Abstract
The Arabidopsis mutation, los2, impairs cold-responsive gene transcription, acquired freezing tolerance and plant resistance to chilling under certain conditions. LOS2 was isolated through positional cloning and shown to encode an enolase in the glycolytic pathway. In animal cells, enolase has also been known to function as a transcription factor that represses the expression of c-myc by binding to the c-myc gene promoter. LOS2 fused to green fluorescent protein is targeted to the nucleus as well as to the cytoplasm. LOS2/enolase protein can bind to the cis-element of the human c-myc gene promoter and to the gene promoter of STZ/ZAT10, a zinc finger transcriptional repressor from Arabidopsis. STZ/ZAT10 expression is induced rapidly and transiently by cold in the wild type, and this induction is stronger and more sustained in the los2 mutant. Furthermore, the expression of a RD29A-LUC reporter gene is repressed significantly by STZ/ZAT10 in transient expression assays in Arabidopsis leaves. Our results demonstrate that cold-responsive gene transcription in plants is controlled by a bi-functional enolase.
Keywords: cold signaling/enolase/LOS2/transcription factor/ZAT10 (STZ)
Introduction
Low temperature is an important environmental factor influencing plant growth and development. Some plants can cold acclimate, i.e. their freezing tolerance can be increased by being exposed to low but non-freezing temperatures (Levitt, 1980). Cold-regulated gene expression is a critical aspect of cold acclimation. Low temperatures induce the expression of a diverse array of plant genes, many of which are known as COR (for cold regulated), KIN (for cold induced), LTI (for low temperature induced) or RD (for responsive to desiccation) genes. Some of the gene products have been shown to contribute functionally to freezing tolerance. For example, the chloroplastic COR15A protein acts as a cryoprotectant by stabilizing membranes against freezing injury (Steponkus et al., 1998).
Substantial progress has been made towards understanding the transcriptional regulation of cold-responsive genes. Many of these genes have in their promoters one or several copies of the DRE/CRT cis-element, which has the core sequence CCGAC (Yamaguchi-Shinozaki and Shinozaki, 1994; Stockinger et al., 1997). A small family of transcription factors known as CBFs or DREB1s (Stockinger et al., 1997) binds to this element and activates transcription of the downstream COR/RD/KIN/LTI genes. Interestingly, the CBF/DREB1 genes are themselves induced by low temperatures, and this induction is transient and precedes that of downstream cold-regulated genes with the DRE/CRT cis-element (Medina et al., 1999). Therefore, there is a transcriptional cascade leading to the expression of the DRE/CRT class of genes under cold stress. Ectopic expression of CBF/DREB1 genes in plants turns on downstream cold-responsive genes even at warm temperatures (Jagglo-Ottosen et al., 1998). In transgenic Arabidopsis plants ectopically expressing CBF3, higher levels of proline and soluble sugars were found, which correlate with increased freezing tolerance (Gilmour et al., 2000).
CBF1 physically interacts with a histone acetyltransferase and a putative transcriptional adaptor protein (Stockinger et al., 2001). Recently, a negative regulator of cold-induced CBF expression was cloned (Lee et al., 2001). This regulator, HOS1, was identified in a genetic screen for Arabidopsis mutants with de-regulated RD29A expression (Ishitani et al., 1998). In homozygous recessive hos1 mutant plants, cold induction of RD29A and its upstream regulators, CBFs, is enhanced. HOS1 encodes a variant RING finger that exists in the cytoplasm at warm temperatures but appears in the nucleus when plants are incubated in the cold (Lee et al., 2001). Since some RING finger proteins can serve as ubiquitin E3 ligases that help degrade specific target proteins, HOS1 has been proposed to function by targeting the transcriptional activator of CBFs for degradation.
In order to have a better understanding of cold sensing and signaling in plants, more regulatory components need to be identified. We report here the characterization and cloning of a mutation, los2, which specifically impairs cold-induced transcript accumulation of stress genes. LOS2 is also critical for cold acclimation, and for chilling resistance under some conditions. LOS2 encodes an enolase (2-phospho-d-glycerate hydrolase, EC 4.2.1.11) that converts 2-phosphoglycerate to phosphoenolpyruvate in the glycolytic pathway. In animal cells, part of the enolase protein has been shown to bind to the promoter element of the c-myc gene and to repress c-myc expression (Feo et al., 2000; Subramanian et al., 2000). The LOS2 protein can bind to the c-myc promoter as well as to the promoter of the zinc finger STZ/ZAT10 from Arabidopsis. STZ/ZAT10 expression is induced rapidly and transiently by cold in wild-type plants, and the induction is significantly enhanced and prolonged by the los2 mutation. These results suggest that LOS2 has a direct regulatory function in controlling gene expression under low temperature stress.
Results
Identification of the LOS2 locus
To facilitate genetic dissection of cold signaling, we previously constructed Arabidopsis plants that emit bioluminescence in response to cold temperatures. The plants express the firefly luciferase reporter gene (LUC) under the control of the RD29A promoter, which contains the cold- and osmotic stress-responsive DRE/CRT element and the abscisic acid (ABA)-responsive ABRE element (Ishitani et al., 1997). The RD29A-LUC plants (referred to herein as wild type) were mutagenized by ethylmethane sulfonate, and mutants with aberrant luminescence responses were screened from the resulting M2 population by luminescence imaging using a cooled CCD camera (Ishitani et al., 1997).
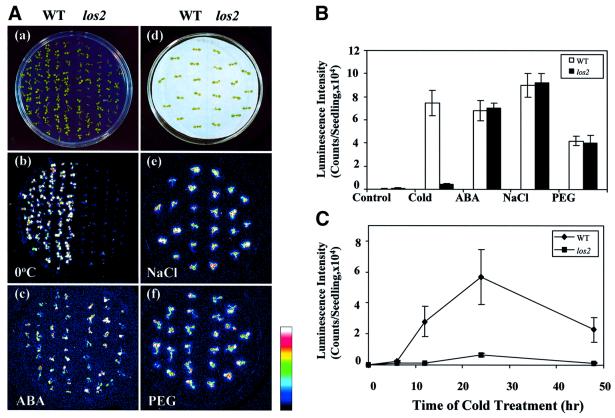
One mutant, designated as los2, shows a substantially reduced luminescence response specifically under cold stress, and was chosen for detailed characterization. Figure 1A shows the RD29A-LUC transgene expression phenotype of los2 and wild-type plants when treated with cold, ABA, NaCl or polyethylene glycol (PEG). Wild-type and mutant plants were grown on the same agar plate and bioluminescence images were taken after the respective stress treatments. Without stress treatment, no RD29A-LUC expression was detected in either the wild-type or los2 plants (data not shown). In response to cold treatment for 24 h, wild-type plants showed strong luminescence emission. In contrast, luminescence emission from cold-treated los2 plants was very weak (Figure 1Ab). ABA, NaCl or PEG treatment induced similar levels of luminescence in wild-type and los2 plants (Figures 1Ac, e and f). These luminescence images indicate that the los2 defect in RD29A-LUC expression is specific to cold stress. A quantification of the luminescence shows clearly that RD29A-LUC expression in los2 is virtually non-responsive to cold, but is still fully responsive to ABA, NaCl or PEG. The cold-induced luminescence was 9.2 times lower in los2 compared with the wild type (Figure 1B).
Fig. 1. The los2 mutation impairs cold-induced luminescence expression. (A) Luminescence images of wild-type and los2 plants. (a) Wild-type and los2 seedlings used for cold or ABA treatment. (b and c) Luminescence of wild-type and los2 seedlings after low temperature (0°C, 24 h) or ABA treatment (100 µM ABA, 3 h). (d) Wild-type and los2 seedlings used for NaCl or PEG treatment. (e and f) Luminescence of wild-type and los2 seedlings after NaCl (300 mM NaCl, 5 h) or PEG treatment (30% PEG 6000, 5 h). The color scale on the right shows the luminescence intensity from dark blue (lowest) to white (highest). (B) Luminescence intensity in wild-type and los2 plants under different stresses. Shown are the mean values ± SE from 20 individual seedlings. Stress treatments were as described in (A). (C) Time course of luminescence expression in wild-type and los2 plants during cold treatment. The wild-type and los2 plants on the same agar plate were incubated at 0°C for the indicated time and luminescence images were taken. Shown are the mean values ± SE from 20 individual seedlings.
To test whether los2 plants display similarly reduced expression of the RD29A-LUC transgene at different time points of cold treatment, 10-day-old seedlings on agar plates were cold treated for 0, 6, 12, 24 or 48 h and then used for luminescence imaging. As shown in Figure 1C, the luminescence intensity of wild-type plants increased quickly in response to cold treatment, and reached a peak level after 24 h of cold treatment. In los2 plants, the luminescence intensity was very low throughout the time course of cold treatment.
To test the dominance of the los2 mutation, F1 plants were obtained by backcrossing los2 plants with the wild type. All F1 plants exhibited the wild-type phenotype of luminescence expression in response to cold treatment (data not shown). The F2 progeny of the selfed F1 showed an ∼3:1 segregation for wild type/mutant, when imaged for luminescence after cold treatment (data not shown). These results indicate that los2 is a recessive mutation in a single nuclear gene.
los2 blocks cold induction of COR/KIN/RD/LTI but not CBF genes
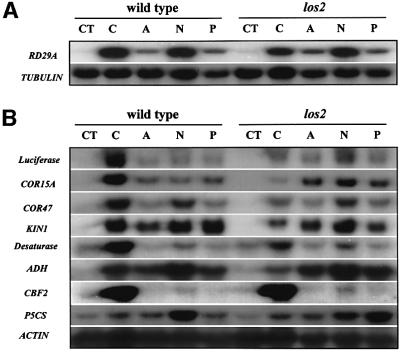
RNA blot analysis was carried out to determine the impact of los2 mutation on the transcript levels of stress-responsive genes. The endogenous RD29A transcript level was greatly induced in the wild-type plants but this induction was lower in los2 mutant plants (Figure 2A). Induction of the RD29A transcript by ABA, NaCl or PEG treatment was not affected by the los2 mutation (Figure 2A). The effect of los2 on the cold induction of endogenous RD29A is less dramatic compared with the effect on RD29A-LUC. This is probably because the RD29A-LUC transgene contains only part of the RD29A promoter (a fragment containing –646 to –1 upstream of the transcription start site), and there may be additional unknown regulatory elements in the endogenous RD29A promoter or even in the untranslated regions or introns of the endogenous RD29A gene. The difference between RD29A-LUC and endogenous RD29A expression levels has also been noticed in other mutants such as hos1 (Ishitani et al., 1998).
Fig. 2. los2 blocks cold induction of COR/KIN/RD/LTI but not CBF genes. (A) Endogenous RD29A expression in wild-type and los2 plants under different stress treatments. (B) The expression of other stress-responsive genes in wild-type and los2 plants. Wild-type and los2 plants were grown in the same agar plates for 3 weeks under continuous light at 22 ± 2°C. The seedlings were then treated with cold (0 ± 0.1°C for 24 h), ABA (100 µM for 3 h), NaCl (300 mM for 3 h) or PEG (30% for 5 h). Genes for β-tubulin or actin were used as loading controls. CT, control; C, cold; A, ABA; N, NaCl; P, PEG.
In contrast to the reduction of cold activation of RD29A expression, the los2 mutation virtually blocked the cold induction of LUC, COR15A, COR47, KIN and ADH (alcohol dehydrogenase) genes (Figure 2B). As with the induction of RD29A, induction of LUC, COR15A, COR47, KIN and ADH by ABA, NaCl or PEG was not significantly affected or not at all affected by the los2 mutation.
The CBF/DREB1 transcription factor genes are induced specifically by cold treatment. We compared the expression of one of them, CBF2, in wild-type and los2 plants (Figure 2B). The los2 mutation had no effect on the cold regulation of CBF2, implying that LOS2 does not function upstream of the CBF genes.
Membrane fluidity is an important determining factor for cell adaptation to both high and low temperature stresses (Levitt, 1980). Plants are known to increase in polyunsaturated phospholipids to maintain proper membrane fluidity under cold stress (Miquel et al., 1993). We determined the expression of one of the desaturase genes, Δ9-desaturase. The result shows that expression of this desaturase gene was highly induced by cold stress in the wild type (Figure 2B). The induction was much diminished in los2 mutant plants, indicating that polyunsaturation of membrane phospholipids may not occur in los2 in response to cold stress.
The compatible osmolyte, proline, plays an important role in freezing tolerance (Delauney et al., 1993). The P5CS gene encodes the rate-limiting enzyme, Δ1-pyrroline-5-carboxylate synthetase, in proline biosynthesis. P5CS expression was induced slightly by cold treatment, and this induction was not affected by the los2 mutation. Curiously, P5CS induction by NaCl appeared to be reduced whereas its induction by PEG was enhanced in the los2 mutant (Figure 2B).
los2 mutant plants are chilling sensitive in the light but not in the dark
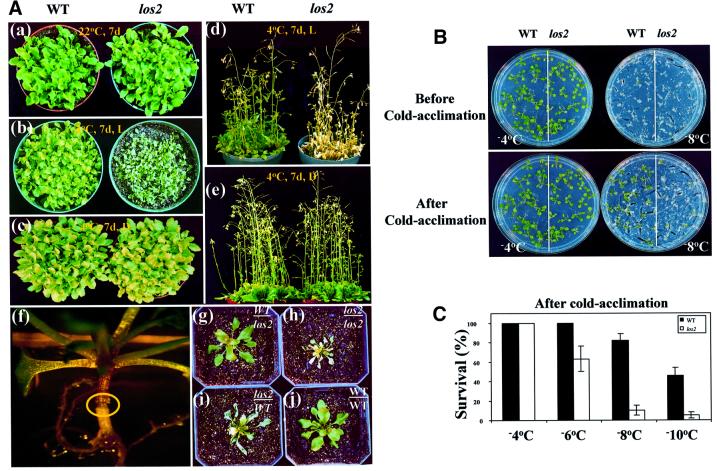
Unless indicated otherwise, all our cold treatments were carried out in the dark. Under the dark cold treatments as well as under normal growth temperatures, los2 plants behaved just like the wild-type plants, and no visible phenotypic changes could be detected. However, when los2 mutant plants were incubated in the cold under light, a striking phenotype was observed: the mutant plants wilted. The wilting of los2 plants first became visible at the leaf tips after 2 days at 4°C and under light. After 1 week, the mutant leaves were entirely desiccated but maintained their green color (Figure 3Ab). With prolonged cold and light exposure, los2 plants became bleached and dead (Figure 3Ad). In comparison, the vigor and survival of wild-type plants were not visibly affected by even prolonged cold and light treatment.
Fig. 3. Chilling and freezing sensitivity in los2 plants. (A) Morphology of wild-type and los2 plants with or without cold treatment and grafting experiment. (a) Wild-type and los2 plants are normal when grown at 22°C. (b and d) los2 but not wild-type plants wilt when incubated under light (20 µmol/m2/s) at 4°C for 7 days. (c and e) Neither los2 nor wild-type plants wilt when incubated in the dark at 4°C for 7 days. (f–j) Grafting experiment showing that wilting is not caused by the los2 root. (f) A successfully grafted plant. The circle highlights the graft junction. (g–j) Chilling sensitivity of grafted plants at 4°C. Grafted plants in soil were incubated in the cold room for 10 days under continuous light (20 µmol/m2/s) before pictures were taken. (g) Seedling with a wild-type shoot and los2 root. (h) Seedling with a los2 shoot and root. (i) Seedling with a los2 shoot and wild-type root. (j) Seedling with a wild-type shoot and root. (B) Morphology of wild-type and los2 plants after freezing stress, with or without cold acclimation treatment. (C) Quantification of the freezing survival rate for wild-type and los2 plants with cold acclimation treatment.
The wilting of los2 plants in response to cold and light treatment appeared to be a result of a water deficit because no wilting occurred when the plants were covered with a plastic film (data not shown). However, the mutant plants did not lose more water than the wild-type plants in the cold (in the light) or at normal growth temperatures (data not shown). Furthermore, the wilting occurred regardless of the amount of water supplied to the roots. We hypothesized that the cold-induced wilty phenotype of los2 plants may be caused by impaired water absorption or conduction by the roots. It has been well documented that some chilling-sensitive plants such as mung bean and tomato experience a sharp decrease in root hydraulic conductance and subsequent wilting when they are exposed to chilling temperatures (Markhart et al., 1979; Bagnall et al., 1983).
We performed a series of grafting experiments to test the ‘water absorption/conduction’ hypothesis. The results show that wilting occurred when the grafted plants had los2 shoots, but no wilting occurred when they had wild-type shoots, regardless of the root genotypes (Figure 3Ag–j). Because the wilting phenotype has nothing to do with the root genotype, the results clearly reject the ‘water absorption/conduction’ hypothesis.
Since los2 mutant plants show light-dependent chilling sensitivity, we performed experiments to determine whether the effect of the mutation on cold-responsive gene expression is also light dependent. Luminescence imaging showed that reduced cold induction of RD29A-LUC expression in los2 seedlings occurred in either light or dark conditions (data not shown). RNA blot analysis revealed that cold-induced expression of RD29A, COR15A and COR47 was diminished by the los2 mutation in either light or dark conditions (data not shown). In contrast, cold-induced CBF2 expression in either light or dark conditions was not reduced by the los2 mutation (data not shown). These results demonstrate that although los2 plants show light-dependent chilling sensitivity, the effect of the mutation on cold-responsive gene expression is not dependent on light.
Chilling-induced cell death and freezing sensitivity in los2 mutant plants
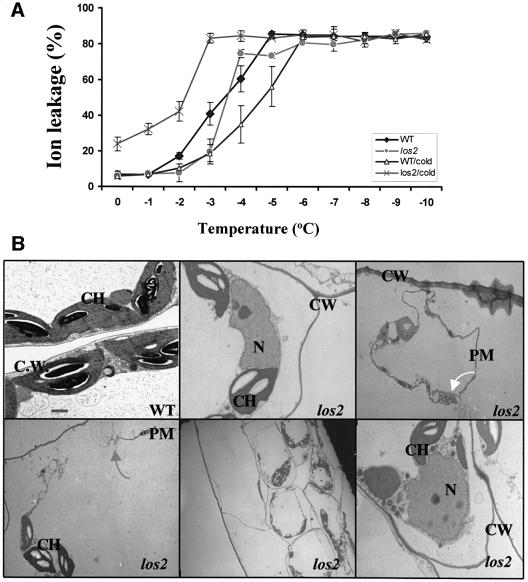
We attempted to examine the effect of cold acclimation on the freezing tolerance of los2 plants. Wild-type and los2 plants were treated at 4°C under light for 2 days, and covered with plastic film to avoid the wilting of the mutant. The leaves were then excised and subjected to a standard electrolyte leakage assay (Ishitani et al., 1998). As expected, the wild-type plants showed an increase in freezing tolerance and a decrease in electrolyte leakage, in response to the cold acclimation treatment (Figure 4A). Surprisingly, los2 leaves showed a dramatic increase in electrolyte leakage after the cold acclimation treatment (Figure 4A). The results suggest that los2 cell membranes became leaky when the mutant plants were exposed to cold and light, consistent with the chilling-sensitive phenotype.
Fig. 4. los2 plants are defective in membrane integrity under light and cold treatment. (A) Ion leakage assay. Wild-type and los2 mutant leaves from 3-week-old plants grown in soil were used for the assay. For cold acclimation (CA), plants were incubated at 4°C for 2 days under light (20 µmol/m2/s) before the ion leakage assay. Shown are the mean values and standard errors from three replicates. (B) Protoplast shrinkage and plasma membrane damage revealed by transmission electron microscopy. Leaves from wild-type and los2 plants incubated at 4°C for 2 days under light (20 µmol/m2/s) are shown. Bar = 1 µm. CW, cell wall; PM, plasma membrane; N, nucleus; CH, chloroplast. The gray arrow indicates broken plasma membrane in los2 leaf cells. The white arrow indicates membrane blebbing in los2 leaf cells.
The los2 leaves that were treated for 2 days at 4°C under light (under a plastic film cover to prevent wilting) were examined under a transmission electron microscope. Under the electron microscope, los2 mesophyll cells were found to have disintegrated cell membranes (Figure 4B). Membrane blebbing was prevalent in los2 cells. These cytological changes indicate that the cold and light treatment induced cell death in los2 leaves.
In order to avoid chilling sensitivity during cold acclimation treatment, wild-type and los2 plants were incubated for 2 days at 4°C in the dark. The method of Xin and Browse (1998) was then used to determine cold-induced freezing tolerance. Without cold acclimation treatment, wild-type and los2 plants had the same level of freezing tolerance (Figure 3B). For example, both the wild type and los2 mutant survived –4°C freezing but neither survived –8°C freezing (Figure 3B). However, after the 2 day cold acclimation treatment in the dark, wild-type plants became substantially more tolerant to freezing than los2 plants (Figure 3B). Most of the los2 plants did not survive freezing at –8°C. On the contrary, ∼81% of the wild-type plants survived the –8°C freezing. Even after freezing at –10°C, ∼43.5% of the wild-type plants survived (Figure 3C). The data indicate that los2 mutant plants are defective in cold acclimation.
Positional cloning reveals that LOS2 encodes an enolase
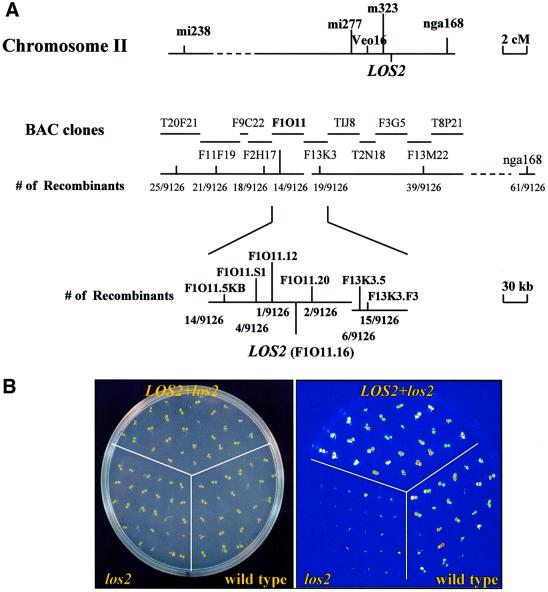
To map the LOS2 gene genetically, los2 plants in the C24 ecotype were crossed with wild-type plants of the Columbia ecotype. The resulting F1 plants were selfed and the F2 progeny were planted and grown on MS agar plates for 10 days. Luminescence images of plants after 24 h cold treatment were obtained to select homozygous los2 plants for genetic mapping. Representative molecular markers that cover five Arabidopsis chromosomes were surveyed, and the LOS2 gene was located to chromosome II. Further mapping positioned LOS2 to the lower arm of chromosome 2, north of nga168. More refined mapping delimited LOS2 to a region covered by 10 bacterial artificial chromosome (BAC) clones T20F21, F11F19, F9C22, F2H17, F1O11, F13K3, TIJ8, T2N18, F3G5 and F13M22 (Figure 5A). For the fine mapping, simple sequence repeats were identified from the BAC sequences, and PCR primer pairs were designed to amplify selected repeats to identify simple sequence length polymorphism (SSLP) markers. Also, sequence polymorphisms between Col and C24 ecotypes identified during candidate gene sequencing to find a los2 mutation were utilized to develop molecular markers (see Materials and methods for details). In summary, with a total of 4563 los2 plants, the los2 mutation was delimited to an ∼28 kb region between the F1O11.12 and F1O11.20 markers (Figure 5A). There are seven predicted open reading frames (ORFs) in this region. All genomic DNA in this region including the seven ORFs was amplified from the wild-type C24 and los2 mutant plants and sequenced. This sequence analysis revealed a single G to A mutation in the F1O11.16 gene that encodes an enolase (Van Der Straeten et al., 1991). The mutation would result in the substitution of Gly325 by serine in the los2 mutant.
Fig. 5. Positional cloning of LOS2 and mutant complementation assay. (A) Positional cloning. Genetic mapping with SSLP markers positioned LOS2 to the BAC clone F1O11. Sequence analysis identified a single nucleotide mutation that would result in an amino acid sequence change (Gly325 to serine) in the ORF F1O11.16. (B) Complementation assay. RD29A-LUC luminescence expression (after 2 days treatment at 0°C) is shown for one representative homozygous T3 line (LOS2+los2) transformed with a construct containing a wild-type F1O11.16 gene. Also included in the agar plate are wild-type and los2 plants as controls.
To confirm that F1O11.16 is the LOS2 gene, the F1O11 BAC clone was digested with restriction enzymes EcoRI and SacI, which yielded a 5157 bp fragment containing the enolase ORF plus 886 bp upstream of ATG and 1386 bp downstream of the stop codon. The enolase fragment was cloned into the binary vector pCAMBIA 1200 and introduced into los2 mutant plants via Agrobacterium-mediated transformation. Forty-five hygromycin-resistant individuals were selected, and their T2 and T3 progeny were subjected to complementation analysis with luminescence imaging. One representative line in the T3 generation was chosen to illustrate its luminescence response to cold treatment. The result is presented in Figure 5B, which shows that the F1O11.16 gene complemented the luminescence phenotype of los2 mutant plants. In addition, all of the 45 transgenic los2 plants expressing the wild-type F1O11.16 transgene were exposed to 4°C under light for a week. None of the transgenic plants wilted, proving that the F1O11.16 gene also complemented the chilling-sensitive phenotype of los2 mutant.
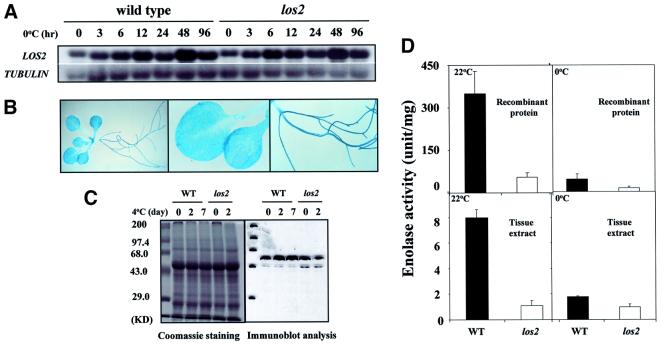
The transcript of the LOS2 gene was constitutively present in Arabidopsis seedlings (Figure 7A). The transcript level appeared to be slightly up-regulated in response to cold treatment. The los2 mutation did not alter this pattern of LOS2 transcript expression (Figure 7A). We fused the LOS2 promoter region (–1055 to –1 from ATG) to the α-glucuronidase reporter gene (GUS), and the resulting construct was introduced into wild-type Arabidopsis plants. GUS expression was detected in all plant tissues examined (Figure 7B). The LOS2 protein level was also up-regulated slightly by cold treatments (Figure 7C). LOS2 protein was present in los2 mutant seedlings, but the level appeared to decline after a 2 day cold treatment (Figure 7C).
Fig. 7. Gene expression pattern and enolase activity of LOS2. (A) Expression of the LOS2 gene transcript. Full-length LOS2 cDNA was used as a probe. The β-tubulin gene was used as a loading control. (B) LOS2 promoter::GUS expression. Wild-type plants containing LOS2 promoter::GUS were stained with X-Gluc for 15 h and visualized under a microscope. (C) Immunoblot analysis. A 20 µg aliquot of total protein from wild-type and los2 leaves was extracted and fractionated in a 12.5% SDS–polyacrylamide gel. Shown are the gel picture of Coomassie staining and immunodetection with polyclonal anti-enolase. Molecular size markers are shown on the left. (D) Wild-type and mutant enolase activity. Shown are the mean values ± SE from three replicates.
The los2 mutation reduces enolase activity at both cold and warm temperatures
Both the wild-type and mutant LOS2 ORFs were expressed in bacteria to produce recombinant enolase proteins. In vitro assays carried out at 22°C showed that the wild-type LOS2 protein had a high enolase activity. In comparison, the los2 mutant protein had a significantly reduced activity at 22°C (Figure 7D). When the assays were carried out at 0°C, both the wild-type and mutant proteins displayed much lower activities.
At 22°C, tissue extracts from los2 mutant also had a significantly lower activity compared with that from wild-type plants (Figure 7D). At 0°C, tissue extracts from both wild-type and los2 mutant plants had low activities, and the difference between the wild-type and los2 plants was reduced (Figure 7D). These results show that LOS2 is indeed an enolase, and the los2 mutation disrupts the enolase activity at both warm and cold temperatures. Therefore, the los2 mutation is not a cold-sensitive allele of LOS2. The fact that los2 tissue extracts had a much lower activity compared with the wild-type extracts suggests LOS2 as the major contributor to enolase activity in Arabidopsis.
LOS2–GFP fusion protein is targeted to the nucleus as well as cytoplasm
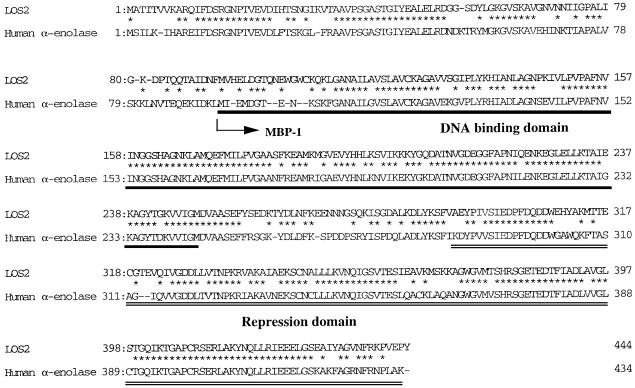
Enolase has been implicated as a multifunctional protein in animals and fungi. For example, in human cells, part of the enolase has been identified as MBP1 that binds to the c-myc promoter and represses c-myc gene expression (Ray and Miller, 1991; Ghosh et al., 1999a; Feo et al., 2000). MBP1 is an alternatively translated form of enolase that simply lacks the first 93 amino acid residues (Feo et al., 2000; Subramanian et al., 2000; Figure 6). As a glycolytic enzyme, enolase is localized in the cytoplasm. As a transcription factor, MBP1 is localized in the nucleus (Feo et al., 2000).
Fig. 6. Amino acid sequence comparison between LOS2 protein and human α-enolase. In human α-enolase, the DNA-binding domain is underlined, and the transcriptional repression domain is double-underlined. The arrow indicates the start site of alternative translation for the c-myc promoter- binding protein (MBP-1).
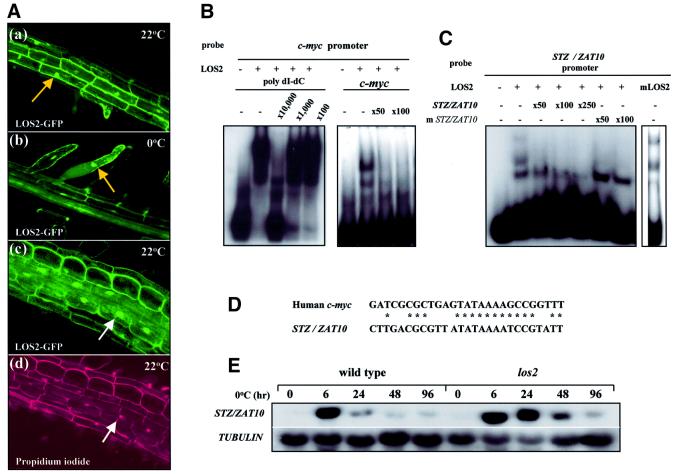
To examine the localization of LOS2 protein, green fluorescent protein (GFP) was fused in-frame to the C-terminus of LOS2, and the fusion protein expressed in Arabidopsis plants under the cauliflower mosaic virus (CaMV) 35S promoter. Fifteen independent T1 lines were selected and examined for GFP expression. Undoubtedly, GFP expression was detected in the cytoplasm (Figure 8A). However, GFP was also clearly detected in the nucleus (Figure 8A). The nuclear GFP expression was detected at both cold and normal growth temperatures (Figure 8A). GFP fusion with several truncated versions of LOS2 (e.g. starting with Met93 or Met187) gave similar localization patterns (data not shown).
Fig. 8. LOS2 subcellular localization and binding to promoter elements. LOS2–GFP localizes in the nucleus as well as in the cytoplasm and binds specifically to the promoter of c-myc and STZ/ZAT10. (A) LOS2–GFP fusion protein is localized in the nucleus as well as in the cytoplasm. (a–c) Confocal images of root tissues expressing LOS2–GFP fusion protein at 22 or 0°C. Arrows point to nuclei. (d) The same root tissue shown in (c) was stained with propidium iodide for nuclei. (B) EMSA with the human c-myc promoter DNA probe. Cold competitors [poly(dI–dC) in the left panel and the c-myc promoter DNA probe in the right panel] were added to determine the binding specificity. (C) EMSA was carried out with the STZ/ZAT10 promoter DNA probe in the presence or absence of wild-type (STZ/ZAT10) or mutated (mSTZ/ZAT10) cold probes as competitors. (D) Sequence alignment between human c-myc and Arabidopsis STZ/ZAT10 promoter elements included in the probes used in the EMSA. (E) Expression of STZ/ZAT10 in response to cold stress in wild-type and los2 plants.
LOS2 binds to the promoter and down-regulates the expression of ZAT10, an early cold-responsive gene encoding a transcriptional repressor
Human α-enolase (ENO1) has been shown to bind to the c-myc promoter and acts as a negative regulator of the c-myc gene (Ray and Miller, 1991; Ghosh et al., 1999a). Comparison of LOS2 and human ENO1 sequences revealed that the putative DNA-binding and repression domains are conserved in LOS2 (Figure 6), suggesting that LOS2 may also be capable of binding to certain DNA sequences, perhaps sequences similar to the animal c-myc promoter element. To address this possibility, we performed an electrophoretic mobility shift assay (EMSA) using His-LOS2 fusion protein. As shown in Figure 8B, several DNA-shifted bands were observed with a DNA probe containing the human enolase-binding site in the c-myc promoter. The binding of LOS2 to the labeled human c-myc probe was competed off by unlabeled cold c-myc probe but not by a 1000-fold excess of poly(dI–dC), indicating the specificity of the LOS2–DNA interaction (Figure 8B).
Since no c-myc homolog is present in Arabidopsis, other genes might be targets for LOS2. The C-terminal region of LOS2 is very similar to that of human enolase (Figure 6), which serves as a repression domain (Ghosh et al., 1999a). To identify possible target genes of LOS2, we surveyed the promoter sequences of Arabidopsis genes encoding putative transcriptional repressors. We found that the region from –774 to –748 upstream of the trans lation initiation site of STZ/ZAT10, which encodes a putative transcriptional repressor (Ohta et al., 2001), has similarity to the enolase/MBP1-binding sequence in the human c-myc promoter (Figure 8D). As shown in Figure 8C, LOS2 was also capable of binding to this STZ/ZAT10 promoter fragment (–775 to –747). Competition experiments with mutated STZ/ZAT10 promoter fragment confirmed specific binding of LOS2 to this sequence. Mutant LOS2 protein could also bind to the fragment (–775 to –747) (Figure 8C).
RNA blot analysis showed that STZ/ZAT10 expression was induced rapidly and transiently by cold stress in the wild type whereas, in the los2 mutant, the cold induction was stronger and more sustained (Figure 8E). These results suggest that LOS2 negatively regulates the expression of STZ/ZAT10 by binding to its promoter.
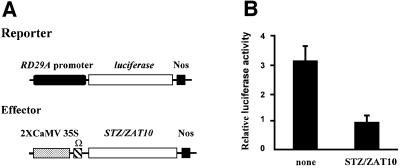
To examine whether STZ/ZAT10 represses the expression of the RD29A gene, transient expression assays were performed in Arabidopsis leaves. The RD29A-LUC transgene, containing an upstream sequence (–646 to –1 from the transcription start site) of the RD29A gene fused to the LUC reporter gene was delivered into Arabidopsis leaves by particle bombardment. When the effector plasmid containing STZ/ZAT10 under the control of the CaMV 35S promoter (Figure 9A) was co-transfected with RD29A-LUC, the LUC activity was reduced to ∼30% (Figure 9B). This observation suggests that STZ/ZAT10 represses the expression of the RD29A gene.
Fig. 9. STZ/ZAT10 can repress the expression of RD29A in Arabidopsis leaves. (A) Schematic representation of the reporter and effector plasmids used in transient expression assays. A portion of the RD29A promoter (black oval) was fused to firefly luciferase. Nos denotes the terminator signal of the gene for nopaline synthase. Ω indicates the translational enhancer of tobacco mosaic virus. (B) Relative luciferase activities after transfection with RD29A-LUC and 2× 35S-STZ/ZAT10. To normalize values obtained after each transfection, pPTRL DNA that contained the CaMV 35S promoter and a gene for luciferase from Renilla was used as internal control. Luciferase activity was expressed in arbitrary units relative to the activity of Renilla luciferase (as described in Ohta et al., 2001). The values are averages of three bombardments, and error bars indicate standard deviations.
Discussion
In the present study, we identified and cloned LOS2, a positive regulator of cold-responsive gene transcription. We identified the los2 mutant by its reduced expression of the RD29A-LUC transgene under cold treatment. Normal responses of los2 plants to ABA, salt and PEG indicate that los2 has a specific defect in cold signaling. Besides its effect on the RD29A-LUC transgene, los2 also specifically impairs cold-regulated expression of the endogenous COR/KIN/RD/LTI genes. The COR/KIN/RD/LTI genes are regulated by cold through the CRT/DRE element in their promoters (Thomashow, 1999). CBF transcription factors bind to the CRT/DRE sequence and activate COR/KIN/RD/LTI expression. However, no difference was detected in the expression of CBF genes between wild-type and los2 plants (Figure 2B). These results suggest that LOS2 has a function downstream of the CBF transcription factors or is required directly or indirectly for CBF activation of the COR/KIN/RD/LTI genes. The effect of los2 on COR/KIN/RD/LTI but not CBF genes is similar to that of sfr6, the only other Arabidopsis mutation known to reduce cold induction of genes (Knight et al., 1999). However, the effect of sfr6 is not cold specific because the mutation also blocks osmotic stress or ABA induction of the COR/KIN/RD/LTI genes (Knight et al., 1999).
LOS2 is not only critical for freezing tolerance, it is also important for chilling resistance. In the light, los2 mutant plants are chilling sensitive. The chilling sensitivity is a consequence of cold- and light-induced cell death. The molecular basis of the induced cell death is unclear. It may be caused by metabolic imbalance because the los2 mutation disrupts enolase activity. It may also have to do with the defective gene regulation in the los2 mutant. As shown in Figure 2B, the transcript level of a desaturase gene is induced by cold in the wild type but this induction is greatly decreased in los2. Membrane fluidity is an important factor for plant survival under cold temperatures (Levitt, 1980). The defect in the desaturase gene induction may contribute to the chilling sensitivity of los2 plants. Interestingly, los2 plants are not chilling sensitive in the dark, although los2 plants are impaired in cold-responsive gene expression in both light and dark. These observations suggest that the chilling sensitivity of los2 plants in the light may also be related to metabolic imbalance caused by photosynthesis. Alternatively, the chilling sensitivity may be a manifestation of programmed cell death. A connection between human enolase and apoptosis has been demonstrated before. Part of human enolase, MBP1, when transfected into fibroblast cells, caused rapid cell death (Ray, 1995). The cold- and light-induced cell death in los2 mutant plants has some hallmarks of programmed cell death, e.g. cell shrinkage, membrane blebbing (Figure 4B), nuclear condensation and DNA fragmentation (data not shown). Therefore, it is possible that the chilling-sensitive phenotype of los2 is a result of programmed cell death caused by the interplay between defective gene regulation, and cold and light signals.
The los2 mutation was found in an enolase gene that converts 2-phosphoglycerate to phosphoenolpyruvate in the glycolytic pathway. This enolase gene was able to complement the los2 mutant phenotypes (Figure 5B). LOS2 transcript can be detected at normal growth temperatures, and cold treatment enhanced transcript accumulation in both wild-type and los2 plants. Since a CCGAC core sequence of CRT/DRE is found in the enolase gene promoter, it is possible that CBF1/DREB1 may regulate the expression of enolase under cold stress.
Interestingly, MBP1, part of human α-enolase, was shown to be targeted to the nucleus and binds to the c-myc promoter (Feo et al., 2000). MBP1 binds just 5′ to the P2 TATA motif and negatively regulates the expression of c-myc by preventing TATA-binding protein (TBP) from binding to the TATA motif (Ray and Miller, 1991). In addition to the inhibition of TBP binding, MBP1 has active repression domains in both its N- and C-terminal regions (Ghosh et al., 1999a; Figure 6). The active repression may be mediated partly through interaction with histone deacetylase (HDAC) (Ghosh et al., 1999b). Consistent with these results on c-myc repression, transfection of human breast carcinoma cells with the MBP1 cDNA inhibits tumor formation (Ray et al., 1995).
We suspected that LOS2/enolase might have a similar function as a transcriptional repressor that regulates cold-responsive gene expression. We found that the LOS2–GFP fusion protein is targeted to the nucleus (Figure 8A). In addition, LOS2 protein can bind to the TATA box region of the c-myc promoter. There is no bona fide c-myc homolog in Arabidopsis. However, we have identified an Arabidopsis zinc finger gene, STZ/ZAT10, which may be a direct target for LOS2/enolase regulation. The STZ/ZAT10 gene promoter contains a similar region to the cis-element in c-myc where MBP1 binds. EMSAs show that LOS2 protein can bind to this STZ/ZAT10 promoter element. The sequence of LOS2 is highly conserved with that of human α-enolase, especially in their C-terminal region that is necessary for active repression (Ghosh et al., 1999a; Figure 6). We showed that both the wild-type and mutant LOS2 proteins could bind to the sequence of the STZ/ZAT10 promoter, indicating that mutation does not affect the binding activity of LOS2. Since the mutation in the los2 allele substitutes Gly325 to serine that is located near the putative repression domain (Ghosh et al., 1999a), the mutation probably causes a defect in repression activity. We hypothesize that LOS2 represses STZ/ZAT10 expression, which in turn may repress the transcription of COR/KIN/RD/LTI genes. STZ/ZAT10 is induced rapidly and transiently by cold treatment, suggesting an important role for STZ/ZAT10 in controlling genes that are induced more slowly by cold (e.g. the COR/KIN/RD/LTI genes). Our finding that STZ/ZAT10 expression is increased in the los2 mutant plants is consistent with LOS2 having a role in repressing STZ/ZAT10 transcription.
STZ/ZAT10 is a TFIIIA/Krüppel-like zinc finger protein that has been shown to repress the trans-activation of genes through an essential repression motif (Ohta et al., 2001). STZ/ZAT10 is similar to other zinc finger proteins such as petunia ZPT2-2 (EPF2-5) (Takatsuji and Matsumoto, 1996). Binding experiments have demonstrated that ZPT2-2 recognizes similar sequences containing the core sequence, AGT (or ACT in a reverse orientation) (Takatsuji and Matsumoto, 1996). In the case of ZPT2-2, the target sequence consists of a tandem repeat of two AGTs, and the spacing between the two AGT sequences is important for ZPT2-2 to distinguish target sequences (Takatsuji and Matsumoto, 1996). Interestingly, we found two tandem AGT sequences with appropriate spacing between them, from –554 to –522 upstream of the transcription initiation site in the RD29A gene promoter (ACTAGTGTA-N13-TCTAGTAAG). This suggests that RD29A may be one of the direct target genes for STZ/ZAT10. All the other genes such as COR15A, COR47, KIN1 and the desaturase that are blocked by the los2 mutation also have in their promoters similar tandem AGT repeats. Moreover, our transient expression assays revealed that indeed STZ/ZAT10 represses RD29A-LUC expression, suggesting that STZ/ZAT10 may interact directly with the sequences in the RD29A promoter. In conclusion, our data provide the first genetic evidence that LOS2/enolase is a positive regulator of the RD29A gene, by controlling the expression of STZ/ZAT10 that represses the RD29A gene. Thus, LOS2/enolase and STZ/ZAT10 along with other transcription factors such as CBFs/DREB1s constitute a fine-tuned transcriptional regulatory circuitry for optimal cellular responses to cold environments.
Materials and methods
Mutant isolation, luminescence imaging and plant growth
Mutant screening, procedures for stress treatment and bioluminescence imaging, and conditions for growing plants in soil were described previously (Ishitani et al., 1997; Lee et al., 2001).
RNA analysis
Two-week-old wild-type and mutant plants grown on standard 0.6% agar medium containing Murashige and Skoog (MS) salt supplemented with 3% sucrose were treated with either low temperature, ABA, NaCl or PEG. Whole seedlings (∼0.3 g) were homogenized and total RNA was extracted as described (Lee et al., 2001). Gene-specific DNA probes were as described (Ishitani et al., 1998; Lee et al., 2001). For the Δ9-desaturase gene, a full-length cDNA was used as the probe.
Grafting experiment
The hypocotyls of 1-week-old seedlings grown vertically on MS agar plates were excised with micro-dissecting scissors (Fisher Scientific) under a dissecting microscope, and wild-type or mutant shoots were placed carefully on top of mutant or wild-type roots, respectively. The re-connected seedlings were incubated in sealed agar plates in order to heal under high humidity. Since the rate of success grafting was very low (∼5%), a large number of seedlings was used. Successfully grafted seedlings were transferred to soil and grown to the rosette stage for the examination of chilling sensitivity.
Electrolyte leakage and freezing tolerance assays
The electrolyte leakage assay was carried out as described previously (Ishitani et al., 1998).
The freezing tolerance assay was carried out as described by Xin et al. (1998). Briefly, seedlings were grown in a standard 0.6% agar medium for 2 weeks under continuous light. Plates with seedlings were placed in a freezing chamber set to –1°C and treated for 16 h prior to decreasing the temperature at 1°C/h. Freezing was initiated by the addition of ice chips to the plates. Plates were removed at the desired temperatures and placed at 4°C overnight for thawing. After recovery for 2 days, the number of surviving plants was scored. For cold acclimation, plates were incubated at 4°C in the dark for 2 days before the freezing treatment.
Genetic mapping and cloning of LOS2
For genetic mapping of the los2 mutation, the los2 mutant of the Arabidopsis C24 ecotype was crossed with wild-type plants of the Columbia ecotype. A total of 4563 homozygous los2 mutants were chosen from the segregating F2 population with the phenotype of low expression of RD29A-LUC after cold treatment. Genetic mapping was carried out as described (Lee et al., 2001).
For los2 complementation analysis, a genomic fragment containing the enolase ORF plus 886 bp of upstream sequence and 1386 bp of downstream sequence was digested with EcoRI and SacI and inserted into the binary vector pCAMBIA 1200. The resulting plasmid was transferred to the los2 mutant via Agrobacterium-mediated transformation. Forty-five hygromycin-resistant T1 transformants were selected, seedlings of T2 and T3 progeny were subjected to cold treatment and luminescence images were taken. In addition, all of the 45 transgenic los2 plants expressing the wild-type F1O11.16 transgene were exposed to 4°C under light for a week to test the complementation of the chilling-sensitive phenotype of los2 plants.
For the LOS2-GUS construct, a LOS2 promoter region from –1055 to –1 (relative to the ATG start codon) was amplified and inserted into SalI–SmaI sites of the binary vector pCAMBIA1391Z. The plasmid was transferred to wild-type C24 plants. Twenty-five hygromycin-resistant T1 transformants were selected, and seedlings of T2 progeny were stained with X-Gluc for 15 h as described (Lee et al., 2001).
Expression of LOS2–GFP fusion protein
Wild-type LOS2 cDNA was amplified by RT–PCR and inserted into the NcoI site of the binary vector pCAMBIA1200/35SP/GFP. Forty lines of hygromycin-resistant T1 transformants were selected and their segregating T2 progeny were examined for GFP expression (Lee et al., 2001).
Enolase activity assay and immunoblot analysis
The wild-type and mutant LOS2 cDNAs were amplified by RT–PCR and inserted into the NcoI site of the expression vector pET14b. The plasmids were transferred to Escherichia coli cells (BL21 DE3, Codon Plus). Expression of His-LOS2 fusion protein was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1.5 mM. Purification of wild-type and mutant His-LOS2 protein was performed according to the manufacturer’s instructions (His-Bind Buffer Kit, Novagen, WI). The enolase activity assay and immunoblot analysis were as described (Van Der Straeten et al., 1991).
DNA binding assay
EMSAs were carried out as described (Hao et al., 1998). The following double-stranded oligonucleotides were used for probes and competitors in EMSAs. Human c-myc, 5′-GGAGGGATCGCGCTGAGTATAAAAGCCGGTTTCGG-3′ (Ray and Miller, 1991); ZAT10, 5′-GGTCTTGACGCGTTATATAAAATCCGTATTA-3′; and m-ZAT10, 5′-GGAGGT CATATGGCGCGCCCCGAATGCGG-3′. These probes were end-labeled with [α-32P]dCTP by Klenow fragment and purified through a Sephadex G-50 column. The labeled probes were incubated with 5 µg of purified His-LOS2 fusion protein in 1× binding buffer (Hao et al., 1998) supplemented with 20 pmol poly(dI–dC) for 20 min at room temperature. The resulting DNA–protein complexes were resolved by electrophoresis on a 6% polyacrylamide gel in 0.5× TBE buffer and visualized by autoradiography. For competition experiments, unlabeled competitors were incubated with the His-LOS2 fusion protein on ice for 30 min prior to adding labeled probes.
Transient expression assay
For the reporter gene construct, a fragment containing –646 to –1 upstream of transcription start site of the RD29A gene was fused transcriptionally to firefly LUC (Promega). A plasmid for the expression of STZ/ZAT10 was constructed as follows. The CaMV 35S promoter and the GUS reporter gene in pBI221was replaced by the 2× CaMV 35S promoter to generate 2× 35S-Nos. The cDNA for STZ/ZAT10 was amplified by PCR, digested with SalI and inserted into SmaI and SalI sites of 2× 35S-Nos. Plasmid DNA was delivered to Arabidopsis leaves using particle bombardment (Ohta et al., 2001).
Acknowledgments
Acknowledgements
We sincerely thank Dr Napoli for assistance with Arabidopsis grafting, Drs Shinshi and Ohme-Takagi for providing pPTRL, 190.LUC and 2× 35S-Nos plasmids for transient expression assays, and Dr Van Der Straten for providing enolase antibodies. This work was supported by USDA NRI grant 2000-00664 and NSF grant DBI-9813360 to J.-K.Z.
References
- Bagnall D., Wolfe,J. and King,R.W. (1983) Chilling-induced wilting and hydraulic recovery in mung bean plants. Plant Cell Environ., 6, 457–464. [Google Scholar]
- Delauney A.J. and Verma,D.P.S. (1993) Proline biosynthesis and osmoregulation in plants. Plant J., 4, 215–223 [Google Scholar]
- Feo S., Arcuri,D., Piddini,E., Passantino,R. and Giallongo,A. (2000) ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett., 473, 47–52 [DOI] [PubMed] [Google Scholar]
- Ghosh A.K., Steele,R. and Ray,R.B. (1999a) Functional domains of c-myc promoter binding protein 1 involved in transcriptional repression and cell growth regulation. Mol. Cell. Biol., 19, 2880–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A.K., Steele,R. and Ray,R.B. (1999b) MBP-1 physically associates with histone deacetylase for transcriptional repression. Biochem. Biophys. Res. Commun., 260, 405–409. [DOI] [PubMed] [Google Scholar]
- Gilmour S.J., Sebolt,A.M., Salazar,M.P., Everard,J.D. and Thomashow,M.F. (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol., 124, 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D., Ohme-Takagi,M. and Sarai,A. (1998) Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem., 273, 26857–26861. [DOI] [PubMed] [Google Scholar]
- Ishitani M., Xiong,L., Stevenson,B. and Zhu,J.K. (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis thaliana: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell, 9, 1935–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M., Xiong,L., Lee,H., Stevenson,B. and Zhu,J.K. (1998) HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis.Plant Cell, 10, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagglo-Ottosen K.R., Gilmour,S.J., Zarka,D.G., Schabenberger,O. and Thomashow,M.F. (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science, 280, 104–106 [DOI] [PubMed] [Google Scholar]
- Knight H., Veale,E.L., Warren,G.J. and Knight,M.R. (1999) The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell, 11, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Xiong,L., Gong,Z., Ishitani,M., Stevenson,B. and Zhu,J.K. (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev., 15, 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt J. (1980) Responses of Plants to Environmental Stress, Vol. 1: Chilling, Freezing and High Temperature Stress. Academic Press, New York, NY, pp. 137–141.
- Markhart A.H. III.,, Fiscus,E.L., Naylor,A.W. and Kramer,P.J. (1979) Effect of temperature on water and ion transport in soybean and broccoli systems. Plant Physiol., 64, 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J., Bargues,M., Terol,J. Perez-Alonso,M. and Salinas,J. (1999) The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol., 119, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M., James,D.,Jr, Dooner,H. and Browse,J. (1993) Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc. Natl Acad. Sci. USA, 90, 6208–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui,K., Hiratsu,K., Shinshi,H. and Ohme-Takagi,M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell, 13, 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R.B. (1995) Induction of cell death in murine fibroblasts by a c-myc promoter binding protein. Cell Growth Differ., 6, 1089–1096. [PubMed] [Google Scholar]
- Ray R. and Miller,D.M. (1991) Cloning and characterization of a human c-myc promoter-binding protein. Mol. Cell. Biol., 11, 2154–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R.B., Steele,R., Seftor,E. and Hendrix,M. (1995) Human breast carcinoma cells transfected with the gene encoding a c-myc promoter-binding protein (MBP-1) inhibits tumors in nude mice. Cancer Res., 55, 3747–3751. [PubMed] [Google Scholar]
- Steponkus P.L., Uemura,M., Joseph,R.A., Gilmour,S.J. and Thomashow,M.F. (1998) Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 95, 14570–14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E.J., Gilmour,S.J. and Thomashow,M.F. (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcription activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl Acad. Sci. USA, 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E.J., Mao,Y., Regier,M.K., Triezenberg,S.J. and Thomashow,M.F. (2001) Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res., 29, 1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. and Miller,D.M. (2000) Structural analysis of α-enolase. Mapping the functional domains involved in down-regulation of the c-myc protooncogene. J. Biol. Chem., 275, 5958–5965. [DOI] [PubMed] [Google Scholar]
- Takatsuji H. and Matsumoto,T. (1996) Target-sequence recognition by separate-type Cys2/His2 zinc finger proteins in plants. J. Biol. Chem., 271, 23368–23373. [DOI] [PubMed] [Google Scholar]
- Thomashow M.F. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol., 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Van Der Straeten D., Rodrigues-Pousada,R.A., Goodman,H.M. and Van Montagu,M. (1991) Plant enolase; gene structure, expression and evolution. Plant Cell, 3, 719–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z. and Browse,J. (1998) eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl Acad. Sci. USA, 95, 7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K. and Shinozaki,K. (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell, 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]