Abstract
XPF–ERCC1 is a structure-specific endonuclease involved in nucleotide excision repair, interstrand crosslink repair and homologous recombination. So far, it has not been shown experimentally which subunit of the heterodimer harbors the nuclease activity and which amino acids contribute to catalysis. We used an affinity cleavage assay and located the active site to amino acids 670–740 of XPF. Point mutations generated in this region were analyzed for their role in nuclease activity, metal coordination and DNA binding. Several acidic and basic residues turned out to be required for nuclease activity, but not DNA binding. The separation of substrate binding and catalysis by XPF–ERCC1 will be invaluable in studying the role of this protein in various DNA repair processes. Alignment of the active site region of XPF with proteins belonging to the Mus81 family and a putative archaeal RNA helicase family reveals that seven of the residues of XPF involved in nuclease activity are absolutely conserved in the three protein families, indicating that they share a common nuclease motif.
Keywords: affinity cleavage/DNA repair/endonuclease/site-directed mutagenesis/XPF–ERCC1
Introduction
DNA lesions formed by UV light and environmental mutagens are mainly removed by nucleotide excision repair (NER) in mammals (Friedberg et al., 1995; Lindahl and Wood, 1999). In humans, NER deficiency is associated with the hereditary syndrome xeroderma pigmentosum (XP), which is characterized by extreme sensitivity to sunlight, a >1000-fold increased frequency of skin cancer and, in some cases, neurological abnormalities (Bootsma et al., 1998). In NER, the damaged DNA strand is incised 5′ and 3′ to the lesion, the damaged oligonucleotide of ∼30 nucleotides in length is removed and the gap is filled by repair synthesis. The excision step involves at least 18 polypeptides, namely the proteins encoded by the XPA to XPG genes representing seven XP complementation groups, ERCC1, hHR23B, the RPA trimer and additional subunits of TFIIH (Sancar, 1996; Wood, 1997; de Laat et al., 1999). The core NER reaction has been reconstituted with purified factors, which has led to considerable progress in the understanding of the biochemical basis of the pathway (Aboussekhra et al., 1995; Mu et al., 1995). Biochemical and cell biological studies suggest that XPC–hHR23B is the first of the known NER factors to detect a damaged site in DNA (Sugasawa et al., 1998; Volker et al., 2001). XPC–hHR23B then recruits TFIIH to the damaged site (Yokoi et al., 2000). The two helicase subunits of TFIIH, XPB and XPD, together with XPA, RPA and XPG, are involved in the formation of a bubble of ∼30 bp around the lesion (Evans et al., 1997; Araujo et al., 2001). XPA and RPA appear to play a role in damage verification and are crucial for the correct positioning of the two endonucleases, XPG and XPF–ERCC1, before incision (de Laat et al., 1998a; Missura et al., 2001). The incision 3′ to the lesion by XPG precedes the 5′ incision by XPF–ERCC1; the latter requires the structural presence, but not the catalytic activity, of XPG (Wakasugi et al., 1997). The incisions by XPG and XPF–ERCC1 are localized at the borders of the double-/single-stranded DNA (ds/ssDNA) in the incision complex, and this structure-specific activity can be reproduced with purified proteins on artificial DNA substrates in the absence of additional factors (Bardwell et al., 1994; O’Donovan et al., 1994; Sijbers et al., 1996a). Observation of the mobility of various NER proteins in living cells suggests that NER proceeds by sequential assembly of the individual factors involved, rather than through the action of a pre-assembled repairosome (Houtsmuller et al., 1999).
A thorough understanding of the NER reaction requires the detailed characterization of the activities of each protein involved and of the interactions between all these proteins. In this context, we have become interested in the characterization of the XPF–ERCC1 endonuclease. XPF and ERCC1 form a stable heterodimer in mammalian cells and in vitro, and various lines of evidence suggest that the two proteins are unstable in the absence of each other (Biggerstaff et al., 1993; Reardon et al., 1993; van Vuuren et al., 1993; Yagi et al., 1997). Apart from its role in NER, XPF–ERCC1 has additional functions in DNA metabolism that are not yet understood. Mammalian and hamster cells defective in XPF or ERCC1 display a much higher sensitivity to interstrand crosslinking (ICL) agents than cells deficient in other NER factors (Hoy et al., 1985). Furthermore, XPF–ERCC1 can make incisions on both sides of an ICL placed near a ds/ssDNA junction (Kuraoka et al., 2000). An involvement of XPF–ERCC1 in homologous recombination was first suggested by studies in the budding yeast Saccharomyces cerevisiae, where the homologous Rad1–Rad10 complex is required for the removal of long non-homologous tails from invading homologous strands (Fishman-Lobell and Haber, 1992; Bardwell et al., 1994). More recently, a role for ERCC1 in homologous recombination has been demonstrated in mammalian cells (Sargent et al., 2000; Niedernhofer et al., 2001). Roles for XPF–ERCC1 outside of NER are also supported by the severe phenotype of ERCC1 knock-out mice, which includes symptoms such as growth retardation, liver and kidney abnormalities, and early death, which cannot be explained by an NER defect alone (McWhir et al., 1993; Weeda et al., 1997).
Some of the functional domains of XPF–ERCC1 have been mapped. The heterodimer is formed through interaction of the very C-terminal domains of XPF and ERCC1 (de Laat et al., 1998b). The C-termini of both XPF and ERCC1 also contain two helix–hairpin–helix motifs, which are frequently found in proteins that bind distorted DNA structures (Sijbers et al., 1996b; Gaillard and Wood, 2001). ERCC1 has been shown to interact with XPA through amino acids 93–120, and this interaction is required for the NER reaction (Li et al., 1994, 1995). Sequence comparisons have linked the XPF protein family to the SF2 superfamily of archaeal RNA helicases and the Mus81 family of proteins, which are part of a complex that functions as a Holliday junction resolvase (Sgouros et al., 1999; Interthal and Heyer, 2000; Boddy et al., 2001; Chen et al., 2001; Kaliraman et al., 2001). The three proteins share the highly conserved V/IERKX3D sequence, which has been predicted to play a role in metal coordination and nuclease activity (Aravind et al., 1999). The aspartate in this motif in Mus81 has recently been shown to be required for nuclease activity (Boddy et al., 2001; Chen et al., 2001).
We used an affinity cleavage approach to identify the metal-binding and active site of XPF–ERCC1 in an unbiased manner. Replacement of the metal cofactor of a protein with divalent iron, which has the ability to undergo a Fenton reaction to generate hydroxyl radicals, results in cleavage of peptide bonds in the immediate vicinity of the metal-binding site. Sequencing of the peptide fragments obtained in this way can yield information on the location of the metal-binding site (Zaychikov et al., 1996; Lykke-Andersen et al., 1997; Cao and Barany, 1998; Kim et al., 1999; Hlavaty et al., 2000). Application of this iron-mediated cleavage technique to XPF–ERCC1 mapped the metal-binding site between amino acids 670 and 740 of the XPF subunit. We generated and characterized a number of mutations in acidic and basic residues in this region, and demonstrate that several of these residues play a role in metal binding and/or catalysis. None of these mutations affects DNA binding, demonstrating that DNA binding and catalysis by XPF–ERCC1 can be separated. Sequence analysis of the experimentally identified active site of XPF–ERCC1, in agreement with theoretical predictions, reveals a new nuclease motif that is widely conserved in eukaryotes and archaea.
Results
Purification of recombinant XPF–ERCC1
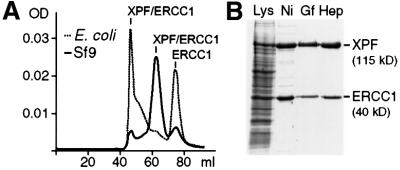
Previous studies have reported the successful purification of XPF–ERCC1 from both Escherichia coli and baculovirus-infected insect cells. We initially evaluated a purification procedure of the complex containing a His6 tag on both subunits overexpressed in E.coli. However, the gel-filtration step in our purification scheme revealed that, in our hands, most of the protein was present in aggregated form and as excess unbound ERCC1 (Figure 1A, dotted line). Only a very small amount of XPF–ERCC1 eluted at a range that could be expected for heterodimeric XPF–ERCC1. Significantly, endonuclease activity was only detected for protein eluting in fractions corresponding to heterodimeric protein (data not shown). We therefore developed a protocol for the purification of XPF–ERCC1 from baculovirus-infected Sf9 insect cells using a His6 tag on ERCC1. We obtained highly purified protein using chromatography on Ni–agarose, gel-filtration and heparin columns (Figure 1B). In contrast to the preparation from E.coli, the profile from the gel-filtration column revealed that the vast majority of XPF–ERCC1 expressed in Sf9 insect cells eluted at the expected range for the heterodimer (Figure 1A, solid line) and displayed the expected nuclease activity.
Fig. 1. Purification of recombinant XPF–ERCC1. (A) Comparison of the gel-filtration profiles of XPF–ERCC1 expressed in E.coli (dotted line) and Sf9 insect cells (solid line). The peak at ∼45 ml corresponds to the void volume of the column with protein in an aggregated state, the peak at ∼65 ml to a molecular weight of ∼200 kDa and the peak at ∼75 ml to a molecular weight of ∼60 kDa. The proteins identified in the peak fractions are indicated. (B) SDS–PAGE gel (10%) of the purification of the XPF–ERCC1 heterodimer from Sf9 cells; proteins were visualized by CBB staining. The cell lysate (Lys) and pooled fractions from after the individual column steps, Ni-NTA–agarose (Ni), gel filtration (Gf) and heparin (Hep), are shown. XPF and ERCC1 have molecular weights of 115 and 40 kDa, respectively.
Mapping of the metal-binding site of XPF–ERCC1 using affinity cleavage
We intended to identify the metal-binding and active site regions of XPF–ERCC1 using an affinity cleavage technique. For this purpose, the metal cofactor (Mg2+ or Mn2+) can be replaced by Fe2+, which has the ability to reduce molecular oxygen to form superoxide and hydroxyl radicals. Such reactive oxygen species can cleave the polypeptide backbone in the vicinity of the metal-binding site to generate peptide fragments that can then be identified by N-terminal sequencing. To determine whether Fe2+ can substitute for Mg2+ or Mn2+ in XPF–ERCC1, we tested the nuclease activity of the protein in the presence of Fe2+. XPF–ERCC1 showed residual nuclease activity with Fe2+ (data not shown). Apparently, Fe2+ is able to enter the active site of the protein, and Fe2+-mediated cleavage experiments could therefore direct us to the active site of XPF–ERCC1. Incubation of XPF–ERCC1 with FeCl2 in the presence of ascorbate (ascorbate reduces Fe3+ that is formed after the reduction of oxygen to regenerate Fe2+) did result in protein cleavage. SDS–PAGE showed a very reproducible cleavage pattern over a wide range of Fe2+ concentrations (10–1000 µM) and incubation times (3–15 h). The most crucial parameter was pH, and best results were obtained at pH 7.0.
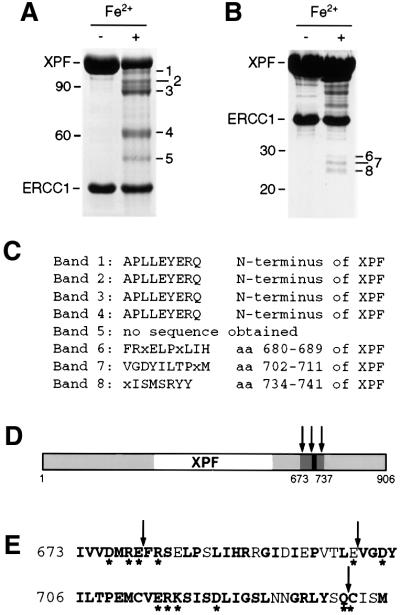
The typical pattern consisted of five bands (bands 1–5, in order of decreasing size), with estimated molecular weights of 102–52 kDa (Figure 2A). To identify the cleavage sites, the gel was blotted on a PVDF membrane and the N-terminus of these fragments was subjected to Edman degradation. Bands 1–4 were readily sequenced and all contained the original N-terminus of XPF (Figure 2C), which suggested that the cleavage sites were located in the C-terminal half of XPF but failed to reveal the exact location of the cleavage sites. Several attempts to sequence band 5 were unsuccessful. Band 5 (52 kDa) may represent the C-terminal half corresponding to band 4 (62 kDa) containing the N-terminal half of XPF, as the sum of their molecular weights adds up to the molecular weight of the full-length protein (115 kDa).
Fig. 2. Iron-induced cleavage of XPF–ERCC1. (A) SDS–PAGE gel (8%) of purified XPF–ERCC1 before and after treatment with 50 µM FeCl2 and 20 mM ascorbate for 15 h at 0°C. The five observed XPF cleavage products were designated bands 1–5, in order of decreasing size and with estimated molecular weights of 102, 95, 88, 62 and 52 kDa, respectively. A 10 kDa protein ladder (Life Technologies) was used as a marker. The positions of the 90 and 60 kDa marker bands are indicated. (B) SDS–PAGE gel (12%) of XPF–ERCC1 treated as in (A). The three bands between 25 and 30 kDa were designated bands 6–8. The positions of the 30 and 20 kDa marker bands are indicated. (C) Results from the N-terminal sequencing of the peptide fragments from bands 1–8 in (A) and (B). (D) Location of the cleavage sites in XPF. Regions of XPF that are conserved between species are shown in gray, the region encompassing the cleavage sites in dark gray and the highly conserved V/IERKX3D motif in black. The cleavage sites are indicated by arrows. (E) The amino acid sequence of region 673–737 of XPF. Conserved residues are in bold, residues selected for site- directed mutagenesis are marked with an asterisk and cleavage sites are marked with arrows.
In order to obtain more information on the cleavage sites that result in the formation of bands 1–3, we sought to obtain the sequences of their C-terminal counterparts. Therefore, we carried out the cleavage reaction on a larger scale and analyzed it on a higher percentage SDS–PAGE gel (Figure 2B). Under these conditions, we observed three additional bands, with molecular weights between 25 and 30 kDa (bands 6–8). In order to obtain N-terminal sequences of the protein fragments in these bands, it was necessary to concentrate the protein before Fe2+/ascorbate treatment and to use ∼200 µg protein per lane and subsequent blotting onto PVDF membranes. Finally, N-terminal sequencing of these three bands revealed that they were indeed XPF derived, with cleavage sites at positions 680, 702 and 734 (Figure 2C–E). Bands 6–8, therefore, most likely constitute the C-terminal counterparts of bands 3, 2 and 1, respectively. The three cleavage sites that we were thus able to map are located in the conserved C-terminal domain of the XPF subunit (Figure 2D and E), on both sides of the highly conserved V/IERKX3D motif. This motif has been predicted to play a role in the nuclease activity of XPF–ERCC1, and the aspartate residue in it has been shown to be involved in the nuclease activity of Mus81 (Aravind et al., 1999; Boddy et al., 2001; Chen et al., 2001). Interestingly, no cleavage of ERCC1 was observed, suggesting that XPF contains the entire metal-binding site.
Generation of XPF–ERCC1 with mutations in the putative active site
Since the three identifiable iron-induced cleavage sites were located between residues 670 and 740 of XPF, we concluded that this region of XPF is important for metal binding and likely contains the active site of XPF–ERCC1. We selected 12 amino acid residues in the XPF subunit for site-directed mutagenesis (Figure 2E). E679, E701, Q733 and C734 were selected because they are located at the cleavage sites. With the exception of E701, these residues are highly conserved. D676, D704, E714 and D720 (and E679) are absolutely conserved acidic residues in this region and might therefore play a role in metal binding and catalysis. The positively charged and highly conserved R678, R681, R715 and K716 were chosen to investigate their roles in catalysis and binding to the DNA backbone. All 12 residues were mutated to alanines and expressed in Sf9 insect cells together with ERCC1. Expression and purification of all 10 proteins with mutations in acidic or basic residues proceeded as for wild-type protein. Importantly, the profiles from the gel-filtration column showed that these mutants were present as heterodimers, indicating that they were properly folded (data not shown). Only the R715A mutant showed a relatively large amount (∼80%) of aggregated protein, but nevertheless a sufficient quantity of heterodimeric protein for biochemical characterization was obtained. By contrast, the Q733A and C734A mutants were present only in the aggregate range of the gel-filtration column (data not shown), indicating that they are not correctly folded. Therefore, we did not characterize these two mutants further.
Mutations in acidic and basic residues near the Fe2+-induced cleavage sites affect endonuclease activity
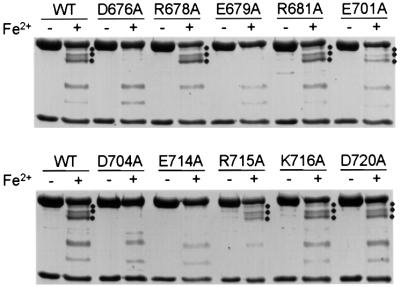
We investigated the endonuclease activity of wild-type and mutant XPF–ERCC1 using a stem–loop substrate described previously (de Laat et al., 1998c). In agreement with earlier studies, we observed optimal activity at metal concentrations of 0.4 mM MnCl2 or 2 mM MgCl2 and a 2-fold excess of protein over substrate. The highest activity was observed when Mn2+ was used as a cofactor, leading to ∼80% cleavage of the stem–loop substrate by XPF–ERCC1 under optimized conditions (Figure 3A, lane b). Of the XPF mutants, only E701A displayed the same level of activity as wild type. R681A displayed ∼70% of wild-type activity, indicating that these two residues are not important for nuclease activity (Figure 3, lanes f and g). E679A and E714A showed 5 and 30% of wild-type activity, respectively, in the presence of MnCl2 and no (E679A) or only residual activity (E714A) in the presence MgCl2 (Figure 3, lanes e and i), suggesting that a mutation in these residues influences the structure of the metal-binding site. Higher MnCl2 or MgCl2 concentrations did not lead to an increase in the activity of these two mutants (data not shown). Substitutions of the two arginine residues at positions 678 and 715 yielded protein with only residual activity (Figure 3, lanes d and j), whereas heterodimers with mutations in D676, D704, K716 and D720 of XPF were devoid of any activity (Figure 3, lanes c, h, k and l), indicating that at least four of the mutated residues are absolutely required for enzyme activity.
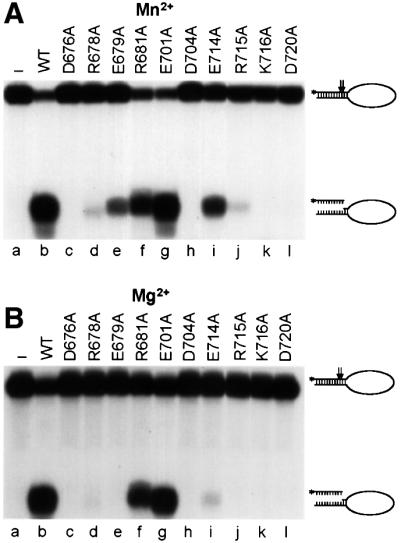
Fig. 3. Nuclease activity of wild-type and mutant XPF–ERCC1. Incision of a stem12–loop22 DNA substrate (100 fmol) by wild-type (WT, lane b) or mutant (lanes c–l) XPF–ERCC1 (200 fmol) in the presence of (A) 0.4 mM MnCl2 or (B) 2 mM MgCl2. Reactions were analyzed on a 15% denaturing polyacrylamide gel. The XPF mutations tested, the 46mer stem–loop substrate (with incision sites indicated by arrows) and the 9/10mer products resulting from cleavage are indicated.
Mutation of most, but not all, acidic residues alters the metal-binding site
Next, we wanted to find out whether the decrease in activity of the mutants is related to the ability of the heterodimer to interact with the metal cofactor. We used the iron-mediated cleavage conditions used to identify the metal-binding site as a measure of the ability of the mutant proteins to interact with the metal. We expected that heterodimers with an intact metal-binding site would display the same cleavage pattern, whereas disruption of the metal-binding site would result in the disappearance of certain characteristic cleavage sites. Since XPF–ERCC1 is active in the presence of both Mg2+ and Mn2+, we expected that only mutations in acidic residues would affect the interaction with the metal, as Mg2+ does not interact favorably with nitrogen ligands. We subjected the XPF–ERCC1 mutants to Fe2+/ascorbate treatment and compared the cleavage pattern formed to the one found for wild-type protein. We observed no change in the cleavage pattern of all the basic mutants and the E701A and D720A mutants (Figure 4). In contrast, bands 1–3 were clearly absent in the cleavage pattern of the D676A, E679A, D704A and E714A mutants, indicating that these substitutions alter or even disrupt the metal-binding site. Mutation of these four acidic residues results in no or reduced nuclease activity, suggesting that they mediate catalysis through metal binding. Bands 4 and 5 were observed in all mutants, indicating either that there are two metal-binding sites or that these bands are formed non-specifically.
Fig. 4. Comparison of iron-induced cleavage of wild-type and mutant XPF–ERCC1. Purified wild-type (WT) and mutant XPF–ERCC1 were incubated with 50 µM FeCl2 and 20 mM ascorbate for 15 h at 0°C and analyzed on an 8% SDS–PAGE gel. Bands 1–3 resulting from iron- induced cleavage, with molecular weights of ∼102, 95 and 88 kDa, respectively, are indicated (where present) by three dots.
Mutations in the active site region of XPF do not influence DNA binding
To examine whether the mutations that resulted in defects in nuclease and metal-binding activities had any influence on DNA binding, we performed electrophoretic mobility shift assays (EMSAs). We initially used the D720A mutant of XPF, which is devoid of nuclease activity but contains an intact metal site, to establish binding conditions (Figure 5A). We investigated the role of divalent metals in this binding interaction by comparing the ability of the wild-type heterodimer, XPF(D676A)–ERCC1 (defective in catalysis and metal coordination) and XPF(D720A)–ERCC1 (defective in catalysis but proficient in metal binding) to bind to the stem–loop substrate in the presence and absence of various metals. As expected, we observed cleavage of the substrate with wild-type protein in the presence of MnCl2 or MgCl2 (Figure 5B, lanes b and c), indicating that the protein–DNA complex observed in the EMSA is in an active conformation. No obvious differences in the binding ability of the two mutant proteins with MnCl2 or MgCl2, or of wild-type and mutant proteins in the presence of CaCl2 and EDTA, were observed (Figure 5B, lanes d– m). Apparently, the presence of divalent metal ions is not a prerequisite for DNA binding, and the identity of the metal cofactor has no significant influence on the binding affinity.
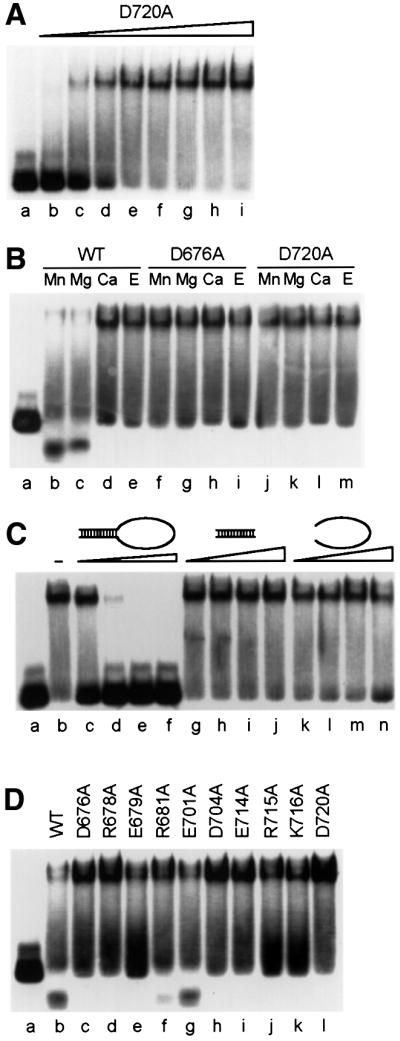
Fig. 5. Catalytically inactive XPF–ERCC1 mutants bind specifically to stem–loop substrates. 5′-32P-labeled stem12–loop22 substrate at a concentration of 17 nM was used for all experiments and was the last component added to the reaction mixture. Unless indicated otherwise, a protein concentration of 68 nM and MnCl2 (0.4 mM) as a cofactor were used. Reactions were incubated for 30 min at room temperature and analyzed on 4% native polyacrylamide gels. (A) Increasing concentrations of XPF(D720A)–ERCC1 were incubated with the substrate to demonstrate binding of XPF–ERCC1 to the stem–loop substrate. Protein concentrations: lane b, 17 nM; lane c, 34 nM; lane d, 51 nM; lane e, 68 nM; lane f, 85 nM; lane g, 102 nM; lane h, 136 nM; lane i, 170 nM. (B) Influence of the presence of divalent metal ions on substrate binding by wild-type and mutant XPF–ERCC1. Wild-type (WT) XPF–ERCC1, lanes b–e; D676A, lanes f–i; D720A, lanes j–m. Concentrations of metal cofactors: Mn2+, 0.4 mM; Mg2+, 2 mM; Ca2+, 2 mM. Concentration of EDTA (E), 5 mM. (C) Influence of increasing amounts of specific and non-specific oligonucleotide competitors on substrate binding by XPF(D720A)–ERCC1. Competitors: lane b, no competitor; lanes c–f, stem12–loop22 (68, 340, 850 and 1700 nM, respectively); lanes g–j, stem12 dsDNA (340, 850, 1700 and 3400 nM, respectively); lanes k–n, loop22 ssDNA (340, 850, 1700 and 3400 nM, respectively). (D) Comparison of substrate binding of wild-type (WT) and mutant XPF–ERCC1.
We went on to investigate the specificity of the binding of XPF–ERCC1 to ds/ssDNA junctions by challenging the binding reaction with specific and non-specific competitors. We used unlabeled stem–loop substrate as our specific competitor and the double- or single-stranded region of this molecule alone as dsDNA and ssDNA competitor, respectively. Addition of a 20-fold excess (340 nM) of specific competitor led to complete inhibition of complex formation, whereas the addition of a 100-fold excess (1.7 µM) of either dsDNA or ssDNA competitor had no effect on the binding reaction (Figure 5C). These observations demonstrate that the binding of XPF–ERCC1 to ds/ssDNA junctions is very specific. Next, we wanted to see whether all the mutants were able to bind to the stem–loop. We observed that all the mutants with impaired nuclease activity showed very similar binding activities (Figure 5D). The R681A and E701A mutants, which displayed near wild-type nuclease activities, were able to partially cleave the substrate under EMSA conditions (which are very similar to nuclease conditions). Our data clearly demonstrate that none of the mutations in XPF affects the DNA-binding ability of the protein.
Discussion
Identification of the active site of XPF–ERCC1
XPF–ERCC1 is a structure-specific endonuclease with roles in NER, ICL repair and homologous recombination. Before this work, the active site of this heterodimer had not been identified. We used an affinity cleavage approach to identify the residues that coordinate the metal ion in the active site of the protein (Zaychikov et al., 1996; Lykke-Andersen et al., 1997; Cao and Barany, 1998; Kim et al., 1999; Hlavaty et al., 2000). This approach takes advantage of the similarity of the mechanism employed by most metal-dependent endonucleases, which involves the coordination of a divalent metal ion (usually Mg2+ or Mn2+) mainly by aspartate or glutamate residues. We replaced the metal cofactor of the enzyme with Fe2+, which can generate hydroxyl radicals in the active site that can cleave the peptide backbone in close proximity. We N-terminally sequenced the fragments obtained in this way and identified three cleavage sites located between amino acids 679/680, 701/702 and 733/734 of XPF. We investigated the role of acidic and basic residues at and around the cleavage sites using site-directed mutagenesis (Figure 2E), and identified several residues that are important or even essential for catalysis. This active site region that we have now biochemically identified agrees well with the predication of Aravind et al. (1999), who suggested that this region comprises a nuclease motif based on multiple sequence alignments (see below). Our results are in contrast, however, to another study that suggested that the N-terminal 378 amino acids of XPF have endonuclease activity (McCutchen-Maloney et al., 1999). This study has already been challenged by Gaillard and Wood (2001), who were not able to detect nuclease activity in XPF alone.
Putative role of residues in the active site
With the exception of the Q733A and C734A mutants, the mutations we introduced into XPF had no effect on protein folding, heterodimer formation with ERCC1 and DNA binding. We examined the nuclease activity of each mutant in the presence of Mg2+ and Mn2+, as well as their ability to undergo iron-mediated cleavage, to see whether they contained an intact metal-binding site. Based on this analysis, we can classify the acidic residues we investigated into three categories. D676 and D704 are absolutely required for both nuclease activity and metal binding, and apparently contribute to catalysis through coordination of the metal ion. D720 is also essential for catalysis but is not involved in metal binding. It may play a role as a general base in activating a water molecule as a nucleophile required for phosphodiester bond hydrolysis. Such a role for acidic residues in nucleases has been demonstrated, for example, for E113 of the restriction endonuclease BamHI (Pingoud and Jeltsch, 2001). Mutations in E679 and E714 resulted in severely reduced nuclease activity in the presence of Mg2+, whereas the use of Mn2+ as a cofactor yielded substantial (5 and 30% of wild type) nuclease activity. Both mutants failed to undergo iron-mediated protein cleavage. Apparently, mutation of these two residues results in an altered metal-binding site, which can still bind Mn2+, but not Mg2+. It is known that Mn2+ has less stringent requirements for ligand coordination and often displays higher affinities for metal-binding sites in proteins than Mg2+ (Cowan, 1998). We generated mutations in four basic residues of XPF. Mutation of R678, R715 and K716 severely impacted nuclease activity, whereas mutation of R681 had no big effect. Since none of these mutations has any apparent influence on the DNA-binding affinity of XPF–ERCC1, R678, R715 and K716 are likely involved in catalysis. The general roles assigned to the residues involved in catalysis are in accordance with what has been observed for other endonucleases (Hosfield et al., 1998; Chevalier and Stoddard, 2001; Pingoud and Jeltsch, 2001). A more precise characterization of the role of these residues awaits structural and more detailed biochemical studies.
A new conserved nuclease motif
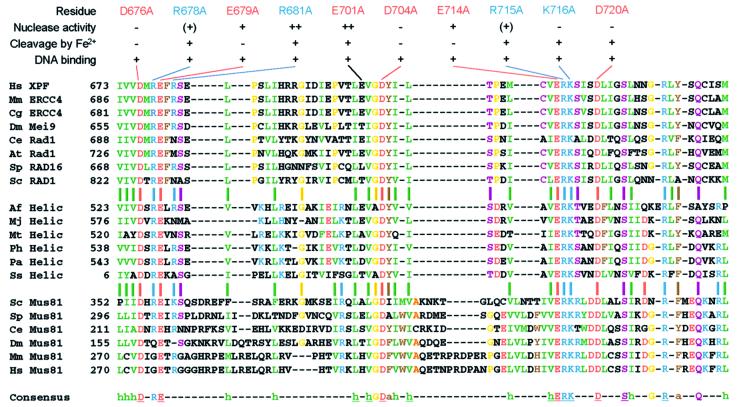
Our study has identified a number of residues that are important or essential for catalysis by XPF–ERCC1. Two classes of proteins have been shown previously to share sequence similarities with XPF: putative archaeal RNA helicases belonging to SF2, and the Mus81 family of Holliday junction resolvases involved in meiotic recombination and restarting of stalled replication forks (Aravind et al., 1999; Sgouros et al., 1999; Interthal and Heyer, 2000; Boddy et al., 2001; Chen et al., 2001; Kaliraman et al., 2001; Mullen et al., 2001). Both families of proteins, like XPF, contain the highly conserved V/IERKX3D motif, which has not been detected yet in other proteins. Inspection of an alignment of proteins from the three families revealed that residues corresponding to D676, E679, D704, E714, R715, K716 and D720 in XPF are absolutely conserved among these proteins and form the core of a new conserved nuclease motif (Figure 6). Further inspection of the sequence alignment reveals that three additional polar residues, S724, R729 and Q733, which may be involved in catalysis, and 15 hydrophobic or aromatic residues, which likely contribute to a conserved overall fold of this endonuclease motif, are also highly to absolutely conserved in this region of the proteins. Mus81, which forms a heterodimer with the ERCC1 homolog Mms4 in S.cerevisiae and Eme1 in Schizosaccharomyces pombe, functions as part of a Holliday junction resolvase and hence has structure-specific endonuclease activity with a slightly different substrate specificity than XPF–ERCC1 (Boddy et al., 2001; Chen et al., 2001; Kaliraman et al., 2001). Consistent with our observations, mutation of two adjacent aspartates in Mus81, corresponding to S719 and D720 in XPF, leads to loss of junction cleavage activity. Furthermore, the corresponding mutant alleles in S.pombe display the phenotype of a Mus81 null allele (Boddy et al., 2001; Chen et al., 2001). To the best of our knowledge, the putative archaeal helicases have not yet been biochemically characterized, but the high conservation of residues involved in catalysis in XPF suggests that they too have nuclease activity.
Fig. 6. Alignment of the endonuclease domain of XPF with its homologs. The alignment shows amino acids 673–737 of human XPF with homologous proteins of the XPF, putative archaeal helicase and Mus81 family. The aligned proteins (DDBJ/EMBL/GenBank accession Nos are shown in parentheses): human (Hs) XPF (Q92889), mouse (Mm) ERCC4 (AAC03240), hamster (Cg) ERCC4 (BAA89299), Drosophila melanogaster (Dm) Mei9 (Q24087), Caenorhabditis elegans (Ce) Rad1 (CAB70217), Arabidopsis thaliana (At) Rad1 (NP_198931), Schizosaccharomyces pombe (Sp) RAD16 (P36617), Saccharomyces cerevisiae (Sc) RAD1 (P06777), Archaeoglobus fulgidus (Af) putative RNA helicase (AAB89786), Methanococcus jannaschii (Mj) putative RNA helicase (AAB99518), Methanothermobacter thermautotrophicus (Mt) putative RNA helicase (AAB85892), Pyrococcus horikoshii (Ph) putative RNA helicase (NP_143722), Pyrococcus abyssi (Pa) putative RNA helicase (NP_125972), Sulfolobus solfataricus (Ss) putative RNA helicase (CAB57574), Sc Mus81 (NP_010674), Sp Mus81 (CAB09772), Ce Mus81 (AAB37627), Dm Mus81 (AAF45571), Mm Mus81 (AAL28066), Hs Mus81 (AAL28065). Conserved residues are indicated in the following colors: green, hydrophobic (I,L,V,M); red, acidic (D,E); blue, basic (K,R,H); brown, aromatic (F,Y,W); yellow (G,P); purple (C,N,Q,S,T); orange (A). Residues that are ≥85% conserved are indicated in the consensus sequence; absolutely conserved residues are underlined. Conserved hydrophobic (h) and aromatic (a) residues are indicated. Results from the biochemical characterization of the mutants generated in this study are summarized above the alignment.
Conclusions
We have characterized the active site of XPF–ERCC1 using affinity cleavage and site-directed mutagenesis. Our studies reveal a conserved nuclease motif that is also present in the Mus81 and archaeal RNA helicase family of proteins. The separation of DNA binding and catalysis by XPF–ERCC1 should be invaluable for the further investigation of the role of this protein in NER, interstrand crosslink repair and homologous recombination.
Materials and methods
Oligonucleotide primers for mutagenesis were synthesized by Microsynth (Switzerland). Oligonucleotides for nuclease and EMSAs were synthesized on an Expedite 8909 nucleic acid synthesis system and purified by denaturing PAGE. All enzymes were purchased from New England Biolabs unless indicated otherwise. [γ-32P]ATP was obtained from Amersham/Pharmacia. All other chemicals and reagents were from Fluka or Sigma. Routine molecular biology manipulations were carried out using standard protocols (Sambrook et al., 1989).
Purification of recombinant XPF–ERCC1 from E.coli
The pET28·XPF/ERCC1 vector containing both XPF and ERCC1 with C-terminal His6 tags was a kind gift from W.de Laat and H.Odijk (Erasmus University, Rotterdam, The Netherlands). Escherichia coli BL21(DE3) cells were transformed with pET28·XPF/ERCC1; a single colony was used to inoculate 1 l of LB medium containing 50 mg of ampicillin and 50 mg of kanamycin, and the culture was grown at 37°C. At an OD600 of 0.6, the culture was chilled to 20°C and IPTG was added to a final concentration of 1 mM, followed by further incubation overnight at 20°C. Cells were collected by low-speed centrifugation and resuspended in 50 ml of lysis buffer (50 mM HEPES pH 7.5, 200 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, 5 mM EDTA and 1 mM PMSF) containing one dissolved protease inhibitor cocktail tablet (Roche). After sonication, the lysate was clarified by centrifugation at 40 000 g for 30 min at 4°C. A streptomycin precipitation was performed by slowly adding a 25% (w/v) solution of streptomycin sulfate to a final concentration of 50 mg/ml lysate. The resulting pellet was removed by centrifigation at 4000 r.p.m. for 30 min at 4°C. Subsequently, an (NH4)2SO4 precipitation was carried out by dropwise addition of 65% (v/v) of a saturated and neutralized (NH4)2SO4 solution to the protein mixture. After an additional incubation for 30 min on ice, the resulting precipitate was harvested by centrifugation at 4000 r.p.m. for 30 min at 4°C. The pellet was resuspended in 15 ml of Ni buffer (50 mM potassium phosphate pH 8.0, 10% glycerol, 5 mM β-mercaptoethanol and 1 mM PMSF) containing 1 mM imidazole, 500 mM NaCl and 1 ml of Ni-NTA–agarose (Qiagen), incubated for 3 h at 4°C and packed in a column. The column was washed twice with, respectively, 10 ml of Ni buffer containing 10 mM imidazole/300 mM NaCl and 10 ml of Ni buffer containing 50 mM imidazole/300 mM NaCl. XPF–ERCC1 was eluted in 4 ml of Ni buffer containing 300 mM imidazole/300 mM NaCl. Subsequently, the protein was loaded onto a HiLoad 16/60 Superdex 200 gel-filtration column (Pharmacia) equilibrated in Gf buffer (25 mM HEPES pH 8.0, 150 mM NaCl, 10% glycerol and 5 mM β-mercapto ethanol). The fractions containing the proteins of interest were flash-frozen and stored at –80°C.
Baculovirus production and purification of the XPF–ERCC1 complexes
Plasmids pFastBac1-XPF and pFastBac1-ERCC1-His were a kind gift from W.de Laat and N.Wijgers, and were used to transform competent DH10Bac cells. Bacmid DNA was isolated and used to transfect Sf9 insect cells, and to amplify the virus according to the manufacturer’s instructions (BAC TO BAC system; Life Technologies). For protein production, 10 T-175 flasks containing 2.5 × 107 cells were co-infected with XPF and ERCC1 viruses at an m.o.i. of 5. Cells were collected 65 h after infection by low-speed centrifugation and resuspended in 40 ml of lysis buffer (50 mM potassium phosphate pH 8.0, 10% glycerol, 0.1% NP-40, 5 mM β-mercaptoethanol and 1 mM PMSF) containing one dissolved EDTA-free protease inhibitor cocktail tablet (Roche). After incubation on ice for 20 min, NaCl was added to a final concentration of 800 mM and the lysate was sonicated. The lysate was clarified by centrifugation at 40 000 g for 30 min at 4°C. The resulting extract was incubated with 1.5 ml Ni-NTA–agarose (Qiagen) for 5 h at 4°C in the presence of 1 mM imidazole. The beads were collected by low-speed centrifugation and resuspended in 15 ml of Ni buffer containing 1 mM imidazole and 800 mM NaCl, and packed in a column. The column was washed twice with, respectively, 10 ml of Ni buffer containing 5 mM imidazole/500 mM NaCl and 10 ml of Ni buffer containing 10 mM imidazole/500 mM NaCl. XPF–ERCC1 was eluted in 5 ml of Ni buffer containing 50 mM imidazole/300 mM NaCl and further purified by gel filtration using a HiLoad 16/60 Superdex 200 column (Pharmacia) equilibrated in Gf buffer. Fractions containing the XPF–ERCC1 heterodimer, eluting from 60 to 70 ml, were pooled and concentrated on a 1 ml HiTrap Heparin column (Pharmacia) equilibrated in Gf buffer. The protein was eluted using a linear, 10-column-volume gradient from 0.15 to 1 M NaCl in Gf buffer. The protein concentration was measured directly at 280 nm, and the purified protein was flash-frozen in aliquots and stored at –80°C. Typically, 0.25–1 mg of purified complex, at a concentration of 0.1–0.6 mg/ml, was obtained.
Fe2+-induced protein affinity cleavage assays
HEPES–NaOH pH 7.0 to a final concentration of 50 mM and freshly prepared ferrous chloride to a final concentration of 50 µM were added to 20–300 µl of purified XPF–ERCC1 protein. The solution was incubated for 15 min on ice, followed by the addition of ascorbate to a final concentration of 20 mM and incubation for another 3–15 h on ice. Cleavage products were separated on 8 or 12% denaturing polyacrylamide gels and either stained immediately with Coomassie Brilliant Blue (CBB) or blotted onto Immobilon-P PVDF membranes (Millipore) for N-terminal sequencing. Electroblotting was performed in 10 mM CAPS pH 11.0 and 10% methanol for 90 min at 100 V. Bands were visualized according to the manufacturer’s instructions. If necessary, protein solutions were TCA precipitated before SDS–PAGE and CBB staining or, if samples were going to be used for N-terminal sequencing, concentrated 5- to 10-fold in centricon tubes (Amicon) before Fe2+/ascorbate treatment. N-terminal sequencing was performed by P.Hunziker and R.Sack on a model G1005A protein sequencer (Agilent Technologies) using standard chemistry protocols as recommended by the manufacturer.
Construction of point mutations in XPF
Site-directed mutagenesis to introduce point mutations in pFastBac1·XPF was performed using the QuikChange site-directed mutagenesis kit according to the manufacturer’s instructions (Stratagene). pFastBac1·XPF served as template and oligonucleotide primers used to generate the mutations contained the desired mutation and, if possible, a marker restriction site for selection. The following primers were used (restriction sites are underlined and indicated, modified nucleotides are shown in italics):
XPF–D676A, GGTACACAGCAAAGCATAGTTGTGGCAATGCGTGAATTTCG (BsrDI); XPF–R678A, GCATAGTTGTGGATATGGCTGAATTCCGAAGTGAGCTTCCATC (EcoRI); XPF–E679A, GTTGTGGATATGCGCGCATTTCGAAGTGAGCTTCC (BssHII); XPF–R681A, GTGGATATGCGTGAATTTGCTAGCGAGCTTCCATCTCTGATCC (NheI); XPF–E701A, GACATTGAACCCGTGACGCTAGCGGTTGGAGATTACATCC (NheI); XPF–D704A, CCCGTGACTTTAGAGGTTGGCGCCTACATCCTCACTCCAG (NarI); XPF–E714A, CCAGAAATGTGCGTGGCGCGCAAGAGTATCAGTG (BssHII); XPF– R715A, CCAGAAATGTGCGTGGAGGCCAAGAGTATCAGTGATTTAATCG (—); XPF–K716A, CCAGAAATGTGCGTGGAGCGCGCTAGCATCAGTGATTTAATCGGC (NheI); XPF–D720A, GAGCGCAAGAGTATCAGCGCGCTAATCGGCTCTTTAAATAACGGC (BssHII); XPF–Q733A, GGCCGCCTCTACAGCGCATGCATCTCCATGTCC (SphI); XPF–C734A, GGCCGCCTCTACAGCCAGGCGATATCCATGTCCCGCTAC (EcoRV).
Positive clones were sequenced to rule out the introduction of additional mutations and were subcloned into the original pFastBac1·XPF vector containing wild-type XPF. Mutants D676A, R678A, E679A and R681A were subcloned using the enzymes XcmI and HpaI, and mutants E701A, D704A, E714A, R715A, K716A, D720A, Q733A and C734A were subcloned using enzymes BstBI and HpaI. Mutant XPF proteins were co-expressed with wild-type ERCC1 in Sf9 insect cells and purified using the same procedure used for the wild-type heterodimer.
Nuclease assays
A stem12–loop22 oligonucleotide (GCCAGCGCTCGGT22CCGAGCGCTGGC) was 5′-32P end-labeled. Nuclease reactions (15 µl) were performed in 25 mM HEPES pH 8.0, 40 mM NaCl, 10% glycerol, 0.5 mM β-mercaptoethanol, 0.1 mg/ml bovine serum albumin (BSA) and 0.4 mM MnCl2 or 2 mM MgCl2. Reaction mixtures contained 100 fmol of DNA substrate and 20–200 fmol of XPF–ERCC1 protein, and were incubated at 30°C for 2 h. Reactions using FeCl2 were performed as above, except that pH 7.0, 10–40 mM NaCl, 20–100 µM FeCl2 and 1 pmol of XPF–ERCC1 were used. Reactions were stopped by adding 15 µl of loading dye (90% formamide/10 mM EDTA) and heating at 95°C for 5 min. Samples were loaded onto 15% (19:1) denaturing polyacrylamide gels containing 0.5× TBE and run for 3 h at 200 V. Reaction products were visualized by autoradiography and quantified on a PhosphorImager (STORM860; Molecular Dynamics).
EMSAs
The same substrate that was used in the nuclease assay was used in all band-shift experiments. Per reaction, 250 fmol of substrate (of which 50 fmol were 5′-32P end-labeled) were incubated with wild-type or mutant XPF–ERCC1 protein in a reaction volume of 15 µl in 25 mM HEPES pH 8.0, 40 mM NaCl, 15% glycerol, 0.5 mM β-mercaptoethanol, 0.1 mg/ml BSA and 0.4 mM MnCl2 or 2 mM MgCl2. The variables that were investigated are described in the legend to Figure 5. After incubation for 30 min at room temperature, samples were loaded onto 4% (37.5:1) native polyacrylamide gels containing 0.5× TBE and run at 100 V for 1 h at 4°C. Gels were dried and the bands visualized by autoradiography.
Acknowledgments
Acknowledgements
We thank Peter Hunziker and Ragna Sack for N-terminal sequencing; Wouter de Laat, Hanny Odijk and Nils Wijgers for providing XPF–ERCC1 expression plasmids; and members of the Schärer laboratory, Josef Jiricny, Primo Schär and Hanspeter Nägeli for continuous support and helpful discussion. This work was supported by the Swiss National Science Foundation, the Human Frontier Science Program and the EMBO Young Investigator Program.
References
- Aboussekhra A. et al. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell, 80, 859–868. [DOI] [PubMed] [Google Scholar]
- Araujo S.J., Nigg,E.A. and Wood,R.D. (2001) Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol., 21, 2281–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Walker,D.R. and Koonin,E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell A.J., Bardwell,L., Tomkinson,A.E. and Friedberg,E.C. (1994) Specific cleavage of model recombination and repair intermediates by the yeast Rad1–Rad10 DNA endonuclease. Science, 265, 2082–2085. [DOI] [PubMed] [Google Scholar]
- Biggerstaff M., Szymkowski,D.E. and Wood,R.D. (1993) Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro. EMBO J., 12, 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard,P.-H.L., McDonald,W.H., Shanahan,P., Yates,J.R.,III and Russell,P. (2001) Mus81–Eme1 are essential components of a Holliday junction resolvase. Cell, 107, 537–548. [DOI] [PubMed] [Google Scholar]
- Bootsma D., Kraemer,K.H., Cleavers,J.E. and Hoeijmakers,J.H.J. (1998) Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. In Vogelstein,B. and Kinzler,K.W. (eds), The Genetic Basis of Human Cancer. McGraw-Hill, New York, NY, pp. 245–274.
- Cao W. and Barany,F. (1998) Identification of TaqI endonuclease active site residues by Fe2+-mediated oxidative cleavage. J. Biol. Chem., 273, 33002–33010. [DOI] [PubMed] [Google Scholar]
- Chen X.-B. et al. (2001) Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell, 8, 1117–1127. [DOI] [PubMed] [Google Scholar]
- Chevalier B.S. and Stoddard,B.L. (2001) Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res., 29, 3757–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan J.A. (1998) Metal activation of enzymes in nucleic acid biochemistry. Chem. Rev., 98, 1067–1087. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Appeldoorn,E., Sugasawa,K., Weterings,E., Jaspers,N.G. and Hoeijmakers,J.H. (1998a) DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev., 12, 2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W.L., Sijbers,A.M., Odijk,H., Jaspers,N.G. and Hoeijmakers,J.H. (1998b) Mapping of interaction domains between human repair proteins ERCC1 and XPF. Nucleic Acids Res., 26, 4146–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W.L., Appeldoorn,E., Jaspers,N.G.J. and Hoeijmakers,J.H.J. (1998c) DNA structural elements required for ERCC1–XPF endonuclease activity. J. Biol. Chem., 273, 7835–7842. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Jaspers,N.G. and Hoeijmakers,J.H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- Evans E., Moggs,J.G., Hwang,J.R., Egly,J.M. and Wood,R.D. (1997) Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J., 16, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J. and Haber,J.E. (1992) Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science, 258, 480–484. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society for Microbiology, Washington, DC.
- Gaillard P.H. and Wood,R.D. (2001) Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res., 29, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavaty J.J., Benner,J.S., Hornstra,L.J. and Schildkraut,I. (2000) Identification of the metal-binding sites of restriction endonucleases by Fe2+-mediated oxidative cleavage. Biochemistry, 39, 3097–3105. [DOI] [PubMed] [Google Scholar]
- Hosfield D.J., Mol,C.D., Shen,B. and Tainer,J.A. (1998) Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell, 95, 135–146. [DOI] [PubMed] [Google Scholar]
- Houtsmuller A.B., Rademakers,S., Nigg,A.L., Hoogstraten,D., Hoeijmakers,J.H. and Vermeulen,W. (1999) Action of DNA repair endonuclease ERCC1/XPF in living cells. Science, 284, 958–961. [DOI] [PubMed] [Google Scholar]
- Hoy C.A., Thompson,L.H., Mooney,C.L. and Salazar,E.P. (1985) Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res., 45, 1737–1743. [PubMed] [Google Scholar]
- Interthal H. and Heyer,W.D. (2000) MUS81 encodes a novel helix–hairpin–helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet., 263, 812–827. [DOI] [PubMed] [Google Scholar]
- Kaliraman V., Mullen,J.R., Fricke,W.M., Bastin-Shanower,S.A. and Brill,S.J. (2001) Functinoal overlap between Sgs1–Top3 and the Mms4–Mus81 endonuclease. Genes Dev., 15, 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.R., Dai,Y., Mundy,C.L., Yang,W. and Oettinger,M.A. (1999) Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev., 13, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka I., Kobertz,W.R., Ariza,R.R., Biggerstaff,M., Essigmann,J.M. and Wood,R.D. (2000) Repair of an interstrand DNA crosslink initiated by ERCC1–XPF repair/recombination nuclease. J. Biol. Chem., 275, 26632–26636 [DOI] [PubMed] [Google Scholar]
- Li L., Elledge,S.J., Peterson,C.A., Bales,E.S. and Legerski,R.J. (1994) Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl Acad. Sci. USA, 91, 5012–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Peterson,C.A., Lu,X. and Legerski,R.J. (1995) Mutations in XPA that prevent association with ERCC1 are defective in nucleotide excision repair. Mol. Cell. Biol., 15, 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Garrett,R.A. and Kjems,J. (1997) Mapping metal ions at the catalytic centres of two intron-encoded endonucleases. EMBO J., 16, 3272–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchen-Maloney S.L., Giannecchini,C.A., Hwang,M.H. and Thelen,M.P. (1999) Domain mapping of the DNA binding, endonuclease, and ERCC1 binding properties of the human DNA repair protein XPF. Biochemistry, 38, 9417–9425. [DOI] [PubMed] [Google Scholar]
- McWhir J., Selfridge,J., Harrison,D.J., Squires,S. and Melton,D.W. (1993) Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nature Genet., 5, 217–224. [DOI] [PubMed] [Google Scholar]
- Missura M., Buterin,T., Hindges,R., Hubscher,U., Kasparkova,J., Brabec,V. and Naegeli,H. (2001) Double-check probing of DNA bending and unwinding by XPA–RPA: an architectural function in DNA repair. EMBO J., 20, 3554–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D., Park,C.H., Matsunaga,T., Hsu,D.S., Reardon,J.T. and Sancar,A. (1995) Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem., 270, 2415–2418. [DOI] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman,V., Ibrahim,S.S. and Brill,S.J. (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics, 157, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L.J., Essers,J., Weeda,G., Berverloo,B., de Wit,J., Muitjens,M., Odijk,H., Hoeijmakers,J.H.J. and Kanaar,R. (2001) The structure-specific endonuclease ERCC1/XPF is rquired for targeted gene replacement in embryonic stem cells. EMBO J., 20, 6540–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan A., Davies,A.A., Moggs,J.G., West,S.C. and Wood,R.D. (1994) XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature, 371, 432–435. [DOI] [PubMed] [Google Scholar]
- Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon J.T., Thompson,L.H. and Sancar,A. (1993) Excision repair in man and the molecular basis of xeroderma pigmentosum syndrome. Cold Spring Harb. Symp. Quant. Biol., 58, 605–617. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- Sargent R.G., Meservy,J.L., Perkins,B.D., Kilburn,A.E., Intody,Z., Adair,G.M., Nairn,R.S. and Wilson,J.H. (2000) Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res., 28, 3771–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgouros J., Gaillard,P.H. and Wood,R.D. (1999) A relationship between a DNA-repair/recombination nuclease family and archaeal helicases. Trends Biochem. Sci., 24, 95–97. [DOI] [PubMed] [Google Scholar]
- Sijbers A.M. et al. (1996a) Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell, 86, 811–822. [DOI] [PubMed] [Google Scholar]
- Sijbers A.M., van der Spek,P.J., Odijk,H., van den Berg,J., van Duin,M., Westerveld,A., Jaspers,N.G., Bootsma,D. and Hoeijmakers,J.H. (1996b) Mutational analysis of the human nucleotide excision repair gene ERCC1. Nucleic Acids Res., 24, 3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Ng,J.M., Masutani,C., Iwai,S., van der Spek,P.J., Eker,A.P., Hanaoka,F., Bootsma,D. and Hoeijmakers,J.H. (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell, 2, 223–232. [DOI] [PubMed] [Google Scholar]
- van Vuuren A.J., Appeldoorn,E., Odijk,H., Yasui,A., Jaspers,N.G., Bootsma,D. and Hoeijmakers,J.H. (1993) Evidence for a repair enzyme complex involving ERCC1 and complementing activities of ERCC4, ERCC11 and xeroderma pigmentosum group F. EMBO J., 12, 3693–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M. et al. (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell, 8, 213–224. [DOI] [PubMed] [Google Scholar]
- Wakasugi M., Reardon,J.T. and Sancar,A. (1997) The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J. Biol. Chem., 272, 16030–16034. [DOI] [PubMed] [Google Scholar]
- Weeda G. et al. (1997) Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol., 7, 427–439. [DOI] [PubMed] [Google Scholar]
- Wood R.D. (1997) Nucleotide excision repair in mammalian cells. J. Biol. Chem., 272, 23465–23468. [DOI] [PubMed] [Google Scholar]
- Yagi T., Wood,R.D. and Takebe,H. (1997) A low content of ERCC1 and a 120 kDa protein is a frequent feature of group F xeroderma pigmentosum fibroblast cells. Mutagenesis, 12, 41–44. [DOI] [PubMed] [Google Scholar]
- Yokoi M., Masutani,C., Maekawa,T., Sugasawa,K., Ohkuma,Y. and Hanaoka,F. (2000) The xeroderma pigmentosum group C protein complex XPC–HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem., 275, 9870–9875. [DOI] [PubMed] [Google Scholar]
- Zaychikov E. et al. (1996) Mapping of catalytic residues in the RNA polymerase active center. Science, 273, 107–109. [DOI] [PubMed] [Google Scholar]