Abstract
Apicomplexan parasites actively secrete proteins at their apical pole as part of the host cell invasion process. The adhesive micronemal proteins are involved in the recognition of host cell receptors. Redistribution of these receptor–ligand complexes toward the posterior pole of the parasites is powered by the actomyosin system of the parasite and is presumed to drive parasite gliding motility and host cell penetration. The microneme protein protease termed MPP1 is responsible for the removal of the C-terminal domain of TgMIC2 and for shedding of the protein during invasion. In this study, we used site-specific mutagenesis to determine the amino acids essential for this cleavage to occur. Mapping of the cleavage site on TgMIC6 established that this processing occurs within the membrane-spanning domain, at a site that is conserved throughout all apicomplexan microneme proteins. The fusion of the surface antigen SAG1 with these transmembrane domains excluded any significant role for the ectodomain in the cleavage site recognition and provided evidence that MPP1 is constitutively active at the surface of the parasites, ready to sustain invasion at any time.
Keywords: Apicomplexa/microneme/MPP1 protease/regulated secretion/Toxoplasma gondii
Introduction
The intracellular protozoan parasite Toxoplasma gondii shares with other members of the Apicomplexa a common set of apical structures involved in host cell invasion. The apical secretory organelles, called micronemes, release numerous soluble and transmembrane proteins at the surface of the parasite during invasion (Tomley and Soldati, 2001). TgMIC2, an adhesin secreted during host cell penetration (Carruthers et al., 1999a), belongs to the family of thrombospondin-related anonymous proteins (TRAPs) previously shown to play an essential role in host cell invasion by Plasmodium sporozoites and ookinetes (Sultan et al., 1997; Dessens et al., 1999; Yuda et al., 1999; Templeton et al., 2000). The C-terminal domain of TgMIC2 was recently demonstrated to functionally replace the equivalent domain of TRAP in Plasmodium (Kappe et al., 1999). TgMIC2 has also been reported recently to form a complex with TgM2AP (Rabenau et al., 2001). Other transmembrane microneme proteins described in T.gondii, such as TgMIC6 and TgMIC8, serve as escorters for the soluble adhesins TgMIC1, TgMIC4 and TgMIC3, respectively (Reiss et al., 2001; Meissner et al., 2002). The stimulation of micronemes discharge is induced by a rise in intracellular Ca2+, and can be mimicked by treatment with Ca2+ ionophore or ethanol (Carruthers and Sibley, 1999; Carruthers et al., 1999b). Microneme proteins undergo complex proteolytic processing events that have been best documented in T.gondii. TgMIC3, TgMIC5, TgMIC6 and TgM2AP are proteolytically cleaved intracellularly during transport to the micronemes (Brydges et al., 2000; Garcia-Reguet et al., 2000; Rabenau et al., 2001; Reiss et al., 2001). In addition, TgMIC2, TgMIC4, TgMIC6 and TgMIC8 are processed post-exocytosis (Carruthers et al., 2000; Brecht et al., 2001; Reiss et al., 2001; Meissner et al., 2002). Two types of proteases, termed MPP1 and MPP2, have been reported to process TgMIC2 upon release at the parasite surface (Carruthers et al., 2000). Recently, the profile of protease inhibitors that prevent the N-terminal processing of TgMIC4 suggested that MPP2 is likely to cleave this protein as well (Brecht et al., 2001). MPP2-dependent cleavage precedes the MPP1 cleavage, and these maturation events on TgMIC2 have been shown to change the adhesive properties of the protein drastically (Carruthers et al., 2000). The C-terminal cleavage of TgMIC2, TgMIC6 and TgMIC8 is expected to cause the release of the adhesin complexes from the parasite surface. Presumably, this event constitutes an essential step during host cell invasion, ensuring the dissociation between the parasite and the host plasma membrane at the end of the penetration process. Little is known about the sequence requirement, the localization and the characteristics of the protease responsible for this activity.
In this study, we have mapped the site of the MPP1-dependent processing at the C-terminus of TgMIC2, TgMIC6 and a newly characterized microneme protein TgMIC12. The cleavage, which appears to be essential for parasite survival, occurs within the transmembrane domain and does not require sequence information from the ectodomain. The protease is constitutively active at the surface of the parasites and conserved throughout the phylum of Apicomplexa, including in Plasmodium.
Results
The amino acid sequences within the membrane-spanning domain of apicomplexan microneme proteins are conserved
An alignment of the transmembrane and cytoplasmic domains of T.gondii, P.falciparum, P.berghei, Eimeria tenella and Sarcocystis muris microneme proteins revealed a striking conservation of the amino acid sequences within their C-terminal membrane-spanning domain (Figure 1A). In T.gondii, TgMIC2 is processed upon secretion, during host cell invasion by an as yet unidentified protease called MPP1 (Carruthers et al., 2000). A very similar processing event occurs on TgMIC6 (Reiss et al., 2001), TgMIC8 (Meissner et al., 2002), TgAMA-1 (Donahue et al., 2000; Hehl et al., 2000) and TgMIC12 (this study), which most probably involves the same protease activity. Taking advantage of the easy accessibility of T.gondii to transfection, we have examined the sequence requirements of this processing event. Some of the most conserved residues across the phylum of Apicomplexa (boxed in grey in Figure 1A) were chosen for the site-specific mutagenesis. The constructs generated and analysed in this study are depicted in Figure 1B.
Fig. 1. (A) Amino acid sequence alignment of the TM domains of the apicomplexan microneme proteins TgMIC2, TgMIC6, TgMIC12, PbTRAP, PfTRAP, EtMIC1, Sm70, TgMIC8, TgAMA1, PfCTRP and EtMIC4. The strictly conserved amino acids are in black boxes, and the conserved residues chosen for the mutagenesis are boxed in grey. (B) Schematic representation of the constructs used in this study: MIC2myc1/2, MIC2myc2mut, MIC6Ty, MIC6Tymut and the chimeric fusions SAG1TyTMCDMIC2, SAG1TyTMCDMIC2mut, SAG1TyTMCDMIC6, SAG1TyTMCDMIC12, SAG1TyTMCDMIC12mut, SAG1TyTMCDMIC12* and SAG1TyTMCDMIC12*mut. c-myc or Ty-1 were used as epitope tags and are boxed. Tg, T.gondii; Pb, P.berghei; Pf, P.falciparum; Et, E.tenella; Sm, S.muris; AMA1, apical membrane antigen 1; CTRP, circumsporozoite TRAP-related protein.
Point mutations within the transmembrane domain of TgMIC2 and TgMIC6 prevent processing and release of these proteins
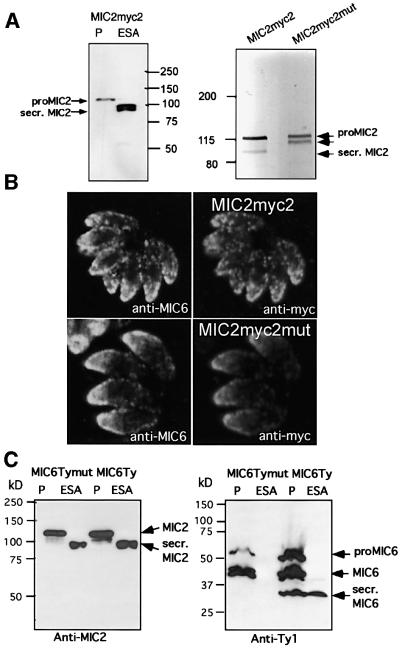
The insertion of a c-myc epitope tag at different positions in the cytoplasmic region of TgMIC2 previously allowed the mapping of sequences relevant for sorting to the micronemes (Di Cristina et al., 2000). The presence of a c-myc tag close to the transmembrane domain, on the luminal side of TgMIC2, compromised the C-terminal processing of MIC2myc1 (data not shown), while a tag inserted a few amino acids further upstream in MIC2myc2 was processed and secreted like wild-type TgMIC2 (Figure 2A). Based on the sequence alignment described above, we decided to insert four point mutations by site-specific mutagenesis within the transmembrane region of TgMIC2 (MIC2myc2), to generate MIC2myc2mut. The replacement of AGGVIGG with VALVIGV in MIC2myc2mut completely hampered the post-exocytotic cleavage at the C-terminus of the protein as analysed by western blot (Figure 2A). Nevertheless, parasites transiently transfected with MIC2myc2mut showed correct targeting of the transgene product to micronemes by indirect immunofluorescence assay (IFA; Figure 2B). While stably transformed clones expressing MIC2myc2 were easily obtained, we failed to isolate recombinant parasites stably expressing MIC2myc2mut, presumably because the lack of processing was deleterious to parasite survival. When five point mutations were introduced in the same region of TgMIC6 (VALVIVL), the corresponding mutant MIC6Tymut was targeted accurately to the micronemes (data not shown) and, as in the case of MIC2myc2mut, no proteolytic cleavage occurred at the C-terminus. In contrast to MIC2myc2mut, stable cell lines expressing MIC6Tymut in the MIC knockout (mic6ko) could be selected over a short period of time, allowing this mutant to be analysed by secretion assay. While TgMIC6Ty behaves like wild-type MIC6, TgMIC6Tymut was not cleaved and consequently not released in the cell culture supernatant (Figure 2C).
Fig. 2. Site-specific mutagenesis within the transmembrane domain of TgMIC2 and TgMIC6 abolished the C-terminal processing but did not affect proper sorting to the micronemes. (A) Western blot analysis of the parasite pellet and ESA of parasites stably transfected with pMMIC2myc2 using mAb 9E10 against the c-myc epitope. Right panel: total cell lysates of parasites 24 h after transfection with pMMIC2myc2 or pMMIC2myc2mut and stimulated for microneme secretion with ethanol treatment. The fully processed form of TgMIC2 with the C-terminal domain released is not detectable in MIC2myc2mut. The partial processing at the N-terminus of TgMIC2 is detectable (see arrow pointing to the band below proMIC2). (B) Double immunofluorescence analysis by confocal microscopy of intracellular parasites transiently expressing MIC2myc2 or MIC2myc2mut and detected with anti-myc antibodies. MIC2myc2mut did co-localize with the microneme marker MIC6 (left panel anti-MIC6). (C) Western blot analysis of secretion assays (parasite pellet and ESA) from stably transformed parasites expressing MIC6Ty or MIC6Tymut. The antibodies used were mAb BB2 recognizing the Ty-1 epitope and T3 4A11 anti-MIC2 as a control. The C-terminal processing of MIC6Tymut did not occur and, consequently, the protein was not released in the ESA.
Mapping of the C-terminal cleavage site of TgMIC6
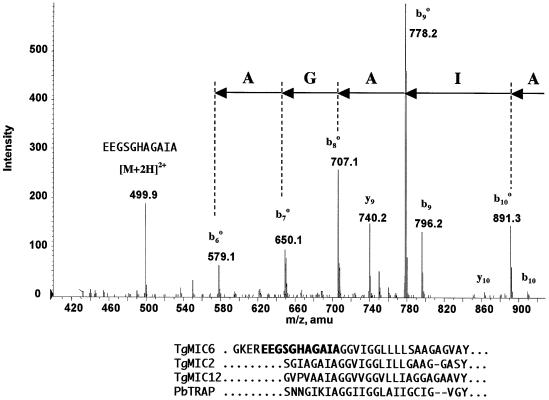
To determine with precision the MPP1 site of cleavage, recombinant parasites expressing TgMIC6Ty were used to purify large amounts of the processed form of TgMIC6 and to analyse it by mass spectrometry. The mature form of TgMIC6 was isolated from 5 × 109 freshly released parasites after stimulation of microneme secretion by ethanol and purification by immunoprecipitation using the monoclonal antibody (mAb) BB2 specific for the Ty-1 epitope. The 25 kDa processed product was separated finally on SDS–PAGE and digested with trypsin. The resulting peptides were analysed by mass spectrometry to identify a peptide whose C-terminus was not generated by trypsin cleavage. A doubly charged peptide ion matching the mass of the peptide EEGSGHAGAIA (m/z = 499.9) was detected. This peptide ion was fragmented by collision-induced dissociation and the fragment ions were recorded under conditions optimized for the mass range 500–1200 to confirm the amino acid sequence. The major fragment peaks are assigned to a bo series with the C-terminal peptide sequence AGAIA (Figure 3). Additionally, the fragments labelled as y9 and y10 are consistent with the two N-terminal amino acids EE of the peptide EEGSGHAGAIA.
Fig. 3. Mapping of the processing site at the C-terminus of TgMIC6 by identification of the peptide EEGSGHAGAIA by MS/ESI. After in-gel digestion of MIC6Ty, the tryptic peptides were bound to Poros R2 and Poros oligoR3 material. In the eluate from the R3 column, a doubly charged peptide ion matching the mass of the peptide EEGSGHAGAIA (m/z = 499.9) was detected. This peptide was fragmented and major fragment peaks are assigned to a bo series with the C-terminal peptide sequence AGAIA. Two additional peaks labelled as y9 and y10 are consistent with the two N-terminal amino acids EE. The amino acid sequence conservation around the cleavage site in TgMIC2, TgMIC6, TgMIC12 and PbTRAP is depicted below, with the tryptic peptide of MIC6 in bold.
MPP1 processing is conserved for micronemal proteins of the Apicomplexa phylum
The precise determination of the MPP1 cleavage site on TgMIC6 revealed the striking conservation of the amino acid sequence flanking this site in all transmembrane micronemal proteins of the Apicomplexa. In Eimeria, EtMIC1 is processed at the C-terminus upon microneme secretion (F.Tomley, personal communication). In Neospora caninum, the homologue of TgMIC2, NcMIC2, also lacks the C-terminal domain (Lovett et al., 2000). In Plasmodium species, both the redistribution of TRAP from the anterior to the posterior pole and the shedding of the protein by parasites, which presumably involves the C-terminal proteolytic cleavage of TRAP, are crucial for sporozoite gliding motility (Kappe et al., 1999). In a previous study, P.berghei TRAP protein was expressed in T.gondii (Di Cristina et al., 2000). We compared the expression of PbTRAP and PbTRAP lacking the transmembrane and cytoplasmic tail (PbTRAPΔTMCD) by transient transfection in T.gondii. The evidence for C-terminal processing of the full-length TRAP was obtained by western blot analysis (Figure 4). Consistent with the removal of the TMCD domain, the processed form co-migrated with the PbTRAPΔTMCD while the deleted mutant remained unprocessed. The C-terminal cleavage of TRAP in T.gondii reinforces the notion that this proteolytic event is ubiquitous in the Apicomplexa and the components of the machinery are functionally conserved.
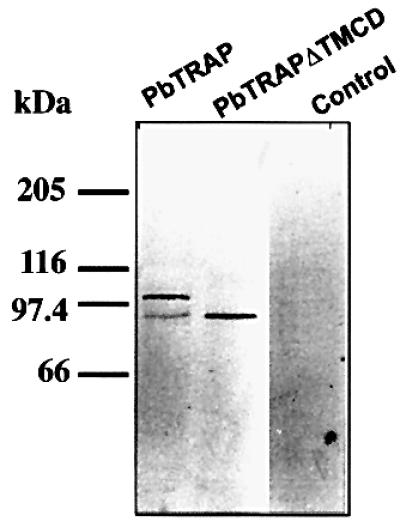
Fig. 4. When expressed in T.gondii, PbTRAP is processed at the C-terminus. Western blot (reducing conditions) analysis of total protein lysates of tachyzoites transiently transformed with PbTRAP wild-type (TRAP), TRAP devoid of the transmembrane domain and cytoplasmic tail (TRAPΔTMCD) and wild-type parasites (control) using mAb anti-PbTRAP.
The minimal amino acid sequence information required for MPP1 cleavage does not include the ectodomains
The C-terminal domains of TgMIC2 and TgMIC6 carry sufficient information for sorting to the appropriate organelles. When the major surface antigen SAG1 lacking its glycosylphosphatidylinositol (GPI) anchor signal was fused to the transmembrane and cytoplasmic domains of TgMIC2 or TgMIC6, the chimeric products were targeted to the micronemes (Di Cristina et al., 2000; Reiss et al., 2001). These SAG1 fusions exhibited a typical type I membrane topology as demonstrated by transient permeabilization followed by proteinase K treatment (Figure 5A). This type I topology was shown previously to be adopted by wild-type TgMIC6 (Meissner et al., 2002) and TgMIC2 (data not shown). Consequently, the sorting signals present in the C-terminal tail are facing the cytosol and are accessible to the adaptor complex machinery implicated in vesicular trafficking (Hoppe et al., 2000). Analysis of the recombinant parasites stably expressing SAG1TMCDMIC2 by secretion assays revealed that SAG1 is secreted upon ethanol stimulation, processed at the C-terminus and released in the cell supernatant in just the same manner as wild-type TgMIC2 (Figure 5B). To demonstrate unambiguously that the same processing event took place on these SAG1 fusions, the point mutations previously introduced in the transmembrane segment of TgMIC2 and TgMIC6 were included in the pSAG1TMCDMIC2 construct. The SAG1TMCDMIC2mut fusion was sorted accurately to the micronemes and, as expected, the C-terminal cleavage of SAG1TMCDMIC2mut was completely abrogated. Identical results were obtained with SAG1TMCDMIC6mut (data not shown). As in the case of MIC2mut, we failed to obtain stable lines expressing SAG1TMCDMIC2mut (Figure 5C). A schematic representation of the products obtained either after proteinase K digestion or upon stimulation of secretion is depicted in Figure 5D.
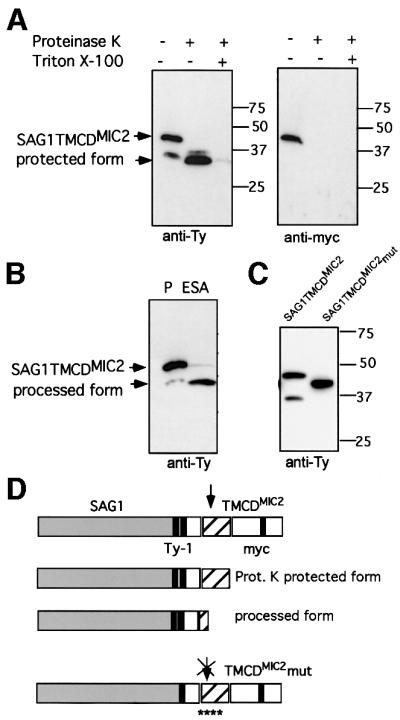
Fig. 5. The minimal requirement for processing to occur involves the luminal region of the MICs. SAG1TyTMCDMIC2 and SAG1TyTMCDMIC6 fusions are targeted to the micronemes and subjected to processing upon release by the micronemes. (A) The type I membrane protein topology of the SAG1TyTMCDMIC2 fusion was determined by partial permeabilization and proteinase K treatment. (B) Western blot analysis of total lysates and ESAs from recombinant parasites expressing SAG1TyTMCDMIC2 or SAG1TyTMCDMIC6. The membrane was probed with anti-Ty-1 antibodies. (C) Western blot analysis of recombinant parasites expressing SAG1TyTMCDMIC2 and SAG1TyTMCDMIC2mut. The presence of the four point mutations prevents the cleavage of the chimeric protein. The membranes were probed with either anti-myc or anti-Ty-1. (D) The construct SAG1TyTMCDMIC2 shown schematically as the full protein form with two Ty-1 epitope tags before the transmembrane domain (TM) and the c-myc epitope tag within the cytoplasmic domain. The smaller form protected from proteinase K treatment and the secreted processed form are shown below. The last cartoon shows the construct SAG1TyTMCDMIC2mut schematically. The localization of only one Ty-1 epitope tag before the TM, as well as the localization of the four point mutations within the TM, is shown.
TgMIC12 is a novel microneme protein, which also contains targeting signals to the organelle in the cytoplasmic tail
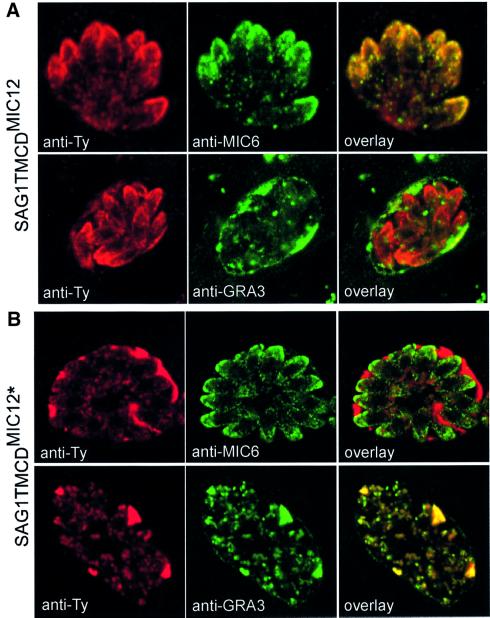
TgMIC12 is a novel microneme protein recently identified in T.gondii and partially sequenced (accession number AAK58479). Antibodies raised against the C-terminus of this protein stained the small apical microneme organelles and detected a product of 50 kDa by western blotting analysis (F.Stavru and D.Soldati, unpublished). The C-terminal region of TgMIC12 exhibited a strong homology to members of the TRAP family (Figure 1) and showed the closest resemblance to the partial sequence of a 70 kDa S.muris microneme protein (Tomley and Soldati, 2001). Consistent with previous observations, the fusion of the TMCD domain of TgMIC12 to the SAG1 ectodomain also localized the chimeric protein to the micronemes (Figure 6A). This SAG1TMCDMIC12 fusion is excluded from the parasitophorous vacuole, as illustrated by the absence of co-localization with the dense granule marker, GRA3. In contrast, an inadvertent mutant generated at the extreme C-terminus of SAG1TMCDMIC12 caused a complete mistargeting of the fusion protein to the dense granules and to the parasitophorous vacuolar space as seen by extensive co-localization with GRA3 (Figure 6B). In this SAG1TMCDMIC12* mutant, the last four amino acids, ADMD, have been replaced by the 10 amino acids QTLLKWIRWH (Figure 1B). The reason for this mistargeting is not understood but might involve an incorrect folding of the cytoplasmic tail, which potentially masks the sorting signals and prevents their interaction with the adaptor complex machinery. Although dense granules were described previously as the default pathway taken only by soluble proteins and not by transmembrane or GPI-anchored proteins in T.gondii (Karsten et al., 1998), SAGTMCDMIC12* is the first example of a transmembrane protein to follow this route by default.
Fig. 6. SAG1TyTMCDMIC12 is targeted to the micronemes while SAG1TyTMCDMIC12* is re-routed to the dense granules and into the parasitophorous vacuole. Double immunofluorescence analysis by confocal microscopy was carried out with anti-MIC6 and anti-GRA3 as microneme and dense granule/parasitophorous vacuole markers, respectively. (A) SAG1TyTMCDMIC12 was detected with anti-Ty-1 (in red) and co-localized with MIC6 (in green) (B) SAG1TyTMCDMIC12*, carrying an out-of-frame mutation changing the sequence at the extreme C-terminus, was detected with anti-Ty-1 (in red) and co-localized with GRA3 (in green).
MPP1 is constitutively active at the plasma membrane of the parasites and does not require the substrate to traffic through the micronemes
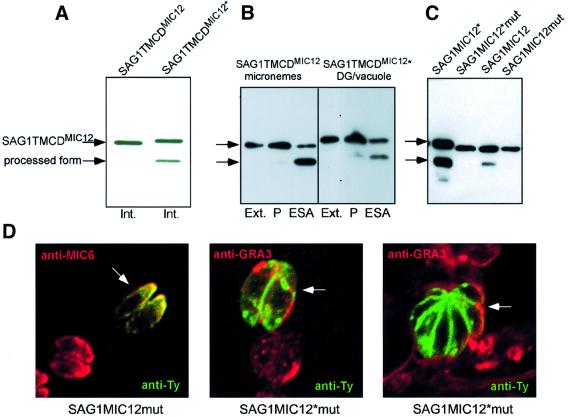
Despite the fact that SAG1TMCDMIC12* accumulated in the vacuole, the protein was processed accurately at its C-terminus (Figure 7A). However, in contrast to the proteins targeted to the micronemes, including SAG1TMCDMIC12, which are only processed after organelle secretion, the cleavage of SAG1TMCDMIC12* occurred constitutively during the intracellular cycle of parasite replication. Since dense granules are constitutively discharging their contents into the vacuole, this result suggested that MPP1-dependent processing occurred either at the parasite surface or in dense granules. To distinguish between the two possibilities, lysates were prepared from extensively washed extracellular parasites and subsequently stimulated for organelle secretion at 37°C for 60 min and analysed by western blot. Both SAG1TMCDMIC12 and SAG1TMCDMIC12* were processed only upon secretion by micronemes and dense granules, respectively (Figure 7B). This result clearly established that the cleavage of SAG1TMCDMIC12* occurred only when the proteins reached the cell surface, and not during storage in the dense granules. Finally, the inhibitory effect of the presence of the five point mutations on SAG1TMCDMIC12mut and SAG1TMCDMIC12*mut confirmed that the cleavage observed on the mistargeted SAG1TMCDMIC12* was identical to that occurring on the same protein targeted to the micronemes (Figure 7C). The subcellular distribution of SAG1TMCDMIC12mut and SAG1TMCDMIC12*mut was examined by immunofluorescence on confocal microscopy after transient transfection. As expected, the mutations introduced in SAG1TMCDMIC12mut did not affect targeting to the micronemes, while in the case of SAG1TMCDMIC12*mut the absence of cleavage leads to the persistence of the protein at the parasite surface (Figure 7D), instead of an accumulation in the vacuolar space (Figure 6B).
Fig. 7. Despite mistargeting to the vacuole, SAG1TyTMCDMIC12* is processed accurately at the C-terminus. (A) Western blot analysis of total lysates from intracellular parasites (Int.) stably transformed with SAG1TyTMCDMIC12 and SAG1TyTMCDMIC12* showed constitutive processing at the C-terminus of SAG1TyTMCDMIC12*. (B) Western blot analysis of total lysates from extracellular (Ext.) parasites as well as pellet (P) and secreted material (ESA). For both SAG1TyTMCDMIC12 and SAG1TyTMCDMIC12*, the processing occurred after release from micronemes and dense granules, respectively. (C) Western blot analysis of parasites transiently transfected with SAG1TyTMCDMIC12, SAG1TyTMCDMIC12mut, SAG1TyTMCDMIC12* and SAG1TyTMCDMIC12*mut established that processing occurred only with the non-mutated constructs and thus they are generated by the MPP1-dependent cleavage. (D) Overlays of double immunofluorecence analysis by confocal microscopy on intracellular parasites transiently expressing SAG1TyTMCDMIC12mut and SAG1TyTMCDMIC12*mut. SAG1TyTMCDMIC12mut detected by anti-Ty (in green) co-localized with anti-MIC6 (in red) while SAG1TyTMCDMIC12*mut is present at the plasma membrane of the parasites (in green) and did not co-localize with GRA3 (in red).
Discussion
Transmembrane microneme proteins are presumed to play a dual essential role during host cell invasion by the Apicomplexa. The adhesin domains present on the luminal side of these proteins or, indirectly, their association with soluble adhesins establish tight and specific interactions with host cell receptors while their cytoplasmic tail presumably is connected to the actomyosin system of the parasite. Only transmembrane proteins are expected to transmit the mechanical force across the plasma membrane and thus to power parasite gliding motion.
Upon stimulation of microneme secretion, some proteins were shown previously to redistribute over the entire surface of the parasites and, as invasion proceeds, these proteins are excluded from the forming vacuole, accumulate at the posterior pole of the parasites and are shed. This has been documented previously for TgMIC2 (Carruthers et al., 1999a) and TgMIC3 (Garcia-Reguet et al., 2000). The proteolytic processing occurring on these proteins and causing their rapid release from the cell surface is anticipated to be an essential step to accomplish the penetration process. In a preliminary experiment, we observed that the insertion of a c-myc tag in close proximity to the transmembrane-spanning domain of TgMIC2 hampered its processing at the C-terminus. Subsequently, we identified by site-specific mutagenesis a few conserved amino acids within the transmembrane domain which were necessary for the cleavage of TgMIC2 to take place. Expression of a non-processed TgMIC2 mutant (MIC2myc2mut, Figures 1B and 2A) appeared to compromise the survival of the parasites as all attempts to generate stable transformants remained unsuccessful. The deleterious effect observed due to the absence of cleavage on TgMIC2 underlines the importance of this event for a successful invasion. In the absence of processing, the exocytosed TgMIC2 might create a persistent bridge between the host cell and parasite surface, thus interfering with the proper sealing of the newly formed parasitophorous vacuole and preventing the parasite from replicating inside the vacuole. The introduction of equivalent point mutations on TgMIC6 provoked the same phenotype and was slightly better tolerated by the parasite, but the stable clones could be maintained for only a few passages. TgMIC6 does not bind directly to the host cell surface receptors but functions as an escorter and, presumably, the lack of processing does not lead to such dramatic consequences. Moreover, we have shown previously that TgMIC6 is dispensable, as we could generate mic6ko by homologous recombination but, in contrast, multiple attempts to disrupt the TgMIC2 gene have been unsuccessful so far. This suggests that unlike TgMIC6, TgMIC2 fulfils a more substantial role. In Plasmodium species, by disruption of the genes coding for several members of the TRAP family it was shown that they are a prerequisite for parasite motility and host cell invasion (Sultan et al., 1997; Dessens et al., 1999; Yuda et al., 1999; Templeton et al., 2000).
Previously, fusions of SAG1 to the TMCD domains of microneme proteins TgMIC2 and TgMIC6 established that the sorting signal to the micronemes are confined within the short cytoplasmic tails. In this study, we provide compelling evidence that the short C-terminal domain of these proteins is sufficient to serve as a substrate for MPP1, excluding an essential role for luminal domains, which contain diverse combinations of adhesive motifs (Tomley and Soldati, 2001). The striking and unexpected conservation within the transmembrane domain combined with the results of the mutagenesis suggested to us that MPP1 functions in the hydrophobic environment of the plasma membrane outer leaflet. This hypothesis was confirmed by mapping of the cleavage site of MPP1 within the membrane-spanning region of TgMIC6. Interestingly, the cleavage site of MPP1 lies within the membrane, apparently beneath the polar head groups of the phospholipids. Enzymes able to cleave polypeptides within the membrane exist and include the site 2 protease (S2P) (Rawson et al., 1997), which is a zinc metalloprotease that cleaves the sterol-regulatory element-binding proteins (SREBPs) at site 2 within transmembrane segments (Duncan et al., 1998). The release of the signal peptide fragments also requires cleavage in the transmembrane region (Weihofen et al., 2000). Additionally, the famous amyloid β precursor protein, involved in Alzheimer’s disease, is cleaved twice in the middle of the membrane-spanning domain (Citron et al., 1996). The enzyme named γ-secretase is responsible for this activity and is likely to be another member of the family of polytopic membrane proteases, displaying pharmacological properties of an aspartyl protease, with presenilin probably acting as a catalytic component (Esler and Wolfe, 2001). So far, only one metalloprotease activity has been reported in T.gondii (Berthonneau et al., 2000).
The chimeric protein SAG1TMCDMIC12* is mistargeted to the surface of the parasite and, nevertheless, is processed very efficiently at the C-terminus. The introduction of point mutations at the cleavage site in SAG1TMCDMIC12* precisely confirmed that the processing is MPP1 dependent. These results indicate that MPP1 is not stored in micronemes but rather is a plasma membrane constitutively active protease. Previous experiments using cytochalasin D treatment established that MPP1 activity was not concentrated at the posterior pole of the parasites but rather distributed all over the parasite surface (Carruthers et al., 1999a). Interestingly, the substrate does not need to traffic through the micronemes in order to be delivered correctly to the enzyme and processed accurately. SAG1TMCDMIC12* is cleaved very efficiently as soon as the molecule reaches the plasma membrane. Similarily, when expressed in T.gondii, PbTRAP appears to be processed accurately at the C-terminus despite the fact that the protein is mistargeted to the vacuole because the Plasmodium tail of TRAP lacks the complete information for sorting to the micronemes of T.gondii (Di Cristina et al., 2000).
The mistargeting of TgMIC4 to the parasitophorous vacuole in mic6ko (Reiss et al., 2001) allowed us to address the similar question with regard to MPP2-dependent processing. Unlike the situation observed here with MPP1, in mic6ko, TgMIC4 protein accumulated in the vacuolar space but could no longer be cleaved by MPP2 even when microneme secretion was stimulated (M.Reiss, U.Jäkle and D.Soldati, unpublished). It is interesting to note that MPP2 processing is sensitive to cysteine protease inhibitors, and recently a member of the subtilisin-like family of cysteine proteases has been localized to the micronemes of T.gondii (Miller et al., 2001).
The compartmentalization of the substrates (transmembrane microneme proteins) and the enzyme (MPP1) appears to be a safe strategy adopted by the parasite to avoid processing at an inappropriate time, while still preserving an active enzyme ready to respond very quickly to stimulation. The strict conservation of the cleavage site among microneme proteins of different species strongly suggests that MPP1 is functionally conserved among the members of the phylum of Apicomplexa. This broad functional conservation across species offers a unique opportunity to study some aspects of the events governing Plasmodium host cell invasion using T.gondii parasites, more easily accessible to rapid transfection and biochemical analysis than the malaria parasites. The presumed crucial role of MPP1 during host invasion and its unique characteristics justify further efforts to identify and characterize this protease as a new potential drug target against a highly relevant group of human and animal pathogens.
Materials and methods
Host cells and Toxoplasma strains growth
Toxoplasma gondii tachyzoites of the RH strain were maintained by growth on monolayers of human foreskin fibroblasts (HFFs) or African green monkey (Vero) cells, grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) containing 5 or 10% fetal calf serum (FCS; Gibco). A clonal isolate of the RHhxgprt– of T.gondii was used as the recipient strain for the experiments described here. The mic6ko mutant generated in the RH strain was described previously (Reiss et al., 2001).
DNA constructs
The cDNA of TgMIC2 was modified by the insertion of a sequence coding for the c-myc epitope tag to generate pMMIC2myc1 and pMMIC2myc2 using a previously described strategy (Di Cristina et al., 2000). The constructs were expressed in T.gondii under the control of the 5′- and 3′-flanking sequences of the TgMIC2 gene. Four point mutations were introduced by site-directed mutagenesis in the transmembrane region of TgMIC2 to create pMMIC2myc2mut. The construct pTMIC6Ty, previously described (Reiss et al., 2001), was used to generate pTMIC6Tymut by inverse PCR using the primers MIC6-1: 5′-CGGGATCCATGCATGTCCACTTCCTTCCTCT-3′ and MIC6-2: 5′-GACATGCATTGGCGATCGTGGCTCTCGTTATTCTCGTTCTTTACTTCTGAGCGC-3′. The vector pMSAG1TyTMCDMIC6 and pMSAG1TyTMCDMIC2 were described previously (Di Cristina et al., 2000; Reiss et al., 2001). A Ty-1 epitope tag was introduced by insertion of double-stranded oligonucleotides into the SalI site using the primers Ty-1/1 (5′-TCGAGCGTCCACACCAACCAGGACCCCCTCGACGGG-3′) and Ty-1/2 (5′-TCGACGTCGAGGGGGTCCTGGTTGGTGTGGACC-3′). The constructs pMSAG1TyTMCDMIC12 and pMSAG1TyTMCDMIC12* were obtained by exchanging the TMCD domain of TgMIC2 with the corresponding domain of TgMIC12 (accession number AY034607) between the SalI and PacI cloning sites. The wild-type TMCDMIC12 and TMCDMIC12* were generated by PCR using the combination of primers MIC12-1 with MIC12-2, and MIC12-1 with MIC12-3, respectively: MIC12-1, 5′-CGGGATCCGTCGACGGCGTTCCAGTCGCAGCGATT-3′; MIC12-2, 5′-CCTTAATTAATTAGTCCATGTCTGCCCATGCGTC-3′; and MIC12-3, 5′-CCTTAATTAACTAGTGATTGCGGATCCACTTTAG-3′. The Ty-1 tag was introduced as described above.
The mutations within the transmembrane domains of TgMIC2 and TgMIC12 in SAG1 fusions were generated by PCR using the sense primers MIC2, 5′-CCGGTCGACAGTTTGGATGGCATAGCTGGGGCTATTGCGGTCGCTCTGGTCATCCTGGTTCTGATACTGCTTGGCGC-3′; and MIC12-4, 5′-CCGGTCGACGGCGTTCCAGTCGCAGCGATTGTCGCTCTGGTCGTCCTGGTTGTGTTGCTCATTGCT-3′, respectively. The mutated TMCDs were cloned in-frame downstream of the SAG1 coding sequence between the SalI and PacI sites.
Transfection and selections
To generate stable transformants, 5 × 107 extracellular RHhxgprt– parasites were transfected and selected using the HXGPRT marker gene as previously described (Donald et al., 1996), with the following modifications. Parasites were transfected with 50–80 µg of linearized plasmid. After 24 h, parasites were subjected to mycophenolic acid/xanthine (MPA/X) exposure and cloned 3 days later by limiting dilution into 96-well microtitre plates, in the presence of MPA/X. Pools of stable transformants were analysed by immunofluorescence assay for the expression of the transgenes.
SDS–PAGE and immunoblots
Freshly released tachyzoites were harvested, washed in phosphate-buffered saline (PBS) and lysed in RIPA solution (150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0, 1 mM EDTA). Polyacrylamide gels (8.5–10%) were run under reducing conditions with 0.1 M dithiothreitol (DTT) in the samples. mAbs 9E10 anti-c-myc; BB2 anti-Ty1 (mouse ascitic fluid; kindly provided by K.Gull, Manchester, UK); T3 4A11anti-TgMIC2 (kindly provided by J.-F.Dubremetz) and rabbit polyclonal antisera were diluted 1:1000 in PBS, 0.05% Tween-20 and 5% non-fat milk powder. After washing, the nitrocellulose membrane was incubated for 1 h with a peroxidase-conjugated goat anti-mouse antibody (Bio-Rad; 1:3000) and bound antibodies were visualized using the enhanced chemiluminescence (ECL) system (Boehringer).
Immunoprecipitation
Parasites (5 × 109) were stimulated to secretion and TgMIC6 was immunoprecipitated according to the protocol described previously (Meissner et al., 2002).
Indirect immunofluorescence microscopy
All manipulations were carried out at room temperature. Tachyzoite-infected HFFs on glass coverslips were fixed with 3% paraformaldehyde/0.05% glutaraldehyde, or 4% paraformaldehyde only, for 20 min, followed by 3 min incubation with 0.1 M glycine in PBS. Fixed cells were permeabilized with 0.2% Triton X-100 in PBS for 20 min and blocked in 2% FCS or bovine serum albumin in PBS for 20 min. The cells were then stained with the primary antibodies followed by Alexa 594 goat anti-rabbit or Alexa 488-conjugated goat anti-mouse antibodies (Molecular Probes, Cappel and Bio-Rad). Confocal images were collected with a Leica laser scanning confocal microscope (TCS-NT DM/IRB) using a 100× Plan-Apo objective with NA 1.30. Single optical sections were recorded with an optimal pinhole of 1.0 (according to Leica instructions) and averaging 16 times. Other micrographs were obtained with a Zeiss Axiophot equipped with a CCD camera (Photometrics Type CH-250). Adobe Photoshop (Adobe Systems, Mountain View, CA) was used for image processing.
Preparation of secreted microneme proteins
For the large-scale preparation of excretory secretory antigens (ESAs), ∼5 × 109 tachyzoites were resuspended in 1 ml of HHE (HBSS containing 10 mM HEPES and 1 mM EGTA). The discharge of micronemes was stimulated by the addition of ethanol to a final concentration of 1.0% and warming to 37°C for 30 min, as previously described (Carruthers et al., 1999b). Cells were removed by centrifugation at 2000 g and the supernatant was kept frozen.
Mass spectrometry analysis
In-gel digestion was performed as described (Rosenfeld et al., 1992; Shevchenko et al., 1996) with minor modifications. Briefly, the excised gel plugs were washed with 100 µl of water, 100 µl of 50% acetonitrile and subsequently shrunk in 100 µl of acetonitrile. Modified trypsin (Promega), 12 ng/µl in 40 mM ammonium bicarbonate buffer, was added and incubated overnight at 37°C. Custom-made chromatographic columns packed with either Poros R2 or Poros oligoR3 (50 µm bead size, PerSeptive Biosystems) were used for desalting the supernatant of the tryptic digest. A 20 µg aliquot of Poros sorbent in 5 µl of 75% methanol/1% acetic acid was packed in a constricted GELoader tip (Eppendorf). After equilibration with 40 µl of 1% acetic acid, an R2 column was aligned with an R3 column to trap hydrophilic peptides not bound by the R2 material (Neubauer and Mann, 1999). The sample was loaded, the columns were washed with 20 µl of 1% acetic acid and peptides were eluted separately from the two columns with 1 µl of 75% methanol/1% acetic acid directly into a pre-coated borosilicate nanoelectrospray needle (MDS Protana, Odense, Denmark) for mass spectrometry (MS) analysis. MS analysis was performed on a Q-TOF Pulsar mass spectrometer (PE SIEX, Weiterstadt, Germany) equipped with a nano-ESI ion source (MDS Protana, Odense, Denmark). A potential of 900 V was applied to the nanoelectrospray needle. Declustering potential and focusing potential were set to 40 and 180, respectively. For the fragmentation of selected peptides (unit resolution), three different collision energies (22, 27 and 35 V) and two parameter sets (without pulsar function: m/z 100–1900; IQ3 = 9.2; IRD = 0; IRP = 9.2; IRW = 5; R02 = 9.2; with pulsar function: m/z 400–1200; IQ3 = 11.2; IRD = 94; IRP = 7.2; IRW = 46; R02 = 7.2) were used. The peptide fragments were labelled according to Biemann (1988).
Acknowledgments
Acknowledgements
We thank Ursula Jaekle and Fabrizia Stavru for their preliminary work on TgMIC12. We are very grateful to Thierry Soldati for his valuable assistance with the confocal imaging and we are indebted to him and other members of the laboratories for critical reading of the manuscript. This work was funded by the Deutsche Forschungsgemeinschaft (DFG grant number SO 366/1-3), the Graduiertenkolleg ‘Pathogene Mikroorganismen: Molekulare Mechanismen und Genome’ and the Howard Hughes Medical Institute Research Scholars programme.
References
- Berthonneau J., Rodier,M.-H., El Moudni,B. and Jacquemin,J.-L. (2000) Toxoplasma gondii: purification and characterization of an immunogenic metallopeptidase. Exp. Parasitol., 95, 158–162. [DOI] [PubMed] [Google Scholar]
- Biemann K. (1988) Contributions of mass spectrometry to peptide and protein structure. Biomed. Environ. Mass Spectrom., 16, 99–111. [DOI] [PubMed] [Google Scholar]
- Brecht S., Carruthers,V.B., Ferguson,D.J., Giddings,O.K., Wang,G., Jaekle,U., Harper,J.M., Sibley,L.D. and Soldati,D. (2001) The toxoplasma micronemal protein MIC4 is an adhesin composed of six conserved apple domains. J. Biol. Chem., 276, 4119–4127. [DOI] [PubMed] [Google Scholar]
- Brydges S.D., Sherman,G.D., Nockemann,S., Loyens,A., Daubener,W., Dubremetz,J.F. and Carruthers,V.B. (2000) Molecular characterization of TgMIC5, a proteolytically processed antigen secreted from the micronemes of Toxoplasma gondii.Mol. Biochem. Parasitol., 111, 51–66. [DOI] [PubMed] [Google Scholar]
- Carruthers V.B. and Sibley,L.D. (1999) Mobilization of intracellular calcium stimulates microneme discharge in Toxoplasma gondii.Mol. Microbiol., 31, 421–428. [DOI] [PubMed] [Google Scholar]
- Carruthers V.B., Giddings,O.K. and Sibley,L.D. (1999a) Secretion of micronemal proteins is associated with toxoplasma invasion of host cells. Cell Microbiol., 1, 225–235. [DOI] [PubMed] [Google Scholar]
- Carruthers V.B., Moreno,S.N. and Sibley,L.D. (1999b) Ethanol and acetaldehyde elevate intracellular [Ca2+] and stimulate microneme discharge in Toxoplasma gondii. Biochem. J., 342, 379–386. [PMC free article] [PubMed] [Google Scholar]
- Carruthers V.B., Sherman,G.D. and Sibley,L.D. (2000) The Toxoplasma adhesive protein MIC2 is proteolytically processed at multiple sites by two parasite-derived proteases. J. Biol. Chem., 275, 14346–14353. [DOI] [PubMed] [Google Scholar]
- Citron M., Diehl,T.S., Gordon,G., Biere,A.L., Seubert,P. and Selkoe,D.J. (1996) Evidence that the 42- and 40-amino acid forms of amyloid β protein are generated from the β-amyloid precursor protein by different protease activities. Proc. Natl Acad. Sci. USA, 93, 13170–13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessens J.T., Beetsma,A.L., Dimopoulos,G., Wengelnik,K., Crisanti,A., Kafatos,F.C. and Sinden,R.E. (1999) CTRP is essential for mosquito infection by malaria ookinetes. EMBO J., 18, 6221–6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristina M., Spaccapelo,R., Soldati,D., Bistoni,B. and Crisanti,A. (2000) Two conserved amino acid motifs mediate protein targeting to the micronemes of the apicomplexan parasite Toxoplasma gondii. Mol. Cell. Biol., 20, 7332–7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue C.G., Carruthers,V.B., Gilk,S.D. and Ward,G.E. (2000) The toxoplasma homolog of plasmodium apical membrane antigen-1 (AMA-1) is a microneme protein secreted in response to elevated intracellular calcium levels. Mol. Biochem. Parasitol., 111, 15–30. [DOI] [PubMed] [Google Scholar]
- Donald R., Carter,D., Ullman,B. and Roos,D.S. (1996) Insertional tagging, cloning and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem., 271, 14010–14019. [DOI] [PubMed] [Google Scholar]
- Duncan E.A., Dave,U.P., Sakai,J., Goldstein,G.L. and Brown,M.S. (1998) Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junctions as determined by cysteine panning. J. Biol. Chem., 273, 17801–17809. [DOI] [PubMed] [Google Scholar]
- Esler W.P. and Wolfe,M.S. (2001) A portrait of Alzheimer secretases—new features and familiar faces. Science, 293, 1449–1454. [DOI] [PubMed] [Google Scholar]
- Garcia-Reguet N. et al. (2000) The microneme protein MIC3 of Toxoplasma gondii is a secretory adhesin that binds to both the surface of the host cells and the surface of the parasite. Cell. Microbiol., 2, 353–364. [DOI] [PubMed] [Google Scholar]
- Hehl A.B., Lekutis,C., Grigg,M.E., Bradley,P.J., Dubremetz,J.F., Ortega-Barria,E. and Boothroyd,J.C. (2000) Toxoplasma gondii homologue of plasmodium apical membrane antigen 1 is involved in invasion of host cells. Infect. Immun., 68, 7078–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe H.C., Ngo,H.M., Yang,M. and Joiner,K.A. (2000) Targeting to rhoptry organelles of Toxoplasma gondii involves evolutionarily conserved mechanisms. Nature Cell Biol., 2, 449–456. [DOI] [PubMed] [Google Scholar]
- Kappe S., Bruderer,T., Gantt,S., Fujioka,H., Nussenzweig,V. and Menard,R. (1999) Conservation of a gliding motility and cell invasion machinery in apicomplexan parasites. J. Cell Biol., 147, 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten V., Qi,H., Beckers,C.J., Reddy,A., Dubremetz,J.F., Webster,P. and Joiner,K.A. (1998) The protozoan parasite Toxoplasma gondii targets proteins to dense granules and the vacuolar space using both conserved and unusual mechanisms. J. Cell Biol., 141, 1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett J.L., Howe,D.K. and Sibley,L.D. (2000) Molecular characterization of a thrombospondin-related anonymous protein homologue in Neospora caninum.Mol. Biochem. Parasitol., 107, 33–43. [DOI] [PubMed] [Google Scholar]
- Meissner M., Reiss,M., Viebig,N., Carruthers,V., Trousel,C., Tomavo,S., Ajioka,J. and Soldati,D. (2002) A family of transmembrane microneme proteins of Toxoplasma gondii contain EGF-like domains and function as escorters. J. Cell Sci., 115, 563–574. [DOI] [PubMed] [Google Scholar]
- Miller S.A., Binder,E.M., Blackman,M.J., Carruthers,V.B. and Kim,K. (2001) A conserved subtilisin-like protein TgSUB1 in microneme organelles of Toxoplasma gondii. J. Biol. Chem., 276, 45341–45348. [DOI] [PubMed] [Google Scholar]
- Neubauer G. and Mann,M. (1999) Mapping of phosphorylation sites of gel-isolated proteins by nanoelectrospray tandem mass spectrometry: potentials and limitations. Anal. Chem., 71, 235–242. [DOI] [PubMed] [Google Scholar]
- Rabenau K.E., Sohrabi,A., Tripathy,A., Reitter,A., Ajioka,J.W., Tomley,F.M. and Carruthers,V.B. (2001) TgM2AP participates in Toxoplasma gondii invasion of host cells and is tightly associated with the adhesive protein TgMIC2. Mol. Microbiol., 41, 1–12. [DOI] [PubMed] [Google Scholar]
- Rawson R.B., Zelenski,N.G., Nijhawan,D., Ye,J., Sakai,J., Hasan,M.T., Chang,T.Y., Brown,M.S. and Goldstein,L.J. (1997) Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell, 1, 47–57. [DOI] [PubMed] [Google Scholar]
- Reiss M., Viebig,N., Brecht,S., Fourmaux,M.-N., Soete,M., Di Cristina,M., Dubremetz,J.F. and Soldati,D. (2001) Identification and characterisation of an escorter for two secretory adhesins in Toxoplasma gondii. J. Cell Biol., 152, 563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J., Capdevielle,J., Guillemot,J.C. and Ferrara,P. (1992) In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem., 203, 173–179. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm,M., Vorm,O. and Mann,M. (1996) Mass spectrometric sequencing of proteins on silver-stained polyacrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Sultan A.A., Thathy,V., Frevert,U., Robson,K.J., Crisanti,A., Nussenzweig,V., Nussenzweig,R.S. and Menard,R. (1997) TRAP is necessary for gliding motility and infectivity of plasmodium sporozoites. Cell, 90, 511–522. [DOI] [PubMed] [Google Scholar]
- Templeton T.J., Kaslow,D.C. and Fidock,D.A. (2000) Developmental arrest of the human malaria parasite Plasmodium falciparum within the mosquito midgut via CTRP gene disruption. Mol. Microbiol., 36, 1–9. [DOI] [PubMed] [Google Scholar]
- Tomley F.M. and Soldati,D.S. (2001) Mix and match modules: structure and function of microneme proteins in apicomplexan parasites. Trends Parasitol., 17, 81–88. [DOI] [PubMed] [Google Scholar]
- Weihofen A., Lemberg,M.K., Ploegh,H.L., Bogyo,M. and Martoglio,B. (2000) Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J. Biol. Chem., 275, 30951–30956. [DOI] [PubMed] [Google Scholar]
- Yuda M., Sakaida,H. and Chinzei,Y. (1999) Targeted disruption of the Plasmodium berghei CTRP gene reveals its essential role in malaria infection of the vector mosquito. J. Exp. Med., 190, 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]