; Descheemaeker et al., 2000). This correlation suggests a role for IdeS both during acute infections and in aseptic sequelae following acute S.pyogenes infections, a notion supported further by a publication where S.pyogenes culture supernatant proteins were analyzed by two-dimensional electrophoresis and N-terminal sequencing (Lei et al., 2000). One of the proteins analyzed, designated SP22, has an N-terminal sequence identical to IdeS. Interestingly, sera from patients with pharyngitis, acute rheumatic fever and convalescent sera from a patient recovering from a toxic and invasive S.pyogenes infection, all contained antibodies reacting with SP22/IdeS (Lei et al., 2000). The enzymatic activity in different bacterial culture supernatants varied considerably also between different M1 strains, which could reflect differences in ideS regulation among different isolates. This could indicate other functions of IdeS, perhaps as an integrin-binding protein, as suggested by Lei et al. (2000).
IgG is the dominant Ig class, and IgGFc has important functions in complement activation and phagocytosis. Moreover, Fcγ receptors are expressed by all immunologically active cells. Therefore, it seems logical that S.pyogenes has evolved a specific IgG-cleaving enzyme, and its specificity underlines a potential role for IdeS in preventing contact between S.pyogenes and phagocytes by cleaving opsonizing IgG in the hinge region. Opsonizing IgG antibodies bind to various S.pyogenes surface structures via the Fab regions. However, most S.pyogenes strains express surface proteins of the M protein family with affinity for IgGFc. The AP1 strain studied here has two such proteins, H and M1, which are structurally closely related (Gomi et al., 1990; Åkesson et al., 1994; Nilson et al., 1995). Large amounts of these IgGFc-binding proteins are present at the bacterial surface, and bind IgG with high affinity (Åkesson et al., 1994; Frick et al., 1994). As a result, AP1 bacteria surrounded by plasma or inflammatory exudate are covered with IgG bound to these proteins through the IgGFc-binding proteins. This IgG population will be present in vast amounts compared with antigen-specific IgG bound to the bacterial surface via Fab. However, the data presented in Figure 6 demonstrate that IdeS cleaves not only opsonizing antibodies, but also IgG bound to the surface via Fc. Whether the Fc and Fab fragments released by IdeS (Figure 7) have biological effects is unclear, but remains an interesting possibility.
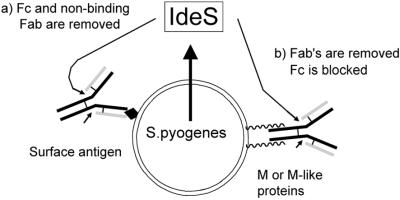
Fig. 7. Streptococcus pyogenes binds, accumulates and presents the IgG substrate to IdeS. Arrows indicate the cleavage sites in IgG. (a) Immune binding of IgG to S.pyogenes surface antigens. Fc (and non-binding Fabs) is removed by IdeS to avoid interaction with Fcγ receptors of phagocytes. (b) M and M-like proteins bind IgG in the Fc region, thereby inhibiting complement activation and binding to Fcγ receptors. IdeS cleaves in the hinge region to release Fabs.
Streptococcus pyogenes has evolved a number of different mechanisms to resist the human immune defense, and M and M-like proteins play a key role in this context. However, in the presence of opsonizing antibodies, the bacteria appear remarkably defenseless. In this study we show that the novel IgG-specific cysteine proteinase, IdeS, has the ability to cleave opsonizing antibodies in the hinge region, causing an increased resistance to killing by phagocytes. Thus, with its unique proteolytic properties, IdeS represents a novel virulence determinant and a potential therapeutic target.
Materials and methods
Bacterial strains and growth conditions
Streptococcus pyogenes strains used in this study are listed in Table I. Streptococci were routinely grown in TH broth (Difco) at 37°C in 5% CO2. Strains BMJ71 and AL1 were grown in the presence of 10 µg/ml tetracycline or 150 µg/ml kanamycin, respectively. In some cases, bacteria were grown in the presence of the cysteine proteinase inhibitor trans-epoxysuccinyl-l-leucylamido-(4-guanidino) butane (E64) (Sigma).
SDS–PAGE analysis and N-terminal sequence determination
Proteins of S.pyogenes growth media were precipitated with trichloroacetic acid (final concentration 5%), washed twice with 1 ml acetone, and resuspended in sample buffer. Proteins were separated by 12% SDS–PAGE (all SDS–PAGE experiments in this work were performed under reducing conditions) and stained with Coomassie Blue. For N-terminal amino acid sequence analysis, proteins were separated by 10% SDS–PAGE and blotted onto a PVDF membrane using 10 mM CAPS buffer and 10% methanol. Proteins were visualized in 0.1% Coomassie Blue R-250 and 50% methanol. After destaining in 50% methanol, membranes were dried, and protein bands were cut out with a scalpel and stored at –20°C until sequencing. Sequencing was performed at the Eurosequence Company (Groningen, The Netherlands).
Purification of IdeS
IdeS was purified by growing bacteria to an OD620 of ∼0.4 and fractionating the culture supernatant with 50% ammonium sulfate. The resulting precipitate was discarded and ammonium sulfate was added to the remaining supernatant to a final concentration of 70%. The second precipitate was resuspended in 1/100 of the starting volume with 20 mM Tris–HCl pH 8.0, and dialyzed against the same buffer. The material was further fractionated by FPLC on a Mono Q column (Pharmacia). Proteins were eluted by a linear NaCl gradient, and a peak eluted at 0.1 M NaCl was found to contain the IdeS activity. Corresponding fractions were collected, analyzed by SDS–PAGE and stored at –20°C until use.
IdeS activity assays
For standard IdeS activity assays, bacterial cultures were grown to an OD620 of 0.4. Bacteria were pelleted by centrifugation and supernatants were sterile-filtered through a 0.22 µm membrane (Millipore) prior to use. For screening of IdeS activity in different S.pyogenes strains, E64 was added to a final concentration of 40 µM. For activity assays, 25 µl of supernatant were mixed with 5 µl of IgG (10 mg/ml, Sigma) and the volume was adjusted with phosphate-buffered saline (PBS) to 100 µl. The mixtures were incubated at 37°C for 30 min and samples were analyzed by 12% SDS–PAGE. For cleavage assays of different classes of Ig, purified IdeS (0.3 µg/ml) was incubated with ∼3 µg Ig for 2 h at 37°C, and analyzed by 12% SDS–PAGE analysis. IgG and IgM were obtained from Sigma; IgA, IgD, IgE and IgG1-4 were from ICN. To investigate cleavage of human plasma proteins, IdeS (1 mg/ml) was incubated with 5% human plasma diluted in PBS.
PCR analysis of genomic DNA for identification of ideS
To analyze the presence of the ideS gene in different streptococcal isolates, PCR template DNA was prepared by boiling S.pyogenes bacteria for 5 min in sterile water. Cell debris was removed by centrifugation and 1 µl of the boiled lysate was used with PCR primers Ide1 (5′-CGTTACTTCCGTTTGGATCCAAGG-3′) and Ide2 (5′-GAAATAGCTACTTCTCGAGCGGAATT-3′). PCR products were analyzed by agarose (1%) gel electrophoresis.
Recombinant expression of IdeS in E.coli
For PCR amplification of ideS, template DNA was prepared by boiling S.pyogenes bacteria (strain AP1) in sterile water. The cell debris was removed by centrifugation and 5 µl of the boiled lysate was used with PCR primers Ide5X (5′-TCGGTAGATCGTGGGATCCTAGCAGATAGT-3′), creating a BamHI restriction site, and Ide3X (5′-CGGAATTCTTAATTGGTCTGATTCCAAC-3′), creating an EcoRI restriction site. A PCR fragment covering base pairs 79–1020 of the intact ideS gene was generated, cleaved with restriction enzymes, and cloned into the corresponding sites of plasmid pGEX-5X-3 (Amersham Pharmacia Biotech). The resulting plasmid was transformed into E.coli strain BL21(DE3) pLysS, according to standard protocols (Sambrook et al., 1989). Protein expression was induced by addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an OD620 of ∼0.2. Growth was continued for 3 h, and lysates were prepared by freezing bacterial pellets at –70°C, followed by resuspension in PBS. Cell debris was removed by centrifugation and 2 µl of supernatant was incubated with 5 µl of IgG (10 mg/ml) in PBS, and separated by 12% SDS–PAGE for analysis of recombinantly expressed IdeS.
Proteinase inhibition assays
Purified IdeS (0.3 µg/ml) or activated SpeB (0.6 µg/ml, ∼60% active) (Berge and Björck, 1995) were incubated with either 20 mM iodoacetic acid, Z-LVG-CHN2 (Björck et al., 1989) at 0.4 mg/ml in 1% dimethyl sulfoxide (DMSO), or E64 (40 µM). The tubes were kept in the dark and incubated for 30 min at room temperature. As controls, IdeS was also kept in PBS or in 1% DMSO, the solvent for Z-LVG-CHN2. After 30 min, 5 µl of 10 mg/ml IgG (Sigma) were added, and the volume was adjusted to 100 µl with PBS. Incubation was continued for 60 min at 37°C. The reaction was stopped by the addition of SDS–PAGE sample buffer and samples were analyzed by 12% SDS–PAGE. For quantification, increasing amounts of Z-LVG-CHN2 were incubated with IdeS at room temperature for 30 min, before adding 10 µl of 5 mg/ml IgG. Z-LVG-CHN2 completely inhibited cleavage of IgG at a molar ratio of approximately 1:1 (data not shown). Thus, the stoichiometric inhibition of IdeS by Z-LVG-CHN2 was used to determine the concentration of IdeS in growth medium. Increasing amounts of the inhibitor (0.2–50 nM) were added to medium samples, taken at different time points. After 15 min, 10 µl of 5 mg/ml IgG (Sigma) were added and incubation was continued for 60 min at 37°C. Inhibition was determined by analysis of IgG cleavage by 12% SDS–PAGE.
Preparation of human neutrophils
Human polymorphonuclear leukocytes were isolated from heparinized blood using polymorphprep™ (Nycomed Pharma, Norway) according to the manufacturer’s instructions. Briefly, whole blood was layered onto polymorphprep medium and centrifuged in a Sigma table-top centrifuge, at 400 g for 30 min. After centrifugation, the neutrophil layer was isolated and washed in PBS. Residual erythrocytes were removed by hypotonic lysis for 20 s. Neutrophils were collected by centrifugation, resuspended in Na medium pH 7.3 (127 mM NaCl, 10.8 mM KCl, 2.4 mM KH2PO4, 1.6 mM MgSO4, 10 mM HEPES, 1.8 mM CaCl2 and 5.6 mM glucose), and counted using a hemocytometer.
Cell culture, infection of eukaryotic cells and bactericidal assays
The murine macrophage-like cell line RAW264.7 was cultured in RPMI 1640 medium (Life Technologies), supplemented with 10% fetal calf serum and antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin, from ICN), at 5% CO2 with 100% relative humidity.
To study phagocytic killing, S.pyogenes strain AP1 was grown at 37°C to an OD620 of ∼0.4. AP1 bacteria were incubated with either immune or non-immune plasma, washed and treated with IdeS or a buffer control, for 1 h at 37°C. Subsequently, bacteria were washed and resuspended in antibiotic-free cell culture medium or Na medium, prior to infection.
Cell lines were carefully washed in antibiotic-free cell culture medium and bacteria were added (0.1–1 bacteria/cell) to confluent RAW264.7 cells. Infections were synchronized by gentle centrifugation at 400 g for 3 min by incubation at 37°C. Ten minutes after infection, the cell cultures were carefully washed in antibiotic-free medium to remove non-adherent bacteria (time 0 h). Control cells were lysed in ice-cold lysis buffer (0.1% Tween), remaining bacteria were diluted, and spread onto TH plates. Parallel cell cultures were incubated at 37°C for 1 h. Subsequently, growth media were removed, and cells were lysed and treated as described above.
For PMN bactericidal assays, bacteria and neutrophils were incubated at 37°C for 1 min, pelleted in an Eppendorf microcentrifuge (20 s), and further incubated at 37°C for 40 s. Subsequently, the bacteria/neutrophil pellet was resuspended and incubated at 37°C. At various time points samples were withdrawn and phagocytosis was stopped by putting samples on ice. Cells were lysed in ice-cold lysis buffer (0.1% Tween), remaining bacteria were diluted, and spread onto TH plates. For analysis of bacterial, survival the number of surviving bacteria after 30 min respectively 1 h, was divided by the number of c.f.u. present at time 0 h.
Acknowledgments
Acknowledgements
We thank Dr Arne Olsén for providing different Ig preparations and for helpful discussions, and Drs Mattias Collin and Magnus Rasmussen for helpful discussions and expert help. Mrs Monica Heidenholm is acknowledged for excellent technical assistance. We acknowledge the Streptococcal Genome Sequencing Project funded by USPHS/NIH grant AI38406 for making the streptococcal genomic sequences available. This work was supported by grants from the Swedish Research Council projects 7480 (to L.B.) and 1055 (to U.v.P.-R.); the Medical Faculty, Lunds University; the foundations of Crafoord (to U.v.P.-R.), Kock, Lundberg, and Österlund; and Hansa Medical AB.
Note added in proof
After the submission of this work, Lei et al. (2001) published a paper describing the streptococcal protein Mac. Streptococcal protein Mac shows homology to the α-subunit of the human β2-integrin Mac-1, and is suggested to function through a process of molecular mimicry to inhibit phagocytosis. Protein Mac is identical to IdeS, a finding that further emphasizes the complexity of bacterial strategies to evade the human immune system.
References
- Abrahamson M., Ritonja,A., Brown,M.A., Grubb,A., Machleidt,W. and Barrett,A.J. (1987) Identification of the probable inhibitory reactive sites of the cysteine proteinase inhibitors human cystatin C and chicken cystatin. J. Biol. Chem., 262, 9688–9694. [PubMed] [Google Scholar]
- Åkesson P., Cooney, J., Kishimoto,F. and Björck,L. (1990) Protein H— a novel IgG binding bacterial protein. Mol. Immunol., 27, 523–531. [DOI] [PubMed] [Google Scholar]
- Åkesson P., Schmidt,K.H., Cooney,J. and Björck,L. (1994) M1 protein and protein H: IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem. J., 300, 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Arnaout M.A., Gupta,S.K., Pierce,M.W. and Tenen,D.G. (1988) Amino acid sequence of the α subunit of human leukocyte adhesion receptor Mo1 (complement receptor type 3). J. Cell Biol., 106, 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashbaugh C.D. and Wessels,M.R. (2001) Absence of a cysteine proteinase effect on bacterial virulence in two murine models of human invasive group A streptococcal infection. Infect. Immun., 69, 6683–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashbaugh C.D., Warren,H.B., Carey,V.J. and Wessels,M.R. (1998) Molecular analysis of the role of group A streptococcal cysteine proteinase, hyaluronic acid capsule and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Invest., 102, 550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge A. and Björck,L. (1995) Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem., 270, 9862–9867. [DOI] [PubMed] [Google Scholar]
- Berge A., Kihlberg,B.M., Sjöholm,A.G. and Björck,L. (1997) Streptococcal protein H forms soluble complement-activating complexes with IgG, but inhibits complement activation by IgG-coated targets. J. Biol. Chem., 272, 20774–20781. [DOI] [PubMed] [Google Scholar]
- Bisno A.L. and Stevens,D.L. (1996) Streptococcal infections of skin and soft tissues. N. Engl. J. Med., 334, 240–245. [DOI] [PubMed] [Google Scholar]
- Björck L. and Kronvall,G. (1984) Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. J. Immunol., 133, 969–974. [PubMed] [Google Scholar]
- Björck L., Bohus,P.M., Trojnar,J., Abrahamson,M., Olafsson,I. and Grubb,A. (1989) Bacterial growth blocked by a synthetic peptide based on the structure of a human proteinase inhibitor. Nature, 337, 385–386. [DOI] [PubMed] [Google Scholar]
- Burns E.H., Marciel,A.M. and Musser,J.M. (1996) Activation of a 66 kDa human endothelial cell matrix metalloprotease by Streptococcus pyogenes extracellular cysteine proteinase. Infect. Immun., 64, 4744–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R. (1985) Immunoglobulin G: functional sites. Mol. Immunol., 22, 161–202. [DOI] [PubMed] [Google Scholar]
- Caparon M.G. and Scott,J.R. (1987) Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc. Natl Acad. Sci. USA, 84, 8677–8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussee M.S., Phillips,E.R. and Ferretti,J.J. (1997) Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect. Immun., 65, 1956–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin M. and Olsén,A. (2001a) EndoS, novel secreted enzyme from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J., 20, 3046–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin M. and Olsén,A. (2001b) Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect. Immun., 69, 7187–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham M.W. (2000) Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev., 13, 470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis N. (1996) Invasive group A streptococcal infection. Curr. Opin. Infect. Dis., 9, 191–202. [Google Scholar]
- Descheemaeker P., Van Loock,F., Hauchecorne,M., Vandamme,P. and Goossens,H. (2000) Molecular characterization of group A streptococci from invasive and non-invasive disease episodes in Belgium during 1993–1994. J. Med. Microbiol., 49, 467–471. [DOI] [PubMed] [Google Scholar]
- Elliott S.D. (1945) A proteolytic enzyme produced by group A streptococci with special reference to its effect on the type-specific M antigen. J. Exp. Med., 81, 573–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti J.J. et al. (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes.Proc. Natl Acad. Sci. USA, 98, 4658–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti V.A. (1989) Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev., 2, 285–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren A. and Sjöquist,J. (1966) ‘Protein A’ from S.aureus. I. Pseudo-immune reaction with human γ-globulin. J. Immunol., 97, 822–827. [PubMed] [Google Scholar]
- Frick I.M., Wikström,M., Forsén,S., Drakenberg,T., Gomi,H., Sjöbring, U. and Björck,L. (1992) Convergent evolution among immunoglobulin G-binding bacterial proteins. Proc. Natl Acad. Sci. USA, 89, 8532–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick I.M. et al. (1994) Protein H-a surface protein of Streptococcus pyogenes with separate binding sites for IgG and albumin. Mol. Microbiol., 12, 143–151. [DOI] [PubMed] [Google Scholar]
- Gomi H., Hozumi,T., Hattori,S., Tagawa,C., Kishimoto,F. and Björck,L. (1990) The gene sequence and some properties of protein H—a novel IgG binding protein. J. Immunol., 144, 4046–4052. [PubMed] [Google Scholar]
- Green G.D. and Shaw,E. (1981) Peptidyl diazomethyl ketones are specific inactivators of thiol proteinases. J. Biol. Chem., 256, 1923–1928. [PubMed] [Google Scholar]
- Hartas J. and Sriprakash,K.S. (1999) Streptococcus pyogenes strains containing emm12 and emm55 possess a novel gene coding for distantly related SIC protein. Microb. Pathog., 26, 25–33. [DOI] [PubMed] [Google Scholar]
- Heath D.G. and Cleary,P.P. (1987) Cloning and expression of the gene for an immunoglobulin G Fc receptor protein from a group A Streptococcus. Infect. Immun., 55, 1233–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwald H., Collin,M., Müller-Ester,W. and Björck,L. (1996) Streptococcal cysteine proteinase releases kinins: a novel virulence mechanism. J. Exp. Med., 184, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S.E., Norrby,A., Bergholm,A.M. and Norgren,M. (1992) Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988–1989. J. Infect. Dis., 166, 31–37. [DOI] [PubMed] [Google Scholar]
- Isberg R.R. and Tran Van Nhieu,G. (1994) Binding and internalization of microorganisms by integrin receptors. Trends Microbiol., 2, 10–14. [DOI] [PubMed] [Google Scholar]
- Janulczyk R. and Rasmussen,M. (2001) Improved pattern for genome-based screening identifies novel cell wall-attached proteins in Gram-positive bacteria. Infect. Immun., 69, 4019–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T.F., Cooney,J.C., Baker,H.M., McSweeney,S., Liu,M., Gubba, S., Musser,J.M. and Baker,E.N. (2000) Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: an integrin-binding cysteine protease. Proc. Natl Acad. Sci. USA, 97, 2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansal R.G., McGeer,A., Low,D.E., Norrby-Teglund,A. and Kotb,M. (2000) Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect. Immun., 68, 6362–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur V., Majesky,M.W., Li,L.L., Black,R.A. and Musser,J.M. (1993) Cleavage of interleukin 1β (IL-1β) precursor to produce active IL-1β by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc. Natl Acad. Sci. USA, 90, 7676–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihlberg B.M., Cooney,J., Caparon,M.G., Olsén,A. and Björck,L. (1995) Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb. Pathog., 19, 299–315. [DOI] [PubMed] [Google Scholar]
- Lancefield R.C. (1962) Current knowledge of type-specific M antigens of group A streptococci. J. Immunol., 89, 307–313. [PubMed] [Google Scholar]
- Lei B., Mackie,S., Lukomski,S. and Musser,J.M. (2000) Identification and immunogenicity of group A Streptococcus culture supernatant proteins. Infect. Immun., 68, 6807–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B. et al. (2001) Evasion of human innate and acquired immunity by a bacterial homolog of CD11b that inhibits opsonophagocytosis. Nature Med., 7, 1298–1305. [DOI] [PubMed] [Google Scholar]
- Lukomski S., Sreevatsan,S., Amberg,C., Reichardt,W., Woischnik,M., Podbielski,A. and Musser,J.M. (1997) Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J. Clin. Invest., 99, 2574–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S., Burns,E.H., Wyde,P.R., Podbielski,A., Rurangirwa,J., Moore-Poveda,D.K. and Musser,J.M. (1998) Genetic inactivation of an extracellular cysteine protease (SpeB) expressed by Streptococcus pyogenes decreases resistance to phagocytosis and dissemination to organs. Infect. Immun., 66, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J.M., Kapur,V., Szeto,J., Pan,X., Swanson,D.S. and Martin,D.R. (1995) Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect. Immun., 63, 994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre W.W. and Schneewind,O. (1999) Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev., 63, 174–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Nilson B.H., Frick,I.M., Åkesson,P., Forsén,S., Björck,L., Åkerström,B. and Wikström,M. (1995) Structure and stability of protein H and the M1 protein from Streptococcus pyogenes. Implications for other surface proteins of Gram-positive bacteria. Biochemistry, 34, 13688–13698. [DOI] [PubMed] [Google Scholar]
- Porter R.R. (1973) Structural studies of immunoglobulins. Science, 180, 713–716. [DOI] [PubMed] [Google Scholar]
- Raeder R.R. and Boyle,M.D (1993) Association of type II immunoglobulin G-binding protein expression and survival of group A streptococci in human blood. Infect. Immun., 61, 3696–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M. and Björck,L. (2002) Proteolysis and its regulation at the surface of Streptococcus pyogenes. Mol. Microbiol., 43, 537–544. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D. and Barrett,A.J. (1994) Families of cysteine peptidases. Methods Enzymol., 244, 461–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis K.J., Ayoub,E.M. and Boyle,M.D.P. (1984) Streptococcal Fc receptors. I. isolation and partial characterization of the receptor from a group C Streptococcus. J. Immunol., 132, 3091–3097. [PubMed] [Google Scholar]
- Roe B.A., Linn,S.P., Song,L., Yuan,X., Clifton,S., McLaughlin,R.E., McShan,M. and Ferretti,J. (2001) Streptococcus pyogenes genome sequencing strain M1 GAS. [Online.] http://www.genome.ou.edu/strep.html, October 17, 2001 (accession date).
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989). Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Stevens D.L. (1995) Streptococcal toxic-shock syndrome: spectrum of disease, pathogenesis and new concepts in treatment. Emerg. Infect. Dis., 1, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D.L. (2000) Streptococcal toxic shock syndrome associated with necrotizing fascitis. Annu. Rev. Med., 51, 271–288. [DOI] [PubMed] [Google Scholar]
- Suvorov A.N. and Ferretti,J.J. (1996) Physical and genetic chromosomal map of an M type 1 strain of Streptococcus pyogenes. J. Bacteriol., 178, 5546–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson M.D., Scaramuzzino,D.A., Sjöbring,U., Olsén,A., Frank,C. and Bessen,D.E. (2000) Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol. Microbiol., 38, 242–253 [DOI] [PubMed] [Google Scholar]
- Wexler D.E., Chenoweth,D.E. and Cleary,P.P. (1985) Mechanism of action of the group A streptococcal C5a inactivator. Proc. Natl Acad. Sci. USA, 82, 8144–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore A.M., Kapur,V., Sullivan,D.J., Musser,J.M. and Kehoe,M.A. (1994) Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol. Microbiol., 14, 619–631. [DOI] [PubMed] [Google Scholar]