Abstract
An investigation was made into the oligomerization, the ability to form pores and the secretion-related properties of the 45 kDa C-terminal domain of the IgA protease (C-IgAP) from Neisseria gonorrhoeae. This protease is the best studied example of the autotransporters (ATs), a large family of exoproteins from Gram-negative bacteria that includes numerous virulence factors from human pathogens. These proteins contain an N-terminal passenger domain that em bodies the secreted polypeptide, while the C-domain inserts into the outer membrane (OM) and trans locates the linked N-module into the extracellular medium. Here we report that purified C-IgAP forms an oligomeric complex of ∼500 kDa with a ring-like structure containing a central cavity of ∼2 nm diameter that is the conduit for the export of the N-domains. These data overcome the previous model for ATs, which postulated the passage of the N-module through the hydrophilic channel of the β-barrel of each monomeric C-domain. Our results advocate a secretion mechanism not unlike other bacterial export systems, such as the secretins or fimbrial ushers, which rely on multimeric complexes assembled in the OM.
Keywords: autotransporters/IgA protease/Neisseria gonorrhoeae/protein secretion
Introduction
Proteins secreted by Gram-negative bacteria must cross the lipid bilayers of the inner (IM) and outer (OM) mem branes as well as the periplasmic space containing the peptidoglycan layer (Nikaido, 1996; Duong et al., 1997; Bernstein, 2000). To achieve this formidable task, different secretion systems have evolved (Lory, 1998; Thanassi and Hultgren, 2000). Some involve the initial translocation of a protein precursor with an N-terminal signal peptide into the periplasmic space (e.g. type II and chaperone/usher pathways) (Russel, 1998; Thanassi et al., 1998a; Soto and Hultgren, 1999; Sandkvist, 2001). The secreted polypeptide, either alone or in association with a periplasmic chaperone, can then be recognized by an OM protein complex that promotes its final translocation to the extracellular medium. Alternatively, some polypeptides are secreted without a periplasmic intermediate by passing directly from the cytoplasm into the extracellular medium through a hydrophilic protein channel connecting the IM and OM (e.g. type I and type III pathways) (Blight and Holland, 1994; Thanabalu et al., 1998; Cheng and Scheneewind, 2000; Buchanan, 2001; Fernández and de Lorenzo, 2001; Plano et al., 2001). A common feature of all these pathways is the participation of several gene products in the assembly of a secretion apparatus, usually some 3–20 polypeptides with at least one being an OM oligomer containing a central hydrophilic pore (Russel, 1998; Stathopoulos et al., 2000).
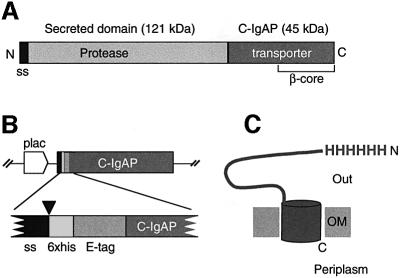
Autotransporters (ATs) are a distinct family of secreted proteins which contain all the necessary elements for translocation across the OM within their own polypeptide sequences (Henderson et al., 1998, 2000). Members of this family include virulence factors of important human pathogens, such as the IgA1 proteases from Neisseria gonorrhoeae, Neisseria meningitidis and Haemophilus influenzae (Pohlner et al., 1987; Lomholt et al., 1995), the actin polymerization factor IcsA from Shigella flexneri (Suzuki et al., 1995), the AIDA-I adhesin from pathogenic Escherichia coli (Benz and Schmidt, 1992; Suhr et al., 1996), the serum-resistant factor BrkA from Bordetella pertussis (Fernandez and Weiss, 1994) and the cytotoxin VacA from Helicobacter pylori (Schmitt and Haas, 1994) among others (Henderson and Nataro, 2001). The fundamental mechanism of AT secretion was outlined by studies of the IgA1 protease (IgAP) from N.gonorroheae (Pohlner et al., 1987). The iga gene encodes a large precursor (∼170 kDa) in which the mature protease (∼106 kDa) is flanked by an N-terminal signal peptide directing the initial export into the periplasm, and an ∼45 kDa (C-IgAP) C-terminal domain starting at Val1124 that inserts into the OM and which is required for translocation of the protease domain toward the bacterial surface (Figure 1A). During this step, autoproteolytic processing releases the mature protease into the extracellular medium. These studies showed that C-IgAP could translocate a heterologous passenger domain lacking disulfide bonds towards the bacterial surface (i.e. the 15 kDa cholera toxin B subunit) (Klauser et al., 1990, 1992). Protection experiments against externally added proteases indicated that the last ∼30 kDa of C-IgAP, the so-called β-core, are embedded in the OM in vivo (Klauser et al., 1993). Predictions of the secondary structure of C-IgAP suggested that this β-core may contain 15 amphipathic β-strands that could fold as a β-barrel similar to those found in outer membrane proteins (OMPs) (e.g. OmpA, OmpF, PhoE, LamB, FhuA, FepA and TolC; for reviews see Koebnik et al., 2000; Tamm et al., 2001).
Fig. 1. Structural organization of the IgA protease and HEβ hybrid. (A) Schematic representation of the IgA protease precursor (∼170 kDa) from N.gonorroheae showing the position of the N-terminal signal sequence (ss), the secreted protease module (∼120 kDa) and the transporter C-domain (∼45 kDa; C-IgAP). The ∼30 kDa β-core region within C-IgAP is also labeled. (B) Organization of the relevant insert of plasmid pHEβ encoding the HEβ hybrid under the control of the IPTG-inducible lac promoter (plac). The heb gene chimera is an in-frame fusion of DNA segments encoding the pelB N-terminal signal sequence (ss), polyhistidine (6×his) and E-tag peptides, and the C-IgAP transporter domain. The site of cleavage of the signal peptidase (just before the His6 peptide) is indicated by an arrowhead. (C) Representation of topology of HEβ in the OM showing the His6 N-passenger domain exposed to the extracellular space (Out) and the C-terminus toward the periplasm. The putative β-barrel of HEβ is depicted as a cylinder embedded in the OM.
The above led to the proposal of an elegant model for AT secretion in which the β-barrel of a monomeric C-domain contained a hydrophilic channel through which an unfolded N-domain could be translocated (Klauser et al., 1990, 1992). However, this model has received no experimental support. Indeed, it has been challenged recently in view of new data showing that homologous and heterologous N-domains can be translocated after folding by the action of periplasmic chaperones (Veiga et al., 1999; Valls et al., 2000; Brandon and Goldberg, 2001). These results raise the question of the actual size and nature of the pore used for secretion in ATs. The present work investigates the structural and functional properties of the transporter C-IgAP domain from N.gonorroheae. The results show that C-IgAP assembles as a ring-like oligomeric complex of ∼500 kDa with a central hydrophilic pore of ∼2 nm diameter. This structure is similar to that of the OM complexes found in other secretion systems (e.g. secretins and fimbrial ushers; Thanassi et al., 1998b; Stathopoulos et al., 2000). Furthermore, evidence was obtained indicating that the central pore of the C-IgAP complex is the site used for the secretion of the N-domains.
Results
Purification of native C-IgAP
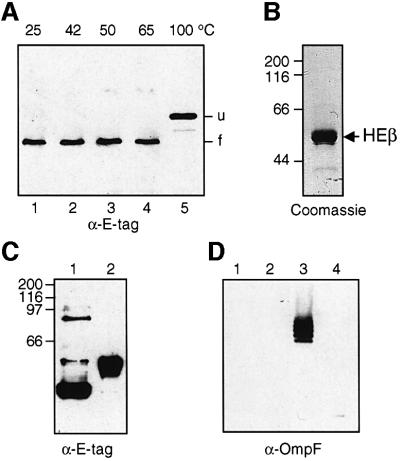
A gene construct encoding a functional polyhistidine-tagged version of the 45 kDa C-IgAP was generated. This hybrid protein, named HEβ (∼50 kDa), contained an N-terminal signal peptide (pelB) followed by six histidines (His6) and a 12 amino acid peptide (E-tag) fused to C-IgAP (Figure 1B and C). When produced into E.coli, HEβ was localized in the OM fraction, with the His6 and E-tag peptides fully exposed toward the bacterial surface [according to whole-cell enzyme-linked immunosorbent assay (ELISA) and digestion with externally added proteases; data not shown]. HEβ showed strong resistance to denaturation in the presence of 1% SDS unless the temperature was increased to 100°C. This was revealed by the two distinct electrophoretic mobilities for HEβ observed in SDS–PAGE, with migration as a faster protein band if the sample was not boiled (Figure 2A). The heat-modifiable electrophoretic mobility of HEβ is a typical feature of OMPs with β-barrel structure, with the folded conformation (more compact) migrating more quickly in polyacrylamide gels than the denatured form (Schnaitman, 1973; Nikaido and Vaara, 1985). Advantage was taken of this behavior to monitor the folding state of HEβ throughout its purification.
Fig. 2. Electrophoretic mobility and purification of HEβ. (A) Western blot developed with anti-E-tag mAb–POD of whole-cell protein extracts from IPTG-induced E.coli UT5600 harboring pHEβ. Before loading onto the 10% polyacrylamide gel, the samples were resuspended in denaturing SDS–PAGE sample buffer and heated for 10 min at the indicated temperatures (25, 42, 50, 65 and 100°C). The faster mobility band of HEβ (f) corresponds to the folded conformation of the hybrid. When unfolded, HEβ migrates as a slower mobility band (u). (B) A sample of purified HEβ is shown after Coomassie Blue staining of a 10% SDS–polyacrylamide gel. (C) Western blot developed with anti- E-tag mAb–POD of purified HEβ samples treated at 25 (lane 1) or 100°C (lane 2) for 10 min before loading onto a 10% SDS– polyacrylamide gel. (D) The possible presence of contaminating OmpF porin in the purified HEβ was evaluated by western blot developed with a polyclonal serum against trimeric OmpF. Excess purified HEβ (10 µg, lanes 1 and 2) and a sample of purified OmpF (0.1 µg, lanes 3 and 4) as a control were loaded onto a 10% SDS–polyacrylamide gel after heating at 25 (lanes 1 and 3) or 100°C (lanes 2 and 4) for 10 min. Only the trimeric OmpF control (lane 3) was detected, ruling out the presence of OmpF in the purified HEβ sample.
To this end, HEβ was produced in E.coli UT5600, a strain lacking the OM protease OmpT (Grodberg and Dunn, 1988). After isolation of the OM fraction from these cells, the OMPs were solubilized in a buffer containing 1% Zwittergent 3–14 and passed through a cobalt-containing resin for immobilized metal affinity chromatography (IMAC). This detergent was instrumental in solubilizing HEβ without affecting its heat-modifiable electrophoretic mobility, even after extended incubations. Solubilized HEβ bound specifically to the IMAC column and was eluted in a buffer containing imidazole and 0.1% Zwittergent 3–14. Finally, the imidazole was removed by dialysis and HEβ was concentrated (see Materials and methods). This purification procedure gave a protein band corresponding to HEβ after Coomassie Blue staining of a denaturing SDS–polyacrylamide gel (Figure 2B) and western blotting with anti-E-tag monoclonal antibody (mAb) (Figure 2C). The folding of purified HEβ was tested by its heat-modifiable mobility in SDS–PAGE (Figure 2C). In this analysis, it was found that the non-boiled sample of HEβ generated bands of high molecular weight (Figure 2C, lane 1). As an additional purity criterion, western blots were used to show that the HEβ preparation contained no detectable trace of the major E.coli porin, OmpF (Figure 2D).
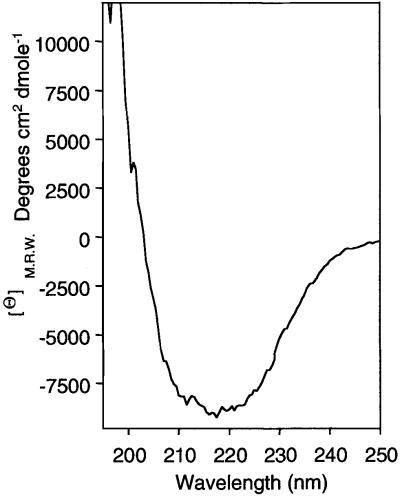
The secondary structure of purified HEβ was investigated using circular dichroism (CD). The CD spectrum obtained using 0.1 mg/ml of HEβ in a buffer containing 0.1% Zwittergent 3–14 is shown in Figure 3. The structural components derived from this spectrum (Perczel et al., 1992) indicate that HEβ contains 30.2% β-sheet conformation, 34.4% α-helix structure, 9.8% turns and 25.6% unordered structure. This analysis is compatible with the presence of 15 amphipathic β-strands (Klauser et al., 1993), which represent 30% of the HEβ sequence, but reveals a rich α-helical content in C-IgAP. In addition, these data confirmed that HEβ remained in a structured state after purification.
Fig. 3. CD spectrum of HEβ. The CD spectrum of purified HEβ (0.1 mg/ml) was monitored at 22°C in TNZ buffer [20 mM Tris–HCl pH 8.0, 10 mM NaCl, 0.1% (w/v) Zwittergent 3–14]. A minimum of four spectra were accumulated and the contribution of the buffer subtracted. Values of mean residue weight ellipticities (Θ) M.R.W. (degrees × cm2 × dmol–1) are indicated.
Biochemical measurement of the pore in C-IgAP
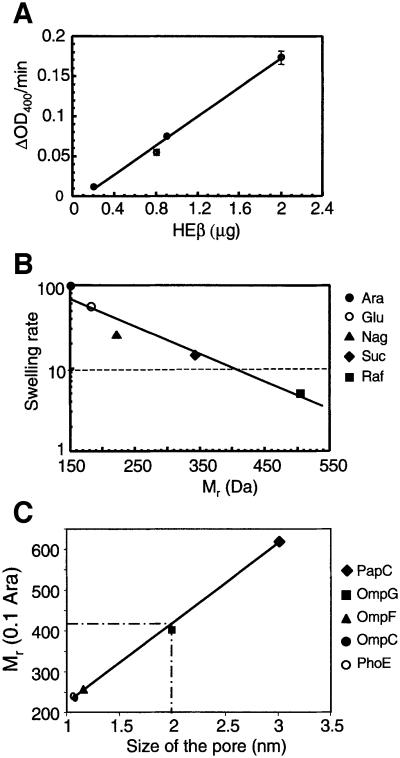
The possible presence of a hydrophilic conduit in HEβ was examined using the proteoliposome swelling assay, which has been used to study the pore-forming activity of porins (Nikaido and Rosenberg, 1981; Nikaido et al., 1991; Nikaido, 1994). Briefly, purified HEβ was embedded into multilamellar liposomes suspended in solutions containing an iso-osmotic concentration of sugars of different Mr. If a hydrophilic pore were present in HEβ, the sugars, concomitant with water, would enter into the liposomes, producing their swelling. This can be monitored as a decrease in the OD400. Pore-forming activity was clearly detected in the proteoliposomes containing HEβ and suspended in arabinose solutions (Mr ∼150 Da). Porin activity was directly proportional to the amount of HEβ used (Figure 4A). Addition of 2 nmol of E.coli lipopolysaccharide (LPS) increased the absolute swelling rate of the liposomes containing HEβ by ∼3-fold (e.g. from 0.14 to 0.4 ΔOD400/min at 1.4 µg of HEβ in arabinose). No swelling was noticed in proteoliposomes reconstituted with bovine serum albumin (BSA; negative control), whereas those with OmpF (positive control) had swelling rates identical to those previously reported (Nikaido and Rosenberg, 1981; Nikaido et al., 1991).
Fig. 4. Pore-forming activity of HEβ. (A) Swelling rates (ΔOD400/min) of proteoliposomes suspended in isosmotic solutions of arabinose and containing the indicated amount of purified HEβ. (B) Swelling rates of proteoliposomes containing HEβ in solutions of sugars with different Mr. The sugars used were arabinose (Ara; 150 Da), glucose (Glu; 180 Da), N-acetylglucosamine (Nag; 221 Da), sucrose (Suc; 342 Da) and raffinose (Raf; 504 Da). The data are shown relative to the swelling in arabinose and are the averages of at least six independent experiments in which a range of amounts of HEβ (from 0.8 to 2.5 µg) was employed. The swelling rate corresponding to 10% of that in arabinose is indicated with a dashed line. (C) The Mr (0.1 Ara) of HEβ (410 Da) was intersected in a plot representing the Mr (0.1 Ara) of OMPs versus their pore size (PhoE, 240 Da and 1.1 nm; OmpC, 236 Da and 1.1 nm; OmpF, 255 Da and 1.2 nm; OmpG, 400 Da and 2.0 nm; PapC, 620 Da and 3 nm) (Nikaido and Rosenberg, 1983; Cowan et al., 1992; Fajardo et al., 1998; Thanassi et al., 1998b). This plot allows an estimation of 2 nm for the size of HEβ.
The size of the channel in HEβ was estimated from the swelling rates with sugars of different Mr in the presence or absence of LPS. When these rates were expressed relative to those obtained with arabinose, the sugar with the lowest Mr, it was apparent that the different sugars had identical relative values in either the presence or absence of LPS (Table I). The relative swelling rates were plotted versus the Mr of the sugars employed to obtain the parameter Mr (0.1 Ara) for HEβ of 410 Da (Figure 4B). This parameter, defined as the Mr of a solute that would diffuse at 10% of the rate of arabinose, has been used extensively to calculate the diameter of channels in OMPs (Nikaido et al., 1991). Comparing the Mr (0.1 Ara) of HEβ with that of other OMPs of known pore size, the hydrophilic channel of the C-IgAP was estimated to have a diameter of ∼2 nm (Figure 4C).
Table I. Relative swelling rates of liposomes containing HEβ (%).
| Sugar | Mr | LPS |
||
|---|---|---|---|---|
| – | + | |||
| Arabinose | 150 | 100 | 100 | |
| Glucose | 180 | 60 | 59 | |
| N-acetylglucosamine | 221 | 20 | 23 | |
| Sucrose | 342 | 15 | 12 | |
| Raffinose | 504 | not tested | 5 |
Thedatapresentedare the average of at least three independent experi mentsinwhichtriplicatesofeachpoint were measured.The typical deviationwasalways<17%oftheaveragevalues.Therateswere normalized to that in arabinose, which was taken as 100%.
To confirm the porin activity detected in HEβ, a different C-IgAP hybrid (Corβ) was used that contained a 10 amino acid viral epitope as the N-domain (Veiga et al., 1999). In addition, it was investigated whether the presence of a sizable N-domain—a recombinant single-chain Fv (scFv) antibody with two disulfide bonds (Mr ∼30 kDa) (Veiga et al., 1999)—influenced the pore-forming activity of the C-IgAP. In these experiments, the hybrid proteins Corβ and FvHβ were expressed in E.coli HN705, a strain lacking the OmpC and OmpF porins (Sugawara and Nikaido, 1992). The OM fractions from these cells were isolated by isopycnic centrifugation and identical amounts of total protein of these OM fractions were used in the liposome swelling assays (see Materials and methods). This E.coli strain reproducibly gave rise to low levels of Corβ and FvHβ hybrids, detected in the OM fractions by western blotting with anti-E-tag mAb. For unknown reasons, the amount of FvHβ was always ∼100-fold higher than that of Corβ in E.coli HN705. Despite these facts, the swelling of the liposomes containing Corβ and FvHβ was readily detectable in arabinose and sucrose, whereas the OM fraction from the non-transformed E.coli HN705 strain induced no significant liposome swelling in sucrose (Table II). Since FvHβ was present in greater amounts than Corβ in these OM fractions, and pore-forming activity was directly proportional to the amount of protein added (Figure 4A), it is very likely that the presence of a bulky scFv N-domain blocked most of the porin activity of C-IgAP. In a previous study, we have shown that a small proportion of FvHβ hybrids (∼15%) fully translocate the folded scFv across the OM, while the remaining hybrids (∼85%) have their N-domain within the OM fraction (Veiga et al., 1999).
Table II. Swelling rates of liposomes containing OM fractions.
| Escherichia coli cells | Arabinose | Sucrose |
|---|---|---|
| HN705 | 0.26 | U |
| HN705/Corβ | 0.4 | 0.04 |
| HN705/FvHβ | 0.47 | 0.05 |
Values presented came from at least three different measurements. OM fractions containing 60 µg of total protein were added to liposomes. The typical deviation was always <20% of the average values.
U = undetectable.
C-IgAP forms an oligomeric complex in the OM
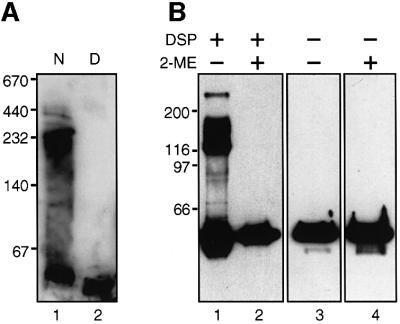
The presence of high Mr bands in the purified sample of HEβ (see Figure 2C) prompted the investigation of whether native C-IgAP forms any sort of oligomers. The first clue pointing in this direction was obtained by electrophoresis of native HEβ in non-denaturing gels (without SDS) with a gradient of 4–15% polyacrylamide. Native HEβ, solubilized in 0.1% Zwittergent 3–14, ran in these gels as a high Mr band with a minimum mass of ∼250 kDa (Figure 5A, lane 1) detected by anti-E-tag mAb. In contrast, after boiling in SDS, HEβ displayed the mobility expected for the monomer (∼50 kDa; Figure 5A, lane 2).
Fig. 5. Oligomeric forms of C-IgAP in polyacrylamide gels. (A) Western blot probed with anti-E-tag mAb–POD of a native gel containing a 4–15% gradient of polyacrylamide. Purified HEβ was resuspended in native PAGE sample buffer (lane 1, N) or the same buffer containing 1% (w/v) SDS (lane 2, D). The sample in lane 2 was boiled for 10 min before loading. (B) Western blot probed with anti-E-tag mAb–POD of total protein extracts from E.coli UT5600 cells expressing HEβ and incubated in vivo (or not) with DSP cross-linker (lanes 1 and 2). When indicated, the protein extracts were resuspended in SDS–PAGE sample buffer containing 5% (v/v) 2-ME (lanes 2 and 4) in order to reduce a disulfide bridge in the cross-linker. All samples were boiled for 10 min before loading.
Evidence of the oligomeric form of HEβ in vivo was gained by cross-linking with the thiol-cleavable, homobifunctional amine-reactive reagent dithiolbis(succinimidyl propionate) (DSP, 12 Å spacer). Escherichia coli cells producing HEβ were incubated with DSP for 30 min at room temperature (see Materials and methods). After this period, the DSP was quenched and the cells boiled in SDS–PAGE sample buffer containing, or not containing, 2-mercaptoethanol (2-ME). Protein bands of HEβ with ∼55, ∼115, ∼175 and >250 kDa were detected in denaturing SDS–polyacrylamide gels by western blot with anti-E-tag mAb (Figure 5B, lane 1), which probably correspond to a monomer, dimer, trimer and at least one tetramer of HEβ. As expected, these high Mr bands appeared after treatment with DSP and were dissociated with 2-ME (Figure 5B, lanes 2–4). This pattern of cross-linking of HEβ in vivo was confirmed with a different reagent, disuccinimidyl glutarate (DSG, 7.7 Å spacer) (data not shown).
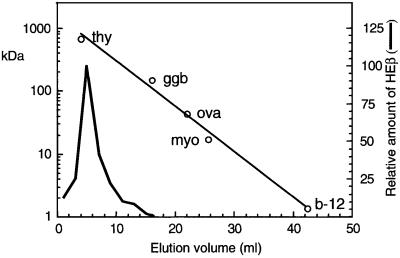
The mass of the oligomeric complex formed by HEβ was assessed by size exclusion chromatography. The elution profile of HEβ in a gel filtration column with an exclusion limit of 1500 kDa (Bio-Gel A-1.5m) is provided in Figure 6. The quantity of HEβ in the different fractions was determined by western blot with anti-E-tag mAb. Proteins of known mass were separated along with HEβ as size markers for generating a standard curve. HEβ eluted as a single peak with an apparent mass of ∼500 kDa. Remarkably, HEβ was not detected in the fractions corresponding to the mass of the monomer (50 kDa). This result was confirmed with three independent preparations of purified HEβ and employing a different chromatographic medium (Macro-Prep SE 1000/40).
Fig. 6. Size exclusion chromatography of purified HEβ. The elution profile in a Bio-Gel A column of HEβ and proteins of known Mr as standards [thyroglobulin (thy; 670 000 Da), bovine γ-globulin (ggb; 158 000 Da), chicken ovalbumin (ova; 44 000 Da), equine myoglobin (myo; 17 000 Da) and vitamin B-12 (b-12; 1350 Da)] is shown. Elution of the Mr standards was monitored by UV absorption at 280 nm (open circles), whereas HEβ was detected by western blotting with anti-E-tag mAb–POD.
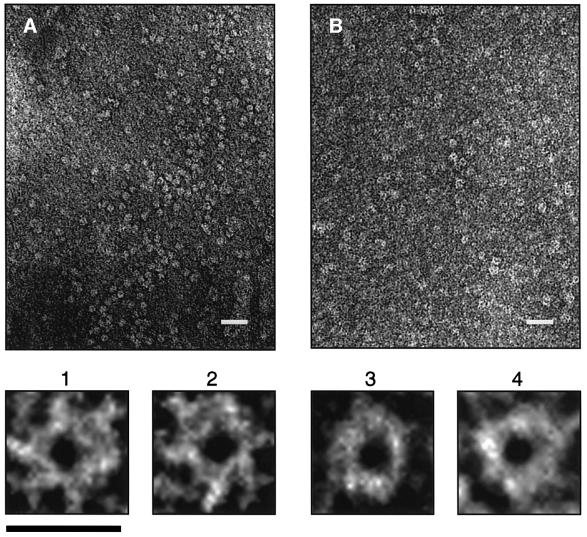
The large size estimated for the HEβ complex raised the possibility that it might be visible by electron microscopy. Thus, samples containing purified HEβ were negatively stained with ammonium molybdate (Figure 7A) or uranyl acetate (Figure 7B) and viewed with an electron microscope. In the unprocessed micrographs obtained with both negative stains, HEβ appeared as ring-shaped complexes with an external diameter of ∼9 nm and a central cavity of ∼2 nm (see magnifications 1–4 in Figure 7).
Fig. 7. Electron microscopy of C-IgAP. Electron micrographs (×60 000) of HEβ samples (0.5 mg/ml) stained with (A) 2% ammonium molybdate or (B) 2% uranyl acetate. The white bar corresponds to 35 nm. Images 1 and 2 were magnified from (A), and images 3 and 4 were magnified from (B). The black bar corresponds to 10 nm.
The N-domains of a complex are secreted through a common channel
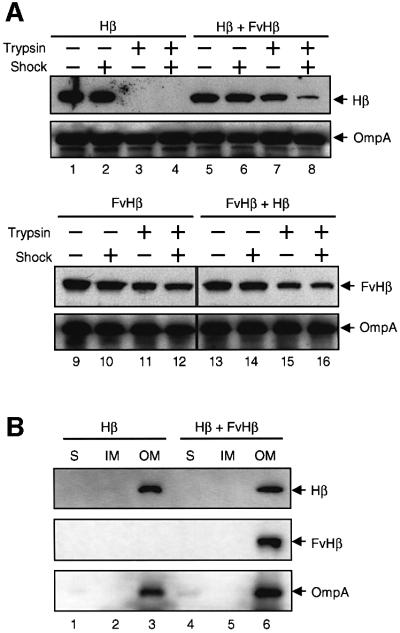
The remarkable consistency between the size of the central pore of the C-IgAP complex observed by electron microscopy and that of the hydrophilic pore measured by liposome swelling suggested that this was the actual site used for the secretion of the N-domains in the complex. To investigate this, it was determined whether the simultaneous production of two distinct N-domains could interfere with their secretion by C-IgAP. The rationale behind this experiment was the assumption that if a common pore is used, a bulky N-domain blocking it would hinder the passage of a small N-domain that would otherwise be secreted efficiently. Thus, two hybrids of C-IgAP, with N-domains with different surface display properties, were co-expressed from compatible plasmids in the same E.coli cell. One of these hybrids was FvHβ, which appeared to block the hydrophilic channel of C-IgAP (see above). The other hybrid selected was Hβ, which completely translocates its N-domain (a His6 epitope) towards the surface of E.coli. Hβ is a derivative of HEβ in which the E-tag was deleted. In this way, FvHβ and Hβ hybrids could be detected independently after their co-expression in E.coli by employing different mAbs, anti-E-tag and anti-His, respectively.
The expression and surface display of the two hybrids in E.coli UT5600 first was analyzed independently. Whole-cell ELISA with anti-His mAb (data not shown) and trypsin accessibility assays (Figure 8A, lanes 1–4) showed that ∼100% of the His6 epitopes were displayed on the surface of E.coli cells producing Hβ. This was clearly revealed by the full accessibility of the His6 peptide to externally added trypsin (compare lanes 1 and 3). In contrast, only 15–20% of the FvHβ produced in E.coli cells exposed the N-scFv on the surface, as judged by whole-cell ELISA with anti-E-tag mAb (Veiga et al., 1999) and trypsin digestion (Figure 8A, lanes 9–12). Most of the N-scFv (∼80%) was protected from proteolysis in intact cells (compare lanes 9 and 11). Importantly, trypsin was not able to degrade the N-scFv fully even after permeabilization of the OM with an EDTA shock (Figure 8A, lanes 10 and 12), indicating that this domain was probably embedded in the OM.
Fig. 8. Interference in the translocation of N-domains. (A) The C-IgAP hybrids Hβ and FvHβ were co-expressed (or expressed independently) in E.coli UT5600 cells. These cells were shocked with 10 mM EDTA or incubated with trypsin (1 µg/ml) as indicated (+). The digestion of Hβ and FvHβ was monitored by western blots using anti-His or anti- E-tag mAbs. The amount of OmpA (detected with rabbit anti-OmpA serum) was used as a loading control. (B) The subcellular location of Hβ and FvHβ in E.coli UT5600 cells producing Hβ alone or in combination with FvHβ is shown. The total protein extracts from these E.coli cells were separated into soluble (S), inner membrane (IM) and outer membrane (OM) protein fractions, as described previously (Veiga et al., 1999). OmpA was used as a control of the OM fractions. The proteins were detected by western blot as in (A).
The surface display properties of the His6 epitope in Hβ changed dramatically when the two hybrids were co-expressed in E.coli UT5600 cells. In this case, the N-passenger of Hβ was almost completely protected (∼95%) from trypsin digestion in intact E.coli cells (Figure 8A, lanes 5 and 7), clearly supporting the hypothesis that the same pore was being used by the translocation of the N-scFv and the His6 N-passenger domains. Indeed, the His6 was partially protected from the protease even when the cells were shocked with EDTA (Figure 8A, lane 8). Three independent induction experiments with E.coli cells carrying either one or both plasmids were performed to confirm these results. OmpA was used as an internal control for the loading of the gels. The subcellular location of the C-IgAP hybrids in the OM was confirmed by cellular fractionation (Figure 8B).
Discussion
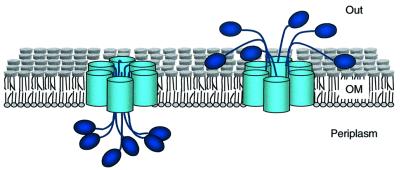
From an initial survey, ATs might appear unrelated to other bacterial secretion systems that use multiple components in the IM, the periplasm and the OM to promote the movement of proteins towards the extracellular medium (Lory, 1998; Thanassi and Hultgren, 2000). The data presented here, however, reveal that the C-domain of ATs share important structural similarities with OM complexes found in most secretion systems (Russel, 1998; Stathopoulos et al., 2000). In the OM, they form a ring-shaped multimeric complex with a central hydrophilic channel. In addition, the present data strongly support the idea that the export of N-passenger domains in ATs occurs through a common channel shared by different subunits assembled into a single complex. A new mechanism for AT secretion can therefore be proposed (summarized in Figure 9). These findings are based on the structural and functional properties obtained with the C-IgAP from N.gonorroheae, a domain used in the past to establish a model for ATs.
Fig. 9. Model for AT secretion. The AT proteins assemble as a ring-shaped oligomeric complex in the bacterial OM. A minimum of six monomers assemble in this complex. The N-terminal passenger domains (P) are depicted as ovals and the C-terminal transporter domain as cylinders anchored in the OM lipid bilayer. The N-domains are translocated from the periplasm (left) to the external medium (right) by passing through the central hydrophilic pore formed by an AT oligomeric complex.
The replacement of the protease module of IgAP with a polyhistidine-linked E-tag in HEβ allowed the purification of C-IgAP in its native form after solubilization of the OMPs of E.coli. C-IgAP showed heat-modifiable electrophoretic mobility and a CD spectrum compatible with the presence of a β-barrel in its folded structure (the β-core). The 15 amphipathic β-strands predicted in C-IgAP (Klauser et al., 1993) represent 30% of HEβ sequence, which fits with the experimental value obtained by CD. This analysis also indicated the presence of a 30% α-helical content for HEβ, which may reflect the existence of a rich α-helical structure in the region joining the N-domain of IgAP with the β-core. In the case of AIDA-I, a similar linker region was shown to be required for the translocation of the N-passenger across the OM (Maurer et al., 1999).
C-IgAP showed a clear porin activity when embedded in artificial liposomes, indicating the existence of a hydrophilic pore of 2 nm. Addition of LPS increased the net diffusion rates of solutes through C-IgAP pores. This phenomenon has also been described for the PapC fimbrial usher, and it was suggested that LPS stabilized the oligomeric state of PapC (Thanassi et al., 1998b). The pore of C-IgAP is larger than those of the typical E.coli porins OmpC (1.1 nm) and OmpF (1.2 nm), and similar to the channel found in the OM components of other protein secretion systems such us the TolC (type I) and PapC oligomers, which have a diameter between 2 and 3 nm (Thanassi et al., 1998b; Koronakis et al., 2000).
A previous report demonstrated the existence of an ion channel in the C-domain of BrkA with an average conductance of 3 nS in 1 M KCl (Shannon and Fernandez, 1999). However, in that study, C-BrkA was refolded in vitro after its solubilization from inclusion bodies in the cytoplasm of E.coli cells. This raises the question of whether this channel was present in the native form of C-BrkA and other ATs. In contrast, the present data demonstrate that a hydrophilic pore of 2 nm exists in the native conformation of C-IgAP, large enough to tolerate the passage of certain protein domains in a folded state. It is important to note that the fimbrial usher PapC, with an ∼3 nm pore, allows the passage of folded fimbrial subunits toward the bacterial surface (Thanassi et al., 1998b). In the case of C-IgAP, a good correlation was found between the size of its pore and the size of the N-domains exported efficiently. For instance, a folded immunoglobulin V-domain with a diameter of ∼2 nm is fully translocated by C-IgAP (unpublished data). However, only ∼15% of the C-IgAP hybrids containing an scFv passenger (FvHβ) export their N-domain toward the extracellular surface (Veiga et al., 1999). In an scFv, the two V-domains of an immunoglobulin are linked by a flexible peptide and associate to produce a polypeptide with an average diameter of ∼4 nm (Ay et al., 2000). Thus, it is conceivable that only a small proportion of FvHβ hybrids are exported through the 2 nm pore of C-IgAP; the rest appear to block the conduit.
The results of the liposome swelling assay show that most of the FvHβ hybrids have their hydrophilic pores blocked. This implies that the channel measured in the liposome swelling assay with C-IgAP is the same as that used for export of the N-domain. Furthermore, since the central cavity found by electron microscopy in the oligomeric C-IgAP is identical in size to the hydrophilic channel measured by liposome swelling, it is tempting to speculate that it is used for the export of the N-domains. This hypothesis was strongly supported by the interference seen in the surface display of Hβ when FvHβ was co-expressed in the same E.coli cell.
The Mr estimated for the C-IgAP complex was >250 kDa by native gel electrophoresis and ∼500 kDa by size exclusion chromatography. In addition, after in vivo cross-linking with 3% (v/v) formaldehyde, it was observed that the C-IgAP complex remained in the stacking gel during 10% SDS–PAGE (data not shown). Given the faster mobility of the native form of C-IgAP in polyacrylamide gels, and the consistency of its behavior in size exclusion chromatography, the Mr of the C-IgAP complex may be close to ∼500 kDa. This size would imply that a minimum of six (but more likely 8–10) monomers of C-IgAP are brought together into a single complex. In the secretins, the number of monomers that form the secretion complex can vary between 12 and 14 (Russel, 1998; Sandkvist, 2001).
The arrangement of C-IgAP as a multimeric ring-shaped complex with a central cavity has obvious similarities to the structure of secretins (Stathopoulos et al., 2000). However, a number of differences, including the lack of sequence homology (Genin and Boucher, 1994), suggest that they are only distantly related. For instance, the central pore of the C-IgAP complex is narrower than those typically found in the secretins of type II and type III systems (between 5 and 7 nm) (Stathopoulos et al., 2000). In addition, the external diameter of the C-IgAP complex (9 nm) is smaller than that of the secretins (i.e. 16.5 nm for PilQ from N.meningitidis (Collins et al., 2001), 19.8 nm for XcpQ from Pseudomonas aeruginosa (Bitter et al., 1998) and 20 nm for the PulD–PulS complex from Klebsiella oxytoca (Nouwen et al., 1999, 2000). Finally, the C-IgAP complex has a lower resistance to denaturation in SDS than that displayed by secretin complexes (Guilvout et al., 1999). Therefore, ATs are not members of the secretin protein family, although these two groups share important structural features. ATs, fimbrial ushers and secretins may be considered analogous secretin-like complexes specialized for the export of polypeptides across the OM. Further work is needed to clarify whether this structural relationship is reflected in their mechanism of function.
Materials and methods
Bacterial strains and growth conditions
The E.coli K-12 strains used were UT5600 (ΔompT proC leu-6 trpE38 entA) (Grodberg and Dunn, 1988), HN705 [Δ(lac-proAB) ΔompC ompF::Tn5 zei-298::Tn10 supE rpsL F′(traD36 proAB+ lacIq lacZΔM15)] (Sugawara and Nikaido, 1992) and XL-1 Blue [endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 lac F′(proAB lacIq lacZΔM15 Tn10)] (Bullock et al., 1987) (Stratagene). Escherichia coli XL-1 Blue was employed for gene cloning. All bacteria were grown at 30°C in LB agar plates (Miller, 1992) containing 2% (w/v) glucose (for repression of the lac promoter) and appropriate antibiotics for plasmid selection. The antibiotics used were chloramphenicol (Cm) at 40 µg/ml and spectinomycin (Sp) at 50 µg/ml. For induction of genes encoding C-IgAP hybrids, single colonies were inoculated in liquid LB or 2×YT medium (Miller, 1992) containing 2% (w/v) glucose and antibiotics, and grown at 30°C until OD600 ∼0.5. At this point, bacteria were harvested by centrifugation, resuspended at the same density in fresh liquid medium (LB or 2×YT) containing antibiotics and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and incubated further at 30°C for 3 h. For purification of HEβ, E.coli UT5600 cells harboring pHEβ were induced with IPTG for 16 h.
Plasmids and oligonucleotides
All DNA was manipulated using standard methods (Ausubel et al., 1994). Plasmids pFvHβ (Cmr) and pCorβ (Cmr) have been described elsewhere (Veiga et al., 1999). pHEβ (Cmr) was constructed by fusing the His6 linker to the 5.6 kb fragment resulting from SfiI digestion of pFvHβ. The His6 linker was annealed with the oligonucleotides HisA (5′-CGGCCATGGCGCATCACCATCACCATCACGCGGCCGCTGCGG-3′) and HisB (5′-CAGCGGCCGCGTGATGGTGATGGTGATGCGCCATGGCCGGCT-3′) obtained from Genemed Synthesis, Inc. Plasmid pHβ (Cmr) was made by removing the DNA sequence coding for the E-tag epitope from pHEβ. To this end, pHEβ was double-digested with EcoRI and SacII, and re-ligated in the presence of the ΔE linker (which has cohesive ends for EcoRI and SacII), allowing the in-frame translation of Hβ. The ΔE linker was made by annealing the oligonucleotides ΔE1 (5′-AATTCGAGCTCCTGC-3′) and ΔE2 (5′-AGGAGCTGC-3′). To co-express FvHβ and Hβ in E.coli, the plasmid pVFvHβ (Spr), a derivative of the low-copy plasmid vector pVLT35 (Spr) (de Lorenzo et al., 1993), was constructed. Plasmid pVFvHβ contained the 2.2 kb XbaI–HindIII DNA fragment from pFvHβ (encoding FvHβ) cloned into the same sites of pVLT35. Plasmids pVFvHβ (Spr) and pHβ (Cmr) have compatible origins of replication from pMMB207 (Morales et al., 1991) and pBR322 (Bolivar et al., 1977), respectively. The expression of FvHβ is under tac promoter control in pVFvHβ.
Purification of HEβ
Escherichia coli UT5600 (pHEβ) cells were grown in 3 l of 2×YT medium at 30°C and induced with 1 mM IPTG for 16 h. Cells were subsequently harvested by centrifugation (4000 g, 10 min) and resuspended in buffer TN (20 mM Tris–HCl pH 8.0, 10 mM NaCl) containing DNase I (0.1 mg/ml; Roche), pancreatic RNase A (0.1 mg/ml; Amresco), 0.1 mM phenylmethylsulfonyl flouride (PMSF) and a cocktail of protease inhibitors (Complete EDTA-free®; Roche). Hereafter, all steps were carried out at 4°C. The suspension of cells was passed through a French press (Sim-Aminco®; Electronic Instruments) at 14 000 p.s.i. followed by a short centrifugation (4000 g, 10 min) to discard non-lysed cells. The supernatant was centrifuged once more (100 000 g, 1 h) in a 50.2Ti rotor (Beckman). The pellet, containing the cellular envelope, was resuspended in 20 ml of TN containing 1.5% (v/v) Triton X-100. After 30 min incubation, this sample was centrifuged (100 000 g, 1 h) in a 90Ti rotor (Beckman) and the subsequent pellet, which corresponded to the OM fraction, resuspended in 10 ml of TN buffer containing 1% (w/v) Zwittergent 3–14 (Calbiochem). After 30 min further incubation, the mixture was centrifuged (100 000 g, 1 h) in a 90Ti rotor (Beckman) and the supernatant, containing the solubilized OMPs, diluted with TN to a final concentration of 0.1% Zwittergent 3–14 (w/v). A 10 ml aliquot of a cobalt-containing agarose resin (50% v/v; Talon®; Clontech) equilibrated in TNZ buffer [TN plus 0.1% (w/v) Zwittergent 3–14] was then added. The resulting suspension was incubated overnight with slow agitation on a gyratory wheel to allow efficient binding of HEβ. The next day, this mixture was passed through a chromatography column (Econopac; Bio-Rad) containing an additional 4 ml of the Talon® resin. This column was washed with 100 ml of TNZ and 50 ml of the same buffer containing 5 mM imidazole. HEβ was eluted in 1 ml fractions with the same buffer containing 100 mM imidazole. Fractions containing HEβ were dialyzed against buffer TNZ. When necessary, the protein was concentrated by ultrafiltration in a Centricon® tube (cut-off Mr 10 000; Millipore).
Electrophoresis and immunoblotting
Denaturing SDS–PAGE was performed in a Miniprotean® system (Bio-Rad) with 4% stacking and 8 or 10% (as indicated) separating gels (acrylamide:bisacrylamide 29:1; Bio-Rad) containing 0.1% (w/v) SDS (Ausubel et al., 1994). For denaturing SDS–PAGE, protein samples were heated at 100°C for 10 min in 60 mM Tris–HCl pH 6.8, 1% (w/v) SDS, 5% (v/v) glycerol, 0.005% (w/v) bromophenol blue and 1% (v/v) 2-ME. For non-denaturing (native) PAGE, protein samples were dissolved in 60 mM Tris–HCl pH 6.8, 5% (v/v) glycerol, 0.005% (w/v) bromophenol blue buffer, and separated in a 4–15% gradient polyacrylamide gel buffered with Tris–HCl pH 8.8 (Ready-gel®; Bio-Rad). For immunoblotting, the gels were transferred onto polyvinylidene difluoride (PVDF) membranes (Immobilon-P®; Millipore) using a semi-dry electrophoresis transfer apparatus (Bio-Rad) or the Miniprotean® wet-transfer device (Bio-Rad). Prior to transfer of the proteins separated under native conditions, the gels were soaked for 10 min in transfer buffer [48 mM Tris, 39 mM glycine, 0.0375% (w/v) SDS, 20% (v/v) methanol]. All membranes were blocked in B-buffer [phosphate-buffered saline (PBS), 0.1% (v/v) Tween-20; 3% (w/v) skimmed milk] for 1 h at room temperature. For the immunodetection of the hybrid proteins, membranes were incubated for 1 h at room temperature in B-buffer with anti-E-tag mAb–peroxidase (POD) conjugate (1:5000; Amersham Bioscience) or anti-His mAb–POD conjugate (1:5000; Clontech). OmpF and OmpA were detected with specific rabbit polyclonal sera raised against one of these proteins. The protein A–POD conjugate (1:5000; Roche) was used for detection of the bound rabbit antibodies. The avidin–POD conjugate (1:5000; Bio-Rad) was used to detect the biotinylated protein markers (Bio-Rad). Membranes were washed four times with 30 ml of PBS containing 0.1% (v/v) Tween-20 to remove unbound antibodies and secondary reagents. In all cases, POD was detected by a chemiluminiscence mixture of 1.25 mM luminol (Sigma), 42 µM luciferin (Roche) and 0.0075% (v/v) H2O2 in 100 mM Tris–HCl pH 8.0. For higher sensitivities, a commercial chemiluminiscence kit for POD was employed (Roche). In all cases, after 1 min incubation at room temperature, the membranes were exposed to an X-ray film (X-OMAT®; Kodak) or to a ChemiDoc® apparatus (Bio-Rad).
Circular dichroism
CD spectra were obtained with a Jasco J-715 spectropolarimeter with a thermostat-equipped cell. The HEβ protein concentration used was 0.1 mg/ml in TNZ buffer (see above). This concentration was estimated from the UV absorption spectrum of purified HEβ (considering ε280 nm = 36 980/M/cm). A minimum of four spectra were accumulated for each sample and the contribution of the buffer always subtracted. Values of mean residual weight ellipticities were calculated on the basis of 110 as the average residue Mr They are reported in terms of the mean residue weight ellipticities (Θ) MRW (degrees × cm2 × dmol–1). The secondary structure of the protein was evaluated by computer fit of the dichroism spectra according to the convex constraint analysis (CCA) algorithm. This method relies on an algorithm that calculates the contribution of the secondary elements that give rise to the original spectral curve without referring to spectra from model systems (Perczel et al., 1992).
In vivo protein cross-linking
The protocol described by Thanabalu et al. (1998) was basically followed. Escherichia coli UT5600 (pHEβ) cells, grown in LB + Cm at 30°C, were harvested after 3 h induction with 0.1 mM IPTG (final OD600 ∼1.0) and resuspended in one-tenth of the original culture volume of PBS containing 0.2 mM DSP (Pierce). Cross-linking was carried out for 30 min at room temperature and quenching with 50 mM Tris–HCl pH 7.5 for 15 min. After this incubation, cells were washed twice with 10 mM Tris–HCl pH 7.5 and resuspended in the same buffer. One volume of SDS–PAGE sample buffer (2×), with or without 5% (v/v) 2-ME, was added to the samples before boiling for 10 min and loading onto gels. The absence of the reducing agent 2-ME in the SDS–PAGE sample buffer allowed the maintenance of a disulfide bridge in the cross-linker.
Size exclusion chromatography
Purified HEβ in TNZ buffer (see above) was passed through a Bio-Gel A 1.5m resin (Bio-Rad) packed in a 1 m long and 1.5 cm wide column (Bio-Rad). Alternatively, the Macrosep 1000/40 resin (Bio-Rad) was employed. The flow-rate of the sample through the column was fixed at 0.1 ml/min using a peristaltic pump (P-1; Amersham Bioscience). The void volume of the column was calculated by the elution of Blue dextran 2000 (Amershan Bioscience). The gel filtration standards (Bio-Rad) were thyroglobulin (Mr 670 000), bovine γ-globulin (Mr 158 000), chicken ovalbumin (Mr 44 000), equine myoglobin (Mr 17 000) and vitamin B-12 (Mr 1350). Elution of the protein standards through the column was monitored by UV light absorption (Uvicord S II; Amersham Bioscience). Fractions of 1 ml were collected (RediFrac collector; Amersham Bioscience) and HEβ detected by western blot using an anti-E-tag mAb–POD (see above). HEβ was quantified in the different fractions by measuring chemiluminiscence signals using Quantity-one® software (Bio-Rad).
Electron microscopy
The HEβ samples, which were concentrated to 0.5 mg/ml in TNZ buffer (see above), were negatively stained with either 2% (w/v) uranyl acetate or 2% (w/v) ammonium molybdate on thin coated collodion grids previously glow-discharged for 15 s. Images of HEβ complexes were taken using a JEOL 1200 EX II electron microscope operated at 100 kV.
Liposome swelling assays
Liposome swelling experiments were performed essentially as described by Nikaido et al. (1991). Liposomes were made from 2.4 µmol egg phosphatidylcholine (Avanti Polar Lipids) and 0.2 µmol of dicetyl phosphate (Sigma). Different amounts of purified HEβ were embedded in the liposomes. When indicated, they also contained 2 nmol LPS isolated from an E.coli Ra mutant (Sigma). The OM fractions from E.coli HN705 cells producing Corβ, FvHβ or none of these C-IgAP hybrids were isolated in a discontinuous sucrose gradient on the basis of their differential buoyant densities (Schnaitman, 1970; Osborn et al., 1972; Smit et al., 1975). The crude extracts of E.coli HN705 cells and their envelope fraction were obtained exactly as described for the purification of HEβ (see above).
Protease accessibility assays
Escherichia coli UT6000 cells harboring pHEβ, pFvHβ or pHEβ and pVFvHβ were grown in 10 ml of LB liquid medium at 30°C and induced with 0.1 mM IPTG for 3 h. Cells were harvested by centrifugation, washed in PBS and resuspended in 1 ml of PBS containing 50 mM glucose (whole cells) or PBS containing 10 mM EDTA, 50 mM glucose and 250 µg/ml lysozyme (Roche) (shocked cells). Where indicated, trypsin (Sigma) was added at 1 µg/ml, and cells incubated for 1 h at 4°C. Subsequently, the trypsin inhibitor (5 µg/ml; Sigma) was added to inhibit further proteolysis. Finally, 50 µl samples were added to 50 µl of denaturing SDS–PAGE sample buffer (2×) and the mixture immediately heated for 10 min at 100°C. SDS–PAGE and western blotting were performed as described above.
Acknowledgments
Acknowledgements
The authors are indebted to Drs Ignacio Rodiguez-Crespo, Jose Luis Carrascosa and Sofía Fraile for their technical help and scientific discussions. This work was supported by EU contracts QLK3-CT2000-00170 and QLK3-CT1999-00041, by grant BIO2001-2274 of the Spanish Comisión Interministerial de Ciencia y Tecnología (CICYT) and by the Strategic Research Groups Program of the Autonomous Community of Madrid. L.A.F. is a holder of the Ramón y Cajal Program for young investigators of the Spanish Ministerio de Ciencia y Tecnología.
References
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1994) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- Ay J., Keitel,T., Kuttner,G., Wessner,H., Scholz,C., Hahn,M. and Hohne,W. (2000) Crystal structure of a phage library-derived single-chain Fv fragment complexed with turkey egg-white lysozyme at 2.0 Å resolution. J. Mol. Biol., 301, 239–246. [DOI] [PubMed] [Google Scholar]
- Benz I. and Schmidt,M.A. (1992) AIDA-I, the adhesin involved in diffuse adherence of diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol. Microbiol., 6, 1539–1546. [DOI] [PubMed] [Google Scholar]
- Bernstein H.D. (2000) The biogenesis and assembly of bacterial membrane proteins. Curr. Opin. Microbiol., 3, 203–209. [DOI] [PubMed] [Google Scholar]
- Bitter W., Koster,M., Latijnhouwers,M., de Cock,H. and Tommassen,J. (1998) Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol., 27, 209–219. [DOI] [PubMed] [Google Scholar]
- Blight M.A. and Holland,I.B. (1994) Heterologous protein secretion and the versatile Escherichia coli haemolysin translocator. Trends Biotechnol., 12, 450–455. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez,R.L., Greene,P.J., Betlach,M.C., Heyneker,H.L. and Boyer,H.W. (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene, 2, 95–113. [PubMed] [Google Scholar]
- Brandon L.D. and Goldberg,M.B. (2001) Periplasmic transit and disulfide bond formation of the autotransported Shigella protein IcsA. J. Bacteriol., 183, 951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan S.K. (2001) Type I secretion and multidrug efflux: transport through the TolC channel-tunnel. Trends Biochem. Sci., 26, 3–6. [DOI] [PubMed] [Google Scholar]
- Bullock W.O., Fernández,J.M. and Short,J.M. (1987) XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. Biotechniques, 4, 376–378. [Google Scholar]
- Cheng L. and Scheneewind,O. (2000) Type III machines of Gram-negative bacteria: delivering the goods. Trends Microbiol., 8, 214–220. [DOI] [PubMed] [Google Scholar]
- Collins R.F., Davidsen,L., Derrick,J.P., Ford,R.C. and Tonjum,T. (2001) Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol., 183, 3825–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan S.W., Schirmer,T., Rummel,G., Steiert,M., Ghosh,R., Pauptit,R.A., Jansonius,J.N. and Rosenbusch,J.P. (1992) Crystal structures explain functional properties of two E.coli porins. Nature, 358, 727–733. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V., Eltis,L., Kessler,B. and Timmis,K.N. (1993) Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene, 123, 17–24. [DOI] [PubMed] [Google Scholar]
- Duong F., Eichler,J., Price,A., Leonard,M.R. and Wickner,W. (1997) Biogenesis of the Gram-negative bacterial envelope. Cell, 91, 567–573. [DOI] [PubMed] [Google Scholar]
- Fajardo D.A., Cheung,J., Ito,C., Sugawara,E., Nikaido,H. and Misra,R. (1998) Biochemistry and regulation of a novel Escherichia coli K-12 porin protein, OmpG, which produces unusually large channels. J. Bacteriol., 180, 4452–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L.A. and de Lorenzo,V. (2001) Formation of disulphide bonds during secretion of proteins through the periplasmic-independent type I pathway. Mol. Microbiol., 40, 332–346. [DOI] [PubMed] [Google Scholar]
- Fernandez R.C. and Weiss,A.A. (1994) Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect. Immun., 62, 4727–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin S. and Boucher,C.A. (1994) A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet., 243, 112–118. [DOI] [PubMed] [Google Scholar]
- Grodberg J. and Dunn,J.J. (1988) OmpT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol., 170, 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilvout I., Hardie,K.R., Sauvonnet,N. and Pugsley,A.P. (1999) Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J. Bacteriol., 181, 7212–7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R. and Nataro,J.P. (2001) Virulence functions of autotransporter proteins. Infect. Immun., 69, 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Navarro-Garcia,F. and Nataro,J.P. (1998) The great escape: structure and function of the autotransporter proteins. Trends Microbiol., 6, 370–378. [DOI] [PubMed] [Google Scholar]
- Henderson I.R., Cappello,R. and Nataro,J.P. (2000) Autotransporter proteins, evolution and redefining protein secretion. Trends Microbiol., 8, 529–532. [DOI] [PubMed] [Google Scholar]
- Klauser T., Pohlner,J. and Meyer,T.F. (1990) Extracellular transport of cholera toxin B subunit using Neisseria IgA protease β-domain: conformation-dependent outer membrane translocation. EMBO J., 9, 1991–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser T., Pohlner,J. and Meyer,T.F. (1992) Selective extracellular release of cholera toxin B subunit by Escherichia coli: dissection of Neisseria Igaβ-mediated outer membrane transport. EMBO J., 11, 2327–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser T., Krämer,J., Otzelberger,K., Pohlner,J. and Meyer,T.F. (1993) Characterization of the Neisseria Igaβ-core. The essential unit for outer membrane targeting and extracellular protein secretion. J. Mol. Biol., 234, 579–593. [DOI] [PubMed] [Google Scholar]
- Koebnik R., Locher,K.P. and Van Gelder,P. (2000) Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol., 37, 239–253. [DOI] [PubMed] [Google Scholar]
- Koronakis V., Sharff,A., Koronakis,E., Luisi,B. and Hughes,C. (2000) Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature, 405, 914–919. [DOI] [PubMed] [Google Scholar]
- Lomholt H., Poulsen,K. and Kilian,M. (1995) Comparative characterization of the iga gene encoding IgA1 protease in Neisseria meningitidis, Neisseria gonorrhoeae and Haemophilus influenzae. Mol. Microbiol., 15, 495–506. [DOI] [PubMed] [Google Scholar]
- Lory S. (1998) Secretion of proteins and assembly of bacterial surface organelles: shared pathways of extracellular protein targeting. Curr. Opin. Microbiol., 1, 27–35. [DOI] [PubMed] [Google Scholar]
- Maurer J., Jose,J. and Meyer,T.F. (1999) Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol., 181, 7014–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1992) A Short Course in Bacterial Ggenetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Morales V.M., Backman,A. and Bagdasarian,M. (1991) A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene, 97, 39–47. [DOI] [PubMed] [Google Scholar]
- Nikaido H. (1994) Porins and specific diffusion channels in bacterial outer membranes. J. Biol. Chem., 269, 3905–3908. [PubMed] [Google Scholar]
- Nikaido H. (1996) Outer membrane. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology, Vol. 1. ASM Press, Washington, DC, pp. 29–47.
- Nikaido H. and Rosenberg,E.Y. (1981) Effect on solute size on diffusion rates through the transmembrane pores of the outer membrane of Escherichia coli. J. Gen. Physiol., 77, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. and Rosenberg,E.Y. (1983) Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J. Bacteriol., 153, 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. and Vaara,M. (1985) Molecular basis of bacterial outer membrane permeability. Microbiol. Rev., 49, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Nikaido,K. and Harayama,S. (1991) Identification and characterization of porins in Pseudomonas aeruginosa. J. Biol. Chem., 266, 770–779. [PubMed] [Google Scholar]
- Nouwen N., Ranson,N., Saibil,H., Wolpensinger,B., Engel,A., Ghazi,A. and Pugsley,A.P. (1999) Secretin PulD: association with pilot PulS, structure and ion-conducting channel formation. Proc. Natl Acad. Sci. USA, 96, 8173–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwen N., Stahlberg,H., Pugsley,A.P. and Engel,A. (2000) Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J., 19, 2229–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M.J., Gander,J.E., Parisi,E. and Carson,J. (1972) Mechanism of assembly of the outer membrane of Salmonella typhimurium. J. Biol. Chem., 247, 3962–3972. [PubMed] [Google Scholar]
- Perczel A., Park,K. and Fasman,G.D. (1992) Analysis of the circular dichroism spectrum of proteins using the convex constraint algorithm: a practical guide. Anal. Biochem., 203, 83–93. [DOI] [PubMed] [Google Scholar]
- Plano G.V., Day,J.B. and Ferracci,F. (2001) Type III export: new uses for an old pathway. Mol. Microbiol., 40, 284–293. [DOI] [PubMed] [Google Scholar]
- Pohlner J., Halter,R., Beyreuther,K. and Meyer,T.F. (1987) Gene structure and extracellular secretion of Neisseria gonorrhoea IgA protease. Nature, 325, 458–462. [DOI] [PubMed] [Google Scholar]
- Russel M. (1998) Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol., 279, 485–499. [DOI] [PubMed] [Google Scholar]
- Sandkvist M. (2001) Biology of type II secretion. Mol. Microbiol., 40, 271–283. [DOI] [PubMed] [Google Scholar]
- Schmitt W. and Haas,R. (1994) Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol. Microbiol., 12, 307–319. [DOI] [PubMed] [Google Scholar]
- Schnaitman C.A. (1970) Protein composition of the cell wall and cytoplasmic membrane of E.coli. J. Bacteriol., 104, 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C.A. (1973) Outer membrane proteins of Escherichia coli. I. Effect of preparative conditions on the migration of protein in polyacrylamide gels. Arch. Biochem. Biophys., 157, 541–552. [DOI] [PubMed] [Google Scholar]
- Shannon J.L. and Fernandez,R.C. (1999) The C-terminal domain of the Bordetella pertussis autotransporter BrkA forms a pore in lipid bilayer membranes. J. Bacteriol., 181, 5838–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J., Kamio,Y. and Nikaido,H. (1975) Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studied with lipopolysaccharide mutants. J. Bacteriol., 124, 942–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto G.E. and Hultgren,S.J. (1999) Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol., 181, 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos C., Hendrixson,D.R., Thanassi,D.G., Hultgren,S.J., St Geme,J.W.,III and Curtiss,R.,III (2000) Secretion of virulence determinants by the general secretory pathway in gram-negative pathogens: an evolving story. Microbes Infect., 2, 1061–1072. [DOI] [PubMed] [Google Scholar]
- Sugawara E. and Nikaido,H. (1992) Pore-forming activity of OmpA protein of Escherichia coli. J. Biol. Chem., 267, 2507–2511. [PubMed] [Google Scholar]
- Suhr M., Benz,I. and Schmidt,M.A. (1996) Processing of the AIDA-I precursor: removal of AIDAc and evidence for the outer membrane anchoring as a β-barrel structure. Mol. Microbiol., 22, 31–42. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Lett,M.C. and Sasakawa,C. (1995) Extracellular transport of VirG protein in Shigella. J. Biol. Chem., 270, 30874–30880. [DOI] [PubMed] [Google Scholar]
- Tamm L.K., Arora,A. and Kleinschmidt,J.H. (2001) Structure and assembly of β-barrel membrane proteins. J. Biol. Chem., 276, 32399–32402. [DOI] [PubMed] [Google Scholar]
- Thanabalu T., Koronakis,E., Hughes,C. and Koronakis,V. (1998) Substrate-induced assembly of a contiguous channel for protein export from E.coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J., 17, 6487–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi D.G. and Hultgren,S.J. (2000) Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol., 12, 420–430. [DOI] [PubMed] [Google Scholar]
- Thanassi D.G., Saulino,E.T. and Hultgren,S.J. (1998a) The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol., 1, 223–231. [DOI] [PubMed] [Google Scholar]
- Thanassi D.G., Saulino,E.T., Lombardo,M.J., Roth,R., Heuser,J. and Hultgren,S.J. (1998b) The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc. Natl Acad. Sci. USA, 95, 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls M., Atrian,S., de Lorenzo,V. and Fernández,L.A. (2000) Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nature Biotechnol., 18, 661–665. [DOI] [PubMed] [Google Scholar]
- Veiga E., de Lorenzo,V. and Fernández,L.A. (1999) Probing secretion and translocation of a β-autotransporter using a reporter single-chain Fv as a cognate passenger domain. Mol. Microbiol., 33, 1232–1243. [DOI] [PubMed] [Google Scholar]