Abstract
The heterotrimeric factor e/aIF2 plays a central role in eukaryotic/archaeal initiation of translation. By delivering the initiator methionyl-tRNA to the ribosome, e/aIF2 ensures specificity of initiation codon selection. The three subunits of aIF2 from the hyperthermophilic archaeon Pyrococcus abyssi could be overproduced in Escherichia coli. The β and γ subunits each contain a tightly bound zinc. The large γ subunit is shown to form the structural core for trimer assembly. The crystal structures of aIF2γ, free or complexed to GDP-Mg2+ or GDPNP-Mg2+, were resolved at resolutions better than 2 Å. aIF2γ displays marked similarities to elongation factors. A distinctive feature of e/aIF2γ is a subdomain containing a zinc-binding knuckle. Examination of the nucleotide-complexed aIF2γ structures suggests mechanisms of action and tRNA binding properties similar to those of an elongation factor. Implications for the mechanism of translation initiation in both eukarya and archaea are discussed. In particular, positioning of the initiator tRNA in the ribosomal A site during the search for the initiation codon is envisaged.
Keywords: G protein/transfer RNA/translation initiation/X-ray structure
Introduction
In all cells, initiation of translation requires precise selection of the start codon on the mRNA. In eubacteria, the ribosome binds directly in the vicinity of the AUG initiation codon through interaction of 16S rRNA with the Shine–Dalgarno sequence on mRNA. Exact positioning of the initiation complex is achieved with the recruitment of a formyl-methionyl-initiator tRNA
![]() and of three initiation factors IF1, IF2 and IF3 (Blanquet et al., 2000; Gualerzi et al., 2000). The main function of IF2 is to stimulate f-Met-tRNA
and of three initiation factors IF1, IF2 and IF3 (Blanquet et al., 2000; Gualerzi et al., 2000). The main function of IF2 is to stimulate f-Met-tRNA
![]() binding to the ribosome. This G protein displays significant sequence similarities to EF-Tu. However, recent biochemical and structural studies have led to the idea that IF2 and EF-Tu have rather different structural and functional organizations (Spurio et al., 2000; Szkaradkiewicz et al., 2000).
binding to the ribosome. This G protein displays significant sequence similarities to EF-Tu. However, recent biochemical and structural studies have led to the idea that IF2 and EF-Tu have rather different structural and functional organizations (Spurio et al., 2000; Szkaradkiewicz et al., 2000).
In eukarya, translation begins at the 5′ capped end of mRNA with the formation of a 43S ribosomal complex containing the small 40S ribosomal subunit, an eIF2–GTP–Met-tRNA
![]() complex and several initiation factors (eIF4A, eIF4B, eIF4F and eIF3; Merrick and Hershey, 1996; Trachsel, 1996; Pestova et al., 2001). Further recruitment of eIF1, eIF1A and eIF5 enables this complex to scan the mRNA until the correct AUG initiation codon is reached (Chaudhuri et al., 1997; Pestova et al., 1998). In the resulting 48S complex, eIF5 induces hydrolysis of GTP bound to eIF2. This allows the release of eIF2–GDP and the joining of the 60S large ribosomal subunit to form the competent elongator 80S ribosome (Chakrabarti and Maitra, 1991; Huang et al., 1997; Choi et al., 1998; Pestova et al., 2000). This step requires the presence of eIF5B, a close structural homologue of bacterial IF2 (Roll-Mecak et al., 2000).
complex and several initiation factors (eIF4A, eIF4B, eIF4F and eIF3; Merrick and Hershey, 1996; Trachsel, 1996; Pestova et al., 2001). Further recruitment of eIF1, eIF1A and eIF5 enables this complex to scan the mRNA until the correct AUG initiation codon is reached (Chaudhuri et al., 1997; Pestova et al., 1998). In the resulting 48S complex, eIF5 induces hydrolysis of GTP bound to eIF2. This allows the release of eIF2–GDP and the joining of the 60S large ribosomal subunit to form the competent elongator 80S ribosome (Chakrabarti and Maitra, 1991; Huang et al., 1997; Choi et al., 1998; Pestova et al., 2000). This step requires the presence of eIF5B, a close structural homologue of bacterial IF2 (Roll-Mecak et al., 2000).
Recent genome analyses suggest that initiation of translation in archaea is a hybrid system between eukarya and eubacteria. On the one hand, archaeal mRNAs are polycistronic, uncapped and lack long poly(A)+ tails (Dennis, 1997; Kyrpides and Woese, 1998a,b). Moreover, archaeal mRNAs most often display Shine– Dalgarno-like sequences. All these features are reminiscent of eubacterial translation. On the other hand, archaeal initiator tRNA does not undergo formylation. It is carried towards the ribosome by a close homologue of eukaryotic eIF2. As in eukarya, a homologue of eubacterial IF2, aIF5B, is present in archaea (Lee et al., 1999). Several other translation factors characteristic of eukarya are also found in archaea. They include the ribosome anti-association factor aIF6, the nucleotide exchange factor aEF1B and the peptide chain release factor ERF1 (Kyrpides and Woese, 1998a,b). However, archaea lack proteins homologous to eIF3, eIF4s or eIF5.
In eukarya and in archaea, the e/aIF2 factor results from the 1:1:1 association of three subunits: eIF2α, eIF2β and eIF2γ. Current knowledge of the contributions of each subunit in the eIF2 function is as follows. The large γ subunit is a G protein sharing sequence similarities with EF-Tu. It is thought to play an important role in initiator tRNA recruitment (Erickson and Hannig, 1996). A regulatory role of eIF2α is suggested by the observation that it is the target of an eIF2-specific protein kinase (Hinnebusch, 1993). The eIF2α subunit has also been implicated recently in the exchange of GDP for GTP (Nika et al., 2001). The function of eIF2β remains unclear. However, a putative zinc binding motif in this protein was proposed to be involved in mRNA binding (Castilho-Valavicius et al., 1992; Laurino et al., 1999).
Until now, biochemical and structural studies of e/aIF2 have been hampered by difficulty in obtaining large amounts of this protein. In the present study, we report the overproduction in Escherichia coli of each of the three subunits of aIF2 from the hyperthermophilic archaeon Pyrococcus abyssi. The subunits could be purified and assembled in vitro to reconstitute a stable aIF2 trimer. Crystals of the large γ subunit were obtained in the presence or absence of guanine nucleotides. The corresponding 3D structures show marked similarities to those of elongation factors EF-Tu and eEF1A. As a result, an aminoacyl-tRNA molecule could be readily docked at the surface of aIF2γ. Such a docking enables recognition of features of the protein that account for the specific recognition of initiator tRNA. Finally, we discuss the possibility that, by mimicking an elongation factor, aIF2 first directs Met-tRNA
![]() to the A site on the ribosome during the search for the initiation codon.
to the A site on the ribosome during the search for the initiation codon.
Results and discussion
Production and assembly of aIF2 subunits
Each subunit of the trimeric aIF2 protein from P.abyssi was produced in E.coli and purified independently. Subunits α (31.9 kDa) and β (16.6 kDa) could be obtained in large amounts reproducibly (>20 mg/l of culture). In the case of the γ subunit (44.8 kDa), a maximum of 3 mg of wild-type protein per litre of culture could be obtained. Possibly, in this case, overproduction was precluded because of some toxic effect in the E.coli context. However, a strain in which the aIF2γ protein accumulated up to 30 mg/l could be isolated. Interestingly, the resulting protein contained a G235D mutation. Such a higher rate of overexpression rendered the G235D mutant useful for biochemical and structural studies.
In vitro assembly of the three purified subunits was assessed by combining HPLC molecular sieving and SDS–PAGE analysis (Figure 1). A stable aIF2 trimer could be obtained in one step upon stoichiometric mixing of the subunits. This assembly was obtained regardless of the presence of the G235D mutation. Such a result indicates that coexpression of the three subunits in vivo is not a prerequisite for trimer formation. Pairwise mixings of subunits were also performed. aIF2γ produced a dimer with either aIF2α or aIF2β (Figure 1), whereas aIF2α did not form any stable dimer with aIF2β. These results indicate that the γ subunit is the core of the trimer. Such a conclusion is in good agreement with data obtained with the yeast system, showing that the β subunit can be removed from the eIF2 trimer by mild proteolysis and that eIF2βγ complexes are obtained in vivo in a strain lacking eIF2α (Flynn et al., 1993; Thompson et al., 2000; Erickson et al., 2001; Nika et al., 2001).
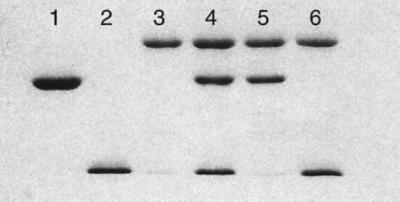
Fig. 1. SDS–PAGE analysis of aIF2 subunits. Lane 1, aIF2α (32 kDa); lane 2, aIF2β (16.6 kDa); lane 3, aIF2γ (45 kDa); lane 4, aIF2αβγ trimer; lane 5, aIF2αγ dimer; lane 6, aIF2βγ dimer. The 12.5% SDS–polyacrylamide gel was stained with Coomassie Blue. The samples in lanes 4–6 were obtained after purification by molecular sieving.
Finally, the zinc content of each subunit was measured by atomic absorption spectroscopy. Whereas the α subunit was devoid of zinc, each of the β and γ proteins contained one tightly bound zinc ion. The two zinc ions were retained in the βγ dimer as well as in the αβγ trimer. It is notable that integrity of a putative zinc binding cysteine cluster in the β subunit (C236, C239, C259 and C262) is essential to eIF2 activity in the yeast system (Donahue et al., 1988).
Overall structure of aIF2γ
Monoclinic crystals of aIF2γ were obtained using PEG8000 as the precipitating agent. The corresponding 3D structure was solved by combining isomorphous derivatives and anomalous scattering data (Table I). In the case of the free protein, two structures were refined. The first one corresponds to the selenomethionylated wild-type protein (1.95 Å resolution) and the second one to the G235D mutant (1.85 Å resolution). In both structures, the final model encompasses 400 of the 410 residues composing the protein. The 10 missing residues are 1–5, 37 and 224–227. Each model also shows one zinc ion and several water molecules (Table I). The two structures are identical within experimental error (r.m.s.d. = 0.7 Å for all 400 Cα atoms), except for the addition of an Asp side chain in the G235D mutant. This demonstrates that the mutation does not cause any conformational change, even locally.
Table I. Native and heavy atom derivative data used in the structure determination of eIF2γ.
| Crystal | G235D native | G235D KPtCl4 | Wild-type Se-Met | G235D GDP-Mg2+ | Wild-type GDP-Mg2+ | G235D GDPNP-Mg2+ | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wavelength (Å) | 0.934 | 1.0721 (peak) | 1.0725 (edge) | 0.9919 (remote) | 0.9795 (peak) | 0.9797 (edge) | 0.9184 (remote) | 0.934 | 0.934 | 0.934 |
| X-ray source | ID14eh1 | ID14eh4 | ID14eh4 | ID14eh4 | ID14eh4 | ID14eh4 | ID14eh4 | ID14eh1 | ID14eh1 | ID14eh1 |
| Unique reflections | 40474 | 11032 | 11039 | 10957 | 34217 | 24180 | 24164 | 27549 | 36551 | 44154 |
| Resolution (Å) | 1.85 | 2.8 | 2.8 | 2.8 | 1.95 | 2.2 | 2.2 | 2.1 | 1.9 | 1.8 |
| Completeness (%) | 99.1 (98.4) | 98.6 (97.9) | 98.6 (97.9) | 98.0 (97.9) | 98.9 (89.4) | 99.9 (100) | 99.9 (100) | 99.8 (99.6) | 97.7 (97) | 99.9 (100) |
| Anomalous data (%) | 98.0 (99.3) | 98.1 (99.4) | 94.8 (99.2) | 93.2 (75.4) | 95.7 (94.5) | 95.6 (94.6) | ||||
| Redundancy | 4.9 | 3.7 | 3.7 | 3.7 | 3.5 | 3.6 | 3.6 | 3.8 | 3.7 | 3.7 |
| Rsym (I) (%)a | 4.9 (32.4) | 2.0 (3.9) | 2.1 (4.4) | 2.6 (5.0) | 4.0 (21.9) | 3.7 (15.4) | 4.0 (23.5) | 5.7 (30.4) | 3.8 (26.4) | 4.3 (30.4) |
| Mean figure of merit | 0.47 (30–2.2 Å) | |||||||||
| R/Free-Rb (%) | 22.5/25.5 | 21.9/25.2 | 21.9/23.9 | 21.5/23.0 | 21.9/24.5 | |||||
| R.m.s.d. bonds (Å)/angles (°) | 0.0051/1.3 | 0.0057/1.5 | 0.0061/1.4 | 0.007/1.5 | 0.0057/1.3 | |||||
| B values (Å2) protein/water/nucleotide | 39.2/43.8/– | 38.0/43.5/– | 35.6/43.2/30.1 | 27.8/36.5/18.8 | 27.3/34.9/19.7 |
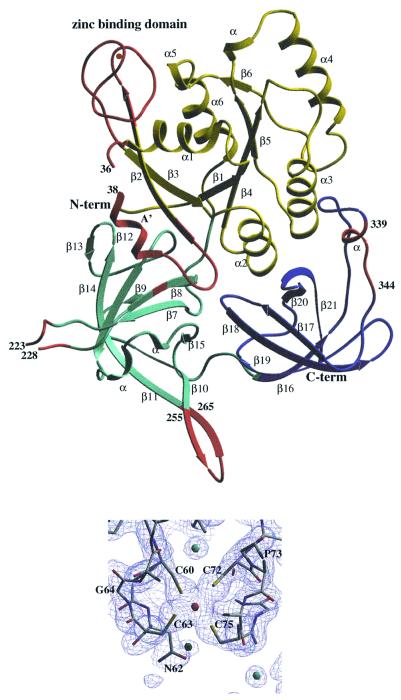
The 3D structure of nucleotide-free aIF2γ shows three domains closely similar to those of EF-Tu or eEF1A (LaCour et al., 1985; Berchtold et al., 1993; Nissen et al., 1995; Andersen et al., 2001). The first domain (coloured yellow in Figure 2), corresponds to the guanine nucleotide binding site. It encompasses residues 1–205. R.m.s. deviation between this domain and the corresponding one in EF-Tu–GDPNP (Berchtold et al., 1993; Nissen et al., 1995) is 1.74 Å for 138 Cα atoms compared. The second domain (light blue in Figure 2) contains residues 206–308. It is superimposable on domain 2 of EF-Tu with an r.m.s.d. of 1.65 Å for 89 Cα atoms compared. The third domain (residues 309–410, dark blue in Figure 2) is comparable to domain 3 of EF-Tu. The r.m.s.d. is 1.50 Å for 74 Cα atoms compared.
Fig. 2. Top: ribbon representation of P.abyssi aIF2γ. The three structural domains are coloured as follows: domain 1 (residues 6–205), yellow; domain 2 (residues 206–308), light blue; and domain 3 (residues 309–410), dark blue. Secondary structure elements are labelled according to Figure 4. The region between β2 and β3 corresponding to the idiosyncratic zinc binding domain (residues 58–80) is coloured in red. Four other regions characteristic of aIF2γ are also coloured in red: switch 1 in domain 1 (residues 32–50, comprising the A′ helix), the mobile loop (residues 223–228), residue 235 within β8 and the β hairpin (residues 255–265) in domain 2 and the small inserted helix in domain 3 (residues 339–344). The figure was drawn with Setor (Evans, 1993). Bottom: close-up view of the 2Fo–Fc electron density map, contoured at 1.2σ, in the region of the zinc binding domain.
Relative arrangement of domains in aIF2γ
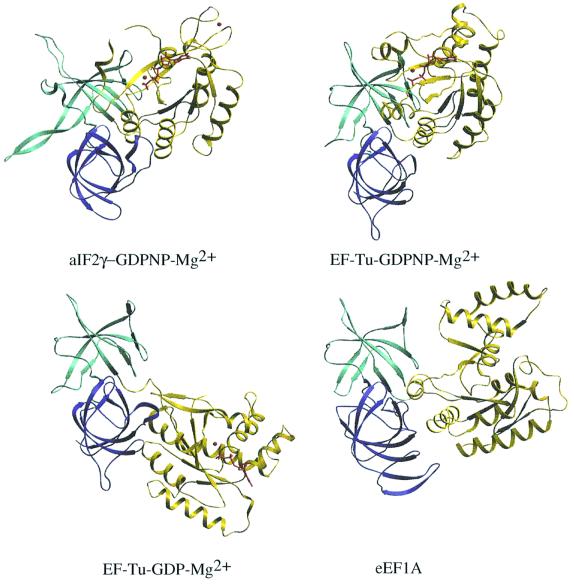
In elongation of translation, a ternary complex composed of EF-Tu (e/aEF1A in eukarya and archaea), GTP and aminoacyl-tRNA is involved. This complex drives the tRNA to the ribosomal A site. After correct codon– anticodon pairing, GTP hydrolysis induces release of EF-Tu–GDP from the ribosome. This cycle results from different conformations of EF-Tu–GDP and EF-Tu–GTP. The conformational changes involve relative motions between domains of the protein, as well as concerted motions of two regions called switch 1 and switch 2. Switch 1 contains the effector region, thought to interact with the ribosome (Zeidler et al., 1996; Clark and Nyborg, 1997). The relative orientations of domains 2 and 3 are identical in the two EF-Tu states, and these two domains can be considered as a single structural unit. On the other hand, the orientation of domain 1 relative to domains 2 and 3 varies strongly in the GTP- and GDP-complexed forms of EF-Tu (LaCour et al., 1985; Berchtold et al., 1993; Polekhina et al., 1996).
Binding of GDP-Mg2+ to aIF2γ was examined in the cases of the wild-type protein and of the G235D mutant. Again, the two refined structures were found to be identical. Finally, the binding of GDPNP-Mg2+ was studied with the G235D mutant (Table I). Notably, in all nucleotide-free or nucleotide-bound aIF2γ structures, the relative orientations of the three domains remained identical (Table I). In the structure of ligand-free aIF2γ as well as in those of the GDP- and GDPNP-complexed proteins, domains 2 and 3 are packed against domain 1. This organization contrasts with that of EF-Tu–GDP, where a hole separates domain 1 from domain 2. Domains 2 and 3 of aIF2γ could be superimposed together on the corresponding domains of EF-Tu (r.m.s.d. = 1.6 Å for 147 Cα atoms). Upon such a superimposition, domain 1 in aIF2γ is rotated by only 14° compared with domain 1 in EF-Tu–GDPNP. A shift of 84° is observed if the comparison is made with domain 1 of EF-Tu–GDP (Figure 3). Therefore, whatever its complexed form, aIF2γ resembles much more an active EF-Tu–GDPNP state than an inactive EF-Tu–GDP state.
Fig. 3. Comparison of domain orientations in GDPNP-Mg2+-complexed aIF2γ and in elongation factors EF-Tu (Berchtold et al., 1993; Polekhina et al., 1996) and eEF1A (Andersen et al., 2001). The two EF-Tu structures shown are those containing GDPNP-Mg2+ or GDP-Mg2+. The structure of eEF1A is drawn from that of the eEF1A–eEF1B complex. Domains 2 and 3 (light and dark blue) in the four structures were superimposed. Each protein is represented according to the resulting orientation. Nucleotides and Mg2+ ions are drawn in red. As described in the text, the aIF2γ–GDPNP-Mg2+ structure has the same domain orientations as those of aIF2γ–GDP-Mg2+or nucleotide-free aIF2γ.
Specific features of the initiation factor 2
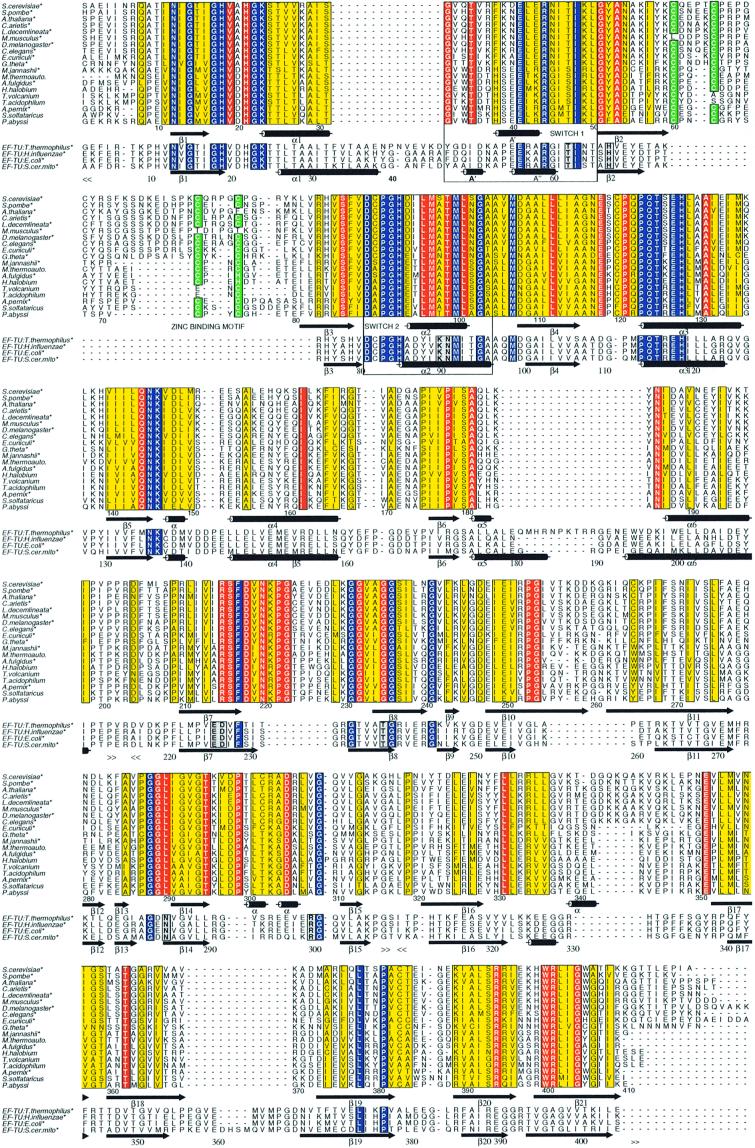
The above superimpositions, together with the sequence alignments deduced from them (Figure 4), enabled us to identify structural features specific to e/aIF2γ proteins or shared with elongation factors. In domain 2, two loops distinguish the e/aIF2γ proteins from EF-Tu. One loop (residues 220–228 between β7 and β8) is disordered from residue 224 to residue 227. It clearly protrudes out of the core of the barrel. The second insertion (residues 255–265 within β10 and β11) is a β-hairpin structure extending outside the barrel (Figures 2 and 4). A role for these two long insertions in the anchoring of the α and/or β subunits can be envisaged. In domain 3, an insertion corresponding to residues 340–343 of aIF2γ is systematically encountered in e/aIF2γ sequences. Another loop characteristic of elongation factors, between β18 and β19 strands, is shortened in e/aIF2γ proteins. The overall topology of the aIF2γ G domain is as described in EF-Tu, with a central β-sheet surrounded by α-helices. α2, α3 and α4 are on one side of the sheet, and α1 and α6 are on the other. In EF-Tu, a third helix, α5, is present on the α1α6 side. Actually, this region, which links β5 to α6, is reduced to a short turn of helix in aIF2γ.
Fig. 4. Multiple sequence alignments of eukaryotic and archaeal eIF2γ and of prokaryotic EF-Tu. The secondary structure elements schematized at the bottom of each block of sequences correspond to either those of P.abyssi aIF2γ or those of the Thermus thermophilus EF-Tu ternary complex (Nissen et al., 1995). The borders of structural domains are indicated by << and >>. Numbering of the upper block of sequence refers to P.abyssi aIF2γ and that of the lower block to T.thermophilus EF-Tu. Residues conserved in all proteins compared are boxed in blue. Strict conservations within the e/aIF2γ sequences only are boxed in red, whereas conservative replacements are boxed in yellow. Cysteines corresponding to the zinc knuckle are boxed in green. The switch 1 and switch 2 regions are boxed and labelled. The positions of residues of elongation factors discussed in the text are in grey boxes. Amino acid sequences were obtained from the SwissProt database. Alignment using the Clustal X program (Thompson et al., 1997) was refined manually. For the sake of clarity, some sequences have been truncated at the N-terminus. These are marked with an asterisk. The sequence of Encephalitozoon cuniculi eIF2γ lacks five residues at the C-terminus. The figure was drawn with Alscript (Barton, 1993).
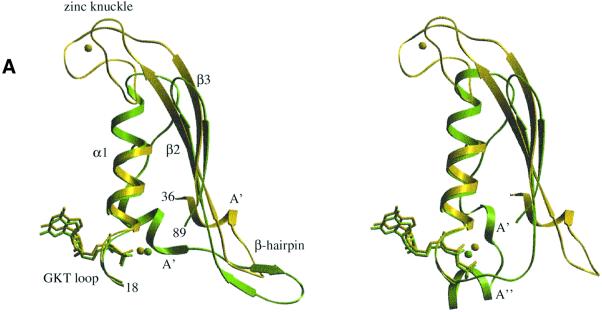
Downstream from the β2 strand, aIF2γ shows a four cysteine cluster (C60, C63, C72 and C75). This motif forms a metal-binding knuckle protruding outside of the G domain (Figures 2 and 5). It occupies the same spatial position as the C-terminal part of helix α1 in EF-Tu. Electronic density clearly showed the presence at this place of a tetra-coordinated metal (Figure 2). Atomic absorption spectroscopy measurements unambiguously identify this metal as a zinc. Moreover, alignment of the e/aIF2γ sequences in this region strongly indicates a zinc knuckle in the initiation factors from most organisms. In some cases, the zinc might have been lost but the knuckle conformation is likely to be conserved (Figure 4).
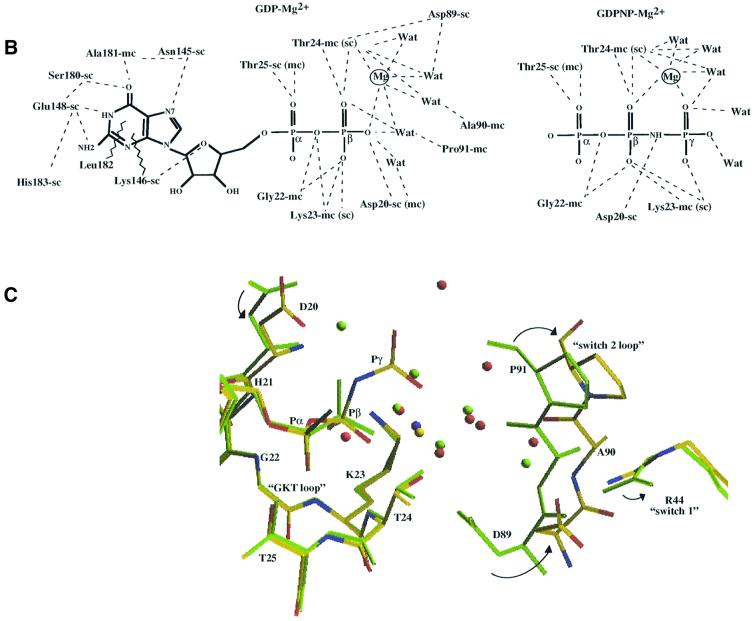
Fig. 5. (A) Left: superimposition of the effector region in aIF2γ–GDP-Mg2+ (yellow) with that in EF-Tu–GDP-Mg2+ (green). Right: same with the GTP-Mg2+ complexes. Secondary structure elements of aIF2γ are labelled according to Figure 4 and numbering refers to aIF2γ. (B) Schematic diagram showing the hydrogen bonding interactions between aIF2γ and GDP-Mg2+ (left) or GDPNP-Mg2+ (right). Only the three phosphates of GDPNP are drawn, since interactions with the nucleoside are identical to those with the guanosine of GDP. An ‘sc’ index after a residue number means that a side-chain atom is involved. ‘mc’ designates the involvement of a main-chain atom. ‘Wat’ indicates a water molecule. For the sake of clarity, only water molecules belonging to the coordination shell of Mg2+ are represented. Broken lines symbolize stacking interactions. (C) Superimposition of aIF2γ–GDPNP (yellow sticks) and aIF2γ–GDP (green sticks) in the region where the phosphates bind. The yellow and blue balls represent Mg2+ bound to GDPNP or GDP, respectively. Red and green balls are water molecules bound to aIF2γ–GDPNP or aIF2γ–GDP structures, respectively. This view illustrates the movement of the switch 2 loop upon GDPNP binding.
The functional importance of this zinc-binding motif has already been highlighted by site-directed mutagenesis experiments. In yeast, substitution by S of C155, the second cysteine ligand of the first motif in the eIF2γ knuckle (C63 according to the P.abyssi numbering), causes extremely slow growth at 30°C as well as temperature sensitivity at 37°C (Erickson et al., 1997). In contrast, mutation of C160 (T68 in P.abyssi), which is not predicted to be a zinc ligand according to the alignment made in the present study (Figure 4), had no effect on growth rate. A role for this zinc-containing specific extension in the binding of aIF2γ to aIF2α and/or aIF2β may be suspected.
Binding of guanine nucleotides to aIF2γ
The binding pocket of GDP-Mg2+ in aIF2γ is superimposable on that in EF-Tu. The regions of aIF2γ involved in the binding of GDP (Figure 5) are the 22GKT24 loop, the switch 2 loop (residues 89–91), the sequences 144QNKIE148 beyond β5 and 180SALH183 corresponding to the small α5 helix. The two latter regions surround the ring of the nucleotide and ensure specificity for guanine (Figure 5). The magnesium ion is held between T24 of the GKT loop and the β phosphate of GDP. Hexa-coordination of the metal is completed by four water molecules. The shell of these water molecules is contributed by the GKT loop and the switch 2 loop. The four regions forming the nucleotide binding cavity move significantly in reaction to GDP-Mg2+ binding. The GKT-loop region becomes better ordered. As a result, the 119–123 region, which interacts with the GKT loop, also changes conformation. The most relevant feature is a 180° switch of the carbonyl group of P91. This switch gives rise to motion of the surrounding region (from D89 to G92). This movement further allows interaction of the carbonyl groups of A90 and P91, as well as of the side chain of D89, with the magnesium ion via water molecules (Figure 5).
When GDPNP-Mg2+ is used instead of GDP-Mg2+, the above switch 2 region recovers its conformation in the nucleotide-free aIF2γ structure (Figure 5). If not, steric hindrance between the carbonyl group of P91 and the γ phosphate of GDPNP-Mg2+ would have occurred. This phosphate replaces a water molecule in the coordination shell of the metal ion. Magnesium therefore bridges the β and γ phosphates and three water molecules.
In EF-Tu, the switch 1 motif is composed of an A′ helix and a β hairpin in the GDP state (Figure 5). In the GTP state, it switches to an A′ plus A′′ helices conformation (Figures 4 and 5). In aIF2γ, the conformation of the switch 1 region is not modified upon GDP or GDPNP binding. It keeps a GDP-like conformation compared with EF-Tu (Figure 5). This behaviour contrasts with the large conformational changes and domain rearrangements observed upon transition between the GDP and GTP states of EF-Tu. In EF-Tu, these movements were proposed to originate from T62 (switch 1) and G84 (switch 2), which both interact with the γ phosphate of GDPNP (Berchtold et al., 1993; Polekhina et al., 1996). In aIF2γ, both residues (T47 and G92) are conserved. However, they are not implicated in GTP binding, and are therefore unable to trigger any movement in the protein.
As noted above, in the ligand-free eIF2γ structure, the three domains are organized in an ‘active state’ conformation. All three are closely packed one to each other (Figure 3). Notably, residues belonging to the switch 1 and 2 regions are involved in this packing. The side chain of T47 (effector region) interacts with T359 (main chain, domain 3). In addition, the side chain of R334 makes contacts with those of H93, E94 (switch 2) and E127. E127 belongs to a conserved sequence (N-terminus of α3), which interacts with the GKT loop when the nucleotide is present. Additional interactions, such as those of R215 (domain 2) with main-chain groups of L42 and R44 (switch 1), contribute to the relative orientations of the three domains. Stability of the resulting network may account for the absence of motion of the switch 1 region upon nucleotide binding. In addition, the involvements of H93 and E94 in interdomain contacts could explain a relatively small displacement of the switch 2 region upon binding of either GTP or GDP.
Considering the limited motions of switch 1 and switch 2, it is difficult to explain the GTP dependence of tRNA binding by an eIF2 trimer, as demonstrated in eukarya (Trachsel, 1996). At this stage, the possibility that either crystal packing interactions or the absence of the two other subunits of aIF2 account for a restricted movement of the two switch regions cannot be excluded. However, we observed that: (i) prolonged (several days) soaking of the native crystals in the presence of the nucleotide did not cause cracking; and (ii) crystal growth in the presence of one or the other nucleotide yielded structures identical to those derived from soaked crystals. To better assess this question, crystallization in another space group would be necessary. At this stage, whatever the answer, it is reasonable to assume that the relatively small conformational change of switch 2 upon GDP binding gets aIF2γ ready for further rearrangements in the reaching of its ‘inactive state’. Full expression of this state might require the presence of the α and/or β subunits.
Functional relationships between aIF2γ and elongation factors
In the EF-Tu–GTP–tRNA complex, two sets of interactions occur between the tRNA molecule and the protein (Nissen et al., 1995). On the one hand, the TΨC stem domain of the polynucleotide is in contact with domain 3 of the protein. However, base-specific interactions are not detectable between these two regions, in agreement with the large diversity of elongator tRNAs accommodated by EF-Tu. On the other hand, the aminoacylated acceptor stem follows a channel made on one side by the effector region and on the other by domain 2 of the protein. Tight interactions involve the 5′-phosphate group of the tRNA and three residues universally conserved in elongator G proteins: K90, N91 and R300 (according to EF-Tu numbering, Figure 4). Base 76 and the esterified amino group fill up a pocket large enough to accommodate any side chain. This pocket is made by residues D227, E226, F229 and T239 (Nissen et al., 1995).
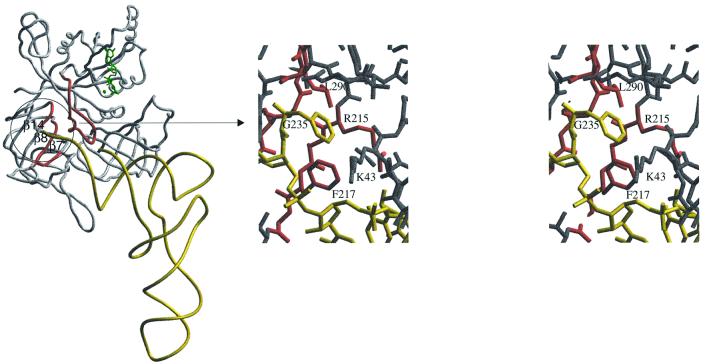
Upon superimposition of domains 2 and 3 of the EF-Tu–GTP–Phe-tRNAPhe complex on the corresponding domains in aIF2γ–GDPNP, the Phe-tRNAPhe molecule could be readily positioned at the surface of the initiation factor subunit. In the resulting model (Figure 6), almost no clash is observed between Phe-tRNAPhe and aIF2γ–GDPNP, although, as noted above, the G domain of aIF2γ differs from that of EF-Tu by a rotation of 14°. A notable exception is a clash between the side chain of the esterified Phe and that of L290 (Figure 6).
Fig. 6. Left: Phe-tRNAPhe from the EF-Tu ternary complex (Nissen et al., 1995) docked on GDPNP-complexed aIF2γ. The nucleotide is shown in green. Three β strands forming the putative methionine binding pocket are labelled. Ribbon regions 215–218, 231–236 and 288–290 containing signature sequences of e/aIF2γ are coloured in red. The switch 1 region, which borders the 3′ end of the docked tRNA, is also coloured in red. Right: close-up stereo view of the putative methionine binding pocket. The aminoacyl moiety of the 3′ end of the docked Phe-tRNAPhe (yellow) lies in a pocket delineated by R215, F217 (β7), G235 (β8) and L290 (β14). Residues coloured in red are strictly conserved in e/aIF2γ sequences (see Figure 4).
One function of eIF2γ is to distinguish only the methionylated initiator tRNAMet among any aminoacylated tRNA. To achieve this goal, eIF2 displays some specificity towards the methionyl moiety of the charged initiator tRNA (Wagner et al., 1984; Drabkin and RajBhandary, 1998). Specific features of the nucleotide sequence of the initiator tRNA itself have also to be recognized by the factor. Accordingly, the first base pair of the acceptor stem (A1–U72), which is almost universally conserved in initiator tRNAs of eukaryotic or archaeal origin, was reported to be important for eIF2 binding (Astrom et al., 1993; Farruggio et al., 1996). The present model indicates that aIF2 may specifically dock Met-tRNA
![]() by using the same scaffold as EF-Tu. In our docking model (Figure 6), the β hairpin in the effector region of aIF2γ comes very close to the end of the acceptor stem. Recognition and possible disruption (Basavappa and Sigler, 1991) of the A1–U72 pair of initiator tRNA by residues contributed by the effector region can therefore be envisaged. Such a hypothesis is compatible with the behaviour of two mutants of yeast eIF2γ, Y142H (F52 within β2 in P.abyssi) and N135K (G45 of switch 1 in P.abyssi). With these mutants, the capacity to bind Met-tRNA
by using the same scaffold as EF-Tu. In our docking model (Figure 6), the β hairpin in the effector region of aIF2γ comes very close to the end of the acceptor stem. Recognition and possible disruption (Basavappa and Sigler, 1991) of the A1–U72 pair of initiator tRNA by residues contributed by the effector region can therefore be envisaged. Such a hypothesis is compatible with the behaviour of two mutants of yeast eIF2γ, Y142H (F52 within β2 in P.abyssi) and N135K (G45 of switch 1 in P.abyssi). With these mutants, the capacity to bind Met-tRNA
![]() is lost (Erickson and Hannig, 1996; Huang et al., 1997). According to the Phe-tRNAPhe docking, the cavity of aIF2γ accommodating the methionine side chain would be formed by the three β7, β8 and β14 strands. In this pocket, stretches of residues specific to initiation factors can be recognized. The first stretch corresponds to 215RSFDΦN220 (β7, where Φ is a hydrophobic residue). Two glycine-rich motifs form the second stretch: 231GGΦΦGG236 in β8 and 288GGLΦ(G,A)Φ(G,A)T295 in β14. Interestingly, the G235D mutation, which allowed higher overproduction of aIF2γ, affects the third glycine in the GGΦΦGG signature. It can be imagined that this mutation decreases the affinity of the initiation factor for Met-tRNA
is lost (Erickson and Hannig, 1996; Huang et al., 1997). According to the Phe-tRNAPhe docking, the cavity of aIF2γ accommodating the methionine side chain would be formed by the three β7, β8 and β14 strands. In this pocket, stretches of residues specific to initiation factors can be recognized. The first stretch corresponds to 215RSFDΦN220 (β7, where Φ is a hydrophobic residue). Two glycine-rich motifs form the second stretch: 231GGΦΦGG236 in β8 and 288GGLΦ(G,A)Φ(G,A)T295 in β14. Interestingly, the G235D mutation, which allowed higher overproduction of aIF2γ, affects the third glycine in the GGΦΦGG signature. It can be imagined that this mutation decreases the affinity of the initiation factor for Met-tRNA
![]() .
Indeed, in the docking model, a D at position 235 would cause steric hindrance or electrostatic repulsion with the esterified amino acid (Figure 6). Under the assumption that the toxicity associated with overexpression of the factor in E.coli reflects non-specific sequestration of some E.coli aminoacylated tRNAs by the archaeal protein, the origin of the G235D mutant now becomes explicable. Indeed, because of a reduced capacity to bind some aminoacyl-tRNAs, possibly the Met-tRNA
.
Indeed, in the docking model, a D at position 235 would cause steric hindrance or electrostatic repulsion with the esterified amino acid (Figure 6). Under the assumption that the toxicity associated with overexpression of the factor in E.coli reflects non-specific sequestration of some E.coli aminoacylated tRNAs by the archaeal protein, the origin of the G235D mutant now becomes explicable. Indeed, because of a reduced capacity to bind some aminoacyl-tRNAs, possibly the Met-tRNA
![]() one, this mutant would have occurred spontaneously.
one, this mutant would have occurred spontaneously.
Two features accounting for the capacity of elongation factors to bind aminoacyl-tRNAs non-specifically are absent from the e/aIF2γ sequences. First, three residues (K90, N91 and R300 according to the Thermus thermophilus EF-Tu numbering) involved in the clamping of the 5′ phosphate group of any elongator tRNA have no equivalent in e/aIF2γ. Instead, electroneutral residues are systematically encountered at corresponding positions in e/aIF2γ proteins (see T98, T99 and A308 in Figure 4). Secondly, region 271–290, which in EF-Tu establishes main-chain contacts with the α amino group and the ester bond of Phe-tRNAPhe, shows a different conformation in aIF2γ. Therefore, aIF2γ possesses distinctive features that should contribute to the exclusion of elongator aminoacyl-tRNAs.
Concluding remarks
In the present study, the three subunits of aIF2 could be obtained separately and mixed in vitro to reconstitute a trimer. By assembling couples of subunits, αγ and βγ dimers were also obtained. The α and β subunits did not react together. Consequently, it can be concluded that the γ subunit forms the structural core of the trimer.
Close 3D structural similarity between aIF2γ and EF-Tu was established. It suggests that inside aIF2 and possibly eIF2, the core γ protein has the capacity to bind both the initiator tRNA and the guanine nucleotide. In agreement with this proposal, yeast can grow in the absence of eIF2α provided that the β and γ subunits as well as the initiator tRNA are all overproduced together (Erickson and Hannig, 1996; Erickson et al., 2001; Nika et al., 2001).
In eukarya, eIF2–GTP–Met-tRNA
![]() plus eIF1, eIF3 and eIF5 make part of a preinitiation complex that scans mRNA to reach the AUG start codon (Asano et al., 2000). It has been proposed that, upon pairing between the tRNA anticodon and the AUG codon, the GTPase activity of eIF2γ is triggered with the help of the eIF5 factor. The ensuing release of eIF2–GDP would eventually leave the initiator tRNA in the ribosomal P site (Huang et al., 1997). Archaeal aIF2 probably plays the same role as that of the eukaryal factor, with the difference however, that its GTPase activity must be exempted from the presence of an eIF5 analogue (Kyrpides and Woese, 1998a; Thompson et al., 2000). Indeed, archaea lack proteins homologous to eIF5 or eIF3. Such a difference is possibly related to the Shine–Dalgarno-driven pre-positioning of the initiation complex in archaea (Dennis, 1997). Such a pre-positioning may be followed by ‘local scanning’ by the ribosome until correct pairing with AUG is established.
plus eIF1, eIF3 and eIF5 make part of a preinitiation complex that scans mRNA to reach the AUG start codon (Asano et al., 2000). It has been proposed that, upon pairing between the tRNA anticodon and the AUG codon, the GTPase activity of eIF2γ is triggered with the help of the eIF5 factor. The ensuing release of eIF2–GDP would eventually leave the initiator tRNA in the ribosomal P site (Huang et al., 1997). Archaeal aIF2 probably plays the same role as that of the eukaryal factor, with the difference however, that its GTPase activity must be exempted from the presence of an eIF5 analogue (Kyrpides and Woese, 1998a; Thompson et al., 2000). Indeed, archaea lack proteins homologous to eIF5 or eIF3. Such a difference is possibly related to the Shine–Dalgarno-driven pre-positioning of the initiation complex in archaea (Dennis, 1997). Such a pre-positioning may be followed by ‘local scanning’ by the ribosome until correct pairing with AUG is established.
Proof-reading of the pairing between the anticodon of a tRNA and a codon on mRNA followed by the triggering of GTPase activity is reminiscent of the functioning of EF-Tu or of e/aEF1A. It is possible that in the decoding of the AUG start codon, because of its resemblance to an elongation factor, e/aIF2 utilizes the same decoding site on the ribosome as the one used by e/aEF1A. This site involves the universally conserved A1492 and A1493 of 16S rRNA (Ogle et al., 2001; Yusupov et al., 2001). A working hypothesis would be that an initiator tRNA carried by e/aIF2–GTP is first positioned at the ribosomal A site while searching for the initiation codon. Upon successful pairing, GTP hydrolysis would be committed and e/aIF2–GDP immediately released. In this context, e/aIF5B should play the role of a translocase by occupying the A site after GTP hydrolysis and shifting the initiator tRNA already paired to the initiation codon towards the P site. Credit is given to this idea by the 3D structure of aIF5B, the overall dimensions of which are very similar to those of EF-G or of an EF-Tu–GTP–tRNA ternary complex (Roll-Mecak et al., 2000). aIF5B lacks the N-terminal extension present in eIF5B. However, it is able to substitute for its yeast homologue both in vivo and in vitro (Lee et al., 1999). At this step, the 60S subunit would join the 48S initiation complex and form the 80S elongator ribosome. Although speculative, this model is supported by the recent proposal that an e/aIF1A–e/aIF5B complex can bind to the A site while stabilizing Met-tRNA
![]() association with the ribosomal P site (Choi et al., 2000; Roll-Mecak et al., 2000). It was also shown that eIF5B is essential to the joining of ribosomal subunits after GTP hydrolysis by eIF2 (Pestova et al., 2000).
association with the ribosomal P site (Choi et al., 2000; Roll-Mecak et al., 2000). It was also shown that eIF5B is essential to the joining of ribosomal subunits after GTP hydrolysis by eIF2 (Pestova et al., 2000).
The above ideas offer an explanation for the observation that both eIF2 and eIF5B are required to ensure efficient initiation of translation. Further studies will be necessary to assess such ideas. In particular, the α and β subunits of the initiation factor will have to be functionally integrated. Hopefully, the 3D model of aIF2γ, as presented here, may stimulate such studies and contribute to the solving of the mechanism of translation initiation in both eukarya and archaea.
Materials and methods
Expression and purification of the aIF2 subunits
The genes encoding the α, β and γ subunits of P.abyssi aIF2 were amplified by PCR and cloned in a modified version of pET3a. The resulting vectors were called, respectively, pET3aαpa, pET3aβpa and pET3aγpa. For the expression of α or β subunit, the vectors were introduced into E.coli BL21 cells together with the tRNA4Arg-overexpressing pSBETa plasmid (Schenk et al., 1995). This tRNA can read the rare AGA and AGG codons. Cultures were 500 ml in 2× TY (1.6% tryptone, 1% yeast extract, 0.5% NaCl pH 7.0) containing 100 µg/ml of ampicillin and 100 µg/ml of kanamycin. Plasmid pET3aγpa was cotransformed with pSJS1240 (Dr S.Sandler), which overexpresses two tRNA species, allowing the decoding of the AGA, AGG (Arg) and ATA (Ile) rare codons. Cultures were 500 ml in 2× TY containing 100 µg/ml of ampicillin and 30 µg/ml of spectinomycin. In all cases, expression was induced by adding 1 mM of isopropyl-β-d-thiogalactopyranoside when OD650 reached 1.6. Culture was then continued for 12 h at room temperature.
Purification of aIF2γ
Overproducing cells corresponding to 500 ml of culture were disrupted by sonication in 40 ml of buffer A (500 mM NaCl, 10 mM MOPS pH 6.7, 10 mM 2-mercaptoethanol). After centrifugation, the supernatant was heated for 10 min at 80°C. Thermostable proteins were treated with 1% w/v streptomycin sulfate and the precipitate removed. Finally, proteins were precipitated with ammonium sulfate to 80% saturation. The pellet was dissolved in 10 ml of buffer A and dialysed for 1 h against the same buffer. This sample was then loaded on to a Q-Hiload column (16 mm × 20 cm; Amersham) equilibrated in buffer A. The protein was recovered in the flow-through fraction (FT) and dialysed against buffer B (100 mM NaCl, 10 mM MOPS pH 6.7, 10 mM 2-mercaptoethanol). The sample was then loaded on to an S-Sepharose column (11 mm × 5 cm; Amersham) equilibrated in buffer B. The protein was recovered in the FT and loaded directly on to a MonoQ column (5 mm × 5 cm; Amersham) equilibrated in buffer B. The FT containing the protein was concentrated by ammonium sulfate to 80% saturation. The pellet was resuspended and dialysed against buffer C (200 mM NaCl, 10 mM MOPS pH 6.7, 10 mM 2-mercaptoethanol). The concentrated sample (5 ml) was finally loaded on to a Superdex75 column (16 mm × 60 cm; Amersham) equilibrated in buffer C. The protein eluted in a single peak and was homogeneous as judged by SDS–PAGE analysis. About 3 mg protein, per litre of culture were obtained in the case of the wild-type proteins, whereas >20 mg/l of culture could be obtained with the G235D mutant. For the preparation of selenomethionylated protein, plasmids pET3aγpa and pSJS1240 were co-transformed into the methionine auxotroph B834-De3 (Novagen). Transformed cells were grown overnight in 30 ml of 2× TY medium. This starter culture was harvested and cells were washed with and resuspended in 30 ml of M9 minimal medium. A 2.5 ml aliquot of this cell suspension were used to inoculate 1 l of M9 minimal medium containing 15 mg of selenomethionine (Sigma), 0.5% glucose, 100 µg/ml of ampicillin and 30 µg/ml of spectinomycin. After 1 week of culture, cells were harvested at an OD650 of 0.3. Purification was performed as above; 100 µg of protein were obtained.
Purification of aIF2α
The protocol used for aIF2α purification was identical to that for aIF2γ until the first chromatographic step. After the Q-Hiload column, the FT was recovered and dialysed against a buffer containing 300 mM NaCl, 10 mM MOPS pH 6.7 and 10 mM 2-mercaptoethanol. The dialysed sample was then loaded on to a MonoS column (5 mm × 5 cm; Amersham) equilibrated in the same buffer. The sample was eluted by a gradient from 300 mM to 0.8 M NaCl in 15 ml. The eluted peak was further loaded on to a Superdex75 column (16 mm × 60 cm; Amersham) equilibrated in buffer A. Routinely, 12 mg of homogeneous protein were obtained from a 500 ml culture (in 2× TY containing 100 µg/ml ampicillin and 25 µg/ml kanamycin).
Purification of aIF2β
The protocol used for aIF2β purification was the same as that used for the γ subunit purification except that buffer B was replaced by buffer C. From a 500 ml culture, 12 mg of purified protein were obtained.
Molecular sieving and atomic absorption spectroscopy
The purified subunits were mixed (in the 10 µM range) pairwise or all three together in 10 mM MOPS pH 6.7, 10 mM 2-mercaptoethanol, 500 mM NaCl. Each mixture (αβ, βγ, αγ, αβγ) was chromatographed on to a TSK3000SWXL column (Tosohaas, Japan, 7.8 × 300 mm2) operated at 0.7 ml/min in the same buffer. From calibration curves, unambiguous assembly was obtained with the βγ, αγ and αβγ combinations, whereas no association of α with β was detectable. Subunit associations were confirmed by SDS–PAGE analysis of the eluted samples (Figure 1).
Zinc atomic absorbancies were followed at a wavelength of 213.9 Å, using a Varian SpectrAA220 spectrophotometer (Mayaux and Blanquet, 1981). Before metal analysis, the protein being studied was dialysed extensively against a buffer containing 20 mM Tris–HCl pH 7.5, 10 mM 2-mercaptoethanol, 500 mM KCl and 0.1 mM EDTA. Zinc contents of the βγ, αγ dimers and of the aIF2 trimer purified from the TSK3000SWXL column were also measured. Values of extinction coefficients deduced from the amino acid compositions of the subunits are: α, 1.2 cm2/mg; β, 0.6 cm2/mg; and γ, 0.6 cm2/mg. However, in the case of the β subunit, a coefficient of 1.1 cm2/mg had to be adopted to be compatible with a 1:1 stoichiometry with either zinc or the γ subunit.
Crystallization of aIF2γ
Initial screening for crystallization conditions of aIF2γ was performed with the hanging drop technique, using protein at 15 mg/ml in buffer C and the commercial Crystal Screen (Hampton Research). Crystals were obtained within 2 days using 20% PEG10K and 0.1 M HEPES pH 7.5. Optimization of crystal growth could be undertaken by varying the mol. wt of PEG, its concentration and the pH. Optimized conditions were eventually defined as 8–18% PEG8000 and 0.1 M Tris–HCl pH 8.5. Microseeding was used to obtain large crystals (1 × 0.3 × 0.1 mm) more reproducibly. Crystals obtained using these conditions belonged to space group P21 with cell parameters of a = 51.5 Å, b = 86.7 Å, c = 58.1 Å and β = 110.4°. Crystals of Se-Met protein were obtained by microseeding using native crystals in the presence of 15% or 18% PEG8000, 0.1 M Tris–HCl pH 8.5, and protein at 5 mg/ml. Binding of guanine nucleotides could be achieved by soaking native crystals in a stabilizing solution containing 10 mM GDPNP-Mg2+ or GDP-Mg2+. Moreover, crystals could also be obtained by microseeding protein solutions in the presence of the chosen nucleotide. Structures of the nucleotide-complexed proteins were found to be identical whichever method was used to obtain the crystals.
Data collection and processing
For data collection, crystals were soaked for 1 min in 18% PEG8000, 0.1 M Tris–HCl pH 8.5 containing 25% glycerol as cryoprotectant and flash-cooled in liquid ethane. A platinum derivative was prepared by soaking native crystals in 15% PEG8000 and 0.1 M Tris–HCl pH 8.5, containing 20 mM potassium tetrachloroplatinate. Data were then collected at 100K using synchrotronic sources at the ESRF (ID14eh1 and ID14eh4 beamlines, Grenoble, France) or at the LURE (DW32 beamline, Orsay, France). Diffraction images were analysed using Mosflm (A.G.W.Leslie, Laboratory of Molecular Biology, Daresbury, UK). Data were processed further with programs of the CCP4 package (Collaborative Computational Project No. 4, 1994).
Structure determination and refinement
Two three-wavelength datasets corresponding to the Se-Met and platinum derivatives were collected at the ID14eh4 beamline (ESRF, Grenoble, France). The native and derivative datasets were then input to the program SOLVE (Terwilliger and Berendzen, 1999) with the help of which the two platinum sites and the expected eight selenium sites were found and refined. These sites were further used to phase the reflections and to compute initial maps with an average figure of merit of 0.47 for data to 2.2 Å resolution. Maps were improved by solvent flattening and maximal likelihood density modification using resolve (Terwilliger, 1999). The quality of the maps were such that >90% of the G235D mutant model could be readily constructed using program O (Jones et al., 1991). The structure was refined by cycles of manual model building and energy minimization using CNS (Brunger et al., 1998). The final working and free R-factors for the G235D protein were 22.5% and 25.5%, respectively. The structure of the wild-type Se-Met protein was refined in parallel with working and free R-factors of 21.9% and 25.1%, respectively. The structures of nucleotide-complexed proteins were solved after positioning of the free protein model using rigid-body refinement. Then, several rounds of positional refinement were performed. Substrate models were built into the difference density. Other modifications of the models were made manually. Results of the refinements are summarized in Table I. The models show good stereochemistry and geometry, as analysed by the program PROCHECK (Laskowski et al., 1993). All residues have φ and ψ angles within the authorized regions of the Ramachandran plot. During the analysis of domain positions, the rotation matrices were converted into polar angles using CONVROT (Urzhumtseva and Urzhumtsev, 1997). Coordinates have been deposited with the Protein Data Bank with IDs 1KJZ, 1KK0, 1KK1, 1KK2 and 1KK3.
Acknowledgments
Acknowledgements
We thank the staff of LURE DW32 and ESRF ID14 beamlines for assistance during data collection. Jean-Claude Thierry and Olivier Poch (IGBMC, Strasbourg) are gratefully acknowledged for the gift of P.abyssi chromosomal DNA and helpful advice.
References
- Andersen G.R., Valente,L., Pedersen,L., Kinzy,T.G. and Nyborg,J. (2001) Crystal structures of nucleotide exchange intermediates in the eEF1A–eEF1Bα complex. Nature Struct. Biol., 8, 531–534. [DOI] [PubMed] [Google Scholar]
- Asano K., Clayton,J., Shalev,A. and Hinnebusch,A.G. (2000) A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev., 14, 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrom S.U., von Pawel-Rammingen,U. and Bystrom,A.S. (1993) The yeast initiator tRNAMet can act as an elongator tRNAMetin vivo. J. Mol. Biol., 233, 43–58. [DOI] [PubMed] [Google Scholar]
- Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Basavappa R. and Sigler,P.B. (1991) The 3 Å crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J., 10, 3105–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold H., Reshtnikova,L., Reiser,C.O.A., Schirmer,N.K., Sprinzl,M. and Hilgenfeld,R. (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature, 365, 126–132. [DOI] [PubMed] [Google Scholar]
- Blanquet S., Mechulam,Y. and Schmitt,E. (2000) The many routes of bacterial transfer RNAs after aminoacylation. Curr. Opin. Struct. Biol., 10, 95–101. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Castilho-Valavicius B., Thompson,G.M. and Donahue,T.F. (1992) Mutation analysis of the Cys-X2-Cys-X19-Cys-X2-Cys motif in the beta subunit of eukaryotic translation initiation factor 2. Gene Exp., 2, 297–309. [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A. and Maitra,U. (1991) Function of eukaryotic initiation factor 5 in the formation of an 80S ribosomal polypeptide chain initiation complex. J. Biol. Chem., 266, 14039–14045. [PubMed] [Google Scholar]
- Chaudhuri J., Si,K. and Maitra,U. (1997) Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J. Biol. Chem., 272, 7883–7891. [DOI] [PubMed] [Google Scholar]
- Choi S.K., Lee,J.H., Zoll,W.L., Merrick,W.C. and Dever,T.E. (1998) Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science, 280, 1757–1760. [DOI] [PubMed] [Google Scholar]
- Choi S.K., Olsen,D.S., Roll-Mecak,A., Martung,A., Remo,K.L., Burley,S.K., Hinnebusch,A.G. and Dever,T.E. (2000) Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol., 20, 7183–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark B.F. and Nyborg,J. (1997) The ternary complex of EF-Tu and its role in protein biosynthesis. Curr. Opin. Struct. Biol., 7, 110–116. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs from protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Dennis P.P. (1997) Ancient ciphers: translation in Archaea. Cell, 89, 1007–1010. [DOI] [PubMed] [Google Scholar]
- Donahue T.F., Cigan,A.M., Pabich,E.K. and Valavicius,B.C. (1988) Mutations at a Zn(II) finger motif in the yeast eIF-2 β gene alter ribosomal start-site selection during the scanning process. Cell, 54, 621–632. [DOI] [PubMed] [Google Scholar]
- Drabkin H.J. and RajBhandary,U.L. (1998) Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol. Cell. Biol., 18, 5140–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson F.L. and Hannig,E.M. (1996) Ligand interactions with eukaryotic translation initiation factor 2: role of the γ-subunit. EMBO J., 15, 6311–6320. [PMC free article] [PubMed] [Google Scholar]
- Erickson F.L., Harding,L.D., Dorris,D.R. and Hannig,E.M. (1997) Functional analysis of homologs of translation initiation factor 2 γ in yeast. Mol. Gen. Genet., 253, 711–719. [DOI] [PubMed] [Google Scholar]
- Erickson F.L., Nika,J., Rippel,S. and Hannig,E.M. (2001) Minimum requirements for the function of eukaryotic translation initiation factor 2. Genetics, 158, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.V. (1993) Setor: hardware lighted three-dimensional solid model representation of macromolecules. J. Mol. Graphics, 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Farruggio D., Chaudhuri,J., Maitra,U. and RajBhandary,U.L. (1996) The A1 × U72 base pair conserved in eukaryotic initiator tRNAs is important specifically for binding to the eukaryotic translation initiation factor eIF2. Mol. Cell. Biol., 16, 4248–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn A., Oldfield,S. and Proud,C.G. (1993) The role of the β-subunit of initiation factor eIF-2 in initiation complex formation. Biochim. Biophys. Acta, 1174, 117–121. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O., Brandi,L., Caserta,E., La Teana,A., Spurio,R., Tomsic,J. and Pon,C.L. (2000) Translation initiation in bacteria. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 477–494.
- Hinnebusch A.G. (1993) Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol. Microbiol., 10, 215–223. [DOI] [PubMed] [Google Scholar]
- Huang H.K., Yoon,H., Hannig,E.M. and Donahue,T.F. (1997) GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev., 11, 2396–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kyrpides N.C. and Woese,C.R. (1998a) Archaeal translation initiation revisited: the initiation factor 2 and eukaryotic initiation factor 2B α-β-δ subunit families. Proc. Natl Acad. Sci. USA, 95, 3726–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides N.C. and Woese,C.R. (1998b) Universally conserved translation initiation factors. Proc. Natl Acad. Sci. USA, 95, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cour T.F., Nyborg,J., Thirup,S. and Clark,B.F. (1985) Structural details of the binding of guanosine diphosphate to elongation factor Tu from E.coli as studied by X-ray crystallography. EMBO J., 4, 2385–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Laurino J.P., Thompson,G.M., Pacheco,E. and Castilho,B.A. (1999) The β subunit of eukaryotic translation initiation factor 2 binds mRNA through the lysine repeats and a region comprising the C2–C2 motif. Mol. Cell. Biol., 19, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Choi,S.K., Roll-Mecak,A., Burley,S.K. and Dever,T.E. (1999) Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc. Natl Acad. Sci. USA, 96, 4342–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayaux J.-F. and Blanquet,S. (1981) Binding of zinc to Escherichia coli phenylalanyl transfer ribonucleic acid synthetase. Comparison with other aminoacyl transfer ribonucleic acid synthetases. Biochemistry, 20, 4647–4654. [DOI] [PubMed] [Google Scholar]
- Merrick W.C. and Hershey,J.W.B. (1996) The pathway and mechanism of eukaryotic protein synthesis. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 31–69.
- Nika J., Rippel,S. and Hannig,E.M. (2001) Biochemical analysis of the eIF2βγ complex reveals a structural function for eIF2α in catalyzed nucleotide exchange. J. Biol. Chem., 276, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikova,L., Clark,B.F.C. and Nyborg,J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Brodersen,D.E., Clemons,W.M., Tarry,M.J., Carter,A.P. and Ramakrishnan,V. (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science, 292, 897–902. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Borukhov,S.I. and Hellen,C.U. (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature, 394, 854–859. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Lomakin,I.B., Lee,J.H., Choi,S.K., Dever,T.E. and Hellen,C.U. (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature, 403, 332–335. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva,V.G., Lomakin,I.B., Pilipenko,E.V., Shatsky,I.N., Agol,V.I. and Hellen,C.U. (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA, 98, 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polekhina G., Thirup,S., Kjeldgaard,M., Nissen,P., Lippmann,C. and Nyborg,J. (1996) Helix unwinding in the effector region of elongation factor EF-Tu–GDP. Structure, 4, 1141–1151. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., Cao,C., Dever,T.E. and Burley,S.K. (2000) X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell, 103, 781–792. [DOI] [PubMed] [Google Scholar]
- Schenk P.M., Baumann,S., Mattes,R. and Steinbiss,H.H. (1995) Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. Biotechniques, 19, 196–200. [PubMed] [Google Scholar]
- Spurio R., Brandi,L., Caserta,E., Pon,C.L., Gualerzi,C.O., Misselwitz,R., Krafft,C., Welfle,K. and Welfle,H. (2000) The C-terminal subdomain (IF2 C-2) contains the entire fMet-tRNA binding site of initiation factor IF2. J. Biol. Chem., 275, 2447–2454. [DOI] [PubMed] [Google Scholar]
- Szkaradkiewicz K., Zuleeg,T., Limmer,S. and Sprinzl,M. (2000) Interaction of fMet-tRNAfMet and fMet-AMP with the C-terminal domain of Thermus thermophilus translation initiation factor 2. Eur. J. Biochem., 267, 4290–4299. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (1999) Reciprocal-space solvent flattening. Acta Crystallogr. D, 55, 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson G.M., Pacheco,E., Melo,E.O. and Castilho,B.A. (2000) Conserved sequences in the β subunit of archaeal and eukaryal translation initiation factor 2 (eIF2), absent from eIF5, mediate interaction with eIF2γ. Biochem. J., 347, 703–709. [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel H. (1996) Binding of initiator methionyl tRNA to ribosomes. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 113–138.
- Urzhumtseva L.M. and Urzhumtsev,A.G. (1997) Tcl/Tk based programs. II. CONVROT: program to recalculate different rotation descriptions. J. Appl. Crystallogr., 30, 402–410. [Google Scholar]
- Wagner T., Gross,M. and Sigler,P.B. (1984) Isoleucyl initiator tRNA does not initiate eucaryotic protein synthesis. J. Biol. Chem., 259, 4706–4709. [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova,G.Z., Baucom,A., Lieberman,K., Earnest,T.N., Cate,J.H. and Noller,H.F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science, 292, 883–896. [DOI] [PubMed] [Google Scholar]
- Zeidler W., Schirmer,N.K., Egle,C., Ribeiro,S., Kreutzer,R. and Sprinzl,M. (1996) Limited proteolysis and amino acid replacements in the effector region of Thermus thermophilus elongation factor Tu. Eur. J. Biochem., 239, 265–271. [DOI] [PubMed] [Google Scholar]