Abstract
Cdc7-related kinases play essential roles in the initiation of yeast DNA replication. We show that mice lacking murine homologs of Cdc7 (muCdc7) genes die between E3.5 and E6.5. We have established a mutant embryonic stem (ES) cell line lacking the muCdc7 genes in the presence of a loxP-flanked transgene expressing muCdc7 cDNA. Upon removal of the transgene by Cre recombinase, mutant ES cells cease DNA synthesis, arresting growth with S-phase DNA content, and generate nuclear Rad51 foci, followed by cell death with concomitant increase in p53 protein levels. Inhibition of p53 leads to partial rescue of muCdc7–/– ES cells from cell death. muCdc7–/–p53–/– embryos survive up to E8.5, and their blastocysts generate inner cell mass of a significant size in vitro, whereas those of the muCdc7–/–p53+/– embryos undergoes complete degeneration. These results demonstrate that, in contrast to cell cycle arrest at the G1/S boundary observed in yeasts, loss of Cdc7 in ES cells results in rapid cessation of DNA synthesis within S phase, triggering checkpoint responses leading to recombinational repair and p53-dependent cell death.
Keywords: Cdc7 kinase/conditional knockout/mouse ES cells/p53-dependent cell death/replication fork arrest
Introduction
G1-to-S transition of mammalian cells is regulated by a series of phosphorylation reactions. Among them, phosphorylation by Cdk4–CyclinD1 plays a crucial role in the decision at the restriction point to commit to the cell cycle (Baldin et al., 1993; Sherr, 1993). Downstream of Cdk4–CyclinD1, Cdk2–CyclinE plays an essential role in the progression from late G1 to S phase (Sherr, 1994; Ohtsubo et al., 1995). Genetic studies in yeast implicated another serine/threonine kinase in this process (Sclafani and Jackson, 1994; Stillman, 1996; Dutta and Bell, 1997; Johnston et al., 1999; Masai and Arai, 2002). Cdc7, which is activated at the G1/S boundary in a complex with Dbf4 (Chapman and Johnston, 1989; Jackson et al., 1993), is essential for initiation of S phase in budding yeast (Hartwell, 1971; Hollingsworth and Sclafani, 1990). Budding yeast cdc7ts cells arrest with dumb-bell forms with 1C DNA content at the non-permissive temperature (Hollingsworth and Sclafani, 1990; Buck et al., 1991), indicative of cell cycle arrest at the onset of DNA synthesis. Similar 1C DNA arrest was also observed in hsk1ts, the fission yeast homolog of CDC7 (Masai et al., 1995; Brown and Kelly, 1998; Takeda et al., 1999). Studies in budding yeast indicated that the Cdc7 functions are required throughout S phase to permit firing of each replication origin on the chromosomes (Bousset and Diffley, 1998; Donaldson et al., 1998; Zou and Stillman, 2000). These results indicate that Cdc7 kinase is required not only for initiation of S phase, but also for initiation of DNA replication at each replication origin.
Genetic and biochemical studies indicate that the minichromosome maintenance (MCM) complex is the primary target of Cdc7 kinases. MCM may function as a cellular replicative DNA helicase (Chong et al., 1996; Kearsey et al., 1996; Aparicio et al., 1997; Ishimi, 1997), and it has been proposed that Cdc7 may activate MCM helicase at the origins (Sclafani, 2000; Labib and Diffley, 2001; Lei and Tye, 2001). Cdc7 phosphorylates the MCM2 component of the MCM complex both in vivo and in vitro (Lei et al., 1997; Sato et al., 1997; Brown and Kelly, 1998; Weinreich and Stillman, 1999), although precise mechanisms of origin activation by Cdc7-mediated phosphorylation of MCM remain elusive.
The structural and functional homologs of Cdc7 kinase have been identified in higher eukaryotes, including Xenopus, mouse and human (Jiang and Hunter, 1997; Sato et al., 1997; Kim et al., 1998). Microinjection of an antibody against ASK, the regulatory subunit for huCdc7, or of that against huCdc7, blocked S-phase initiation in mammalian cells (Jiang et al., 1999; Kumagai et al., 1999), and immunodepletion of Cdc7 from Xenopus egg extracts inhibited DNA replication (Roberts et al., 1999; Jares and Blow, 2000; Walter, 2000), indicating the requirement of Cdc7 kinase for metazoan DNA replication. However, the precise understanding of functions of mammalian Cdc7 kinase in cell proliferation requires genetic characterization.
In this study, we attempt to genetically dissect the roles of mammalian Cdc7 kinase in control of cell proliferation and differentiation. We first generated murine Cdc7 (muCdc7) knockout mice and showed that they are early embryonic lethal. We then generated conditional muCdc7-deficient embryonic stem (ES) cell lines and showed that muCdc7 is essential for DNA replication of ES cells. Furthermore, our results revealed unexpected phenotypes of Cdc7 knockout ES cells, namely S-phase arrest and p53-dependent cell death, indicating that S phase is being monitored very strictly by the p53-dependent checkpoint pathway to ensure high-fidelity genome inheritance in ES cells.
Results
Disruption of the muCdc7 gene
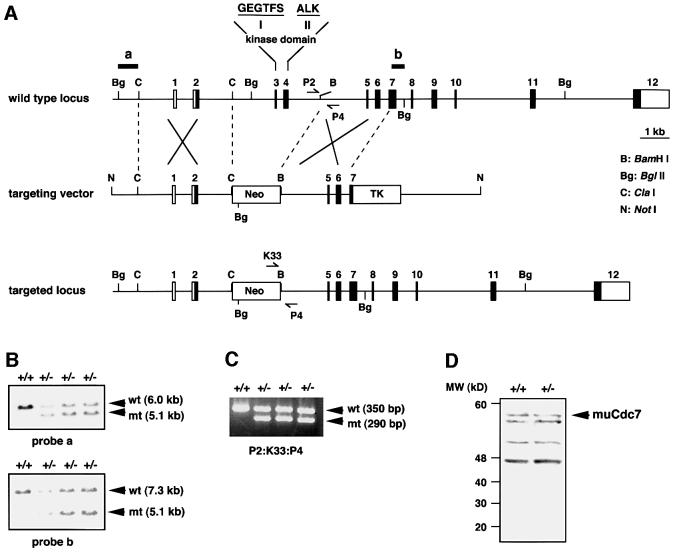
We previously reported that the muCdc7 gene, located on the mouse chromosome 5E5, consists of 12 exons (Kim et al., 1998). In order to genetically dissect the functions of mammalian Cdc7 kinase, we attempted to disrupt the muCdc7 gene through homologous recombination. Our strategy was to replace exons 3 and 4, encoding kinase domains I and II, with a neor resistance cassette (Figure 1A). This manipulation was expected to completely inactivate the kinase activity of muCdc7. Cells from two independent ES lines containing a disrupted muCdc7 allele were injected into C57BL/6 blastocysts, and the resulting chimeras were backcrossed to C57BL/6 wild-type animals to generate heterozygous animals. The successful disruption of the muCdc7 locus was confirmed by Southern blotting and PCR (Figure 1B and C). Western blotting of the extracts prepared from muCdc7+/+ and muCdc7+/– ES cells indicated the absence of aberrant forms of muCdc7 generated from the knockout locus (Figure 1D). No homozygous null animals were observed in a total of >1500 live births from Cdc7+/– heterozygous intercrosses using either of the two founder lines (Table I). Wild-type and heterozygous mice were born at the expected frequencies, and the heterozygous mice appeared normal, healthy and fertile.
Fig. 1. Targeted disruption of the muCdc7 gene. (A) Restriction maps of the wild-type muCdc7 gene, the targeting vector and the mutant locus resulting from homologous recombination. The numbered boxes indicate exons. Genomic fragments used as probes for Southern blotting are indicated by horizontal bars labelled a and b. The positions of the primers used for genotyping mice and embryos are shown by converging arrows P2/P4 and K33/P4. (B) Identification of muCdc7+/– ES clones by Southern blot analysis. Genomic DNA was digested with BglII and hybridized with either a 5′ (a) or 3′ (b) probe. The hybridizing DNA fragments of wild-type (wt) and mutant (mt) alleles are shown by arrows. (C) PCR genotyping of mice and embryos from a muCdc7 heterozygous breeding. Three primers (P2/K33/P4) were included in PCRs to amplify a 350 or 290 bp fragment specific for the wild type or a mutant allele, respectively. (D) Western blotting of whole-cell extracts from muCdc7+/+ and muCdc7+/– ES cells using anti-muCdc7 antibody as described previously (Kim et al., 1998). The arrow indicates the endogenous 532 amino acid muCdc7 polypeptide expressed in ES cells. There are extra bands that react non-specifically with the antibody.
Table I. Genotypes of offspring from muCdc7 heterozygous intercrosses.
| Age | +/+ | +/– | –/– | Total No. |
|---|---|---|---|---|
| E3.5 | 6 | 54 | 15 | 75 |
| E6.5 | 8 | 27 | (1)a | 36 |
| E7.5 | 12 | 40 | 0 | 52 |
| E8.5 | 26 | 60 | 0 | 86 |
| Weaning | 512 | 1080 | 0 | 1592 |
aNumber in parentheses indicates the embryo that was about to be resorbed.
To determine at what embryonic stage muCdc7–/– null embryos die, we analyzed embryos from heterozygous intercrosses at different stages of gestation. Null blastocysts (E3.5) were identified, and their morphology was indistinguishable from that of the wild-type or heterozygous blastocysts (data not shown). However, among 174 embryos between E6.5 and E8.5, only one muCdc7 null embryo (E6.5) was detected that was about to be resorbed (Table I). These results indicate that muCdc7 null embryos are viable and normal until E3.5, but die between E3.5 and E6.5. The apparent death of the null embryos after E3.5 may be due to run-out of the maternal stock and/or to active cell death caused by unknown mechanisms.
The muCdc7 mutation causes degeneration of in vitro cultured blastocyst outgrowths
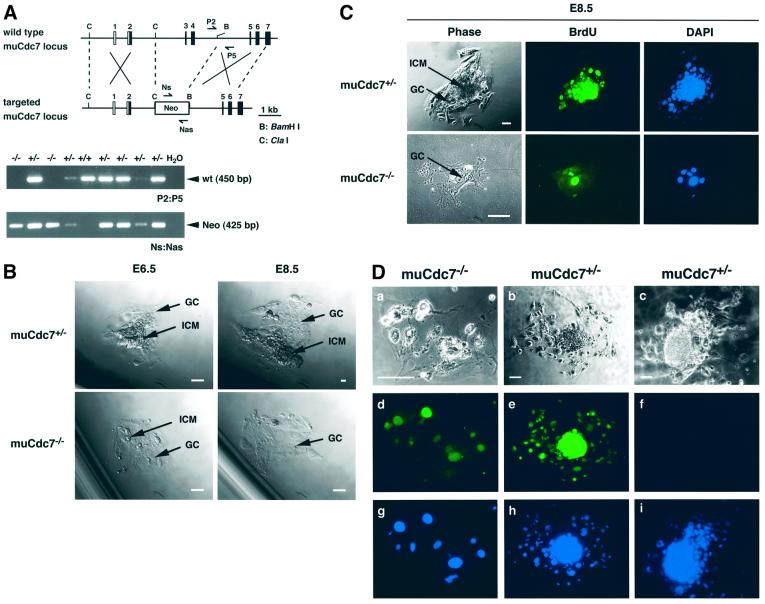
To examine growth of blastocysts in vitro, blastocysts from muCdc7+/– heterozygous intercrosses were collected, cultured in vitro, and subsequently genotyped by PCR (Figure 2A). After 24–48 h in culture, the spherical blastocysts flattened onto the culture dish and the inner cell mass (ICM) grew a mound on top of the trophoblast giant cells (GCs) (Figure 2B). When muCdc7 null and heterozygous embryos were cultured in vitro, they grew at similar rates through E5.5. However, the sizes of the ICM of muCdc7 null blastocysts were smaller than those of the wild-type counterpart at E6.0 (data not shown). Subsequently, muCdc7–/– ICM diminished further in size and completely disappeared by E8.5, leaving behind a monolayer of GCs with large nuclei (Figure 2B). Analysis of Cdc7 null GCs by phase-contrast microscopy and BrdU labeling demonstrated that they were alive and undergoing DNA replication at E8.5 (Figure 2C). GCs are mitotically inactive and undergo repeated DNA replication, generating a polyploid nucleus and a large cytoplasm (Rugh, 1990). Immunostaining of in vitro cultured blastocysts with a muCdc7-specific antibody indicates the presence of muCdc7 protein in muCdc7–/– GCs (Figure 2D). Thus, the survival of muCdc7–/– GCs is likely to be due to the presence of residual Cdc7 protein that persists in the undividing cells.
Fig. 2. Growth and DNA synthesis of muCdc7-deficient embryos in culture. (A) The converging arrows, P2/P5 and Ns/Nas, indicate the primers used for genotyping blastocyst outgrowths, which detect the wild-type (wt) and mutant (Neo) alleles, respectively. (B) Blastocysts from muCdc7 heterozygous intercrosses were cultured in vitro for 5 days and photographed using phase-contrast microscopy. Arrows point to the ICM and the trophoblast GC. Scale bar = 50 µm. (C) Blastocysts from muCdc7 heterozygous intercrosses were cultured in vitro until E8.5, and were incubated in the presence of 10 µM BrdU for 3 h. Blastocyst outgrowths were processed for indirect immunofluorescence using anti-BrdU antibody (green) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI; blue). Scale bars = 100 µm. (D) Immunohistochemical analysis of muCdc7–/– GCs. Blastocysts grown in culture as in (C) were fixed and stained with anti-muCdc7 antibody (d and e, green) or control IgG (f). Nuclei were detected by DAPI staining (g, h and i; blue). a, b and c, phase-contrast images of the cells. The genotype of each embryo was determined by PCR. Scale bars = 100 µm.
Generation of conditional muCdc7-deficient ES cell lines
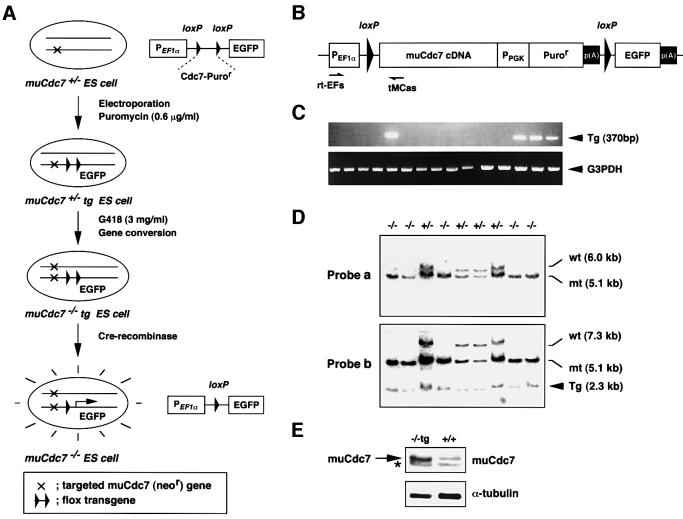
The above results indicate that the muCdc7 functions are essential for early embryogenesis or cell proliferation per se, or for both. In order to determine whether muCdc7 functions are required for cell proliferation per se, we decided to generate mutant ES cell lines in which inactivation of muCdc7 can be conditionally induced (Figure 3A). For this purpose, we constructed a flox muCdc7 transgene, which is flanked by the loxP sites and can be excised later with Cre recombinase. The cDNA used for the transgene was derived from one of the muCdc7 alternatively spliced forms (Kim et al., 1998) and encodes a 532 amino acid polypeptide which is fully active for phosphorylation of the MCM complex in vitro (J.M.Kim and H.Masai, unpublished data). The transgene plasmid also contains a promoterless EGFP gene downstream of the transgene, which is expressed only after excision of the loxP-flanked transgene and therefore can serve as an indicator for the efficiency of Cre-mediated excision (Figure 3B). The transgene was transfected into muCdc7+/– heterozygous ES cells by electroporation, and puromycin-resistant muCdc7+/–tg ES clones carrying the transgene were identified by RT–PCR (Figure 3C). Fifteen out of 48 puromycin-resistant clones were shown to express transgene-derived mRNA. Four out of 15 clones carried a single copy of the transgene. We then converted the endogenous wild-type allele to the disrupted one containing the neomycin marker through gene conversion. This was accomplished simply by elevating the concentration of G418 to 3 mg/ml (Mortensen et al., 1992). It caused the selection of ES cells homozygous for the targeted allele, which are expected to display resistance to a higher concentration of G418 due to the presence of two copies of the neor gene. Using one of the muCdc7+/–tg ES cells carrying a single copy transgene, 38 surviving clones were isolated. Southern blot analysis indicated that 11 had undergone expected gene conversion, generating conditional muCdc7-deficient (muCdc7–/–tg) ES cell lines (Figure 3D). Western blotting showed that the expression level of the transgene-encoded muCdc7 is ∼3–5 times higher than that of endogenous protein (Figure 3E). The homozygous muCdc7 null ES cell lines could not be established in the absence of the transgene (data not shown), suggesting that muCdc7 functions are essential for cell proliferation of ES cells.
Fig. 3. Generation of conditional muCdc7-deficient ES cell lines. (A) Strategy for the generation of conditional muCdc7–/– ES cell lines carrying a Cre-removable (flox) muCdc7 transgene. (B) A schematic drawing of the structure of muCdc7 flox transgene. p(A) indicates a polyadenylation signal. (C) RT–PCR analysis of total RNA isolated from muCdc7+/–tg ES clones. The primer pair, rtEFs/tMCas, shown in (B), was used to detect the expression of a transgene (upper panel). Expression of G3PDH was also examined as a control (lower panel). (D) Southern blotting analysis of conditional muCdc7-deficient ES clones. BglII-digested genomic DNA was hybridized with the probe a or b (see Figure 1A). Tg indicates the bands derived from the transgene. (E) Expression of the transgene-encoded muCdc7 protein in conditional muCdc7-deficient ES clones. Whole-cell extracts prepared from the indicated cells were analyzed by immunoblotting with anti-muCdc7 (upper) or anti-α-tubulin (lower) antibody. A non-specifically reacting band is indicated by the asterisk.
muCdc7 is essential for ES cell viability
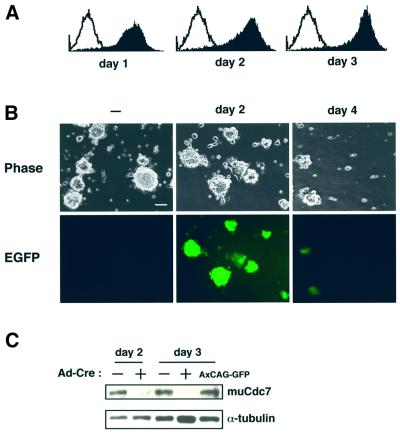
We subsequently infected these muCdc7–/–tg ES cells with adenoviruses expressing Cre recombinase (Ad-Cre) to excise the transgene. FACS analysis indicated that nearly 100% of the cells were positive for EGFP as early as day 1 after infection and its extent reached a maximum at day 3 (Figure 4A). This was also confirmed by examining the cells under a microscope (Figure 4B). At day 4 after infection, Ad-Cre-infected muCdc7–/–tg ES cells had started to detach from the plates (see below), and EGFP expression also decreased. We showed that muCdc7 protein expressed from the transgene disappeared at day 2 after infection (Figure 4C). Thus, we were able to establish mutant ES cell lines in which inactivation of the muCdc7 gene could be conditionally induced.
Fig. 4. Excision of the transgene in muCdc7–/–tg ES cells by infection of Ad-Cre. (A) Flow cytometry analysis of green fluorescent protein (GFP) expression in muCdc7–/–tg ES cells at indicated times after infection of Ad-Cre. The open and filled histograms indicate the fluorescence profile of uninfected and infected muCdc7–/–tg ES cells, respectively. (B) Phase-contrast and fluorescence microscope evaluation of GFP expression in muCdc7–/–tg ES cells infected with Ad-Cre. Left, uninfected; middle, 2 days after infection; right, 4 days after infection. Upper, phase contrast; lower, fluorescence microscope. Scale bar = 40 µm. (C) Expression of the transgene-encoded muCdc7 protein was detected by western blotting at days 2 and 3 after infection of muCdc7–/–tg ES cells with Ad-Cre. Upper, anti-muCdc7 antibody (Kim et al., 1998); lower, anti-α-tubulin antibody. ‘–’ and ‘+’ indicate uninfected and infected cells, respectively. The rightmost lane represents the same cells infected with a control adenovirus, AxCAG–GFP (kindly provided by Dr Yumiko Kamogawa), for 3 days.
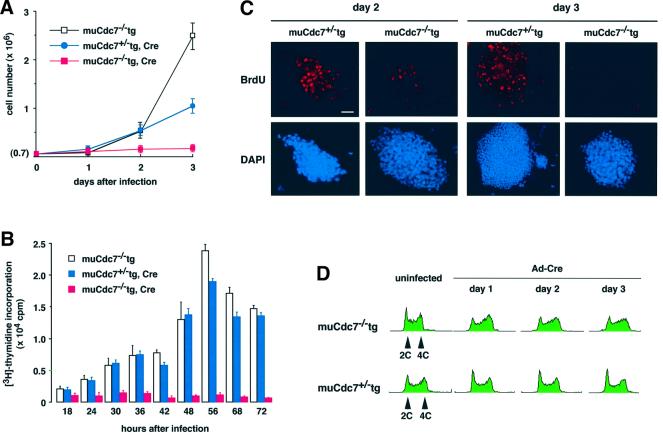
We next examined the growth properties of the muCdc7–/–tg ES cells after infection with Ad-Cre. The increase in cell numbers stopped almost immediately (Figure 5A). As the control, muCdc7+/–tg ES cells carrying the same transgene kept increasing in number under the same condition, although the rate decreased compared with non-infected cells, presumably due to the effect of adenovirus infection. [3H]thymidine incorporation was measured in these cell lines to determine the level of DNA synthesis. In muCdc7–/–tg ES cells, DNA synthesis was almost completely inactivated as early as day 1 after infection with Ad-Cre (Figure 5B). BrdU incorporation was also significantly reduced at day 2 after infection and was completely lost at day 3 (Figure 5C). These results indicate that loss of muCdc7 results in rapid cessation of DNA synthesis.
Fig. 5. Cell proliferation, DNA synthesis and cell cycle profile of muCdc7–/–tg ES cells after removal of the transgene. muCdc7–/–tg and muCdc7+/–tg ES cells were infected with Ad-Cre at a m.o.i. of 50. (A) The numbers of viable cells, as indicated in the figure, were counted at various time points (0–3 days) after infection. (B) The same set of cells were incubated for 1 h in the presence of 1 µCi of [3H]thymidine at different times after infection, and its incorporation was measured. In uninfected muCdc7–/–tg or Ad-Cre-infected muCdc7+/–tg ES cells, the drops in [3H]thymidine incorporation at 68 and 72 h after infection are the results of overgrowth. (C) At days 2 or 3 after infection, cells were pulse-labeled with 10 µM BrdU for 20 min, fixed and incubated with anti-BrdU antibody to detect BrdU incorporation (upper, red). DNA was detected by DAPI staining (lower, blue). Scale bar = 20 µm. (D) Flow cytometry analysis of DNA content of muCdc7–/–tg (upper) or muCdc7+/–tg (lower) at indicated times after infection of Ad-Cre. In (A) and (B), the values represent average of three independent experiments.
Depletion of muCdc7 in ES cells results in accumulation of S-phase cells and formation of nuclear Rad51 foci
We examined cell cycle distribution of Ad-Cre-infected muCdc7–/–tg ES cells. Wild-type ES cells proliferating in an undifferentiated state are characterized by unique cell cycle distribution, with ∼20% of the cells being in G1 phase and ∼80% in S and G2/M phases (Savatier et al., 1994). Prior to infection, muCdc7–/–tg ES cells essentially exhibited the same pattern as the wild-type ES cells (Figure 5D; data not shown). After infection of Ad-Cre, G1 phase peak decreased and the population of S/G2 phase cells increased in muCdc7–/–tg ES cells (Figure 5D). This is expected from the results in the previous section, since rather quick arrest of DNA synthesis after excision of the transgene would lead to accumulation of S-phase cells.
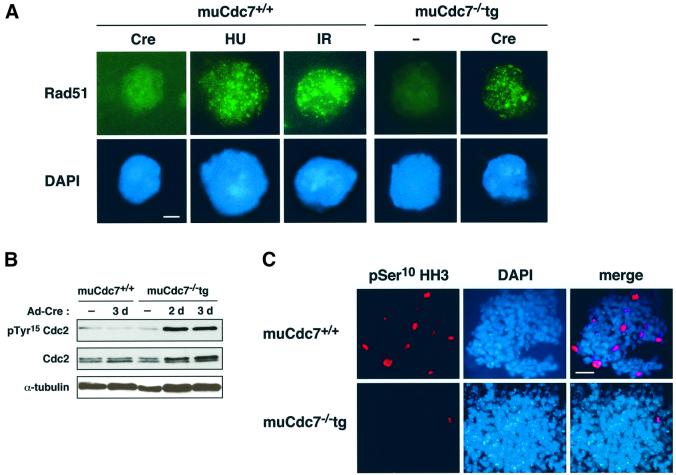
The above results indicate that muCdc7 inactivation in ES cells induces the arrest of replication forks, generating signals for DNA damage or DNA structure checkpoint. Upon DNA damage, recombination-dependent repair processes are induced, and this leads to the appearance of Rad51 foci in the nuclei (Haaf et al., 1995). We investigated whether Rad51 protein generates foci in cells depleted for muCdc7. Ad-Cre-infected muCdc7–/–tg ES cells exhibited typical Rad51 nuclear foci, whereas wild-type ES cells showed no nuclear foci under the same condition (Figure 6A). This is consistent with the idea that the arrested replication forks are being repaired in a recombination-dependent manner. Similar focus formation was also observed in hydroxyurea (HU)-treated or γ-ray-irradiated wild-type ES cells (Figure 6A).
Fig. 6. Activation of G2/M checkpoint in muCdc7–/– ES cells. muCdc7+/+ and muCdc7–/–tg ES cells were infected with Ad-Cre at a m.o.i. of 50. (A) The cells were infected with Ad-Cre for 2.5 days (Cre), treated with HU for 12 h (2 mM, HU), γ-irradiated (10 Gy, IR) or untreated (–). Rad51 nuclear foci were detected by immunostaining using anti-Rad51 antibody (upper, green), and the nucleus was localized by DAPI (lower, blue). Scale bar = 1 µm. (B) muCdc7+/+ and muCdc7–/–tg ES cells were harvested at the indicated times after infection, and the cell extracts were analyzed by immunoblotting with antibodies against Cdc2, phosphorylated Tyr15 of Cdc2 and α-tubulin. (C) Cells infected for 2.5 days were stained with an antibody specific to Ser10 phosphohistone H3 (pSer10 HH3, red) to detect those in mitosis. Cells were counterstained with DAPI to localize the nucleus (blue). Merge: merged images of pSer10 HH3 and DAPI staining. Scale bar = 10 µm.
Activation of G2/M checkpoint in muCdc7–/– ES cells
The block of replication forks by DNA damage or HU is known to induce G2/M blocks through inactivation of Cdc25 phosphatase (Furnari et al., 1997; Sanchez et al., 1997). We investigated whether muCdc7 inactivation in ES cells induces the G2/M checkpoint by examining the phosphorylation on tyrosine 15 of Cdc2 kinase, a target site of Cdc25 phosphatase. We observed a sharp increase in tyrosine 15 phosphorylation upon inactivation of muCdc7 (Figure 6B). We also conducted immunostaining with an antibody that detects the phosphorylated serine 10 of histone H3. The H3 phosphorylation has been reported to be an excellent marker for chromosome condensation during mitotic prophase in mammalian cells (Bradbury, 1992). Ad-Cre-infected muCdc7–/–tg ES cells were virtually negative with this antibody, whilst muCdc7+/+ ES cells treated similarly showed significant fractions of immunopositive cells (Figure 6C). These results indicate that loss of muCdc7 induces G2/M checkpoint signals.
Induction of cell death and increased p53 expression in muCdc7–/– ES cells
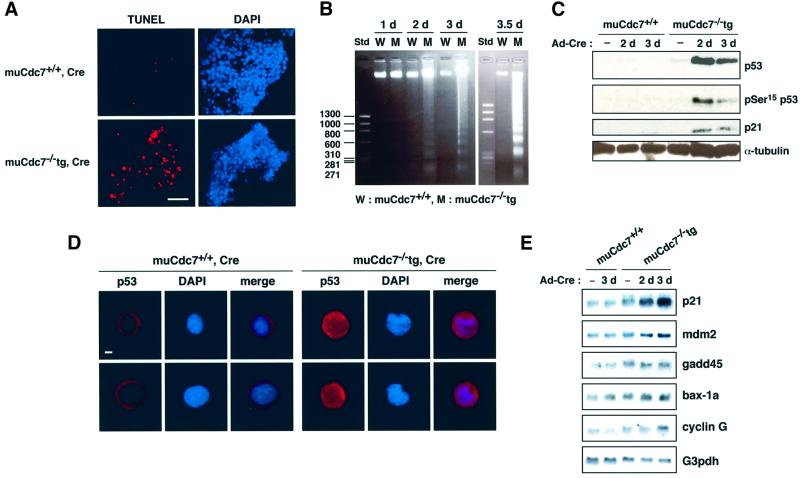
At day 4 after infection, the Ad-Cre-infected muCdc7–/–tg ES cells started to detach from the plates and lose viability. We therefore examined whether infected muCdc7–/–tg ES cells were undergoing apoptosis using terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assays. Figure 7A shows that most of the Ad-Cre-infected muCdc7–/–tg ES cells at day 3.5 were TUNEL positive. These observations indicate that the depletion of muCdc7 protein in ES cells results in growth arrest in S phase and eventually leads to cell death. The occurrence of cell death was also indicated by the appearance of DNA fragmentation in muCdc7–/– ES cells (Figure 7B).
Fig. 7. Induction of cell death with concomitant activation of p53 protein in muCdc7–/– ES cells. muCdc7+/+ and muCdc7–/–tg ES cells were infected with Ad-Cre at a m.o.i. of 50. (A) TUNEL assays (red) and DAPI staining (blue) of ES cells at 3.5 days after infection. Scale bar = 20 µm. (see Supplementary figure 1 for positive control of TUNEL assay.) (B) DNA was extracted from infected ES cells at the indicated times and was analyzed on 1.5% agarose gel. Std, DNA size marker. (C) Western blotting analysis of p53 and p21 in muCdc7–/–tg ES cells. Cells were harvested at the indicated times after infection and cell extracts were analyzed by immunoblotting using antibodies against p53, phosphorylated Ser15 of p53, p21 and α-tubulin. (D) Cellular localization of p53 at 2.5 days after infection. Cells were immunostained with an antibody against p53 (red) and counterstained with DAPI (blue). Merge: merged images of p53 and DAPI staining. Scale bar = 1 µm. (E) Northern blot analysis of some of the p53 target genes. Fifteen micrograms of total RNA isolated from the ES cells at the indicated times after infection was run on agarose gel and blotted with the probes indicated. (See also Supplementary figure 2 for responses of the wild-type ES cells to HU and γ-ray irradiation.)
The observed cell death could simply be due to permanent cell cycle arrest at S phase caused by lack of muCdc7 protein. Alternatively, active cell death may be responsible for this. In response to DNA damage induced by UV or γ-ray irradiation, p53 is stabilized and accumulates in the nucleus, where it induces the expression of genes involved in cell cycle arrest or apoptosis (Carr, 1996; Hansen and Oren, 1997; Levine, 1997). We have noted that levels of p53 and phosphorylation on serine 15 of p53 (Banin et al., 1998; Canman et al., 1998; Tibbetts et al., 1999; Shieh et al., 2000) increased in infected muCdc7–/–tg ES cells, whereas the wild-type ES cells showed no increase under the same conditions (Figure 7C).
We then conducted immunostaining to determine whether increased p53 protein localized in the nucleus. Previous studies demonstrated that embryonic carcinoma cells and undifferentiated ES cells contain cytoplasmic wild-type p53 protein under normal growing conditions (Lutzker and Levine, 1996; Sabapathy et al., 1997; Aladjem et al., 1998). In uninfected or Ad-Cre-infected wild-type ES cells, p53 protein was almost exclusively localized in the cytoplasm. In contrast, Ad-Cre-infected muCdc7–/–tg ES cells exhibited staining of higher intensity, which was detected in both the cytoplasm and nuclei (Figure 7D). These results indicate that muCdc7 inactivation in ES cells resulted in induction and nuclear localization of p53 protein. In order to examine p53-mediated transcriptional activation in Ad-Cre-infected muCdc7–/–tg ES cells, we conducted northern blot analysis of endogenous p53 target genes (Figure 7E). p21, mdm2 and cyclinG mRNA levels increased, while bax-1a and gadd45 levels stayed relatively constant. These data indicate that p53 is transcriptionally active in muCdc7-deficient ES cells. Western blot analysis revealed that the level of p21 protein increased in infected muCdc7–/–tg ES cells (Figure 7C).
Our results suggest a possibility that aberrant replication structures resulting from S-phase arrest in muCdc7–/– cells may be sensed as checkpoint-inducing signals, eventually leading to p53-dependent apoptosis.
Cell death in muCdc7–/– ES cells requires p53 function
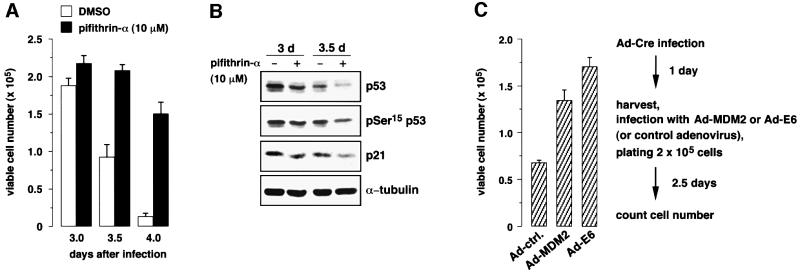
In order to address whether cell death in muCdc7–/– ES cells depends on p53 function, we have used pifithrin-α, which is known to prevent nuclear accumulation of p53 (Komarov et al., 1999). Treatment of Ad-Cre-infected muCdc7–/–tg ES cells with pifithrin-α led to a significant increase of surviving cells (Figure 8A). Under this condition, the levels of p53 and p21 proteins decreased (Figure 8B), indicating that the drug decreased stability of the p53 protein. We also examined the effect of expression of human papilloma virus E6 protein, which inactivates p53 (Thompson et al., 1997), and the Mdm2 protein, which promotes degradation of p53 (Haupt et al., 1997; Kubbutat et al., 1997). Both E6 and Mdm2 proteins partially prevented the cell death of muCdc7–/– ES cells (Figure 8C), whereas they did not affect the viable cell numbers in muCdc7+/+ cells under the same condition (data not shown). These results indicate that cell death observed in muCdc7–/– ES cells requires p53 function.
Fig. 8. p53 is required for apoptosis observed in muCdc7–/– ES cells. muCdc7+/+ and muCdc7–/–tg ES cells were infected with Ad-Cre at a m.o.i. of 50. (A) Pifithrin-α (10 µM), a specific inhibitor of p53, or dimethylsulfoxide was (DMSO) added at day 1 after infection. Cells were collected at the indicated times after infection and the cell number was determined by Trypan Blue staining. (B) ES cells treated as in (A) were harvested at the indicated times. Cell extracts were analyzed by immunoblotting with antibodies against p53, phosphorylated Ser15 of p53, p21 and α-tubulin. (C) Mdm2 or E6-expressing adenovirus was infected at day 1 after Ad-Cre infection. Cell number was determined at 3.5 days after Ad-Cre infection. In (A) and (C), the values are averages of three independent experiments.
Partial rescue of muCdc7–/– embryonic lethality by p53 null mutation
To examine whether the early embryonic lethality of the muCdc7-deficient embryos would be rescued by the loss of p53 function, 190 pups from double heterozygous crosses (muCdc7+/–p53+/– × muCdc7+/–p53+/–) were genotyped by PCR. None of them was homozygous for muCdc7 (data not shown), indicating that the lethal phenotype could not be rescued in a p53 null background. Next, 94 embryos at E7.5, 96 embryos at E8.5 and 65 embryos at E9.5 from double heterozygous crosses were isolated and genotyped by PCR (Figure 9A). Interestingly, four E7.5 and two E8.5 embryos were homozygous for both muCdc7 and p53 (Figure 9A; Table II), although they were growth retarded in development. No double knockout embryos survived past E9.5. These results indicate that loss of p53, leading, presumably, to reduced apoptosis, can partially rescue the early embryonic lethality of muCdc7-deficient embryos.
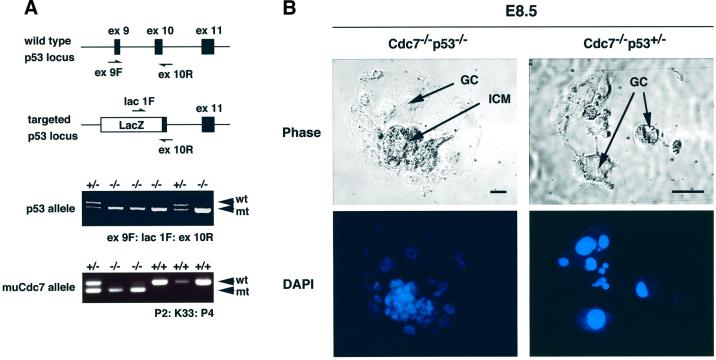
Fig. 9. Partial rescue of ICM proliferation of muCdc7–/– blastocysts by p53 null mutation. (A) PCR genotyping of embryos isolated from muCdc7+/–p53+/– intercrosses. The top illustrations show the locations of the primers (ex9F/lacIF/ex10R) used for the analysis of p53 alleles (Gondo et al., 1994). Filled and open boxes indicate exons and inserted lacZ, respectively. muCdc7 allele was detected by P2/K33/P4 as in Figure 1C. (B) Blastocysts isolated from muCdc7+/–p53+/– intercrosses, with the genotype indicated, were cultured in vitro until E8.5. Arrows indicate ICM and GCs. Upper, phase-contrast microscopy; lower, DAPI staining. Scale bars = 50 µm.
Table II. Partial rescue of early embryonic lethality of muCdc7-deficient mice by p53 null mutation.
| Genotypes of p53 | p53+/+ |
p53+/– |
p53–/– |
Total No. | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes of muCdc7 | +/+ | +/– | –/– | +/+ | +/– | –/– | +/+ | +/– | –/– | |
| No. of E7.5 embryos | 5 | 18 | 0 | 13 | 38 | 0 | 6 | 10 | 4 | 94 |
| No. of E8.5 embryos | 6 | 18 | 0 | 16 | 40 | 0 | 4 | 10 | 2 | 96 |
To further confirm this notion, we examined muCdc7–/– blastocysts. E3.5 blastocysts were collected from double heterozygous crosses, cultured in vitro until E8.5, and subsequently genotyped. muCdc7–/– outgrowth in the p53 null background developed ICM, although its size was smaller than that of the wild type, whereas muCdc7–/– outgrowths in either a wild-type or heterozygous p53 background developed no ICM at E8.5 (Figure 9B), as was shown above (Figure 2B). This indicates that loss of p53 can partially rescue survival of muCdc7–/– ICM in vitro.
Taken together, our observations strongly indicate that loss of Cdc7 functions results in p53-dependent cell death in ES cells and in early embryos.
Discussion
The machinery of DNA replication is strikingly conserved throughout evolution (Stillman, 1996). In eukaryotes, initiation and progression of DNA replication need to be under strict regulation during normal cell cycle progression as well as in an emergency (such as in the presence of DNA-damaging agents). To achieve this exquisite regulation, protein kinases play crucial roles. Cdc7, originally discovered in budding yeast, is expected to play conserved essential functions in DNA replication (Sclafani, 2000; Masai and Arai, 2002). In the present study, we have attempted to genetically define the roles of Cdc7 kinase in mammalian development and cell proliferation. We have done this by generating muCdc7-deficient mice as well as by exploiting a conditionally inducible gene inactivation system in ES cells.
muCdc7 is essential for embryonic development
We have shown that muCdc7 deficiency leads to early embryonic lethality in mice (Table I), and that muCdc7 null blastocysts cultured in vitro until E8.5 did not develop any ICM (Figure 2B). However, muCdc7–/– GCs survived and synthesized DNA under the same conditions, and this is likely to be caused by remaining maternal stock of muCdc7 protein in undividing GCs (Figure 2C and D). Similarly, survival of the null embryos until E3.5 (blastocysts) may be due to the maternal muCdc7 stock. It is also possible that DNA replication of fertilized eggs may not require Cdc7 functions for initiation, and may be regulated by mechanisms entirely different from those of somatic cells.
Strategy for generation of conditional muCdc7-deficient ES cell lines
Since our initial attempts to generate muCdc7 knockout mice indicated that muCdc7 deficiency results in early embryonic lethality, we needed to manipulate the genome so that we can conditionally induce inactivation of the muCdc7 gene. Our strategy was to induce complete knockout of the endogenous muCdc7 in the presence of a single copy of the transgene, which encodes a muCdc7 cDNA and could support the growth of the knockout cells (Figure 3A and B). The transgene is flanked by the loxP sites and, therefore, can be excised. We inserted a promoterless EGFP gene downstream of the transgene (outside the loxP cassette) so that it was expressed only after successful excision of the transgene and we could monitor the efficiency of excision (Figure 4A and B). We used the EF1α promoter, which was expected to be ubiquitously active, to express the transgene, since Cdc7 expression is relatively constant during the cell cycle (Kim et al., 1998; Kumagai et al., 1999). The complete loss of the endogenous muCdc7 alleles could be efficiently achieved by screening for the survivors in the presence of higher concentration of G418. When this was conducted on the heterozygous muCdc7+/– ES cells in the absence of a transgene, very few survivors were obtained and they did not have expected chromosome structures at the muCdc7 locus (data not shown). In contrast, in the presence of the transgene, the cell lines with complete disruption of both alleles were readily obtained (Figure 3D), and these mutant cell lines were indistinguishable from the wild-type cells in terms of growth properties (data not shown). This indicates that constitutive expression of the 532 amino acid form of muCdc7 can fully support normal growth of ES cells.
Upon infection of these cell lines with adenoviruses encoding the Cre recombinase, EGFP expression was induced within 1 day and almost 100% of the cell population was positive for EGFP (Figure 4A and B), indicating that excision of the transgene took place efficiently. We obtained 11 independent clones expressing a transgene and in all cases the transgene was efficiently excised by Cre recombinase, indicating that the method could generally work in generating mutant ES cells, in which essential genes are conditionally inactivated.
Loss of muCdc7 leads to S-phase arrest, induction of recombinational repair and the G2/M checkpoint
The mutant ES cells rapidly ceased the increase in cell numbers and DNA synthesis following inactivation of muCdc7 (Figure 5A–C). Unexpectedly, analysis of DNA content revealed S-phase arrest of Cdc7-depleted ES cells (Figure 5D), rather than G1/S arrest with DNA content of unreplicated cells, which is observed in yeasts under the equivalent condition. In budding yeast, replication forks derived from early firing origins can complete the replication of the entire chromosomes in the absence of firing of late origins. In ES cells, in contrast, the loss of firing from late origins causes rather immediate cessation of DNA synthesis and results in suspended S phase, indicating that the chromosomal DNA replication cannot be completed in the absence of continuous firing of origins. This difference may be due to much larger sizes of the mammalian chromosomes, the replication of which would require firing of thousands of origins under normal growing conditions. The replication forks from the early firing origins would continue to travel along the chromosome, but would be likely to encounter obstacles along the way, which would halt their progression or disintegrate the replicative complexes at the forks. Alternatively, collision of ongoing replication forks with unfired prereplicative complex may arrest the forks in ES cells. It is also possible that mammalian Cdc7 may target molecules essential for DNA chain elongation.
Whatever the case, the arrested replication forks generate checkpoint signals so that the cell cycle progression is temporally suspended and the forks are rescued (Hartwell and Weinert, 1989; Elledge, 1996; Chan et al., 2000). We have shown that this is indeed the case in muCdc7–/– ES cells by showing induction of phosphorylation of Cdc2 on tyrosine 15 and the absence of histone H3 phosphorylation, as well as formation of nuclear Rad51 foci (Figure 6).
p53-dependent cell death induced by replication fork block in ES cells
We have shown that inactivation of muCdc7 in ES cells eventually induces cell death, and that it depends on the p53 function. Extension of the lifespan of muCdc7–/– embryos by p53–/– mutation also suggests the occurrence of p53-dependent cell death in early embryogenesis upon loss of muCdc7.
The wild-type ES cells undergo rapid cell death upon treatment with HU as well, with concomitant induction of nuclear Rad51 foci and p53 protein (Figure 6A; Supplementary figure 2 available at The EMBO Journal Online). Thus, ES cells appear to be particularly sensitive to S-phase arrest. Transcription of p21, mdm2 and cyclinG target genes of p53, is induced in ES cells after inactivation of muCdc7 or treatment with HU (Figure 7E; Supplementary figure 2). This is in contrast to differentiated mammalian cells, in which p53 protein is stabilized but target genes are generally not induced in response to replication block (Gottifredi et al., 2001). We consider the following possibilities to explain this difference. (i) A unique signal transduction pathway is induced by replication fork block in undifferentiated ES cells, which generates transcriptionally active p53, whereas, in differentiated cells, p53 protein, although stabilized, is somehow rendered inactive after S-phase arrest. (ii) The number of arrested replication forks is larger in undifferentiated ES cells, resulting in a higher level of checkpoint signals and hyperactivation of p53. In fact, the numbers of Rad51 foci of ES cells arrested in S phase by inactivation of muCdc7 as well as by HU are larger than that of γ-ray-irradiated ES cells (Figure 6A), in which cells are arrested at G2/M phase without significant cell death (see Supplementary figure 2). (iii) It could simply be due to the level of endogenous p53, which, in an unperturbed state, is higher in undifferentiated ES cells than in differentiated cells. Whatever the mechanism, the tendency of ES cells to undergo apoptosis in response to replication fork block would facilitate the maintenance of high genomic integrity as a population, and this characteristic would be highly desirable for those cells with tautipotency.
In summary, our results demonstrate the essential roles of Cdc7 kinase in DNA synthesis and cell proliferation of mammalian cells, and that the absence of Cdc7 kinase leads to S-phase arrest, which generates DNA structure checkpoint signals resulting in recombinational repair and eventual p53-dependent cell death.
Materials and methods
Cells
The parental ES cell line used in this study was CCE 28. ES cells were maintained on a layer of mitomycin C-treated feeder cells in Dulbecco’s modified Eagle’s medium, supplemented with 20% fetal bovine serum, 2 mM l-glutamine, 0.1 mM non-essential amino acids and 0.1 mM β-mercaptoethanol in the presence of 103 U/ml murine leukemia inhibitory factor (LIF).
Generation of muCdc7 knockout mice
The targeting vector plasmid, in which the 4.3 kb DNA fragment containing exons 3 and 4 (encoding essential kinase domains I and II) of muCdc7 was replaced with a 1.1 kb pMC1-neomycin resistance cassette (neor), was linearized and electroporated into CCE 28 ES cells. Transformants were selected by resistance to G418 at 250 µg/ml for the presence of the neor genes, and by resistance to ganciclovir at 5 µM for the absence of the tk gene.
Construction of a flox transgene vector
The flox transgene plasmid containing a hEF1α promoter-driven wild-type muCdc7 cDNA flanked by the loxP sites (Figure 3B) was constructed as follows. First, pEF1α-loxP-p(A)-loxP-EGFP was constructed by inserting the EGFP coding region at the SwaI site of pEF1α-loxP-p(A)-loxP, in which the original chicken β-actin gene (CAG) promoter of pCAG-loxP-p(A)-loxP was replaced with hEF1α promoter (kindly provided by Dr Sumio Sugano). The muCdc7-PGKpuror cassette, constructed by inserting a muCdc7 cDNA fragment into the pPGK-Puro-p(A) plasmid, was cloned at the NotI site present between the two loxP sites, resulting in the flox transgene vector pEF1α-loxP-muCdc7-PGKpuror-loxP-EGFP.
Generation of muCdc7–/– ES cells carrying a muCdc7 transgene
The flox muCdc7 transgene pEF1α-loxP-muCdc7-PGKpuror-loxP-EGFP was introduced into one of the muCdc7+/– ES clones by electroporation, and transfected cells were cultured in the presence of 0.6 µg/ml puromycin for 8 days. Southern blotting and RT–PCR were conducted to identify four puromycin-resistant clones (muCdc7+/–tg ES clones), which carried a single copy of the transgene on a chromosome and transcribed the transgene-derived mRNA. The primer set, ‘rt-EFs’ and ‘tMCas’, amplified a 370 bp fragment from the mRNA product of the transgene. We then generated muCdc7–/–tg ES clones by culturing one of the muCdc7+/–tg ES clones in the presence of an increased concentration of G418 (3 mg/ml).
Infection of ES cells with adenoviruses
AxCANCre (Ad-Cre; Kanegae et al., 1995) was amplified on 293 cells and purified by two rounds of CsCl density centrifugation. Ad-Mdm2, Ad-E6 and Ad-ctrl were gifts from Cyrus Vaziri (Nghiem et al., 2001). Added to ES cells, resuspended in a small volume of ES medium, were recombinant adenoviruses at a m.o.i. of 50 (Ad-Cre) or a m.o.i. of 200 (Ad-ctrl, Ad-Mdm2 and Ad-E6). After incubation for 1 h, cells were diluted with fresh ES medium containing murine LIF (104 U/ml) and plated in 6-well gelatinized plates. The cultures were maintained with daily change of the media.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Cyrus Vaziri for generous gifts of recombinant adenoviruses. We also thank Miho Nagoya for assistance in experiments with mice, and Naofumi Takemoto for critical reading of manuscript. We thank the members of the Katsuki laboratory and our laboratory for help and helpful discussion.
References
- Aladjem M.I., Spike,B.T., Rodewald,L.W., Hope,T.J., Klemm,M., Jaenisch,R. and Wahl.G.M. (1998) ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol., 8, 145–155. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Baldin V., Lukas,J., Marcote,M.J., Pagano,M. and Draetta,G. (1993) Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev., 7, 812–821. [DOI] [PubMed] [Google Scholar]
- Banin S. et al. (1998) Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science, 281, 1674–1677. [DOI] [PubMed] [Google Scholar]
- Bousset K. and Diffley,J.F. (1998) The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev., 12, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury E.M. (1992) Reversible histone modifications and the chromosome cell cycle. BioEssays, 14, 9–16. [DOI] [PubMed] [Google Scholar]
- Brown G.W. and Kelly,T.J. (1998) Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J. Biol. Chem., 273, 22083–22090. [DOI] [PubMed] [Google Scholar]
- Buck V., White,A. and Rosamond,J. (1991) CDC7 protein kinase activity is required for mitosis and meiosis in Saccharomyces cerevisiae. Mol. Gen. Genet., 227, 452–457. [DOI] [PubMed] [Google Scholar]
- Canman C.E., Lim,D.S., Cimprich,K.A., Taya,Y., Tamai,K., Sakaguchi,K., Appella,E., Kastan,M.B. and Siliciano,J.D. (1998) Activation of ATM kinase by ionizing radiation and phosphorylation of p53. Science, 281, 1677–1679. [DOI] [PubMed] [Google Scholar]
- Carr A.M. (1996) Checkpoints take the next step. Science, 271, 314–315. [DOI] [PubMed] [Google Scholar]
- Chan T.A., Hwang,P.M., Hermeking,H., Kinzler,K.W. and Vogelstein,B. (2000) Cooperative effects of genes controlling the G2/M checkpoint. Genes Dev., 14, 1584–1588. [PMC free article] [PubMed] [Google Scholar]
- Chapman J.W. and Johnston,L.H. (1989) The yeast gene, DBF4, essential for entry into S phase is cell cycle regulated. Exp. Cell Res., 180, 419–428. [DOI] [PubMed] [Google Scholar]
- Chong J.P., Thommes,P. and Blow,J.J. (1996) The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem. Sci., 21, 102–106. [PubMed] [Google Scholar]
- Donaldson A.D., Fangman,W.L. and Brewer,B.J. (1998) Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev., 12, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A. and Bell,S.P. (1997) Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell Dev. Biol., 13, 293–332. [DOI] [PubMed] [Google Scholar]
- Elledge S.J. (1996) Cell cycle checkpoints: preventing an identity crisis. Science, 274, 1664–1672. [DOI] [PubMed] [Google Scholar]
- Furnari B., Rhind,N. and Russell,P. (1997) Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science, 277, 1495–1497. [DOI] [PubMed] [Google Scholar]
- Gondo Y., Nakamura,K., Nakao,K., Sasaoka,T., Ito,K., Kimura,M. and Katsuki,M. (1994) Gene replacement of the p53 gene with the lacZ gene in mouse embryonic stem cells and mice by using two steps of homologous recombination. Biochem. Biophys. Res. Commun., 202, 830–837. [DOI] [PubMed] [Google Scholar]
- Gottifredi V., Shieh,S.Y., Taya,Y. and Prives,C. (2001) p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc. Natl Acad. Sci. USA, 98, 1036–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T., Golub,E.I., Reddy,G., Radding,C.M. and Ward,D.C. (1995) Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl Acad. Sci. USA, 92, 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R. and Oren,M. (1997) p53: from inductive signal to cellular effect. Curr. Opin. Genet. Dev., 7, 46–51. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. (1971) Genetic control of the cell cycle in yeast II. Genes controlling DNA replication and its initiation. J. Mol. Biol., 59, 183–194. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. and Weinert,T.A. (1989) Checkpoints: controls that ensure the order of cell cycle events. Science, 246, 629–634. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya,R., Kazaz,A. and Oren,M. (1997) Mdm2 promotes the rapid degradation of p53. Nature, 387, 296–299. [DOI] [PubMed] [Google Scholar]
- Hollingsworth J.R.E. and Sclafani,R. (1990) DNA metabolism gene CDC7 from yeast encodes a serine/threonine protein kinase. Proc. Natl Acad. Sci. USA, 87, 6272–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Pahl,P.M.B., Harrison,K., Rosamond,J. and Sclafani,R.A. (1993) Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol. Cell. Biol., 13, 2899–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P. and Blow,J.J. (2000) Xenopus Cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev., 14, 1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Jiang W. and Hunter,T. (1997) Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc. Natl Acad. Sci. USA, 94, 14320–14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., McDonald,D., Hope,T.J. and Hunter,T. (1999) Mammalian Cdc7–Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J., 18, 5703–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L.H., Masai,H. and Sugino,A. (1999) First the CDKs, now the DDKs. Trends Cell Biol., 9, 249–252. [DOI] [PubMed] [Google Scholar]
- Kanegae Y., Lee,G., Sato,Y., Tanaka,M., Nakai,M., Sakaki,T., Sugano,S. and Saito,I. (1995) Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res., 23, 3816–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey S.E., Labib,K. and Maiorano,D. (1996) Cell cycle control of eukaryotic DNA replication. Curr. Opin. Genet. Dev., 6, 208–214. [DOI] [PubMed] [Google Scholar]
- Kim J.M., Sato,N., Yamada,M., Arai,K. and Masai,H. (1998) Growth regulation of the expression of mouse cDNA and gene encoding a serine/threonine kinase related to Saccharomyces cerevisiae CDC7 essential for G1/S transition. J. Biol. Chem., 273, 23248–23257. [DOI] [PubMed] [Google Scholar]
- Komarov P.G., Komarova,E., Kondratov,R.V., Christov-Tselkov,K., Coon,J.S., Chernov,M.V. and Gudkov,A.V. (1999) A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science, 285, 1733–1737. [DOI] [PubMed] [Google Scholar]
- Kubbutat M.H., Jones,S.N. and Vousden,K.H. (1997) Regulation of p53 stability by Mdm2. Nature, 387, 299–303. [DOI] [PubMed] [Google Scholar]
- Kumagai H., Sato,N., Yamada,M., Mahony,D., Seghezzi,W., Lees,E., Arai,K. and Masai,H. (1999) A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol. Cell. Biol., 19, 5083–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. and Diffley,J.F. (2001) Is the MCM2–7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev., 11, 64–70. [DOI] [PubMed] [Google Scholar]
- Lei M. and Tye,B.K. (2001) Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci., 114, 1447–1454. [DOI] [PubMed] [Google Scholar]
- Lei M., Kawasaki,Y., Young,M.R., Kihara,M., Sugino,A. and Tye,B.K. (1997) Mcm2 is a target of regulation by Cdc7–Dbf4 during the initiation of DNA synthesis. Genes Dev., 11, 3365–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.J. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- Lutzker S.G. and Levine,A.J. (1996) A functionally inactive p53 protein in teratocarcinoma cells is activated by either DNA damage or cellular differentiation. Nature Med., 2, 804–810. [DOI] [PubMed] [Google Scholar]
- Masai H. and Arai,K. (2002) Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J. Cell Physiol., 190, 287–296. [DOI] [PubMed] [Google Scholar]
- Masai H., Miyake,T. and Arai,K. (1995) hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J., 14, 3094–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen R.M., Conner,D.A., Chao,S., Geisterfer-Lowrance,A.A. and Seidman,J.G. (1992) Production of homozygous mutant ES cells with a single targeting construct. Mol. Cell. Biol., 12, 2391–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem P., Park,P.K., Kim,Y.S., Vaziri,C. and Schreiber,S.L. (2001) ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc. Natl Acad. Sci. USA, 98, 9092–9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M., Theodoras,A.M., Schumacher,J., Roberts,J.M. and Pagano,M. (1995) Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol., 15, 2612–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B.T., Ying,C.Y., Gautier,J. and Maller,J.L. (1999) DNA replication in vertebrates requires a homolog of the Cdc7 protein kinase. Proc. Natl Acad. Sci. USA, 96, 2800–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugh R. (1990) Normal development of the mouse. The Mouse. Oxford University Press, London, UK, pp. 44–101.
- Sabapathy K., Klemm,M., Jaenisch,R. and Wagner,E.F. (1997) Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J., 16, 6217–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y., Wong,C., Thoma,R.S., Richman,R., Wu,Z., Piwnica-Worms,H. and Elledge,S.J. (1997) Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science, 277, 1497–1501. [DOI] [PubMed] [Google Scholar]
- Sato N., Arai,K. and Masai,H. (1997) Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J., 16, 4340–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatier P., Huang,S., Szekely,L., Wiman,K.G. and Samarut,J. (1994) Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene, 9, 809–818. [PubMed] [Google Scholar]
- Sclafani R.A. (2000) Cdc7p–Dbf4p becomes famous in the cell cycle. J. Cell Sci., 113, 2111–2117. [DOI] [PubMed] [Google Scholar]
- Sclafani R.A. and Jackson,A.L. (1994) Cdc7 protein kinase for DNA metabolism comes of age. Mol. Microbiol., 11, 805–810. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. (1993) Mammalian G1 cyclins. Cell, 73, 1059–1065. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. (1994) G1 phase progression: cycling on cue. Cell, 79, 551–555. [DOI] [PubMed] [Google Scholar]
- Shieh S.Y., Ahn,J., Tamai,K., Taya,Y. and Prives,C. (2000) The human homologs of checkpoint kinases Chk1 and Cds1(Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev., 14, 1439–1447. [PMC free article] [PubMed] [Google Scholar]
- Stillman B. (1996) Cell cycle control of DNA replication. Science, 274, 1659–1664. [DOI] [PubMed] [Google Scholar]
- Takeda T., Ogino,K., Matsui,E., Cho,M.K., Kumagai,H., Miyake,T., Arai,K. and Masai,H. (1999) A fission yeast gene, him1+/dfp1+, encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol. Cell. Biol., 19, 5535–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D.A., Belinsky,G., Chang,T.H., Jones,D.L., Schlegel,R. and Munger,K. (1997) The human papillomavirus-16 E6 oncoprotein decreases the vigilance of mitotic checkpoints. Oncogene, 15, 3025–3035. [DOI] [PubMed] [Google Scholar]
- Tibbetts R.S., Brumbaugh,K.M., Williams,J.M., Sarkaria,J.N., Cliby, W.A., Shieh,S.Y., Taya,Y., Prives,C. and Abraham,R.T. (1999) A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev., 13, 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J.C. (2000) Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem., 275, 39773–39778. [DOI] [PubMed] [Google Scholar]
- Weinreich M. and Stillman,B. (1999) Cdc7p–Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J., 18, 5334–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. and Stillman,B. (2000) Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p–Dbf4p kinase. Mol. Cell. Biol., 20, 3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]